Introduction
Iron is a major nutrient and an essential component
of hemoglobin, which is present in red blood cells and carries
oxygen from the lungs to the rest of the body. Iron is also
involved in metabolism redox regulation and reactive oxygen species
(ROS) production. To support these mechanisms, a comprehensive
association between iron transport and homeostasis is required
(1). The balance between iron as a
nutritional source and a toxicant is sensitive and delicate, and
iron transport and homeostasis are closely associated with
inflammation (2,3). Macrophages are essential in balancing
iron transport and inflammation. M1 macrophages are involved in
iron uptake when iron is accumulated through ferroportin (FPN) and
heme oxygenase (HO)-1 activities; this directs M2 macrophages,
along with ferritin, to export excess iron and to inhibit
proinflammatory cytokines (4).
Iron is a major contributor in the biosynthesis of
heme, which is a porphyrin molecule containing a ferrous ion
(Fe2+). Notably, iron-dependent ferritin, carbon
monoxide and bilirubin are formed through heme degradation
(5), and heme is involved in
various physiological processes, including inflammation that is
induced by excess heme production. However, heme is an
iron-containing complex that is essential for a number of
biological processes, including oxygen transport and storage,
electron transfer and protein synthesis. Therefore, heme is
considered a double-edged sword because it can be toxic by
mediating inflammation (6). It
also participates in oxygen transport, signal transduction and
mitochondrial function, and an imbalance in iron/heme metabolism
may cause tumorigenesis (7). Free
heme molecules and heme-containing proteins are generated under
various pathological conditions, such as ischemia/reperfusion,
hemorrhage and muscle injuries (8). An excess amount of heme also
contributes to renal failure because of local inflammation and
promotes the expression of proinflammatory cytokines, such as IL-1β
and TNF-α (9).
Lipopolysaccharide (LPS) is a vital component of the
outer layer of gram-negative bacteria, which induces innate
immunity and initiates the inflammatory response in other cells
(10). LPS is a pathogenic
stimulator and can induce an inflammatory response by regulating
pathogen-associated molecular patterns (11). Toll-like receptor (TLR)4 recognizes
innate LPS and controls the innate immune response by neutralizing
proinflammatory factors (12). LPS
also activates various inflammatory signaling molecules, such as
inducible nitric oxide synthase (iNOS), COX-2, NF-κB and p38-MAP
kinase pathways (13). When
exposed to LPS, TLRs activate innate immunity by activating NF-κB
through canonical or noncanonical pathways and proinflammatory
cytokines, such as TNF-α and various ILs, in response to
inflammation (14).
Sulfur is an essential element in amino acids, such
as cysteine and methionine, as well as in some sulfur-containing
natural compounds that exert anti-inflammatory activities (15). Sulfur intake is possible through
the diet, for example via the consumption of garlic, onion and duck
meat (16). Methylsulfonylmethane
(MSM) is a natural sulfur compound with various capabilities, such
as anti-ketotic (17), antioxidant
(18,19) and anticancer activities (20,21).
Studies on MSM have demonstrated its anti-inflammatory activity
against various inflammatory conditions (22,23).
MSM is also found in cardiac cells, where it inhibits the
inflammatory response by mediating TNF-α (24). In some instances, sulfur can be
toxic; however, it can be coated with various nontoxic substances
to enhance the effect of sulfur compounds against various
conditions, including inflammation (25). Nontoxic sulfur (NTS) is a sulfur
compound coated with a nontoxic substance, which when used to
enrich livestock feed can enhance meat quality and boost immunity
(26). In a previous study, rats
orally administered NTS did not experience cell death (27). Furthermore, our previous study on
NTS indicated that it can enhance the signaling of growth hormone
in C2C12 mouse myoblasts (28). In
addition, NTS inhibits inflammatory responses in C2C12 cells by
mediating TLR4 and JAK2/STAT3 signaling to regulate IL-6 expression
(29). Our previous study also
demonstrated the anti-inflammatory effects of MSM and NTS against
high glucose-induced inflammatory conditions by regulating NF-κB
signaling in THP-1 human monocytes (30).
The present study aimed to determine the effects of
NTS and MSM on LPS-induced inflammation, and to assess the role of
iron/heme metabolism in the anti-inflammatory activity of these
sulfur compounds. Molecular analyses of the TLR and NF-κB signaling
pathways, iron homeostasis and heme biosynthesis were also
performed under LPS-induced conditions.
Materials and methods
Reagents and antibodies
MSM (cat. no. PHR1346) and LPS (cat. no. L2630) were
obtained from MilliporeSigma. The following primary antibodies were
purchased from Santa Cruz Biotechnology, Inc.: Ferrochelatase
(FECH; cat. no. sc-377377), phosphorylated (p)-IκBα (cat. no.
sc-8404), TLR2 (cat. no. sc-21759), TLR4 (cat. no. sc-293072),
COX-1 (cat. no. sc-19998), COX-2 (cat. no. sc-19999, IKKα/β (cat.
no. sc-7607), and β-actin (cat. no. sc-47778); from Cell Signaling
Technology, Inc.: IL-1β (cat. no. 12703), p-ERK (cat. no. 9101),
ERK (cat. no. 9102), IκBα (cat. no. 9242), p-IKKα/β (cat. no.
2697), NF-κB (cat. no. 8242), p-p38 (cat. no. 4511), p38 (cat. no.
8690), ATR (cat. no. 2790), ATM (cat. no. 2873), Chk2 (cat. no.
2662), BRCA1 (cat. no. 9010), p53 (cat. no. 9282), p-MDM2 (cat. no.
3521), MDM2 (cat. no. 86934) and the DNA Damage Antibody Sampler
Kit (cat. no. 9947; containing p-ATM, p-ATR, p-Chk2, p-BRCA1 and
p-p53 antibodies); from Abcam: Divalent metal transporter (DMT)1
(cat. no. ab55735), 5′-aminolevulinate synthase (ALAS)1 (cat. no.
ab84962), STEAP3 (cat. no. ab151566), HO-1 (cat. no. ab137749),
transferrin receptor (TfR; cat. no. ab84036), PKCα (cat. no.
ab179523), p-PKCα (cat. no. ab59411), TNF-α (cat. no. ab183218),
IL-6 (cat. no. ab6672) and TATA-binding protein (TBP; cat. no.
ab818); from LifeSpan BioSciences, Inc.: ABCB10 (cat. no.
LS-C381841) and FLVCR1 (cat. no. LS-C750126); from LS Bio; Vector
Laboratories, Inc.: FPN (cat. no. NBP1-21502) and iNOS (cat. no.
NB300-605); from Novus Biologicals; Bio-Techne: Mitoferrin (MFRN;
cat. no. MBS6013473); and from Boster Biological Technology: ABCB6
(cat. no. PA1723). Horseradish peroxidase (HRP)-conjugated
anti-rabbit (cat. no. 7074) and anti-mouse (cat. no. 7076)
secondary antibodies were purchased from Cell Signaling Technology,
Inc. NTS was provided by the NARA Bioetch.
Cell culture and treatment
Human THP-1 cells were purchased from the Korean
Cell Line Bank; Korean Cell Line Research Foundation and cultured
in RPMI-1640 medium (cat. no. L0498; Biowest) containing 10% fetal
bovine serum (cat. no. A5670801; Gibco; Thermo Fisher Scientific,
Inc.) and 1% penicillin/streptomycin at 37°C in 5% CO2.
The cells (1×106 cells/ml) were cultured for 72 h with
10 ng/ml LPS, with or without NTS, MSM or TLR4-C34 (cat. no. S0822;
Selleck Chemicals).
Fe2+ determination
assay
The cells were stained with 5 µM FerroFarRed
solution (cat. no. GC903-01; Goryo Chemical, Inc.) and incubated in
a CO2 incubator at 37°C for 30–40 min. After staining,
the cells were washed with 1 ml pre-warmed serum-free RPMI-1640
medium and used for flow cytometric analysis (FACSCalibur; BD
Biosciences). FlowJo v10 software (FlowJo; BD Biosciences) was used
for analysis.
Western blotting
Whole-cell lysates were prepared by incubating
untreated or LPS-treated THP-1 cells on ice with
radioimmunoprecipitation lysis buffer (cat. no. 20-188;
MilliporeSigma) containing protease and phosphatase inhibitors.
Protein concentrations were measured using the Bradford method
(Thermo Fisher Scientific, Inc.). The same amounts of protein (30
µg/well) were then separated by SDS-PAGE on 6–15% gels and the
separated proteins were transferred onto nitrocellulose membranes.
The blots were blocked for 1 h at room temperature with 5% skim
milk (BD Biosciences) in TBS-Tween-20 (TBS-T) buffer [20 mM
Tris-HCl (MilliporeSigma), pH 7.6; 137 mM NaCl (Formedium Limited);
0.1X (0.1%) Tween-20 (Scientific Sales, Inc.)]. The membranes were
then incubated with primary antibodies diluted in 5% skim milk
(1:1,000 dilution) overnight at 4°C with agitation. Subsequently,
the membranes were washed with TBS-T and incubated for 1 h at room
temperature with HRP-conjugated secondary antibodies (1:5,000).
Detection was performed using a West-Q Pico ECL Solution (cat. no.
W3652-020; GenDEPOT, LLC) and a LAS-4000 imaging device (FUJIFILM
Wako Pure Chemical Corporation).
Reverse transcription-quantitative PCR
(RT-qPCR) assay
Total cellular RNA was extracted using an RNeasy
Mini Kit (Qiagen GmbH) according to the manufacturer's protocol.
The isolated RNA was quantified spectrophotometrically at 260 nm,
and cDNA was synthesized at 42°C for 1 h and 95°C for 5 min with
oligo d(T) primers and a first-strand cDNA synthesis kit (cat. no.
K-2041; Bioneer Corporation). qPCR was then conducted using a
thermal cycler (C1000 Thermal Cycler; Bio-Rad Laboratories, Inc.)
as follows: 2 µl diluted cDNA was added to diluted forward and
reverse primers (1 µl each, 100 pM) and 10 µl TB Green Advantage
Premix (Takara Bio, Inc.). The thermocycling conditions were as
follows: Initial denaturation at 95°C for 5 min; followed by 40
cycles of denaturation at 95°C for 40 sec, annealing at 58°C for 40
sec and extension at 72°C for 40 sec; and a final extension step at
72°C for 5 min. All measurements were performed in triplicate.
Relative target gene expression was normalized to GAPDH. The
calculations were performed using the 2−ΔΔCq values
obtained (31). The primer
sequences are provided in Table
I.
 | Table I.Primer sequences used for reverse
transcription-quantitative PCR analysis. |
Table I.
Primer sequences used for reverse
transcription-quantitative PCR analysis.
| Gene | Sequence |
|---|
| TLR2 | Sense:
5′-TGCAAGTACGAACTGGACTTCT-3′ |
|
| Antisense:
5′-CCAGGTAGGTCTTGGTGTTCATT-3′ |
| TLR4 | Sense:
5′-TAGCCATTGCTGCCAACATCAT-3′ |
|
| Antisense:
5′-AAGATACACCAACGGCTCTGAA-3′ |
| iNOS | Sense:
5′-TGCTCAGCTCATCCGCTATG-3′ |
|
| Antisense:
5′-GATGTTCCATGGCCACCTCA-3′ |
| NF-κB | Sense:
5′-GAAATTCCTGATCCAGACAAAAAC-3′ |
|
| Antisense:
5′-ATCACTTCAATGGCCTCTGTGTAG-3′ |
| COX-1 | Sense:
5′-GGCAGCAGAGTTGGAGGAAT-3′ |
|
| Antisense:
5′-CTTCTTCAGTGTGGCCGTCT-3′ |
| COX-2 | Sense:
5′-TTGCATTCTTTGCCCAGCAC-3′ |
|
| Antisense:
5′-ACCGTAGATGCTCAGGGACT-3′ |
| IL-1β | Sense:
5′-ATTGCCTCTTCCAGCAGCTT-3′ |
|
| Antisense:
5′-GGCTTCACTGAGGTTGCCTT-3′ |
| IL-6 | Sense:
5′-GCTGATCCTGCCTCTGCC-3′ |
|
| Antisense:
5′-GACCCTCAAACCCACCCG-3′ |
| TNF-α | Sense:
5′-TGGTGAGACAGAAAGAGCGG-3′ |
|
| Antisense:
5′-AGCCCTGAGGTGTCTGGT-3′ |
| GAPDH | Sense:
5′-ACCCACTCCTCCACCTTTGA-3′ |
|
| Antisense:
5′-CATACCAGGAAATGAGCTTGACAA-3′ |
Flow cytometric analysis
After cultured cells were washed with pre-chilled
PBS, cell pellets were incubated with 10% BSA (cat. no. A3311;
MilliporeSigma) on ice for 20 min. PE-conjugated anti-human CD284
(TLR4) antibody (1:200; cat. no. 312806; Biolegend, Inc.) and
APC-conjugated anti-human CD282 (TLR2) antibody (1:200; cat. no.
309720; Biolegend, Inc.) was used to stain the cells on ice for 30
min. Stained cells were then washed with pre-chilled PBS. Cells
were also stained with CM-H2DCFDA (5 µM; cat. no. C6827;
Invitrogen; Thermo Fisher Scientific, Inc.) for cellular ROS and
placed in a CO2 incubator at 37°C for 30 min. The
stained cells were washed with 1 ml prewarmed staining buffer (cat.
no. 554656; BD Bioscience). Flow cytometric analysis was performed
using a FACSCalibur flow cytometer. FlowJo v10 software was used
for analysis.
Comet assay
The comet assay kit (cat. no. ab238544; Abcam) was
used according to the manufacturer's protocol for measuring
cellular DNA damage. A base layer of comet agarose was used to coat
the slide, and a layer of cells (70% confluence) treated with 10
ng/ml LPS with the indicated concentrations of NTS or MSM was
added, followed by another layer of 100 cells and agarose, followed
by lysis. Electrophoresis was performed under neutral conditions,
and cells were stained with DNA dye. Cell morphology was observed
by fluorescence microscopy (IX71/DP72; Olympus Corporation).
Chromatin immunoprecipitation (ChIP)
assay
A ChIP assay was performed using an Imprint
Chromatin Immunoprecipitation Kit (cat. no. 17-295; MilliporeSigma)
according to the manufacturer's protocol. THP-1 cells were fixed
with 1% formaldehyde for 10 min at 37°C and quenched with 1.25 M
glycine at room temperature for 30 min at 37°C. After washing with
PBS, cells were suspended in a nucleus preparation buffer and
sonicated in a shearing buffer under optimized conditions at 4°C
for 10 times (20 kHz, 10 sec on/30 sec off). This sheared DNA was
diluted with a dilution buffer (1:1 ratio), and 5 µl diluted sample
was removed as an internal control. The diluted supernatant was
incubated for 5 min at room temperature in wells pre-coated with
antibodies (1:50 dilution) specific for IL-1β (cat. no. sc-12742;
Santa Cruz Biotechnology, Inc.), TNF-α (cat. no. 6945; Cell
Signaling Technology, Inc.) or IL-6 (cat. no. sc-57315; Santa Cruz
Biotechnology, Inc.) at room temperature for 90 min. Normal mouse
IgG (1:10 dilution; cat. no. M8695; MilliporeSigma) and anti-RNA
polymerase II (1:10 dilution; cat. no. R1530; MilliporeSigma) were
used as negative and positive controls, respectively. The unbound
DNA was washed off with wash buffer, and the bound DNA was
collected by cross-link reversal using a DNA release buffer
containing proteinase K. The released DNA and the DNA from the
internal controls were purified with GenElute Binding Column G. DNA
was then quantified and RT-qPCR was performed using the
aforementioned reagents; the thermocycling conditions were as
follows: Initial denaturation at 95°C for 5 min; followed by 50
cycles of denaturation at 95°C for 20 sec, annealing at 58°C for 20
sec and extension at 72°C for 20 sec. All measurements were
performed in triplicate. Relative target gene expression was
normalized to GAPDH. The calculations were performed using the
2−ΔΔCq values obtained.
Nuclear extraction assay and NF-κB
detection
Nuclear protein extracts were prepared with the
Nuclear Extract Kit (cat. no. ab113474; Abcam). After harvesting
the cells, the extraction buffer was prepared by adding DTT
solution and a protease inhibitor cocktail at a 1:1,000 dilution.
The cell pellet was treated with the prepared extraction buffer at
a volume of 10 µl/106 cells, and then incubated on ice
for 15 min, briefly vortexing for ~5 sec every 3 min for mixing.
Subsequently, the mixture was centrifuged at 18,500 × g for 10 min
at 4°C. The resulting supernatant was carefully transferred to a
new microcentrifuge tube; this contained the nuclear protein
extract. The nuclear extract was then used for western blot
analysis iof NF-κB. Equal amounts of proteins (150 µg/well) were
separated by SDS-PAGE on 10% gels and were then transferred onto
nitrocellulose membranes. Subsequently, he blots were blocked for 1
h at room temperature with 5% skim milk. The membranes were then
incubated with NF-κB and TBP antibodies diluted in 5% skim milk
overnight at 4°C with agitations. The membranes were then washed
with TBS-T and incubated for 1 h with HRP-conjugated secondary
antibodies at room temperature. Detection was performed as
aforementioned.
Statistical analyses
All experiments were performed in triplicate. Data
are presented as the mean ± SEM of three independent experiments
conducted in triplicate (n=3). The control group was set to 100 for
RT-qPCR results. A one-way ANOVA followed by Tukey's post hoc test
was used for the statistical analysis. The analyses were performed
using SAS 9.3 software (SAS Institute, Inc.). P<0.05 was
considered to indicate a statistically significant difference.
Results
Sulfur compounds inhibit LPS-induced
iron metabolism and heme biosynthesis in THP-1 human monocytes
We previously demonstrated that LPS-induced
inflammation enhances iron metabolism through increased iron
production and transport, which is responsible for iron homeostasis
(5). To determine the effect of
sulfur compounds on LPS-induced iron metabolism, the amount of
Fe2+ was estimated following the treatment of THP-1
cells with NTS, MSM or TLR4-C34, a specific TLR4 inhibitor
(Fig. 1A). Flow cytometry revealed
increased Fe2+ levels after LPS treatment, whereas NTS
and MSM exhibited decreased Fe2+ levels. A slightly
different phenomenon was observed in TLR4-C34-treated cells,
indicating the activity of sulfur compounds to inhibit LPS-induced
inflammation by attenuating TLR4-independent iron metabolism. Next,
western blot analysis was performed to identify the proteins
responsible for iron transport and the molecular mechanisms
associated with sulfur compound-regulated iron homeostasis. The
results indicated upregulation of the TfR and FPN proteins by LPS,
which was suppressed by treatment with NTS, MSM or TLR4-C34
(Fig. 1B). In addition, the
expression levels of DMT1 and STEAP3, which are key factors in iron
metabolism, were increased by LPS treatment. The regulation of iron
metabolism may lead to heme synthesis. To confirm the induction of
heme biosynthesis by LPS, the expression levels of proteins
responsible for heme synthesis were examined through western blot
analysis (Fig. 1C). The results
indicated notable LPS-induced upregulation of HO-1, MFRN, ABCB6,
ABCB10, ALAS1, FECH and FLVCR in THP-1 cells; however, these
protein levels were markedly decreased following NTS or MSM
treatment, but increased by TLR4-C34 treatment. These results
indicated that iron/heme metabolism is pivotal in the
anti-inflammatory activity of sulfur compounds, independent of
TLR4.
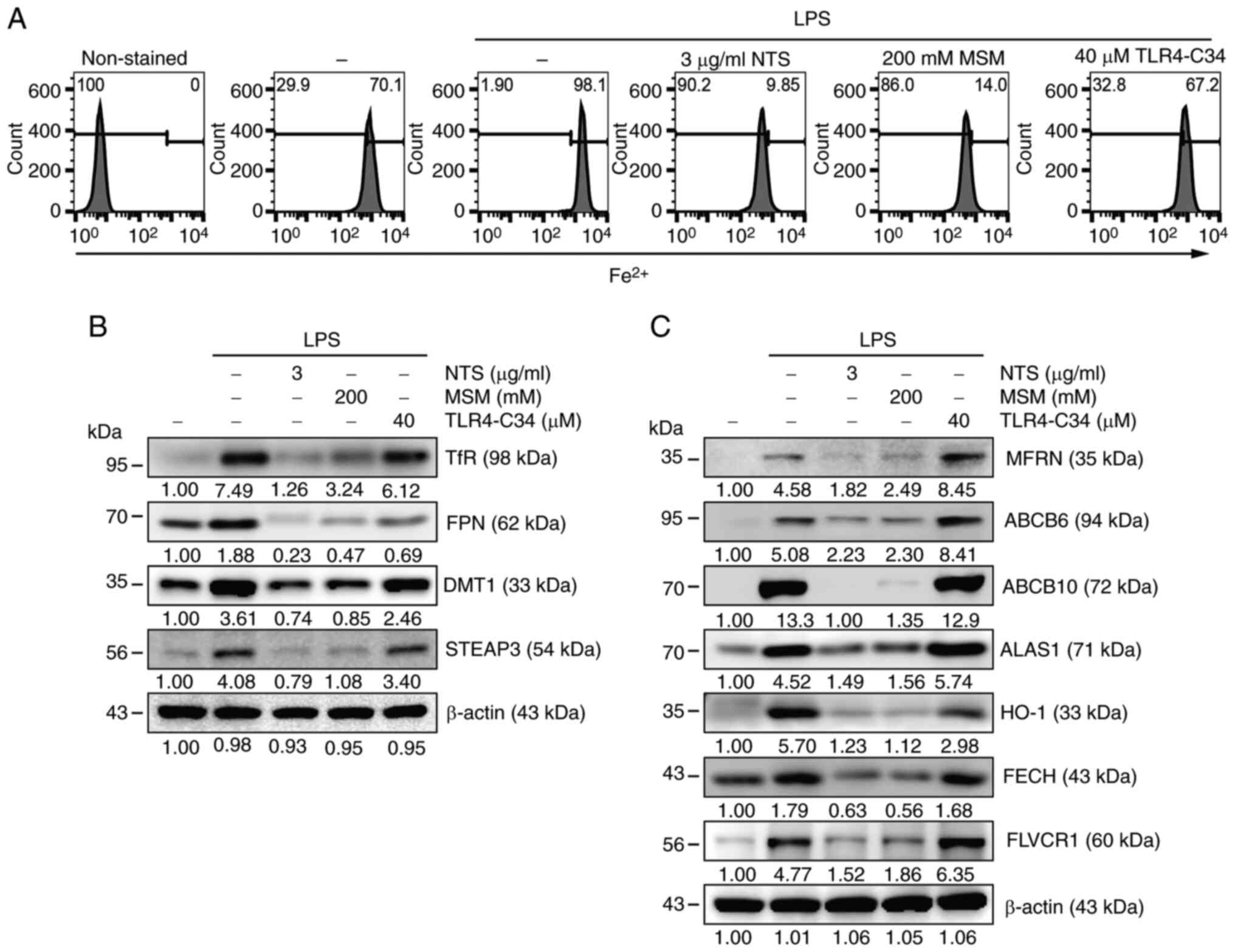 | Figure 1.Sulfur compounds inhibit LPS-induced
inflammation and iron/heme metabolism. (A) Flow cytometry showing
Fe2+ levels in THP-1 cells following treatment with LPS (10 ng/ml)
+ NTS (3 µg/ml), MSM (200 mM) or TLR-C34 (40 µM) for 48 h. Western
blot analysis of (B) transferrin receptor, ferroportin, DMT1 and
STEAP3; and (C) MFRN, ABCB6, ABCB10, ALAS1, HO-1, FECH and FLVCR
proteins in THP-1 cells following treatment with LPS (10 ng/ml) +
NTS (3 µg/ml), MSM (200 mM) or TLR-C34 (40 µM) for 48 h. ALAS1,
5′-aminolevulinate synthase 1; DMT1, divalent metal transporter 1;
Fe2+, ferrous ion; FECH, ferrochelatase; FPN, ferroportin; HO-1,
heme oxygenase-1; LPS, lipopolysaccharide; MFRN, mitoferrin; MSM,
methylsulfonylmethane; NTS, nontoxic sulfur; TfR, transferrin
receptor; TLR, Toll-like receptor. |
Sulfur compounds downregulate the
LPS-induced expression of TLRs in THP-1 cells
TLRs act as receptors for the inflammatory response
in immune cells. Thus, the current study analyzed the interaction
of sulfur compounds with TLRs upon LPS-induced inflammation. First,
the expression pattern of TLRs (TLR2/4) was examined in response to
two sulfur compounds during LPS-induced inflammation. The results
indicated that LPS increased the expression levels of TLR2/4
proteins, which were markedly downregulated by the addition of 3
µg/ml NTS, 200 mM MSM or 40 µM TLR4-C34 (Fig. 2A). The present study also analyzed
the mRNA expression levels of TLR2/4 after LPS treatment in the
presence of NTS, MSM or TLR4-C34 in THP-1 cells. The results
confirmed the inhibition of LPS-induced expression of TLR2/4 by
NTS, MSM and the TLR4 inhibitor (Fig.
2B). Flow cytometric analysis also confirmed that TLR2/4 was
downregulated by NTS, MSM or TLR4-C34 (Fig. 2C). These results suggested that the
anti-inflammatory effects of natural sulfurs depend on the
inhibition of TLR2/4 during LPS-induced inflammation.
Sulfur molecules decrease LPS-induced
ROS generation in THP-1 cells
Generally, LPS is known to induce inflammation by
generating ROS. The present study analyzed the effects of sulfur
compounds on ROS generation in LPS-induced inflammation. First, the
expression levels of the iNOS protein, which serves an important
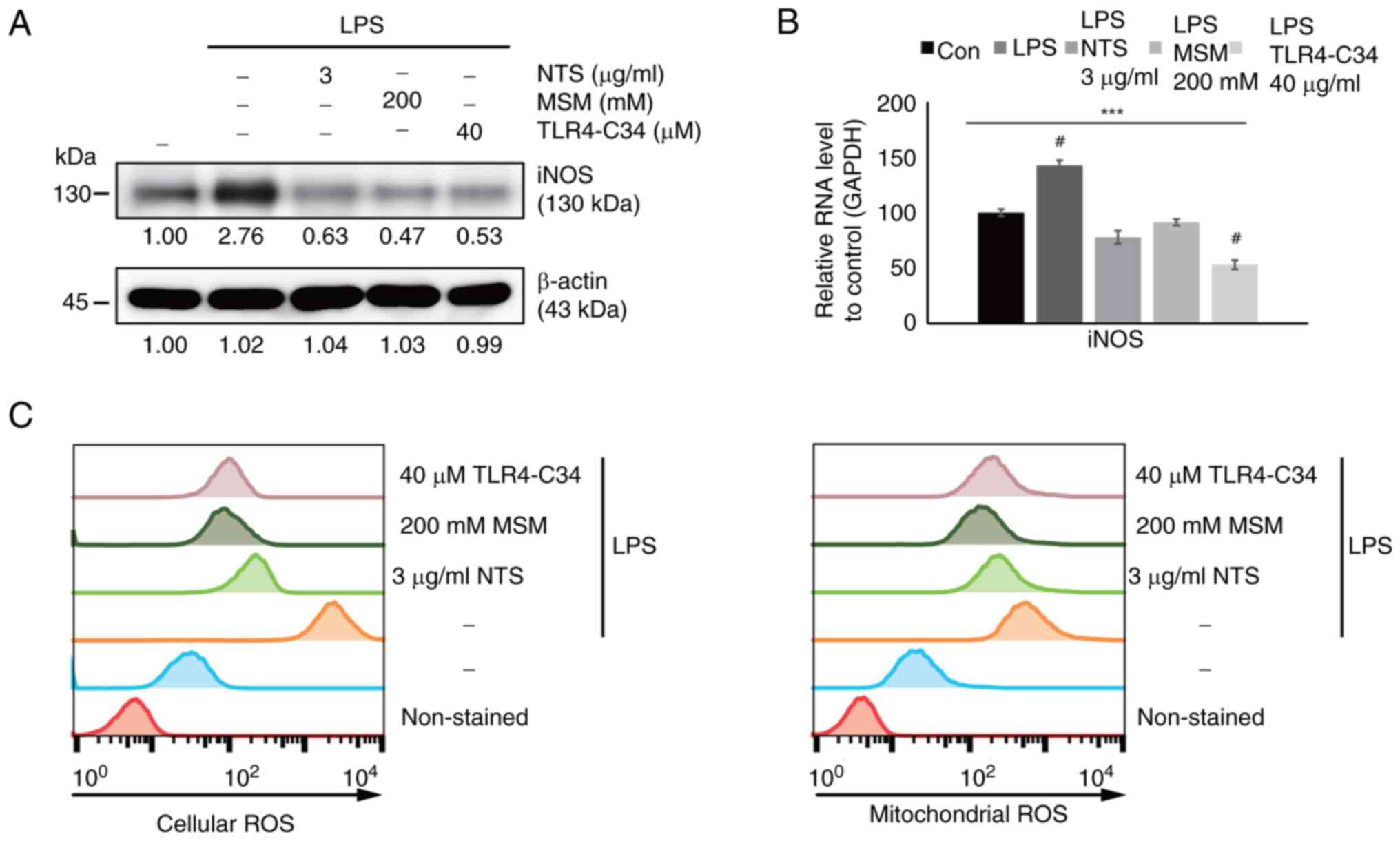
role in ROS generation, were measured. Western blot analysis
revealed increased expression levels of iNOS in response to LPS,
which were inhibited by NTS, MSM, and TLR4-C34 (Fig. 3A). These effects were also
confirmed at the transcriptional level; increased iNOS mRNA
expression levels were observed following LPS treatment, which were
suppressed by NTS, MSM, and TLR4-C34 treatment (Fig. 3B). These results suggested that
sulfur molecules exhibit similar effects to TLR4-C34, thus
indicating the anti-inflammatory effects of NTS and MSM against
LPS-induced inflammation. Finally, ROS generation was measured in
response to treatment with sulfur compounds at the cellular and
mitochondrial levels (Fig. 3C).
Increased ROS levels were observed following LPS treatment, which
were significantly reduced by treatment with natural sulfur
compounds and a TLR4 inhibitor. These results indicated the
cytoprotective nature of NTS and MSM against inflammation.
Sulfur molecules inhibit PKC-mediated
inflammation and canonical NF-κB pathways against LPS-induced
inflammation in THP-1 cells
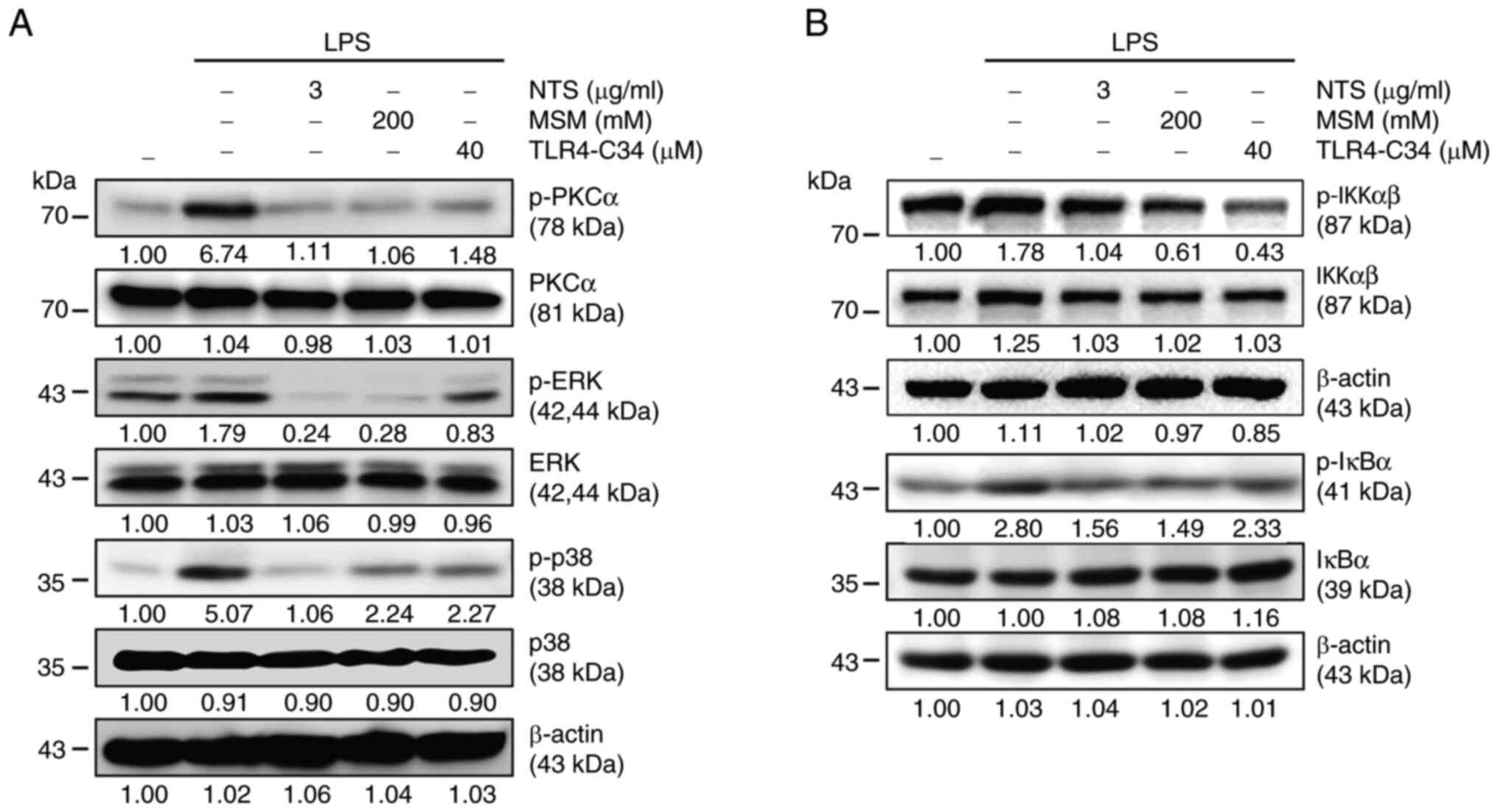
It was hypothesized that NF-κB signaling may be
responsible for the anti-inflammatory response induced by NTS and
MSM; therefore, the PKC-mediated upstream targets of NF-κB were
examined. Increased expression levels of p-PKCα, p-ERK, and p-p38
were observed in LPS-treated THP-1 cells (Fig. 4A). By contrast, NTS, MSM and
TLR4-C34 markedly reduced LPS-induced expression of PKC-mediated
signaling factors, suggesting the possible involvement of NF-κB
activity in the inflammatory response. Next, the present study
assessed the expression levels of canonical NF-κB pathway elements.
Treatment with LPS upregulated the expression levels of p-IKKα/β
and p-IκBα, which were decreased by NTS, MSM, and TLR4-C34
treatment (Fig. 4B). These results
indicated the involvement of NF-κB in the anti-inflammatory effects
of sulfur compounds against LPS-induced inflammation.
Sulfur molecules alleviate LPS-induced
DNA damage in THP-1 cells
DNA damage is a possible outcome of prolonged ROS
production (32). In the present
study, LPS-induced ROS generation was inhibited by treatment with
sulfur compounds. Subsequently, a comet assay was performed to
determine whether LPS induced DNA damage and if sulfur compounds
could revert such an effect or induce a DNA damage response (DDR).
The analysis of THP-1 cells by fluorescence microscopy revealed an
increase in comet length and the number of comet-positive cells
following LPS exposure, whereas these effects were reversed by NTS,
MSM, and TLR4-C34 treatment (Fig.
5A). These results suggested the possible induction of DDR by
sulfur compounds. Next, the current study analyzed the expression
levels of several molecular signaling proteins responsible for DDR.
LPS induced the expression of DNA damage markers, including p-ATR,
p-ATM, p-Chk2, p-BRCA1 and p-p53, and DNA damage by LPS stabilized
p53 and p-p53 by inhibiting MDM2-mediated degradation (Fig. 5B); however, NTS, MSM, and TLR4-C34
notably decreased their expression levels, indicating the ability
of natural sulfurs to induce DDR in response to LPS-induced
inflammation by regulating TLR4 expression.
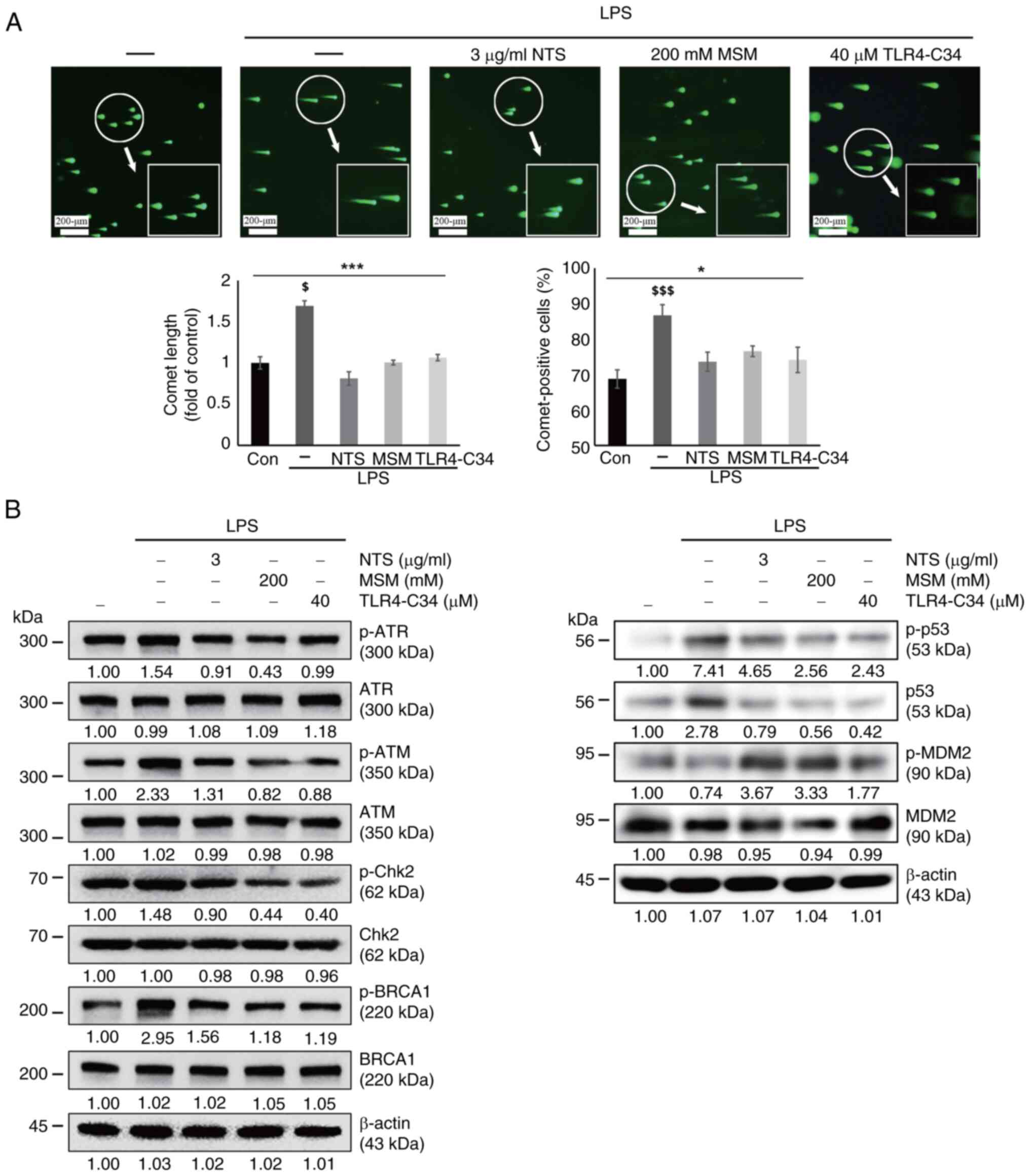 | Figure 5.Sulfur compounds induce DNA damage
response following LPS-induced DNA damage. (A) Images of the comet
assay were captured by fluorescence microscopy at ×10 and ×40
magnification levels, showing the fragmented DNA migrating out of
the nucleoid body, which formed a comet tail following treatment
with LPS (10 ng/ml) + NTS (3 µg/ml), MSM (200 mM) or TLR-C34 (40
µM) for 48 h. *P<0.05 and ***P<0.001; $P<0.05
vs. non-treated control; and $$$P<0.001 vs.
non-treated control (one-way ANOVA and Tukey's test). (B) Western
blot analysis of THP-1 cells; NTS (3 µg/ml) and MSM (200 mM)
inhibited the LPS-induced expression of p-ATM, p-ATR, p-Chk2,
p-BRCA1, and p-p53. However, the expression levels of p-MDM2 were
suppressed by LPS treatment, and were increased by NTS (3 µg/ml),
MSM (200 mM) or TLR4-C34 (40 µM). LPS, lipopolysaccharide; MSM,
methylsulfonylmethane; NTS, nontoxic sulfur; p-, phosphorylated;
TLR, Toll-like receptor. |
Sulfur molecules suppress the
expression of LPS-induced NF-κB and proinflammatory cytokines in
THP-1 cells
Two sulfur compounds downregulated the upstream
targets of NF-κB during LPS-induced inflammation; therefore, the
current analyzed the regulation of NF-κB under the same conditions.
The results indicated increased protein expression levels of COX-2
and NF-κB, but not COX-1, in response to LPS, whereas treatment
with MSM, NTS, and TLR4-C34 markedy reduced their expression levels
(Fig. 6A). The mRNA expression
levels of NF-κB and COX-2 were consistent with their protein
levels; sulfur molecules inhibited the LPS-induced expression of
NF-κB and COX-2, but not COX-1 (Fig.
6B). The present study also analyzed the expression of
proinflammatory cytokines, including IL-6, IL-1β and TNF-α, at the
protein and mRNA levels (Fig. 6C and
D); these were consistently upregulated by LPS and
downregulated by NTS, MSM and the TLR4 inhibitor. Next, the current
study evaluated the binding of NF-κB to the promoter region of
proinflammatory cytokines using ChIP assay, and a significant
inhibition of NF-κB binding to proinflammatory cytokines was
observed by natural sulfur compounds (Fig. 6E). The inhibition of the
LPS-induced nuclear translocation of NF-κB by NTS or MSM also
strongly supported the inhibitory mechanism against the LPS-induced
NF-κB-dependent inflammatory response (Fig. 6F). These results suggested that
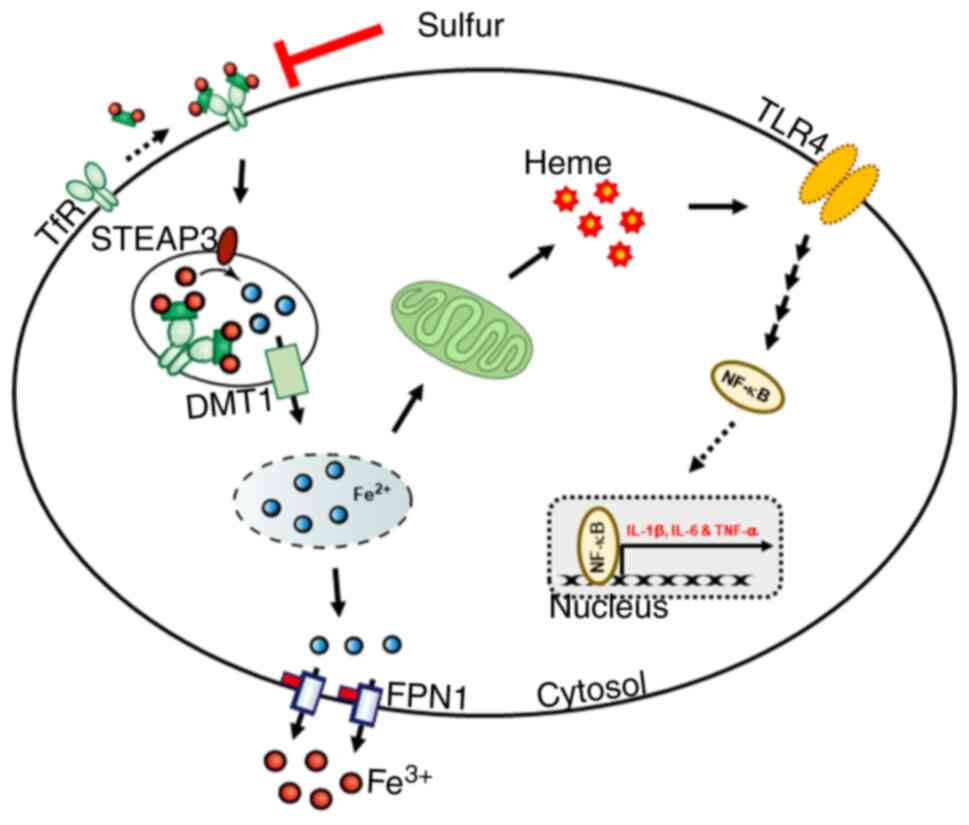
sulfur compounds may suppress LPS-induced inflammatory responses by
inhibiting iron homeostasis and heme biosynthesis in THP-1 human
monocytes (Fig. 7).
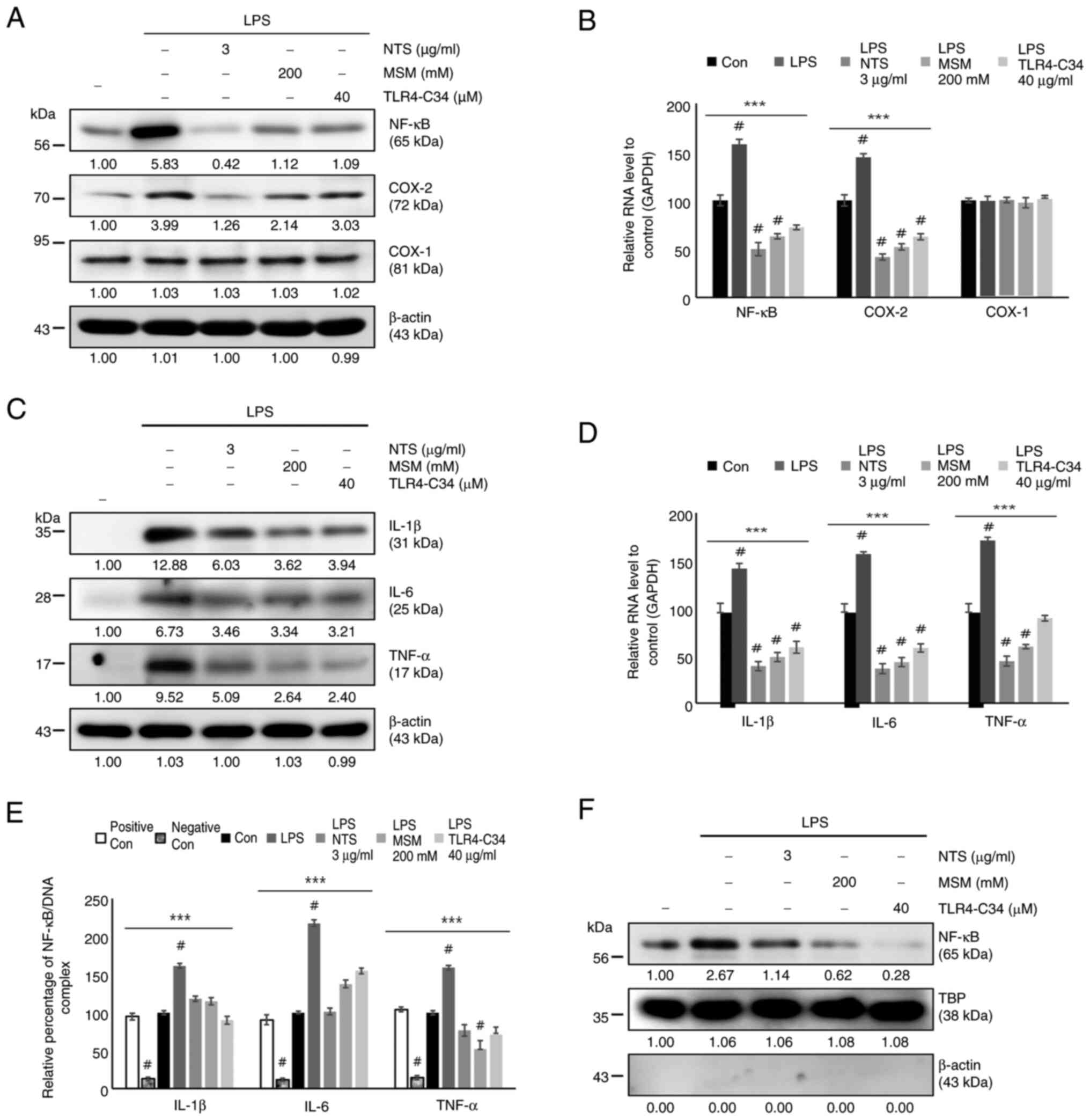 | Figure 6.Sulfur compounds inhibit LPS-induced
NF-κB, COX-2 and proinflammatory cytokine expression. (A) Western
blot analysis of the expression levels of NF-κB, COX-1 and COX-2 in
THP-1 cells treated with LPS (10 ng/ml) + NTS (3 µg/ml), MSM (200
mM) or TLR-C34 (40 µM) for 48 h. (B) RT-qPCR analysis of the
relative expression levels of NF-κB, COX-1 and COX-2 normalized to
GAPDH following treatment with LPS (10 ng/ml) + NTS (3 µg/ml), MSM
(200 mM) or TLR-C34 (40 µM) for 48 h. ***P<0.001;
#P<0.001 vs. non-treated control (one-way ANOVA and
Tukey's test). (C) Western blot analysis of the expression levels
of IL-1β, IL-6 and TNF-α in THP-1 cells treated with LPS (10 ng/ml)
+ NTS (3 µg/ml), MSM (200 mM) or TLR-C34 (40 µM) for 48 h. (D)
RT-qPCR analysis of the relative expression levels of IL-1β, IL-6
and TNF-α normalized to GAPDH in cells following treatment with LPS
(10 ng/ml) + NTS (3 µg/ml), MSM (200 mM) or TLR-C34 (40 µM) for 48
h. ***P<0.001; #P<0.001 vs. non-treated control
(one-way ANOVA and Tukey's test). (E) Chromatin immunoprecipitation
assay of THP-1 cells treated with LPS (10 ng/ml) + NTS (3 µg/ml),
MSM (200 mM) or TLR-C34 (40 µM) for 48 h showing the relative
binding of NF-κB to the promoters of IL-1β, IL-6 and TNF-α.
***P<0.001; #P<0.001 vs. non-treated control
(one-way ANOVA and Tukey's test). (F) Nuclear protein extract
analysis of THP-1 cells treated with LPS (10 ng/ml) + NTS (3
µg/ml), MSM (200 mM) or TLR-C34 (40 µM) for 48 h showing the
protein expression levels of nuclear NF-κB. TBP was used as the
housekeeping protein for the nuclear extract and β-actin was used
to show the efficacy of nuclear protein extraction. LPS,
lipopolysaccharide; MSM, methylsulfonylmethane; NTS, nontoxic
sulfur; RT-qPCR, reverse transcription-quantitative PCR; TBP,
TATA-binding protein; TLR, Toll-like receptor. |
Discussion
Natural compounds can effectively ameliorate
inflammatory responses as they generally lack side effects; they
also exhibit anti-inflammatory properties against LPS-induced
inflammation (33). MSM and NTS
exert anti-inflammatory effects, and these compounds may be
suitable candidates for anti-inflammatory treatment, similar to
artemisinin or artesunate, if they can inhibit the effects of
LPS-induced inflammation. In the present study, it was demonstrated
that NTS and MSM inhibited LPS-induced inflammation in THP-1 human
monocytes and that iron/heme metabolism may serve an important role
in this mechanism.
Iron metabolism is important to the inflammatory
process and is essential for human health. Notably, an imbalance in
iron transport can cause inflammation (34). LPS-treated THP-1 cells exhibit
elevated Fe2+ levels, suggesting an increased amount of
iron that must be oxidized. A failure in converting Fe2+
into Fe3+ for oxidation results in inflammation
(35). The present study observed
decreased Fe2+ levels following NTS and MSM treatment,
suggesting that these sulfur compounds can regulate iron transport
and inflammation. Generally, iron balance depends on FPN and TfR,
which mediate iron homeostasis through iron transport; whereas FPN
exports iron, TfR takes up Fe3+ ion for metabolism
(36). DMT1 is another membrane
transporter that transports iron from the endosomal system to the
cytosol and functions in intestinal iron absorption by oxidizing
Fe3+ before the translocation of Fe2+
(37,38). Inside the endosome, STEAP3 controls
iron transport by reducing Fe3+ to Fe2+
(39). Therefore, dysregulation in
the expression of these receptors may contribute to inflammation.
The present study observed the increased expression of TfR, FPN,
DMT1 and STEAP3 following LPS treatment, suggesting increased iron
transport. NTS and MSM inhibited the expression of these receptors,
which in turn, ameliorated iron transport and exerted
anti-inflammatory effects.
Iron is an important molecule for heme biosynthesis
and in the mitochondria, where it is essential for energy
production, antioxidant defense and signal transduction (40). Generally, ALAS1 is important for
heme synthesis, and combines glycine and succinyl-CoA to form
aminolaevulinic acid. The FECH enzyme is also involved in heme
synthesis by combining Fe2+ and protoporphyrin IX
(41). ABCB6 is a putative
promoter that contributes to the transition of ALA, and with the
help of ALAS1, FECH and MFRN, promotes iron uptake for its
interaction with FECH and ABCB10, which leads to heme synthesis
(42,43). The resulting heme is then
transferred to the cytoplasm with the help of a heme exporter,
FLVCR1 (44). Similar to the
present study, in a previous study, LPS has been shown to
upregulate the expression of HO-1, which catalyzes the degradation
of heme, and the expression of the proteins responsible for heme
synthesis, including ALAS1, FECH, ABCB6, MFRN, ABCB10 and FLVCR
(5). These results indicated that
LPS-induced inflammation may promote heme production. NTS and MSM
inhibited the expression of these proteins, which indicated an
inhibition of heme synthesis by these sulfur compounds. The TLR4
inhibitor did not alter the expression of these proteins,
indicating that heme synthesis may be independent of TLR4
expression.
TLR4 is a transmembrane receptor that mediates a
signaling response to inflammation and represents a major part of
the LPS-induced inflammatory response (45,46).
TLR4 transduces signals to the major downstream target NF-κB, which
is translocated into the nucleus upon activation to upregulate
proinflammatory cytokines (47,48).
The current study observed increased expression levels of TLR2 and
TLR4 during LPS treatment, which were significantly reduced by NTS
or MSM. These findings indicated that the anti-inflammatory effect
of these sulfur compounds may be mediated by regulating TLR
expression. A similar effect was also observed on NF-κB expression.
Furthermore, the inhibition of NF-κB expression by TLR4-C34
treatment indicated that these sulfur compounds act through
TLR4/NF-κB signaling.
Activation of NF-κB occurs through a canonical
pathway (49) or a PKC-dependent
pathway (50). The canonical
pathway depends on IκBα and IKKα/β, whereas the PKC-dependent
pathway signals through the ERK/p38 signaling pathway (11). Similar to the present study, in a
previous study, LPS has been reported to upregulate p-IKKαβ and
IκBα levels in the canonical pathway, whereas NTS or MSM
downregulated the expression of these molecules without affecting
the expression of total IκBα (11). In the PKC-dependent pathway,
LPS-induced inflammation increased the expression of p-PKCα, p-ERK
and p-p38 without altering the expression levels of their total
forms. By contrast, the natural sulfur compounds NTS and MSM
suppressed the LPS-induced expression of p-PKCα, p-ERK and p-p38
during TLR4-independent signaling. Upon inflammation, NF-κB is
translocated into the nucleus and induces the transcription of
genes encoding proinflammatory cytokines to elicit an immune
response (51). The current study
demonstrated that LPS-induced inflammation promoted the
translocation of NF-κB into the nucleus, where it may bind to the
promoters of the proinflammatory cytokines, IL-6, IL-1β and TNF-α.
Notably, NTS and MSM successfully inhibited the translocation of
NF-κB into the nucleus and blocked its binding to these
proinflammatory cytokine gene promoters. Furthermore, the
LPS-induced COX-2 expression was reversed by the two sulfur
compounds without affecting the expression of COX-1, indicating the
inhibition of the inflammatory response by NTS and MSM in a
COX-1-independent and TLR4-dependent manner.
LPS-induced inflammation generates ROS that leads to
oxidative damage (52). iNOS
induction also results in ROS generation, and LPS can induce iNOS
expression and ROS generation (53). The present results confirmed the
increase in iNOS mRNA and protein expression, as well as cellular
and mitochondrial ROS generation. Moreover, NTS and MSM suppressed
the expression levels of iNOS and ROS generation at the cellular
and mitochondrial levels. These results indicated the role of
oxidative stress in LPS-dependent inflammation and the effect of
sulfur compounds, which markedly reduced the inflammatory response
by suppressing oxidative stress. This prolonged oxidative stress
can cause DNA damage, and LPS has been shown to induce DNA
double-strand breaks to promote tumorigenesis (32). Anti-inflammatory drugs may inhibit
DNA damage by inducing DDR. In the present study, NTS and MSM
suppressed the formation of DNA strand breaks, as determined using
the comet assay, suggesting the induction of DDR by these sulfur
molecules. The downregulation of LPS-induced expression levels of
the proteins responsible for DDR by NTS or MSM also strongly
supported the anti-inflammatory effects of these sulfur molecules
by inhibiting LPS-induced oxidative stress. The present study also
observed that the natural sulfur molecules could decrease the
expression levels of MFRN, ABCB6, ABCB10, ALAS1, FECH and FLVCR,
which are associated with the iron/heme metabolism in THP1 cells,
whereas TLR4-C34 (TLR4 inhibitor) did not affect these expression
levels. In addition, it was confirmed that natural sulfur molecules
inhibited the TLR4/NF-κB-mediated inflammatory response. Although
the direct interaction between sulfurs and TLR4-C34 has not yet
been assessed, it may be hypothesized that sulfur molecules
decrease the inflammatory response by inhibiting iron/heme
metabolism prior to suppressing the TLR4/NF-κB pathway.
In conclusion, the natural sulfur compounds NTS and
MSM inhibited the LPS-induced inflammatory response in THP-1 human
monocytes. Iron/heme metabolism is important to the
anti-inflammatory activity of these sulfur compounds, which
includes the inhibition of LPS-induced expression of TLR4 and NF-κB
through canonical and PKC-dependent pathways, thus suppressing the
production of the proinflammatory cytokines, COX-2, IL-1β and IL-6.
In addition, NTS and MSM suppressed LPS-induced ROS production and
induced DDR in THP-1 human monocytes. Therefore, NTS and MSM may
have potential as therapeutic candidates for inflammatory diseases
caused by LPS, similar to artemisinin or artesunate.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Research Foundation of
Korea (NRF) grant funded by the Korean government (Ministry of
Science and ICT) to KJJ (grant no. RS-2024-00450676). This work was
also supported by the Basic Science Research Program to the
Research Institute for Basic Sciences of Jeju National University
through the NRF funded by the Ministry of Education to SWB (grant
no. 2019R1A6A1A10072987). Additionally, this research was supported
by a Korea Basic Science Institute (National Research Facilities
and Equipment Center) grant funded by the Ministry of Education
(grant no. 2023R1A6C101A045).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
KJJ designed the experiments and wrote the
manuscript. DYK and SWB performed all the experiments and analyzed
the data. KJJ, DYK and SWB confirm the authenticity of all the raw
data. All authors helped to revise the manuscript, and read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wessling-Resnick M: Iron homeostasis and
the inflammatory response. Annu Rev Nutr. 30:105–122. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Martins AC, Almeida JI, Lima IS, Kapitao
AS and Gozzelino R: Iron metabolism and the inflammatory response.
IUBMB Life. 69:442–450. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Alam Z, Devalaraja S, Li M, To TKJ,
Folkert IW, Mitchell-Velasquez E, Dang MT, Young P, Wilbur CJ,
Silverman MA, et al: Counter regulation of spic by NF-kappaB and
STAT signaling controls inflammation and iron metabolism in
macrophages. Cell Rep. 31:1078252020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang L, Harrington L, Trebicka E, Shi HN,
Kagan JC, Hong CC, Lin HY, Babitt JL and Cherayil BJ: Selective
modulation of TLR4-activated inflammatory responses by altered iron
homeostasis in mice. J Clin Invest. 119:3322–3328. 2009.PubMed/NCBI
|
|
5
|
Kang DY, Sp N, Jo ES, Lee JM and Jang KJ:
New insights into the pivotal role of iron/Heme metabolism in
TLR4/NF-κB Signaling-mediated inflammatory responses in human
monocytes. Cells. 10:25492021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wagener FA, Volk HD, Willis D, Abraham NG,
Soares MP, Adema GJ and Figdor CG: Different faces of the heme-heme
oxygenase system in inflammation. Pharmacol Rev. 55:551–571. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sp N, Kang DY, Jo ES, Lee JM, Bae SW and
Jang KJ: Pivotal role of iron homeostasis in the induction of
mitochondrial apoptosis by 6-Gingerol through PTEN regulated PD-L1
expression in embryonic cancer cells. Front Oncol. 11:7817202021.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nath KA, Vercellotti GM, Grande JP,
Miyoshi H, Paya CV, Manivel JC, Haggard JJ, Croatt AJ, Payne WD and
Alam J: Heme protein-induced chronic renal inflammation:
Suppressive effect of induced heme oxygenase-1. Kidney Int.
59:106–117. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wagener FADTG, Eggert A, Boerman OC, Oyen
WJ, Verhofstad A, Abraham NG, Adema G, van Kooyk Y, de Witte T and
Figdor CG: Heme is a potent inducer of inflammation in mice and is
counteracted by heme oxygenase. Blood. 98:1802–1811. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yucel G, Zhao Z, El-Battrawy I, Lan H,
Lang S, Li X, Buljubasic F, Zimmermann WH, Cyganek L, Utikal J, et
al: Lipopolysaccharides induced inflammatory responses and
electrophysiological dysfunctions in human-induced pluripotent stem
cell derived cardiomyocytes. Sci Rep. 7:29352017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sp N, Kang DY, Kim HD, Rugamba A, Jo ES,
Park JC, Bae SW, Lee JM and Jang KJ: Natural sulfurs inhibit
LPS-induced inflammatory responses through NF-κB signaling in
CCD-986Sk skin fibroblasts. Life-Basel. 11:4272021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Piktel E, Wnorowska U, Ciesluk M, Deptula
P, Pogoda K, Misztalewska-Turkowicz I, Paprocka P,
Niemirowicz-Laskowska K, Wilczewska AZ, Janmey PA and Bucki R:
Inhibition of inflammatory response in human keratinocytes by
magnetic nanoparticles functionalized with PBP10 peptide derived
from the PIP2-binding site of human plasma gelsolin. J
Nanobiotechnology. 17:222019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rafi MM, Yadav PN and Rossi AO:
Glucosamine inhibits LPS-induced COX-2 and iNOS expression in mouse
macrophage cells (RAW 264.7) by inhibition of p38-MAP kinase and
transcription factor NF-kappaB. Mol Nutr Food Res. 51:587–593.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bjorkbacka H, Kunjathoor VV, Moore KJ,
Koehn S, Ordija CM, Lee MA, Means T, Halmen K, Luster AD, Golenbock
DT and Freeman MW: Reduced atherosclerosis in MyD88-null mice links
elevated serum cholesterol levels to activation of innate immunity
signaling pathways. Nat Med. 10:416–421. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
van der Merwe M and Bloomer RJ: The
influence of methylsulfonylmethane on inflammation-associated
cytokine release before and following strenuous exercise. J Sports
Med (Hindawi Publ Corp). 2016:74983592016.PubMed/NCBI
|
|
16
|
Koh E and Surh J: Influence of sulfur
fertilization on the antioxidant activities of onion juices
prepared by thermal treatment. Prev Nutr Food Sci. 21:160–164.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Preetha NS, Kang DY and Darvin P:
Induction of ketosis condition and suppression using
methylsulfonylmethane by altering ANGPTL3 expression through STAT5b
signaling mechanism. Anim Cells Syst. 19:30–38. 2015. View Article : Google Scholar
|
|
18
|
P NS, Kang DY, Kim BJ, Joung YH, Darvin P,
Byun HJ, Kim JG, Park JU and Yang YM: Methylsulfonylmethane induces
G1 arrest and mitochondrial apoptosis in YD-38 gingival cancer
cells. Anticancer Res. 37:1637–1646. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kang DY, Sp N, Bae SW and Jang KJ:
Methylsulfonylmethane relieves cobalt chloride-induced hypoxic
toxicity in C2C12 myoblasts. Life Sci. 301:1206192022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
S PN, Darvin P, Yoo YB, Joung YH, Kang DY,
Kim DN, Hwang TS, Kim SY, Kim WS, Lee HK, et al: The combination of
methylsulfonylmethane and tamoxifen inhibits the Jak2/STAT5b
pathway and synergistically inhibits tumor growth and metastasis in
ER-positive breast cancer xenografts. BMC Cancer. 15:4742015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kim DH, Nipin SP, Kang DY, Jo ES, Rugamba
A, Jang KJ and Yang YM: Effect of methylsulfonylmethane on
proliferation and apoptosis of A549 lung cancer cells through G/M
Cell-cycle arrest and intrinsic cell death pathway. Anticancer Res.
40:1905–1913. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sousa-Lima I, Park SY, Chung M, Jung HJ,
Kang MC, Gaspar JM, Seo JA, Macedo MP, Park KS, Mantzoros C, et al:
Methylsulfonylmethane (MSM), an organosulfur compound, is effective
against obesity-induced metabolic disorders in mice. Metabolism.
65:1508–1521. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Miller L, Thompson K, Pavlenco C, Mettu
VS, Haverkamp H, Skaufel S, Basit A, Prasad B and Larsen J: The
effect of daily methylsulfonylmethane (MSM) consumption on
High-density lipoprotein cholesterol in healthy overweight and
obese adults: A randomized controlled trial. Nutrients.
13:36202021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Miller LE: Methylsulfonylmethane decreases
inflammatory response to tumor necrosis factor-alpha in cardiac
cells. Am J Cardiovasc Dis. 8:31–38. 2018.PubMed/NCBI
|
|
25
|
Caron JM, Bannon M, Rosshirt L, Luis J,
Monteagudo L, Caron JM and Sternstein GM: Methyl sulfone induces
loss of metastatic properties and reemergence of normal phenotypes
in a metastatic cloudman S-91 (M3) murine melanoma cell line. PLoS
One. 5:e117882010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kim YB, Lee SH, Kim DH, Lee HG, Choi Y,
Lee SD and Lee KW: Effects of dietary organic and inorganic sulfur
on laying performance, egg quality, ileal morphology, and
antioxidant capacity in laying hens. Animals (Basel). 12:872021.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lee JS, Kwon JK, Han SH, An IJ, Kim SJ,
Lee SH, Park YS, Park BK, Kim BS, Kim S, et al: Toxicity study of
detoxication sulphur at 3 months post-treatment in rats. J Food
Hygiene Safety. 25:263–268. 2010.
|
|
28
|
Kang DY, Sp N, Jo ES, Kim HD, Kim IH, Bae
SW, Jang KJ and Yang YM: Non-toxic sulfur enhances growth hormone
signaling through the JAK2/STAT5b/IGF-1 pathway in C2C12 cells. Int
J Mol Med. 45:931–938. 2020.PubMed/NCBI
|
|
29
|
Kang DY, Sp N, Jo ES, Rugamba A, Kim HD,
Kim IH, Park JC, Bae SW, Jang KJ and Yang YM: Non-toxic sulfur
inhibits LPS-induced inflammation by regulating TLR-4 and
JAK2/STAT3 through IL-6 signaling. Mol Med Rep. 24:4852021.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jo ES, Sp N, Kang DY, Rugamba A, Kim IH,
Bae SW, Liu Q, Jang KJ and Yang YM: Sulfur compounds inhibit high
Glucose-induced inflammation by regulating NF-κB signaling in human
monocytes. Molecules. 25:23422020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Qiao W, Huang Y, Bian Z, Sun X, Wang X,
Gao Q, Peng Y and Meng L: Lipopolysaccharide-induced DNA damage
response activates nuclear factor κB signalling pathway via GATA4
in dental pulp cells. Int Endod J. 52:1704–1715. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lee DY, Li H, Lim HJ, Lee HJ, Jeon R and
Ryu JH: Anti-inflammatory activity of sulfur-containing compounds
from garlic. J Med Food. 15:992–999. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Soares P, Silva C, Chavarria D, Silva FSG,
Oliveira PJ and Borges F: Drug discovery and amyotrophic lateral
sclerosis: Emerging challenges and therapeutic opportunities.
Ageing Res Rev. 83:1017902023. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Osterholm EA and Georgieff MK: Chronic
inflammation and iron metabolism. J Pediatr. 166:1351–1357.e1.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang MW, Yang G, Zhou YF, Qian C, Mu MD,
Ke Y and Qian ZM: Regulating ferroportin-1 and transferrin
receptor-1 expression: A novel function of hydrogen sulfide. J Cell
Physiol. 234:3158–3169. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Latunde-Dada GO, Van der Westhuizen J,
Vulpe CD, Anderson GJ, Simpson RJ and McKie AT: Molecular and
functional roles of duodenal cytochrome B (Dcytb) in iron
metabolism. Blood Cells Mol Dis. 29:356–360. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fleming MD, Romano MA, Su MA, Garrick LM,
Garrick MD and Andrews NC: Nramp2 is mutated in the anemic Belgrade
(b) rat: Evidence of a role for Nramp2 in endosomal iron transport.
Proc Natl Acad Sci USA. 95:1148–1153. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Byrne SL, Krishnamurthy D and
Wessling-Resnick M: Pharmacology of iron transport. Annu Rev
Pharmacol Toxicol. 53:17–36. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Richardson DR, Lane DJR, Becker EM, Huang
ML, Whitnall M, Suryo Rahmanto Y, Sheftel AD and Ponka P:
Mitochondrial iron trafficking and the integration of iron
metabolism between the mitochondrion and cytosol. Proc Natl Acad
Sci USA. 107:10775–10782. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chiabrando D, Mercurio S and Tolosano E:
Heme and erythropoieis: More than a structural role. Haematologica.
99:973–983. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen W, Paradkar PN, Li LT, Pierce EL,
Langer NB, Takahashi-Makise N, Hyde BB, Shirihai OS, Ward DM,
Kaplan J and Paw BH: Abcb10 physically interacts with mitoferrin-1
(Slc25a37) to enhance its stability and function in the erythroid
mitochondria. Proc Natl Acad Sci USA. 106:16263–16268. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen W, Dailey HA and Paw BH:
Ferrochelatase forms an oligomeric complex with mitoferrin-1 and
Abcb10 for erythroid heme biosynthesis. Blood. 116:628–630. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chiabrando D, Marro S, Mercurio S, Giorgi
C, Petrillo S, Vinchi F, Fiorito V, Fagoonee S, Camporeale A, Turco
E, et al: The mitochondrial heme exporter FLVCR1b mediates
erythroid differentiation. J Clin Invest. 122:4569–4579. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yu B, Li Q and Zhou M: LPS-induced
upregulation of the TLR4 signaling pathway inhibits osteogenic
differentiation of human periodontal ligament stem cells under
inflammatory conditions. Int J Mol Med. 43:2341–2351.
2019.PubMed/NCBI
|
|
46
|
Ngkelo A, Meja K, Yeadon M, Adcock I and
Kirkham PA: LPS induced inflammatory responses in human peripheral
blood mononuclear cells is mediated through NOX4 and Giα dependent
PI-3kinase signalling. J Inflamm (Lond). 9:12012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xiang PJ, Chen T, Mou Y, Wu H, Xie P, Lu
G, Gong X, Hu Q, Zhang Y and Ji H: NZ suppresses TLR4/NF-κB
signalings and NLRP3 inflammasome activation in LPS-induced
RAW264.7 macrophages. Inflamm Res. 64:799–808. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zusso M, Lunardi V, Franceschini D,
Pagetta A, Lo R, Stifani S, Frigo AC, Giusti P and Moro S:
Ciprofloxacin and levofloxacin attenuate microglia inflammatory
response via TLR4/NF-kB pathway. J Neuroinflammation. 16:1482019.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Sun SC: The non-canonical NF-κB pathway in
immunity and inflammation. Nat Rev Immunol. 17:545–558. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Dasu MR, Devaraj S and Jialal I: High
glucose induces IL-1beta expression in human monocytes: Mechanistic
insights. Am J Physiol Endocrinol Metab. 293:E337–E346. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Lee JS and Surh YJ: Nrf2 as a novel
molecular target for chemoprevention. Cancer Lett. 224:171–184.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhang XQ, Wang CH, Shan S, Liu XY, Jiang
ZM and Ren T: TLR4/ROS/miRNA-21 pathway underlies
lipopolysaccharide instructed primary tumor outgrowth in lung
cancer patients. Oncotarget. 7:42172–42182. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Lee CW, Kim SC, Kwak TW, Lee JR, Jo MJ,
Ahn YT, Kim JM and An WG: Anti-Inflammatory effects of
Bangpungtongsung-San, a traditional herbal prescription. Evid Based
Complement Alternat Med. 2012:8929432012. View Article : Google Scholar : PubMed/NCBI
|