Introduction
The occurrence and development of gastric cancer
(GC) are predominantly associated with Helicobacter pylori
infection (1–3). Recent advances in medical detection
technologies, along with innovative approaches to GC treatment,
have resulted in a 5-year survival rate approaching 95% for
individuals diagnosed with early-stage cancer; however, the
majority of patients with GC are diagnosed with advanced stage
cancer, resulting in an unfavorable prognosis and a low 5-year
survival rate (1). The primary
challenge hindering timely diagnoses lies in the absence of
screening tools possessing suitable sensitivity, specificity and
practicality (1). The management
of advanced GC primarily encompasses targeted drug therapy,
immunotherapy and neoadjuvant chemotherapy (2). Unlike directly targeting the tumor
itself, the objective of immunotherapy is to overcome
immunosuppression induced by the tumor microenvironment to allow
the innate immune system to target and eliminate cancer cells
(2). Stromal cells, immune cells,
vascular endothelium and intravascular blood cells are all present
in tumor tissues, along with tumor cells. Immune cells contribute
to the tumor microenvironment and perform a crucial function, known
as tumor immune infiltration (3).
The presence of immune cells infiltrating into the
tumor microenvironment is associated with the proliferation,
invasion and spread of tumor cells. The various forms of immune
cells that infiltrate into tumor tissues have a strong association
with tumor classification and have a substantial impact on the
response to immunotherapy, as well as the survival and prognosis of
patients (4). The immune cells
that infiltrate the tumor microenvironment not only attack cancer
cells to prevent their growth, but also filter out which cancer
cells are capable of surviving in the environment, potentially
facilitating the progression of the tumor. Tumor immunotherapy is a
crucial area of current research in cancer therapeutics.
Distinguishing the specific type of immune infiltration in tumors
can potentially indicate the efficacy of tumor immunotherapy
(5). Immunotherapy based on immune
checkpoint inhibitors has resulted in a new era of tumor treatment;
therefore, identifying biomarkers to predict which patients are
likely to benefit from tumor immunotherapy is important (6,7).
Nucleotide-binding oligomerization domain, leucine
rich repeat and pyrin domain containing (NLRP) proteins form
inflammasomes, which serve a crucial role in innate immunity and
inflammatory responses (8). As an
inflammatory response sensor signaling protein, NLRP mediates the
activation of proteases to induce cytokine maturation and
contributes to pyroptosis (9).
Different NLRP members are expressed in various organs and tissues,
participating in the occurrence and progression of different types
of cancer by regulating innate and adaptive immune responses, cell
death and proliferation (10).
Among the inflammasome family members, NLRP3 is the most important
member and is involved in a range of regulatory mechanisms
(9). Certain cytokines, such as
IL-1β precursors, require caspase activation by NLRP inflammasomes
in the cytoplasm, particularly NLRP3 (11), which activates caspase enzymes via
adaptor protein ASC for cleaving and activating IL-1β/IL-18
precursor secretion. In a previous study, the CagA virulence factor
from H. pylori was introduced into gastric mucosal cells,
altering the Wnt/β-catenin signaling pathway, thus enhancing the
expression of related transcription factors and promoting the
sustained high expression of NLRP3 (12). Notably, the expression and
activation of the NLRP3 inflammasome can facilitate the
proliferation, invasion and metastasis of cancer cells, thereby
exerting a notable impact on the initiation and progression of
several types of cancer (13,14).
NLRP3 is a potent tumor suppressor in colitis-induced colorectal
cancer, effectively inhibiting liver metastasis by increasing the
anticancer activity of natural killer (NK) cells (15,16).
Understanding the potential role of the NLRP3 inflammasome in
tumorigenesis and immunomodulation, and its functional mechanism,
may reveal relationships with specific types of tumors.
The present study aimed to examine the differential
gene expression caused by H. pylori infection and the
distribution patterns of the NLRP3 inflammasome in GC tissues. The
correlation between NLRP3 status and immune cell composition in the
tumor microenvironment of GC was also examined, and the effect of
NLRP3 on immune infiltration was analyzed to better understand the
mechanisms taking place during GC immunotherapy. Additionally, the
association between NLRP3 expression levels, immune infiltration
and the clinical prognosis of patients with GC was analyzed by
integrating data from online public databases and clinical
specimens.
Materials and methods
Datasets
The Cancer Genome Atlas (TCGA; http://portal.gdc.cancer.gov/) and Gene
Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/gds/) databases were used
to obtain GC data. The data obtained included gene sequencing and
RNA expression data, and relevant clinical information, such as
clinical stage, pathological classification and survival status.
Cases with incomplete information were excluded from the analysis.
A total of 412 sets of clinical data for patients with GC and data
from 36 normal tissues were downloaded from TCGA. Among these
cases, 157 exhibited H. pylori infection, whereas 20 cases
showed no evidence of H. pylori infection. The primary
dataset included patient characteristics such as age, sex, tumor
stage, histological grade, initial treatment response,
progression-free survival time and overall survival (OS) time. To
complement the bioinformatics analysis, a transcriptome dataset
from the GEO was obtained (GSE36968) (17) and GSE26253 (18). The majority of GC cases analyzed in
the databases mentioned were H. pylori-associated GC. The
levels of NLRP3 expression among various types of tumors were
assessed using TIMER2.0 datasets (19). The association between NLRP3
expression and infiltration of immune cell types in GC was also
assessed using NLRP3 expression data obtained from the TIMER2.0
database.
Clinical GC specimens
A total of 65 GC tissues and their adjacent normal
tissues were collected from patients diagnosed with GC between May
2018 and June 2023. These patients were recruited from the
Gastroscopy Room, Department of Oncology and Radiotherapy, and
Department of General Surgery at the Affiliated Changshu Hospital
of Nantong University (Changshu, China). GC cases obtained before
2019 were sourced from the biobank established by the hospital. The
remaining GC specimens were directly collected from the clinical
departments of the hospital. All the patients had provided written
informed consent for medical research and were suitable for the
study. Out of the 65 patients, 34 were male and 31 were female. The
median age was 65.7 years (40.2–81.3 years). All patients with
superficial gastritis were identified using gastroscopy. The
presence of H. pylori infection was verified using a rapid
urease test (RUT) (20). GC or
precancerous lesions were surgically extracted; two clinically
experienced pathologists assessed the histological subtypes and
clinical stages based on the World Health Organization
classification criteria (21) and
pathological test results. Patients with incomplete standard
clinical data, multiple organ failure, another primary tumor or
those who had incomplete follow-up data were excluded from the
study. Written informed consent for the use of samples for research
purposes was obtained from each participant and the Ethics
Committee of the Affiliated Changshu Hospital of Nantong University
(Changshu Second People's Hospital) approved the study (approval
no. 2019-KY-049). Pathological characteristics, including tumor
size, tumor location and pathological type were recorded after the
initial diagnosis. GC and paracancerous tissues from surgically
obtained samples were promptly frozen in liquid nitrogen
containers.
NLRP3 mRNA expression levels
NLRP3 mRNA expression levels in clinical samples
were quantified by reverse transcription-quantitative PCR. Briefly,
total RNA was extracted from GC tissues using TRIzol® Reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocols. The obtained RNA was reverse-transcribed
into cDNA using an RT kit (Cat #. C11027-2; Guangzhou RiboBio Co.,
Ltd.) at 42°C for 30 min. TaqMan Gene Expression Assay primers
(cat. no. 4331182; Thermo Fisher Scientific, Inc.) were used for
NLRP3 (assay ID Hs00918080_g1) and GAPDH (assay ID Hs02786624_g1)
expression. In the Hs00918080_g1 assay kit, the forward and reverse
primers for NLRP3 were provided as 5′-AGTGGCTGGTGGTGGCTGGT-3′ and
5′-CAGGTGCTGCAGGTGCTGC-3′, respectively, and qPCR amplification was
performed using a Taq Pro Universal SYBR qPCR Master Mix kit (cat.
no. Q712-02; Vazyme Biotech Co., Ltd.) under the following
conditions: Initial denaturation at 95°C for 5 min, followed by 35
cycles of denaturation at 95°C for 30 sec, annealing at 58°C for 30
sec and extension at 72°C for 30 sec, with a final extension step
at 72°C for 5 min. All of the reactions were run in triplicate. The
cycle quantification (Cq) data were determined using fixed
threshold settings. A comparative Cq was used to compare each
condition to the control. The relative mRNA expression levels of
NLRP3 normalized to the control were calculated with the
2−ΔΔCq method (22).
Association between NLRP3 and
pathological characteristics
Kaplan-Meier Plotter (http://www.kmplot.com; n=875 patients with GC) was
used to explore the association between NLRP3 expression levels and
clinical survival. The results are presented as survival curves
showing the hazard ratios (HRs) and P-values were calculated using
the log-rank test. The association between NLRP3 expression and the
clinical characteristics of patients with GC was conducted using
data from the clinical patients.
Acquisition of hub genes
A protein-protein interaction (PPI) network of NLRP3
was constructed using the STRING database (https://www.string-db.org/).
Western blotting
Total protein was extracted from GC specimens using
Enhanced RIPA Lysis buffer (cat. no. C5029; BIOSS) and protein
concentration was measured using a BCA assay (cat. no. P00105;
Beyotime Institute of Biotechnology). Protein samples (40 µg/lane)
were separated by SDS-PAGE on an 8% SDS-gel (cat. no. P0012AC;
Beyotime Institute of Biotechnology) and were transferred to PVDF
membranes (Immobilon®-P; MilliporeSigma). The membranes were then
blocked using blocking solution (cat. no. P0023B; Beyotime
Institute of Biotechnology) for 1 h at room temperature and then
incubated with primary antibodies at 4°C overnight. The following
primary antibodies were used in the present study: Rabbit
anti-NLRP3 (1:500; cat. no. ab263899; Abcam) and anti-β-actin
(1:1,000; ab8226; Abcam). Subsequently, membranes were washed with
TBS-Tween (0.1%) and were incubated with horseradish peroxidase
(HRP)-conjugated secondary antibodies (1:5,000; cat. no. TA373083;
OriGene Technologies, Inc.) for 1 h at room temperature. Finally,
signals were visualized using a BeyoECL Plus kit (cat. no. P00185;
Beyotime Institute of Biotechnology) using a ChemiDoc™ MP Imaging
system (Bio-Rad Laboratories, Inc.).
Immunohistochemistry (IHC)
GC tissues and adjacent normal tissues were fixed in
4% paraformaldehyde at room temperature for 24 h, dehydrated
through a graded ethanol series (70, 80, 95 and 100%), cleared in
xylene, embedded in paraffin and sectioned into ~4-µm slices. To
dewax the tissues, they were immersed in xylene twice (10 min
each), followed by hydration for 5 min using a decreasing series of
alcohol solutions. Following antigen retrieval using EDTA,
endogenous peroxidase activity was quenched using 3% hydrogen
peroxide. The cells were then blocked using 1% BSA (cat. no. 0332;
Amresco, LLC) at 37°C for 1 h. Primary anti-human antibodies [NLRP3
(1:500; cat. no. ab263899), CD3 (1:100; cat. no. ab25109), CD4
(1:200; cat. no. ab317787), CD19 (1:500; cat. no. ab245235), CD21
(1:100; cat. no. ab227662) and CD206 (1:500; cat. no. ab64693); all
from Abcam] were then added to the tissues and incubated at 4°C
overnight. Subsequently, the tissues were washed three times with
PBS (5 min each) and were incubated with HRP-labeled goat
anti-rabbit IgG (1:500; cat. no. TA373083; OriGene Technologies,
Inc.) and HRP-labeled goat anti-mouse IgG (1:500; cat. no.
TA373082; OriGene Technologies, Inc.) at 37°C for 1 h. The tissues
were washed a further three times with PBS (5 min each) and DAB
chromogenic solution was used for color development. The nuclei
were stained with hematoxylin at 25°C for 30 min. The staining
results were observed under a light microscope, and the areas of
positive cell staining were subjected to grayscale analysis
(Quantity One 4.62; Bio-Rad Laboratories, Inc.).
RUT
Biopsy specimens of the gastric antrum mucosa,
obtained 3–5 cm away from the pylorus during gastroscopy
examination, were promptly immersed in RUT detection reagent
(Shandong Biomedia Laboratories Co., Ltd.) for 5 min at 37°C. The
change in color of the reagent was directly observed and compared
against the corresponding standard. If the phenol red in the
reagent transitioned from yellow to red, indicating an elevated pH
level, this suggested the presence of H. pylori in the
tissue, whereas no change in color indicated the absence of H.
pylori.
Gene enrichment in GC
The R software clusterProfiler package (23) was used to perform Gene Ontology
(GO) and KEGG pathway enrichment analysis for TCGA GC database. The
median NLRP3 expression levels were used as a threshold to examine
the differences in gene enrichment between the groups with high and
low NLRP3 expression based on TCGA database. Gene Set Enrichment
Analysis (GSEA; http://www.gsea-msigdb.org/) was performed using the
clusterProfiler software package (23). The immune cell gene symbol file
(h.all.v2023.2.Hs.symbols.gmt) was obtained from GSEA. Meanwhile
KEGG (https://www.kegg.jp/) enrichment
analysis was performed on the proteins in the PPI network to
identify the pathways associated with the direct interaction
targets in the network. Statistical analysis was performed on the
functional clustering results and a KEGG pathway enrichment bar
chart was generated to visualize the enriched pathways.
Immune cell infiltration and
co-expression analysis
The CIBERSORT tool (https://ciberxortx.stanford.edu/) in R studio (version
4.3.1) (24) enables calculations
for 22 immune cell proportions in tumor tissues using the inverse
convolution approach. Immune cell proportions with P<0.05,
indicating statistical significance, were included in the
subsequent analysis The gastric tumors in TCGA database were
classified into high- and low-expression groups using the median
NLRP3 mRNA expression level as the threshold. Subsequently, the
potential impact of NLRP3 gene expression on the infiltration of
immune cells in GC tissues was analyzed. The TIMER2.0 database was
used to examine the association between NLRP3 expression and the
prevalence of four types of tumor-associated macrophages
(macrophage, monocyte, M1 macrophage and M2 macrophage) that
infiltrated GC tumors. Co-expression Pearson correlation analysis
was performed between NLRP3 and immune checkpoint genes using GC
RNA-sequencing (RNA-seq) data from TCGA. The data were analyzed
using the R graph visualization software ggplot2 (https://cran.r-project.org/).
Immune infiltration survival
analysis
GC RNA-seq data from TCGA were combined with
clinical GC tissue data to analyze the relationship between
different immune cell infiltrates and patient survival based on the
immune cell infiltrates that were significantly influenced by
NLRP3. Survival analysis was performed by plotting survival curves
for TCGA GC using the survminer package (https://cran.r-project.org/web/packages/survminer/index.html).
Statistical analysis and
visualization
R version 4.1.2 was used for bioinformatics
analysis. A Wilcoxon rank-sum test was used to evaluate the
proportions of immune cell infiltrates in GC tissues between the
NLRP3 high-expression and low-expression groups. The NLRP3
expression data were ranked from low to high, and the median of the
data was used for statistical calculation of P-values and HR values
in survival and survminer packages. All data met the minimum value
required for statistical testing (i.e., for data that conforms to a
normal distribution, the minimum sample size for each group should
be >5). Spearman's correlation analysis was performed to assess
the correlation between NLRP3 expression and immune cell
infiltrates. GSEA and visualization was performed using
clusterProfiler and ggplot2 packages. The Fisher's exact test was
performed to analyze the relationship between gene expression
levels and clinicopathological parameters. Data are presented as
the mean ± SD. Intergroup comparisons were performed using a paired
or unpaired Student's t-test, depending on whether the groups were
paired or independent. P<0.05 was considered to indicate a
statistically significant difference.
Results
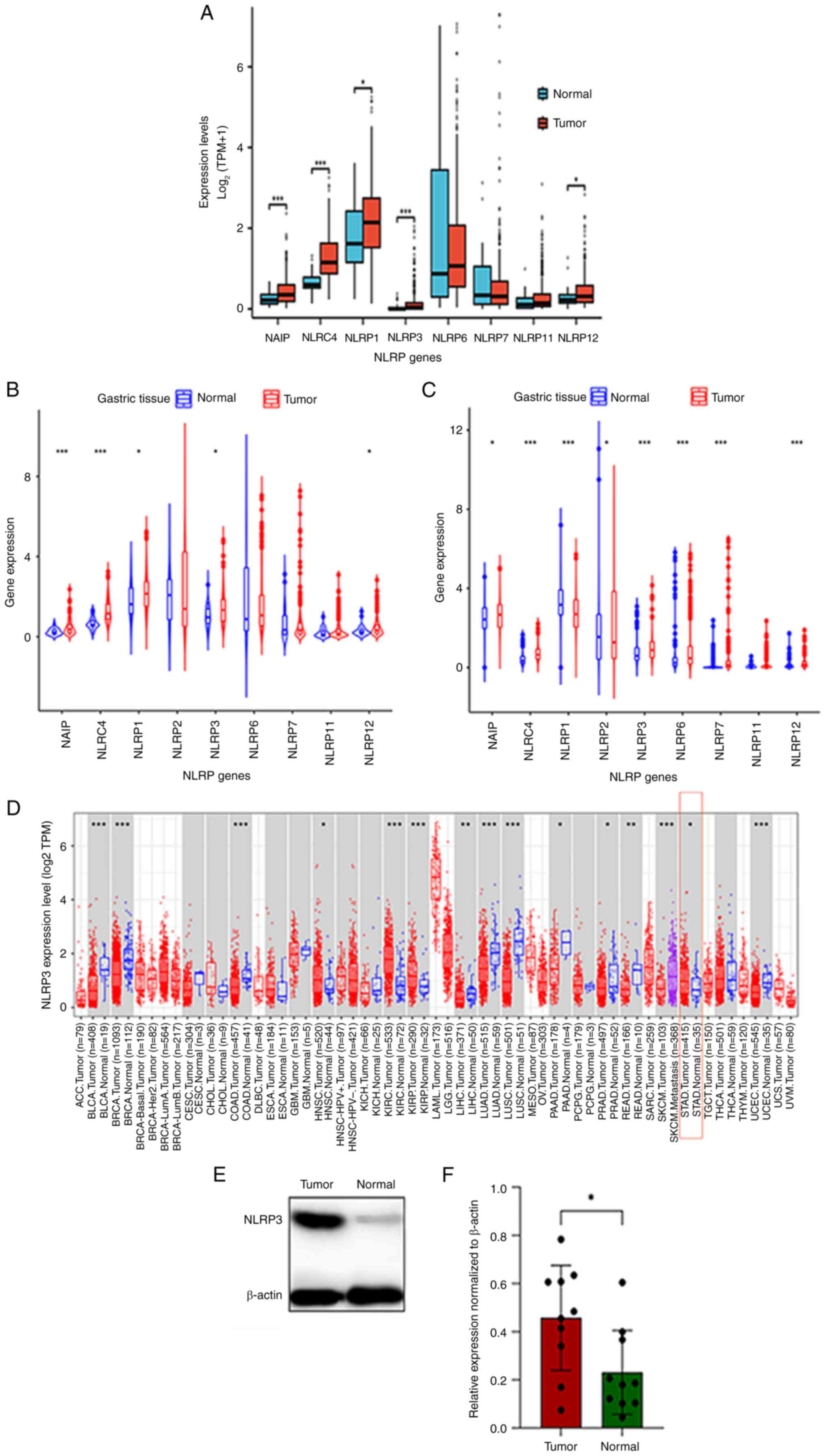
Expression profile of the NLRP
inflammasome in GC
The NLRP family in TCGA datasets consists of a wide
variety of inflammasomes. In TCGA, the expression of several NLRP
inflammasomes was assessed; specifically, NAIP, NLRC4, NLRP1,
NLRP3, NLRP6, NLRP7, NLRP11 and NLRP12. The expression levels of
NAIP, NLRC4, NLRP1, NLRP3 and NLRP12 in GC tissues were
significantly higher than those in adjacent normal tissues
(Fig. 1A). GC datasets (GSE36968
and GSE26253) from the GEO were also analyzed (Fig. 1B and C). GSE36968 included 407 GC
tissues and 32 paracancerous tissues, and GSE26253 consisted of 414
GC tissues and 36 paracancerous tissues. Compared with in the
paracancerous normal tissues, the expression levels of NLRP3 were
significantly greater in GC tissues. Although NLRP3 was not the
only upregulated NLRP protein, only NAIP, NLRC4 and NLRP3 were
upregulated in GC tissues in both GEO datasets and TCGA datasets.
To thoroughly investigate the relationship between NLRP3 and the
development of tumors, NLRP3 expression levels in different types
of tumors were analyzed. Fig. 1D
shows that there were differences in the levels of NLRP3 expression
among various types of tumors in TIMER 2.0 datasets. Downregulation
of NLRP3 was observed in nine tumor types, namely, bladder
urothelial carcinoma, bladder urothelial carcinoma, colon
adenocarcinoma, liver hepatocellular carcinoma, pancreatic
adenocarcinoma, prostate adenocarcinoma, rectum adenocarcinoma and
uterine corpus endometrial carcinoma, compared with in the
paracancerous tissues from the patients with cancer. By contrast,
increased expression of NLRP3 was observed in head and neck
squamous cell carcinoma, renal clear cell carcinoma, kidney renal
papillary cell carcinoma and stomach adenocarcinoma (STAD). Further
analysis of NLRP3 expression in 10/65 sets of clinical GC tissues
and surrounding normal tissues was subsequently performed. Using
western blotting, it was demonstrated that the protein expression
levels of NLRP3 in GC tissues were higher than in those in normal
tissues (Fig. 1E and F). These
results indicated that NLRP3 may be significantly upregulated in
GC, and could therefore be involved in the development and/or
progression of GC.
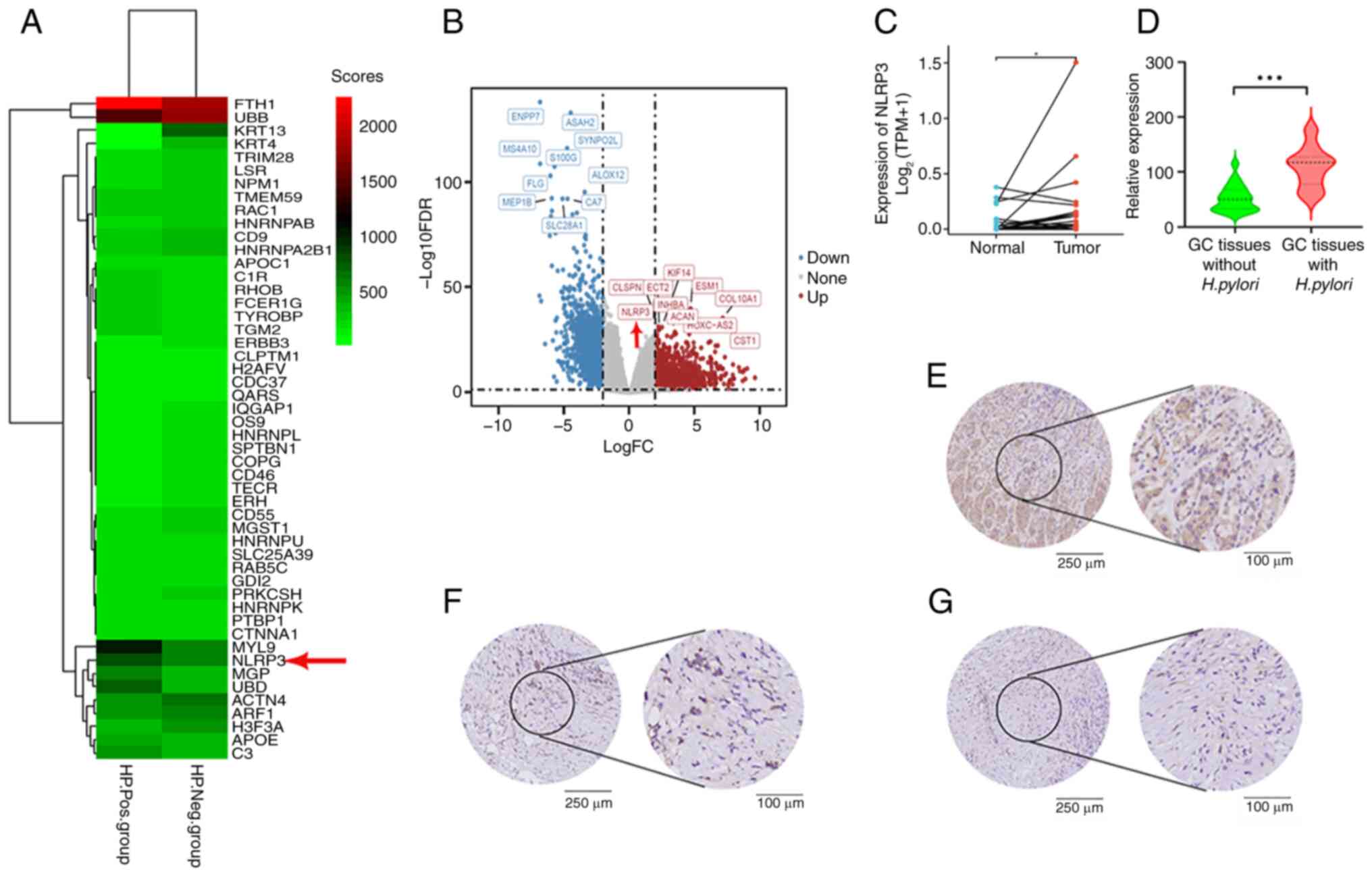
H. pylori infection promotes NLRP3
expression in GC
A total of 157 cases of GC that tested positive for
H. pylori infection and 20 cases of GC that tested negative
were obtained from the RNA-seq database of TCGA. Fig. 2A shows a comparison and analysis of
the differential gene expression between these two groups. Some of
the top 50 genes displayed considerable variations in their
expression levels. Specifically, certain GC tissues presented
increased expression of the NLRP3 gene as a result of H.
pylori infection. A volcano plot revealed that among the top 10
highly expressed genes in GC tissues positive for H. pylori
infection, NLRP3 was enriched (Fig.
2B). Furthermore, IHC revealed high expression of NLRP3 in
selected clinical GC tissues with H. pylori infection
(Fig. 2D). Notably, NLRP3
expression was elevated in H. pylori-infected GC tissues
compared with in uninfected tissues, indicating that H.
pylori infection might not be the only factor responsible for
its upregulation (Fig. 2E-G).
Furthermore, examination of NLRP3 mRNA transcription levels in GC
tissues revealed a significant increase compared with that in
normal gastric mucosa tissues from the same patients (Fig. 2C). The results of IHC consistently
revealed considerably high NLRP3 expression in GC tissues infected
with H. pylori (Fig.
2D).
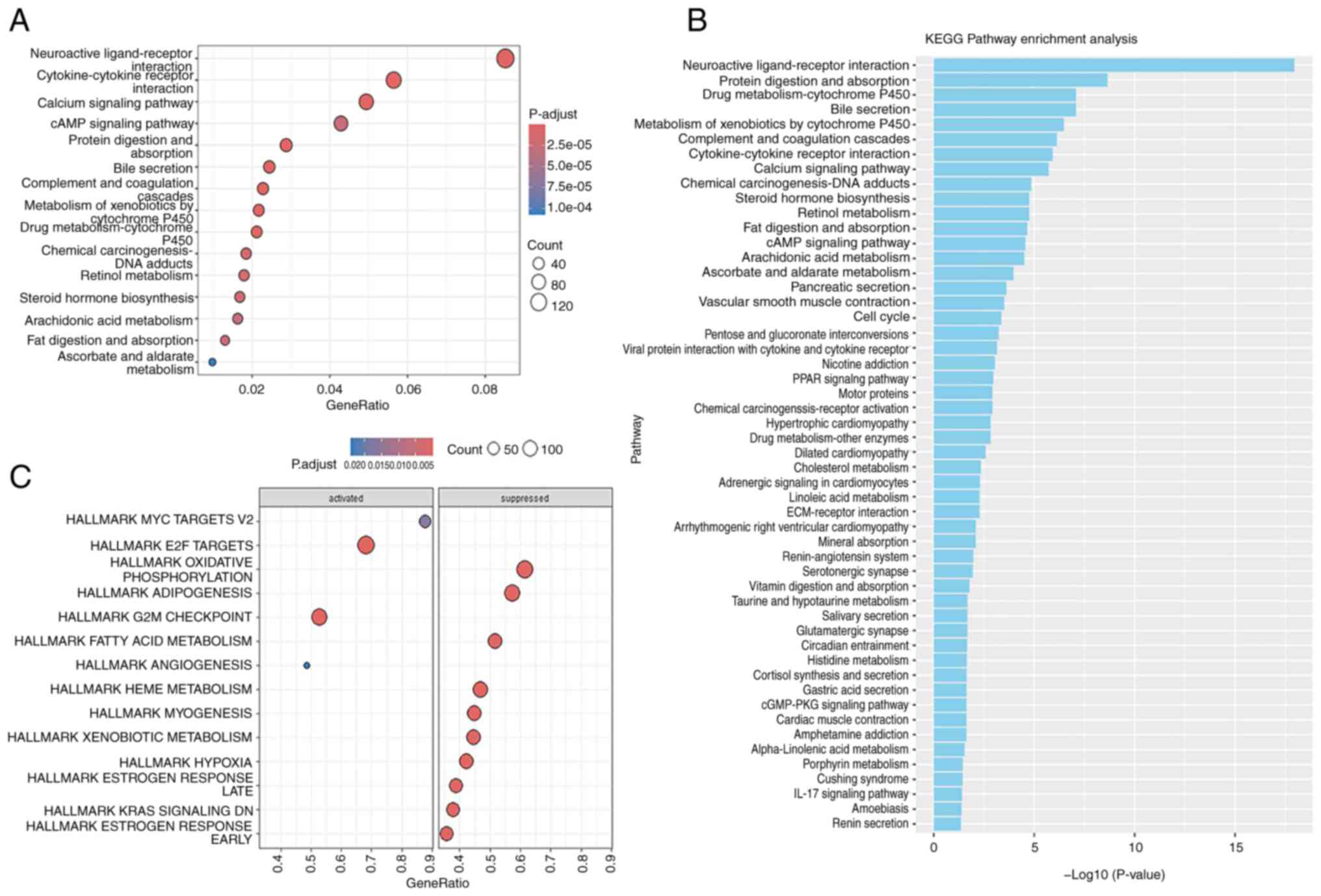
Functional enrichment analysis of
expressed genes in GC
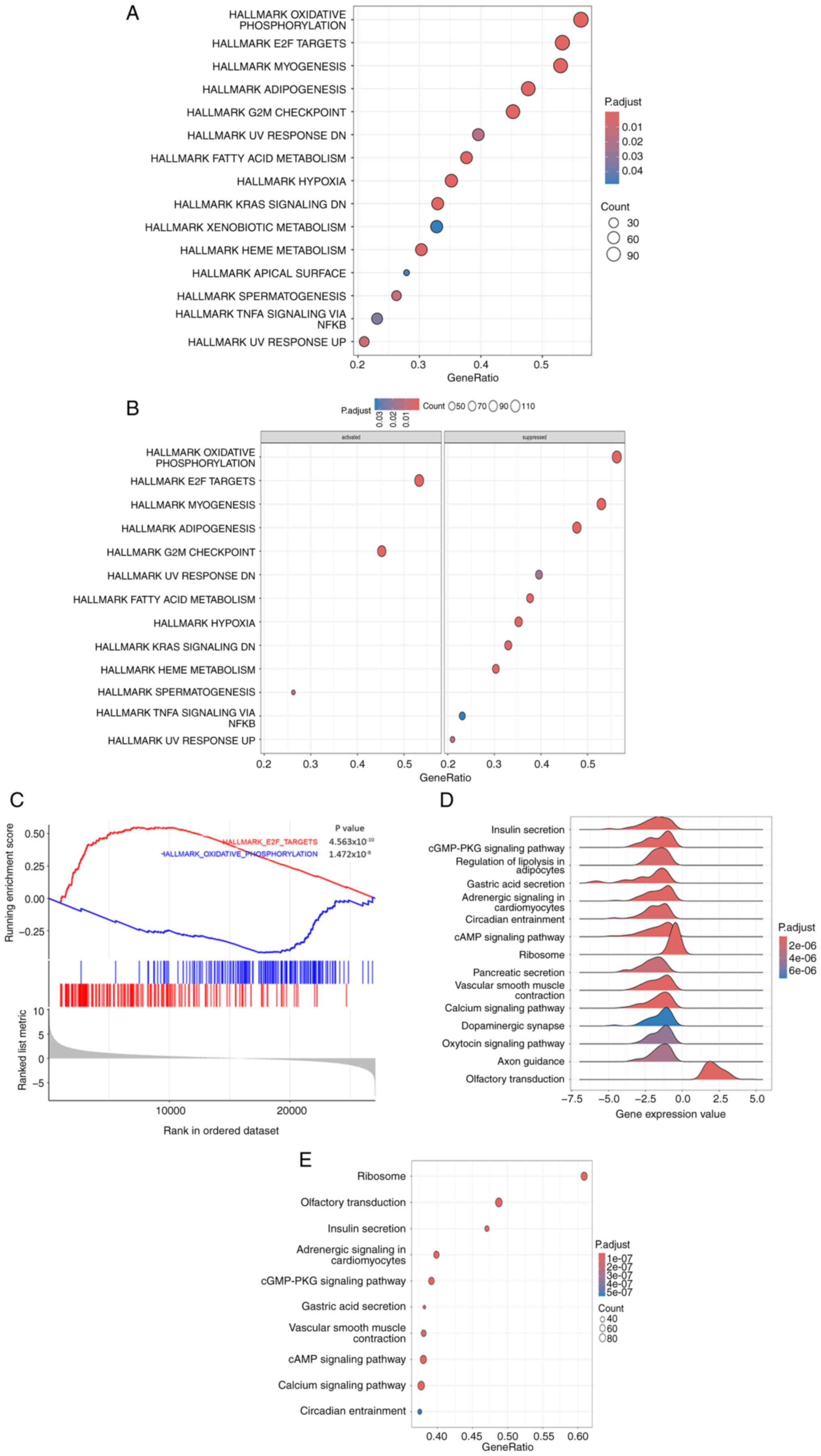
GO analysis revealed that the genes that were
expressed in the STAD cohort of patients in TCGA were enriched
primarily in pathways related to the interaction between
‘Neuroactive ligand-receptor interaction’, ‘Cytokine-cytokine
receptor interaction’, ‘Calcium signaling pathway’ and ‘cAMP
signaling pathway’ (Fig. 3A). GSEA
of the differentially expressed genes in GC revealed that the genes
were enriched in the MYC, which targets V2, E2F, G2M checkpoint and
angiogenesis pathways. Conversely, the top three inhibited pathways
included oxidative phosphorylation, adipogenesis and fatty acid
metabolism (Fig. 3C). In addition,
the KEGG pathway analysis revealed notable enrichment of genes
associated with ‘Neuroactive ligand-receptor interaction’, ‘Protein
digestion and absorption’, ‘Drug metabolism-cytochrome P450’ and
the ‘Bile secretion’ pathways (Fig.
3B).
Acquisition of the hub genes
The top seven directly related proteins with the
highest degree scores were obtained; these were, IL-1B, PYCARD,
MAVS, HSP90AA1, TXNIP, NEK7 and BRCC3 (Fig. 4A). These proteins are involved
primarily in physiological functions, including infection,
inflammation, apoptosis, pyroptosis, innate immunity, transcription
regulation, and kinase and cell cycle regulation (25,26).
Therefore, NLRP3 may be involved in these important functions. The
expression patterns of these proteins in GC in TCGA are shown in
Fig. 4B. The expression levels of
PYCARD, MAVS, HSP90AA1 and BRCC3 were upregulated in GC tissues.
Survival analysis revealed that these seven hub genes were not
individually significantly associated with GC prognosis in TCGA
dataset (Fig. 4C-I).
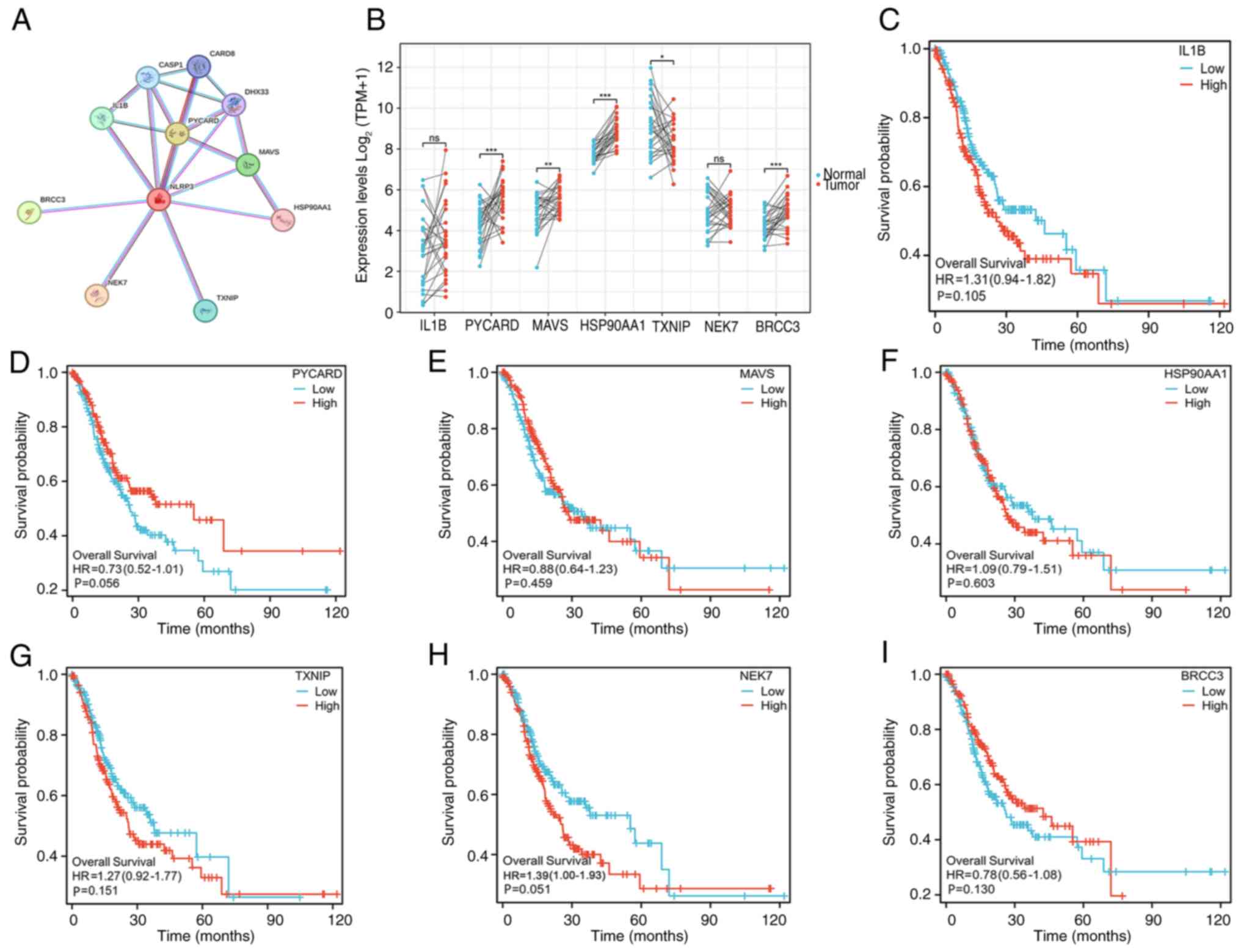 | Figure 4.Analysis of hub genes. (A)
Protein-protein interaction network of NLRP3. (B) Expression levels
of seven related genes in patients with gastric cancer in The
Cancer Genome Atlas database. *P<0.05, **P<0.01,
***P<0.001. Survival curve analyses of (C) IL-1B, (D) PYCARD,
(E) MAVS, (F) HSP90AA1, (G) TXNIP, (H) NEK7 and (I) BRCC3 genes in
gastric cancer. NLRP, nucleotide-binding oligomerization domain,
leucine rich repeat and pyrin domain containing; ns, not
significant. |
Association between NLRP3 expression
and clinicopathological parameters
The median of data were calculated for the optimal
cut-off value, which was used to split patients into high and low
NLRP3 expression. Patients with GC were divided into high
expression and low expression groups according to the cut-off
value. The Fisher's exact test was conducted to assess the
association of NLRP3 expression with clinicopathological
characteristics, and no statistically significant differences were
identified among patients based on age, sex, pathological type or
tumor location (Table I). By
contrast, high NLRP3 mRNA expression was significantly associated
with poorly differentiated tumors, lymph node metastasis and
Tumor-Node-Metastasis (TNM) stage III + IV. These findings
indicated that NLRP3 expression may increase as TNM stage
progresses with enhanced invasion and metastasis, suggesting an
association with the malignant advancement of clinical GC.
 | Table I.Relationships between NLRP3 mRNA
expression levels and the clinicopathologic features of patients
with gastric cancer. |
Table I.
Relationships between NLRP3 mRNA
expression levels and the clinicopathologic features of patients
with gastric cancer.
| Clinical
feature | Number (%) | NLRP3 mRNA high
expression (n=31) | NLRP3 mRNA low
expression (n=34) | P-value |
|---|
| Age |
|
|
| 0.788 |
| <55
years | 18 (27.7) | 8 | 10 |
|
| ≥55
years | 47 (72.3) | 23 | 24 |
|
| Sex |
|
|
| 0.216 |
|
Male | 34 (52.3) | 19 | 15 |
|
|
Female | 31 (47.7) | 12 | 19 |
|
| Tumor size |
|
|
| >0.999 |
| <5
cm | 41 (63.1) | 20 | 21 |
|
| ≥5
cm | 24 (36.9) | 11 | 13 |
|
| Lymph node
metastasis |
|
|
| 0.021a |
| No | 38 (58.5) | 17 | 21 |
|
|
Yes | 27 (41.5) | 14 | 13 |
|
| TNM stage |
|
|
|
<0.0001a |
| I +
II | 40 (61.5) | 11 | 29 |
|
| III +
IV | 25 (38.5) | 20 | 5 |
|
| Pathological
type |
|
|
| 0.074 |
|
Adenocarcinoma | 51 (78.5) | 20 | 31 |
|
|
Others | 14 (21.5) | 11 | 3 |
|
| Degree of
differentiation |
|
|
| 0.032a |
| High
differentiation | 8 (12.3) | 2 | 6 |
|
|
Moderatedifferentiation | 29 (44.6) | 11 | 18 |
|
| Low
differentiation | 28 (43.1) | 18 | 10 |
|
| Tumor location |
|
|
| 0.116 |
| Gastric
antrum | 36 (55.4) | 16 | 20 |
|
| Entire
gastric area | 16 (24.6) | 11 | 5 |
|
|
Others | 13 (20.0) | 4 | 9 |
|
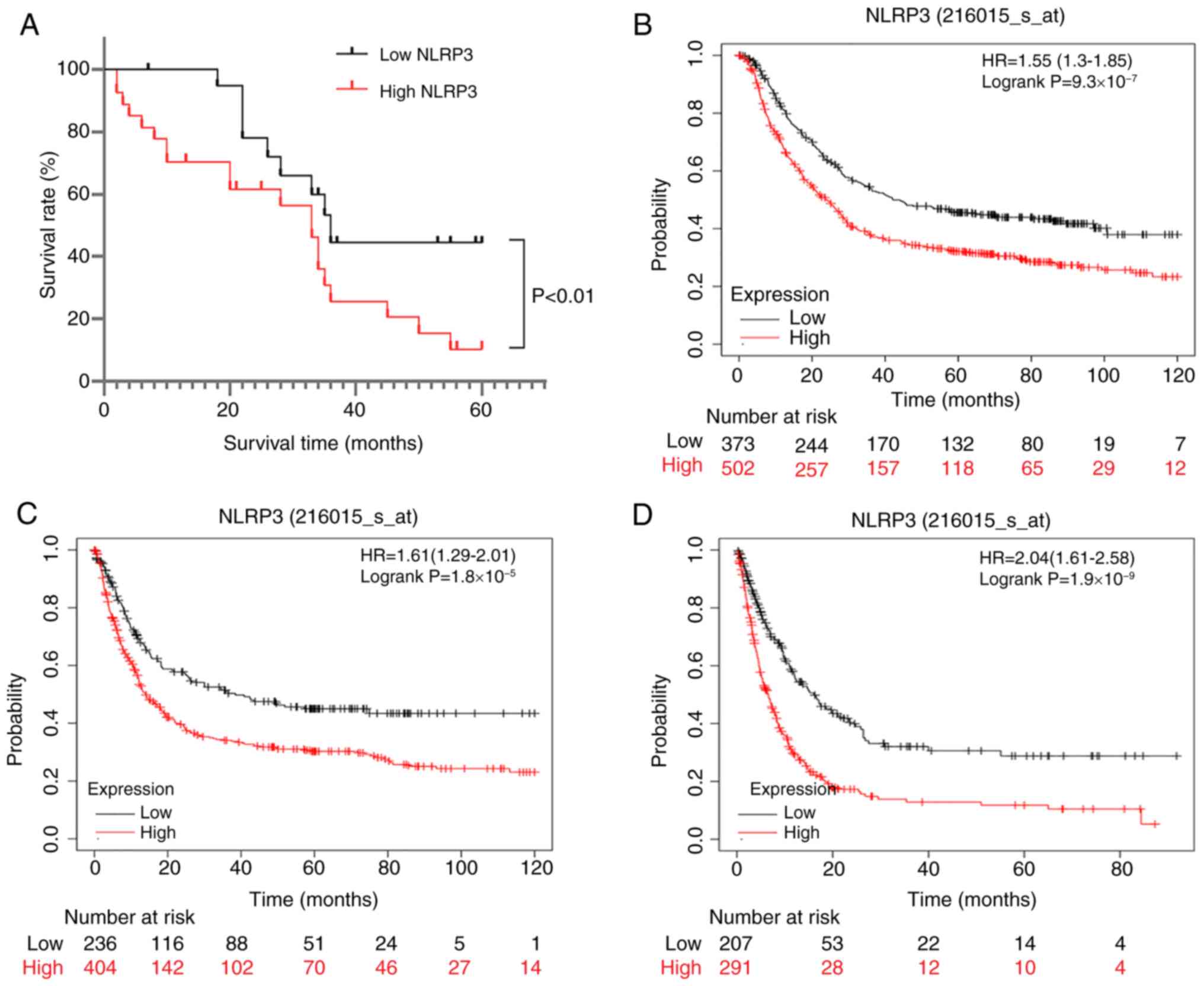
Clinical survival and prognostic
significance of NLRP3 in GC
The median expression of NLRP3 in GC was used to
split patients into high and low NLRP3 expression groups.
Kaplan-Meier survival analysis of 875 patients with GC in TCGA
database revealed that patients with high levels of NLRP3
expression had significantly lower 5-year OS, progression-free
survival and post-progression survival (PPS) rates than those with
low expression levels (Fig. 5B-D;
OS, HR=1.55, P=9.3×10−7; FP, HR=1.61,
P=1.8×10−5; PPS, HR=2.04, P=1.9×10−9). These
data indicated a possible link between increased NLRP3 expression
and lower survival rates in GC. To verify these findings follow-up
data from clinical samples were collected to perform a survival
study on a cohort of 65 individuals with GC. The findings revealed
a reduced survival rate in individuals with higher NLRP3 expression
(Fig. 5A), in agreement with the
aforementioned results. Thus, constitutively high levels of NLRP3
expression may be associated with a worse prognosis in patients
with GC.
NLRP3 affects the infiltration of
different types of immune cells
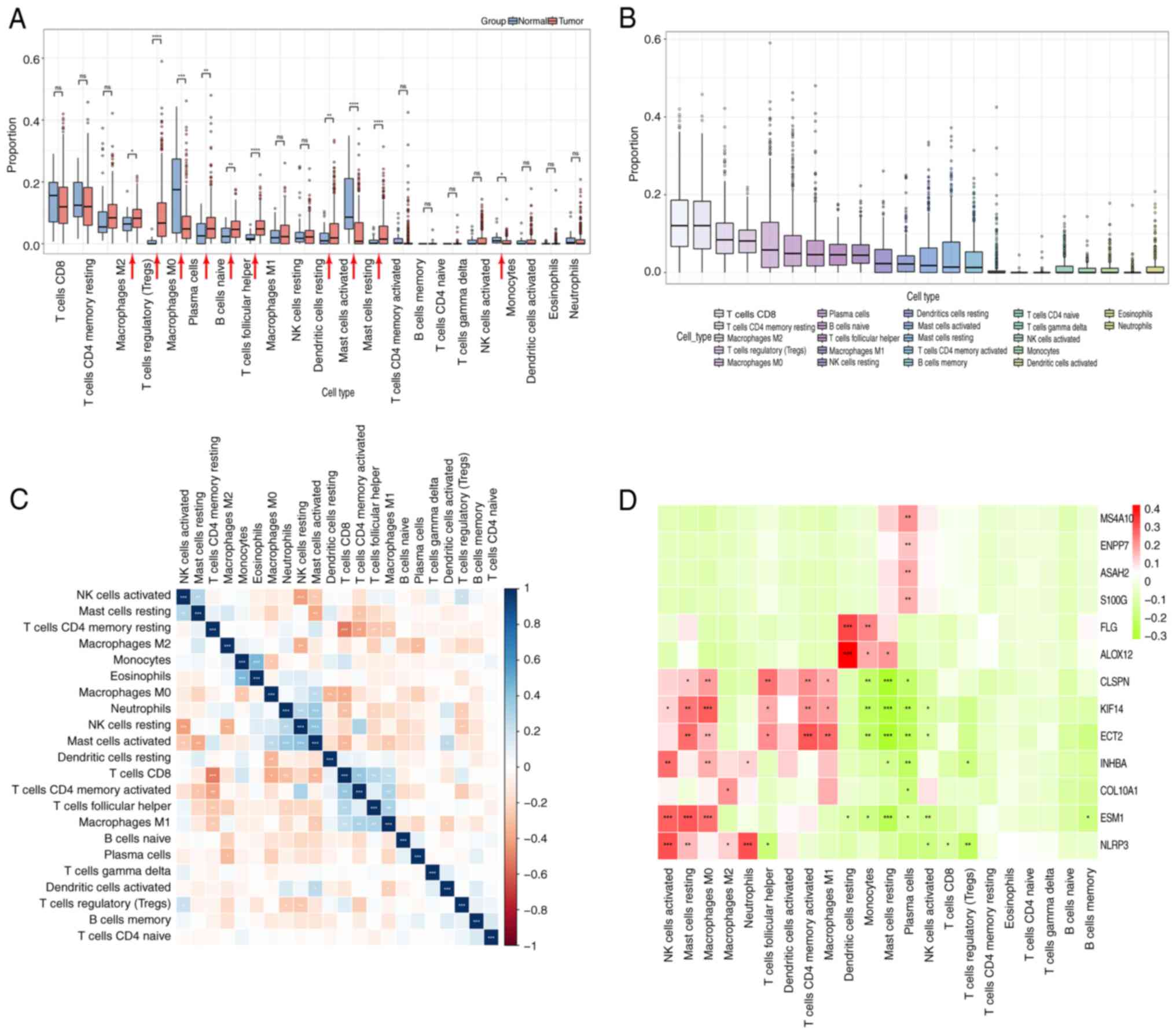
CIBERSORT was used to examine the presence of 22
immune cell types in tissues. Analysis revealed that in the GC
group, the presence of regulatory T cells (Tregs), M0 macrophages,
naïve B cells, T helper (Th) cells, M1 macrophages, activated mast
cells and memory-activated CD4+ T cells was
significantly higher in GC compared with in normal tissues
(Fig. 6A). By contrast, the
presence of plasma cells, resting mast cells and monocytes was
significantly reduced in tumor tissues. Regarding the distribution
of immune cells within the tumor, the majority of immune cells were
CD8+ T cells and resting memory CD4+ T cells,
followed by M2 macrophages, Tregs, M0 macrophages, plasma cells and
naïve B cells (Fig. 6B).
Correlation analysis was performed to examine the relationship
between immune cell ratios in GC tissues. The results revealed a
strong negative association between CD8+ T cells and
resting memory CD4+ T cells (Fig. 6C). Conversely, there was a positive
correlation between M1 macrophages and each of the following:
Active memory CD4+ T cells, CD8+ T cells and
T cells follicular helper (Fig.
6C). Furthermore, high expression of NLRP3 in GC was positively
correlated with activated mast cells, resting NK cells, M2
macrophages and neutrophils, whereas it exhibited negative
correlation with Tregs, CD8 T cells, activated NK cells and
follicular helper T cells (Fig.
6D). Additionally, there was a positive correlation between NK
cells and M2 macrophages; conversely, there was a negative
correlation with Tregs and CD8+ T cells (Fig. 6C).
GSEA of GC immune signaling pathways
associated with NLRP3
GSEA was used to examine the immune gene expression
sets specifically associated with NLRP3 in GC. The data were split
into two categories (high and low) based on median NLRP3 expression
levels. GSEA revealed that GC tissues with high NLRP3 expression
exhibited a notable enrichment in the oxidative phosphorylation
signaling pathway, E2F signaling pathway (Fig. 7A). The expression of NLRP3
suppressed oxidative phosphorylation, with the primary activation
routes being E2F signaling and G2M checkpoint signaling (Fig. 7B); the activating and inhibitory
effects of NLRP3 in the two signaling pathways are shown in
Fig. 7C (the red line represents
the E2F signaling pathway activated by NLRP3, and the blue line
represents oxidative phosphorylation signaling inhibited by NLRP3).
The E2F target gene set was activated, whereas the oxidative
phosphorylation gene set was inhibited. The mountain range map
(Fig. 7D) and bubble plot
(Fig. 7E) show that NLRP3 mostly
affected gene activities related to ribosomes, olfactory
transduction and insulin secretion.
Relationship between NLRP3 and immune
cell checkpoints
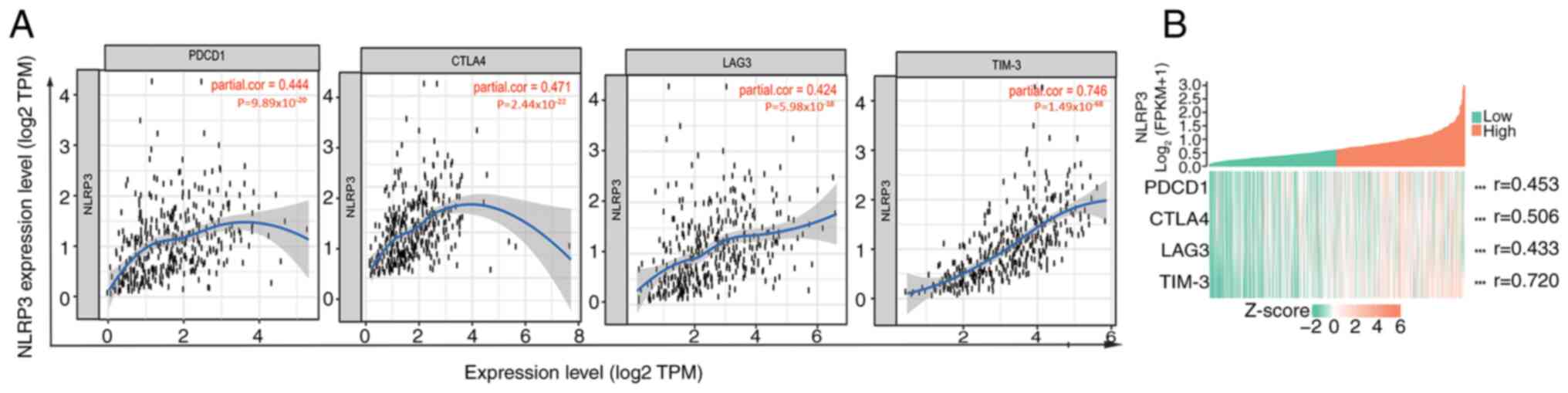
The relationship between NLRP3 expression and 57
immune cell markers in GC was analyzed to investigate the possible
mechanisms by which NLRP3 regulated the infiltration of immune
cells. The expression of NLRP3 in innate immune cells was strongly
positively associated with various indicators of monocytes,
tumor-associated macrophages (TAMs), M2 macrophages, neutrophils
and dendritic cells. The correlation coefficients for all these
markers were between 0.424 and 0.746 and the P-values were all
<0.0001 (Fig. 8A).
Specifically, the expression levels of NLRP3 were strongly
positively associated with the expression levels of programmed cell
death-1 (PDCD1) (r=0.453), cytotoxic T-lymphocyte antigen 4 (CTLA4;
r=0.506), lymphocyte activation 3 (LAG3) (r=0.433), and T-cell
immunoglobulin, and mucin structural domain-containing protein 3
(TIM-3; r=0.720) (P<0.0001; Fig.
8B); These markers serve a role in immune checkpoints. The
co-expression heatmap of each single gene revealed consistent
findings (Fig. 8B).
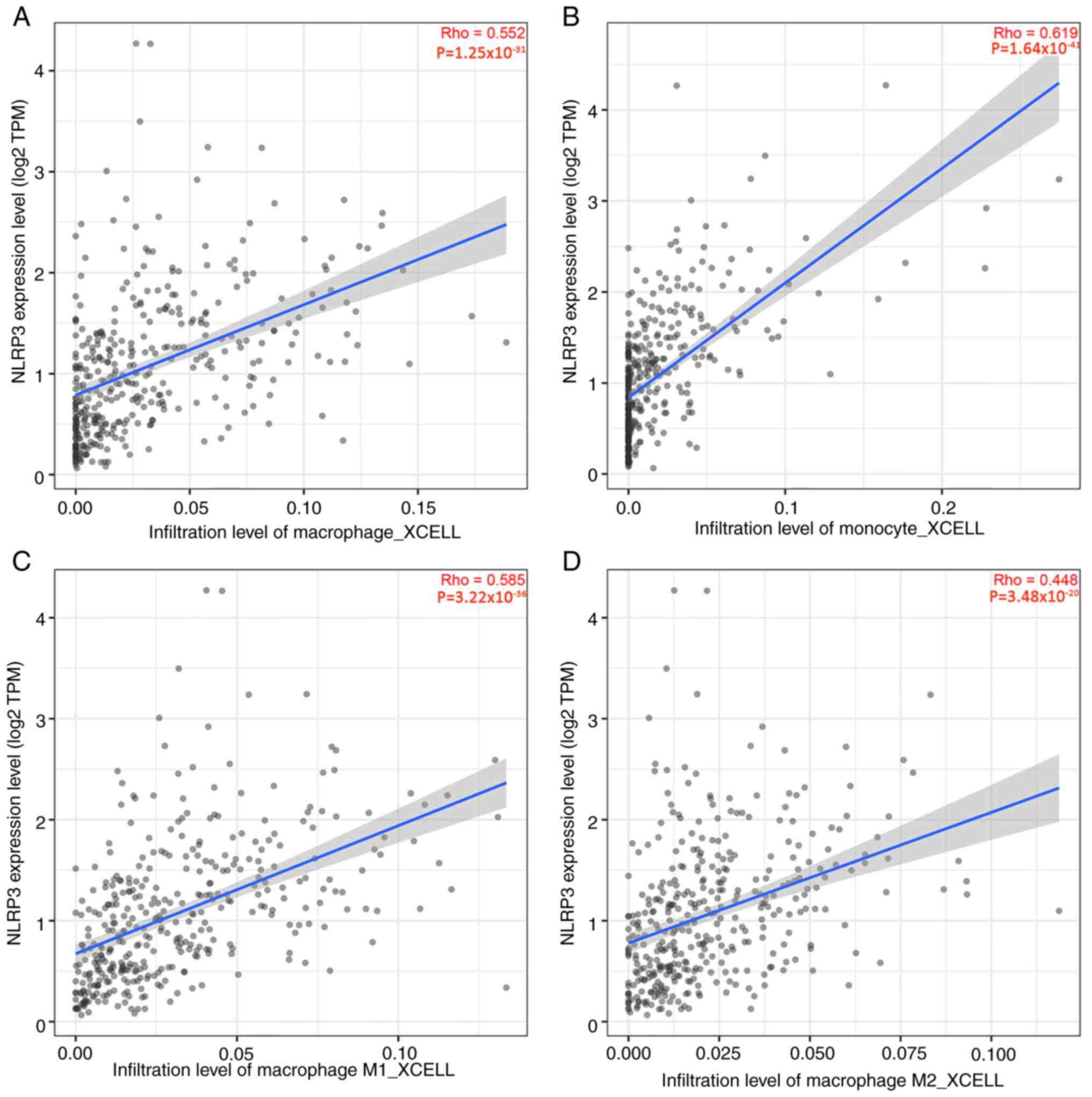
NLRP3 is involved in macrophage
polarization
The levels of NLRP3 expression were weakly
correlated with M1 macrophages in GC tissues, and strongly
correlated with M2 macrophages, TAMs and monocytes (Fig. 9A-D). This suggested that NLRP3 may
serve a role in promoting the polarization of TAMs towards the M2
phenotype. The aforementioned findings strongly indicated that
NLRP3 may have a role in the regulation of immune cell infiltration
in GC.
Immune infiltration of T lymphocytes
and macrophages in clinical GC tissues
Examination of immune infiltration revealed that
increased expression of NLRP3 led to a higher presence of
CD8+ T cells, CD4+ resting memory T cells, M2
macrophages and Tregs in GC tissues (Fig. 6B). Conversely, the levels of Tregs,
follicular Th cells and M1 macrophages were higher in GC tissues
compared with in normal tissues (Fig.
6A). Based on the median NLRP3 score, the 65 cases of GC were
categorized into high and low expression groups. Comparing the
immune cell counts in the two groups revealed that the GC tissues
with high NLRP3 expression had significantly higher counts of
CD3+, CD4+ and CD206+ cells
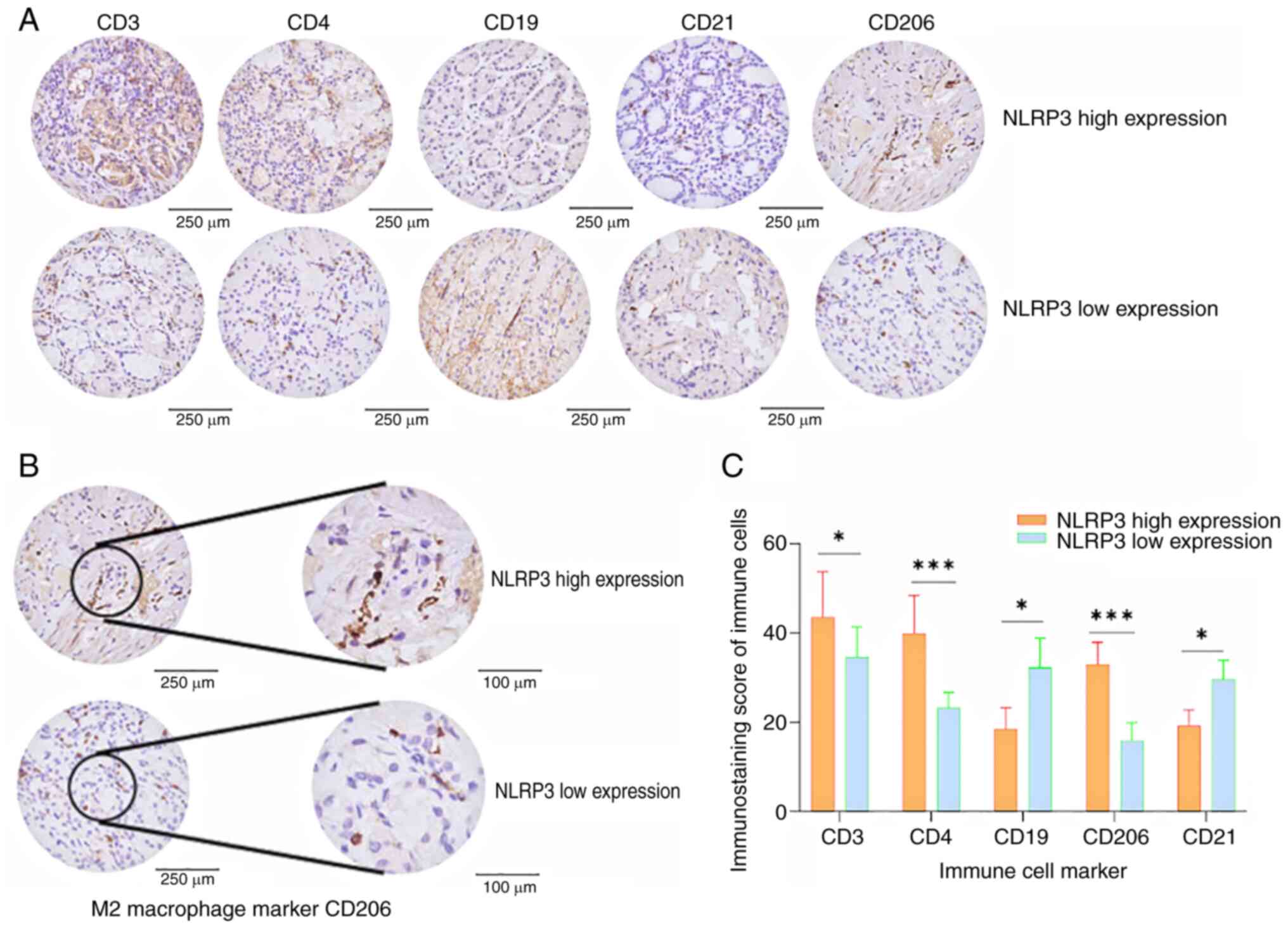
compared with those in the low NLRP3 expression group (Fig. 10). Conversely, the counts of
CD19+ and CD21+ cells were lower in the high
NLRP3 expression group. Representative IHC images are shown in
Fig. 10A and B; and comparative
analysis between the target immune cell marker scores is shown in
Fig. 10C.
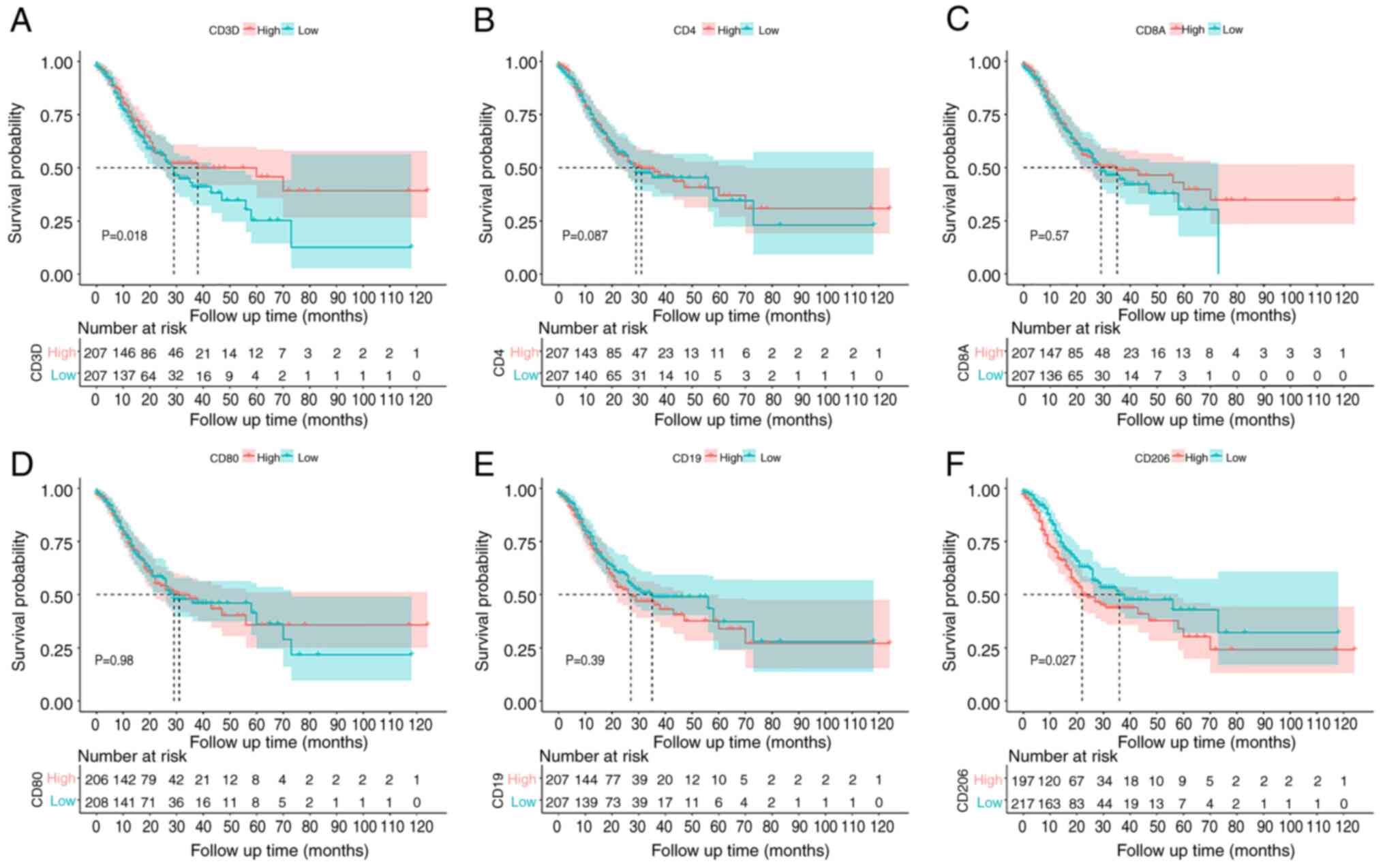
Immune infiltration survival
analysis
The survminer software package was used to plot and
analyze survival curves. The proportion of CD3D (a component of the
CD3 complex) and CD206 cell infiltration had a significant effect
on the survival of patients with GC. Patients with a high
percentage of T lymphocytes with high CD3D expression had a
significantly greater survival rate compared with those with a low
percentage (Fig. 11A). However,
patients with a high percentage of M2 macrophage infiltration (high
CD206 expression) had a lower survival rate (Fig. 11F). There was no significant
relationship between the proportion of infiltration of other types
of immune cells (CD4, CD8, CD80 and CD19) and survival rate
(Fig. 11B-E).
Discussion
In the present study, NLRP3 expression exhibited
marked variation based on cancer type. The NLRP proteins are
implicated in various cellular physiological functions, with
distinct mechanisms attributed to different members across
different tissues and/or diseases (27,28).
The protein expression of NLRPs varies across different types of
tumors, necessitating further investigation into the underlying
mechanisms (14). In the current
study, the aberrant upregulation of NLRP3 has been observed in GC
tissues, which could be attributed to persistent H. pylori
infection. NLRP3 upregulation was observed in GC tissues collected
from patients, and this upregulation was associated with the
malignant degree of GC. In addition, elevated NLRP3 expression was
associated with distant metastases. Upregulated expression of NLRP3
was also predictive of an unfavorable prognosis for patients with
GC. Notably, there was a positive association between NLRP3
expression and lymph node metastasis, infiltration and distant
metastasis, which is in agreement with a previous pan-cancer
analysis of NLRP3 (29).
NLRP3 may participate in the regulation of
infection-induced inflammatory receptor ligands and the modulation
of cellular inflammatory factors. The tumor microenvironment
secretes various factors that regulate tumor behavior (30), and inflammation may act as a
defense mechanism against cancer cells, with inflammasomes serving
a pivotal role in tumor progression, prognosis and treatment
response (31,32). Research has demonstrated the
benefits of utilizing the immune system to combat tumors, as
opposed to conventional treatments (33). The immune cell makeup in cancer
tissues can reveal promising targets for cancer therapy that may be
utilized to modulate the immune system and effectively combat
cancer cells (34,35).
In the present study, patients with GC with a high
degree of macrophage infiltration experienced a significant
decrease in survival. Furthermore, NLRP3 was positively correlated
with Tregs, and it has been suggested that NLRP3 may stimulate the
production of Tregs. It could be hypothesized that increased NLRP3
expression may lead to enhanced infiltration of T lymphocytes,
particularly Tregs, and can promote the transformation of
macrophages from an M1 to an M2 phenotype. CD3+ T cells
are widely hypothesized to possess antitumor properties, and
substantial infiltration of CD3+ T cells is indicative
of a favorable prognosis for patients with GC (36). CD4+ T lymphocytes
consist of four distinct groups: Th1, Th2, Th17 and Treg cells
(37). Th1 and Th2 cells have the
potential to contribute to the suppression of tumor cells, and the
extent of their infiltration is directly proportional to the
benefits of immunotherapy for patients with GC (37,38).
In addition, CD4+ T cells primarily exert antitumor
actions; however, the impact of CD8+ T cells on tumor
cells is contingent upon the specific condition (39). A previous study demonstrated that
the presence of Tregs and M2 macrophages in GC tissues can
positively influence the effectiveness of antitumor treatment
(40). In the current study,
patients with GC who exhibited high expression levels of CD3D in
their tumor tissues demonstrated a significantly higher survival
rate, whereas those with elevated levels of CD206 and a high
infiltration of M2 macrophages showed a lower survival rate.
TAMs are mostly identified using CD68 as their major
marker (41). The distribution of
TAMs differs across different types of cancer, and different types
of TAMs can have either anticancer or pro-cancer effects (42). Research has indicated that TAMs may
function as promoters of cancer during tumor growth (43). Upregulation of NLRP3 in GC tissues
has been shown to lead to the polarization of macrophages towards
an M2 phenotype, which promotes the development of cancer. NLRP3
may also lead to the activation of caspase-1, and the production of
IL-1β and IL-18 cytokines, which may facilitate the transformation
of immune cell infiltration (44).
B cells have been hypothesized to develop a range of
antibodies and to enhance the function of T cells, which allows the
body to combat foreign antigens from viral and bacterial infections
(45). The association between the
functional enrichment of genes and immune cell infiltration in
cancer is intricate. Upregulated expression of NLRP3 was shown to
be associated with the enrichment of cell cycle-related gene sets
(E2F targets and G2M checkpoint), which are associated with
ribosomes, olfactory transduction and insulin secretion.
Tumor immunotherapy employs immune checkpoint
inhibitors to restore antitumor immune responses by activating
co-suppressive T-cell signaling. This therapy has demonstrated
significant clinical effectiveness in several types of cancer
(46,47). In the present study, a favorable
correlation between NLRP3 and the immune checkpoint genes TIM-3,
PDCD1, CTLA4 and LAG3 was identified. TIM-3 has a crucial role in
controlling the function of dendritic cells and limiting the
immunological response to tumors by regulating activation of the
NLRP3 inflammasome (48). The
NLRP3 inflammasome increases the expression of PD-L1 and
contributes to the inhibition of the immune response in lymphoma
(49). Thus, based on the findings
of previous studies, NLRP3 may regulate immunological checkpoints
and facilitate the immune system evasion of tumor cells.
In the present study, the function of NLRP3 as a
prognostic marker of GC was assessed and the results showed an
association between NLRP3 and immune cell infiltration in GC. NLRP3
exhibits contrasting effects in the tumor microenvironment of GC,
exhibiting distinct pro- and anticancer effects through immune cell
infiltration. By reducing the infiltration ratio of M1/M2
macrophages, cancer progression is promoted, whereas increasing the
M1/M2 ratio can exert anticancer effects. By targeting NLRP3, the
immune cell infiltration pattern in GC tissues may be manipulated
to improve patient outcomes. By stimulating the infiltration of
immune cells to shift from inhibiting tumor immune evasion to
promoting antitumor effects, it could be possible to suppress the
proliferation and migration of GC cells. Modulating NLRP3 levels by
interfering with protein expression, such as via antibody-based
drug inhibition, genetic interference and inflammation control,
could prevent NLRP3 inflammation-related diseases, including GC.
However, completely blocking NLRP3 may also lead to notable side
effects, as it would eliminate the beneficial roles of NLRP3
inflammation in eliminating harmful pathogens (50). Developing precise and efficient
methods that appropriately block the inflammatory activity of the
NLRP3 inflammasome may serve as a novel strategy and approach for
preventing and treating GC.
The present study has some limitations. Databases
provided by different research centers may have differences in
research methodologies and experimental outcomes. Although
statistical methods minimized the bias, the heterogeneity of the
data can hinder the reproducibility of the findings to some extent.
Increasing the sample size and extending the follow-up period may
allow for a more accurate assessment of prognostic value. The
detailed mechanisms by which NLRP3 regulates immune infiltration in
GC require further research. NLRP3 may serve as a prognostic marker
for GC; however, the practicality and effectiveness of assessing
its expression in different clinical contexts remain to be
confirmed. The heterogeneity among individuals with GC may also
affect its general applicability as a prognostic marker.
In conclusion, the results of the present study
showed that NLRP3 expression was upregulated in GC tissues compared
with in normal tissues from TCGA dataset; this finding was
confirmed in clinical samples. Higher NLRP3 expression was
demonstrated to promote the progression of larger tumors with lymph
node metastasis, advanced TNM stages or poor differentiation. NLRP3
was revealed to be associated with NK cells and M2 macrophages, and
to be inversely correlated with Tregs and CD8+ T cells.
Furthermore, high NLRP3 expression led to increased
CD3+, CD4+ and CD206+ cell
infiltration, which may regulate cancer cell proliferation and
migration, ultimately leading to an unfavorable prognosis in
patients with GC.
Acknowledgements
Not applicable.
Funding
This study was supported by grants from the Medical Research
Support Project of Changshu Health Committee (grant no.
CSWS202106), the Changshu Science and Technology Program Project
(grant no. CS202018), the Basic Research Program-Medical
Application Basic Research Project (grant no. CY202330) and Suzhou
Science and Technology Bureau Clinical Trial Institution Capacity
Improvement Project (SLT2023007).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
CW, YX and YG conceived the project. PW, YZ and XC
collected specimens and performed experiments. CW, YX and PW
conducted the experiments. YX, PW and YZ analyzed the data. CW and
PW drafted the manuscript. PW, YX and YG confirm the authenticity
of all the raw data. All authors read and approved the final
version of the manuscript.
Ethics approval and consent to
participate
The study was conducted in accordance with The
Declaration of Helsinki, and was approved by the Ethics Committee
of Affiliated Changshu Hospital of Nantong University (approval no.
2019-KY-049). Written informed consent for the use of samples for
research purposes was obtained from each participant.
Patient consent for publication
All patients provided written informed consent for
the publication of medical research findings.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Heuvers ME, Wisnivesky J, Stricker BH and
Aerts JG: Generalizability of results from the national lung
screening trial. Eur J Epidemiol. 27:669–672. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Billan S, Kaidar-Person O and Gil Z:
Treatment after progression in the era of immunotherapy. Lancet
Oncol. 21:e463–e476. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shaopeng Z, Yang Z, Yuan F, Chen H and
Zhengjun Q: Regulation of regulatory T cells and tumor-associated
macrophages in gastric cancer tumor microenvironment. Cancer Med.
13:e69592024. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhai Y, Liu X, Huang Z, Zhang J, Stalin A,
Tan Y, Zhang F, Chen M, Shi R, Huang J, et al: Data mining combines
bioinformatics discover immunoinfiltration-related gene SERPINE1 as
a biomarker for diagnosis and prognosis of stomach adenocarcinoma.
Sci Rep. 13:13732023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kao KC, Vilbois S, Tsai CH and Ho PC:
Metabolic communication in the tumour-immune microenvironment. Nat
Cell Biol. 24:1574–1583. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen F, Chen N, Gao Y, Jia L, Lyu Z and
Cui J: Clinical progress of PD-1/L1 inhibitors in breast cancer
immunotherapy. Front Oncol. 11:7244242022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen X, Zhang W, Yang W, Zhou M and Liu F:
Acquired resistance for immune checkpoint inhibitors in cancer
immunotherapy: Challenges and prospects. Aging (Albany NY).
14:1048–1064. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wen H, Miao EA and Ting JP: Mechanisms of
NOD-like receptor-associated inflammasome activation. Immunity.
39:432–441. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mitchell PS, Sandstrom A and Vance RE: The
NLRP1 inflammasome: New mechanistic insights and unresolved
mysteries. Curr Opin Immunol. 60:37–45. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kufer TA and Sansonetti PJ: NLR functions
beyond pathogen recognition. Nat Immunol. 12:121–128. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Arfsten H, Cho A, Prausmüller S, Spinka G,
Novak J, Goliasch G, Bartko PE, Raderer M, Gisslinger H, Kornek G,
et al: Inflammation-based scores as a common tool for prognostic
assessment in heart failure or cancer. Front Cardiovasc Med.
8:7259032021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yong X, Tang B, Li BS, Xie R, Hu CJ, Luo
G, Qin Y, Dong H and Yang SM: Helicobacter pylori virulence
factor CagA promotes tumorigenesis of gastric cancer via multiple
signaling pathways. Cell Commun Signal. 13:302015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hu Q, Zhao F, Guo F, Wang C and Fu Z:
Polymeric nanoparticles induce NLRP3 inflammasome activation and
promote breast cancer metastasis. Macromol Biosci. 17:17002732017.
View Article : Google Scholar
|
|
14
|
Zhang X, Li C, Chen D, He X, Zhao Y, Bao
L, Wang Q, Zhou J and Xie Y: H. pylori CagA activates the
NLRP3 inflammasome to promote gastric cancer cell migration and
invasion. Inflamm Res. 71:141–155. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Allen IC, TeKippe EM, Woodford RM, Uronis
JM, Holl EK, Rogers AB, Herfarth HH, Jobin C and Ting JP: The NLRP3
inflammasome functions as a negative regulator of tumorigenesis
during colitis-associated cancer. J Exp Med. 207:1045–1056. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dupaul-Chicoine J, Arabzadeh A, Dagenais
M, Douglas T, Champagne C, Morizot A, Rodrigue-Gervais IG, Breton
V, Colpitts SL, Beauchemin N and Saleh M: The Nlrp3 inflammasome
suppresses colorectal cancer metastatic growth in the liver by
promoting natural killer cell tumoricidal activity. Immunity.
43:751–763. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chang HR, Nam S, Kook MC, Kim KT, Liu X,
Yao H, Jung HR, Lemos R Jr, Seo HH, Park HS, et al: HNF4α is a
therapeutic target that links AMPK to WNT signalling in early-stage
gastric cancer. Gut. 65:19–32. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Oh SC, Sohn BH, Cheong JH, Kim SB, Lee JE,
Park KC, Lee SH, Park JL, Park YY, Lee HS, et al: Clinical and
genomic landscape of gastric cancer with a mesenchymal phenotype.
Nat Commun. 9:17772018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu
JS, Li B and Liu XS: TIMER: A web server for comprehensive analysis
of tumor-infiltrating immune cells. Cancer Res. 77:e108–e110. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Elbehiry A, Marzouk E, Aldubaib M,
Abalkhail A, Anagreyyah S, Anajirih N, Almuzaini AM, Rawway M,
Alfadhel A, Draz A and Abu-Okail A: Helicobacter pylori
infection: Current status and future prospects on diagnostic,
therapeutic and control challenges. Antibiotics (Basel).
12:1912023. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kushima R: The updated WHO classification
of digestive system tumours gastric adeno-carcinoma and dysplasia.
Pathologe. 43:8–15. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
RStudio Team R Studio. Integrated
Development for R. RStudio, Inc.; Boston, MA: 2015, http://www.rstudio.com/
|
|
25
|
Ghazi BK, Bangash MH, Razzaq AA, Kiyani M,
Girmay S, Chaudhary WR, Zahid U, Hussain U, Mujahid H, Parvaiz U,
et al: In silico structural and functional analyses of NLRP3
inflammasomes to provide insights for treating neurodegenerative
diseases. Biomed Res Int. 23:98190052023. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
An S, Li X, Li B and Li Y: Comprehensive
analysis of epigenetic associated genes with differential gene
expression and prognosis in gastric cancer. Comb Chem High
Throughput Screen. 26:527–538. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shen Y, Qian L, Luo H, Li X, Ruan Y, Fan
R, Si Z, Chen Y, Li L and Liu Y: The significance of NLRP
inflammasome in neuropsychiatric disorders. Brain Sci. 12:10572022.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Moon SW, Son HJ, Mo HY, Yoo NJ and Lee SH:
Somatic mutation of NLRP genes in gastric and colonic cancers.
Pathol Oncol Res. 27:6073852021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shadab A, Mahjoor M, Abbasi-Kolli M,
Afkhami H, Moeinian P and Safdarian AR: Divergent functions of
NLRP3 inflammasomes in cancer: A review. Cell Commun Signal.
21:2322023. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kim J and Bae JS: Tumor-associated
macrophages and neutrophils in tumor microenvironment. Mediators
Inflamm. 2016:60581472016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bruchard M, Mignot G, Derangère V, Chalmin
F, Chevriaux A, Végran F, Boireau W, Simon B, Ryffel B, Connat L,
et al: Chemotherapy-triggered cathepsin B release in
myeloid-derived suppressor cells activates the Nlrp3 inflammasome
and promotes tumor growth. Nat Med. 19:57–64. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Karki R and Kanneganti TD: Diverging
inflammasome signals in tumorigenesis and potential targeting. Nat
Rev Cancer. 19:197–214. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lin C, He H, Liu H, Li R, Chen Y, Qi Y,
Jiang Q, Chen L, Zhang P, Zhang P, et al: Tumour-associated
macrophages-derived CXCL8 determines immune evasion through
autonomous PD-L1 expression in gastric cancer. Gut. 68:1764–1773.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Huo J, Wu L and Zang Y: Development and
validation of a robust immune-related prognostic signature for
gastric cancer. J Immunol Res. 2021:55543422021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tang X, Guo T, Wu X, Gan X, Wang Y, Jia F,
Zhang Y, Xing X, Gao X and Li Z: Clinical significance and immune
infiltration analyses of the cuproptosis-related human copper
proteome in gastric cancer. Biomolecules. 12:14592022. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Du Z, Xiao Y, Deng G and Song H, Xue Y and
Song H: CD3+/CD4+ cells combined with myosteatosis predict the
prognosis in patients who underwent gastric cancer surgery. J
Cachexia Sarcopenia Muscle. 15:1587–1600. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shi W, Wang Y, Xu C, Li Y, Ge S, Bai B,
Zhang K, Wang Y, Zheng N, Wang J, et al: Multilevel proteomic
analyses reveal molecular diversity between diffuse-type and
intestinal-type gastric cancer. Nat Commun. 14:8352023. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Khan M, Lin J, Wang B, Chen C, Huang Z,
Tian Y, Yuan Y and Bu J: A novel necroptosis-related gene index for
predicting prognosis and a cold tumor immune microenvironment in
stomach adenocarcinoma. Front Immunol. 13:9681652022. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhao S, Wu Y, Wei Y, Xu X and Zheng J:
Identification of biomarkers associated with CD8+ T cells in
coronary artery disease and their pan-cancer analysis. Front
Immunol. 13:8766162022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang F and Luo H: Diosmetin inhibits the
growth and invasion of gastric cancer by interfering with M2
phenotype macrophage polarization. J Biochem Mol Toxicol.
37:e234312023. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Qu Y, Wang X, Bai S, Niu L, Zhao G, Yao Y,
Li B and Li H: The effects of TNF-α/TNFR2 in regulatory T cells on
the microenvironment and progression of gastric cancer. Int J
Cancer. 150:1373–1391. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jian F, Yanhong J, Limeng W, Guoping N,
Yiqing T, Hao L and Zhaoji P: TIMP2 is associated with prognosis
and immune infiltrates of gastric and colon cancer. Int
Immunopharmacol. 110:1090082022. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wei C, Chen M, Deng W, Bie L, Ma Y, Zhang
C, Liu K, Shen W, Wang S, Yang C, et al: Characterization of
gastric cancer stem-like molecular features, immune and
pharmacogenomic landscapes. Brief Bioinform 23: bbab386, 2022. Wang
P, Gu Y, Yang J, Qiu J, Xu Y, Xu Z, Gao J and Wan C: The prognostic
value of NLRP1/NLRP3 and its relationship with immune infiltration
in human gastric cancer. Aging (Albany NY). 14:9980–10008.
2022.PubMed/NCBI
|
|
44
|
Hamarsheh S and Zeiser R: NLRP3
inflammasome activation in cancer: A double-edged sword. Front
Immunol. 11:14442020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ohnishi N, Yuasa H, Tanaka S, Sawa H,
Miura M, Matsui A, Higashi H, Musashi M, Iwabuchi K, Suzuki M, et
al: Transgenic expression of Helicobacter pylori CagA
induces gastrointestinal and hematopoietic neoplasms in mouse. Proc
Natl Acad Sci USA. 105:1003–1008. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Marangoni F, Zhakyp A, Corsini M, Geels
SN, Carrizosa E, Thelen M, Mani V, Prüßmann JN, Warner RD, Ozga AJ,
et al: Expansion of tumor-associated Treg cells upon disruption of
a CTLA-4-dependent feedback loop. Cell. 184:3998–4015.e19. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang W, Zhang Q, Yang N, Shi Q, Su H, Lin
T, He Z, Wang W, Guo H and Shen P: Crosstalk between
IL-15Rα+ tumor-associated macrophages and breast cancer
cells reduces CD8+ T cell recruitment. Cancer Commun
(Lond). 42:536–557. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Dixon KO, Tabaka M, Schramm MA, Xiao S,
Tang R, Dionne D, Anderson AC, Rozenblatt-Rosen O, Regev A and
Kuchroo VK: TIM-3 restrains anti-tumour immunity by regulating
inflammasome activation. Nature. 595:101–106. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lu F, Zhao Y, Pang Y, Ji M, Sun Y, Wang H,
Zou J, Wang Y, Li G, Sun T, et al: NLRP3 inflammasome upregulates
PD-L1 expression and contributes to immune suppression in lymphoma.
Cancer Lett. 497:178–189. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chen Y, Ye X, Escames G, Lei W, Zhang X,
Li M, Jing T, Yao Y, Qiu Z, Wang Z, et al: The NLRP3 inflammasome:
Contributions to inflammation-related diseases. Cell Mol Biol Lett.
28:512023. View Article : Google Scholar : PubMed/NCBI
|