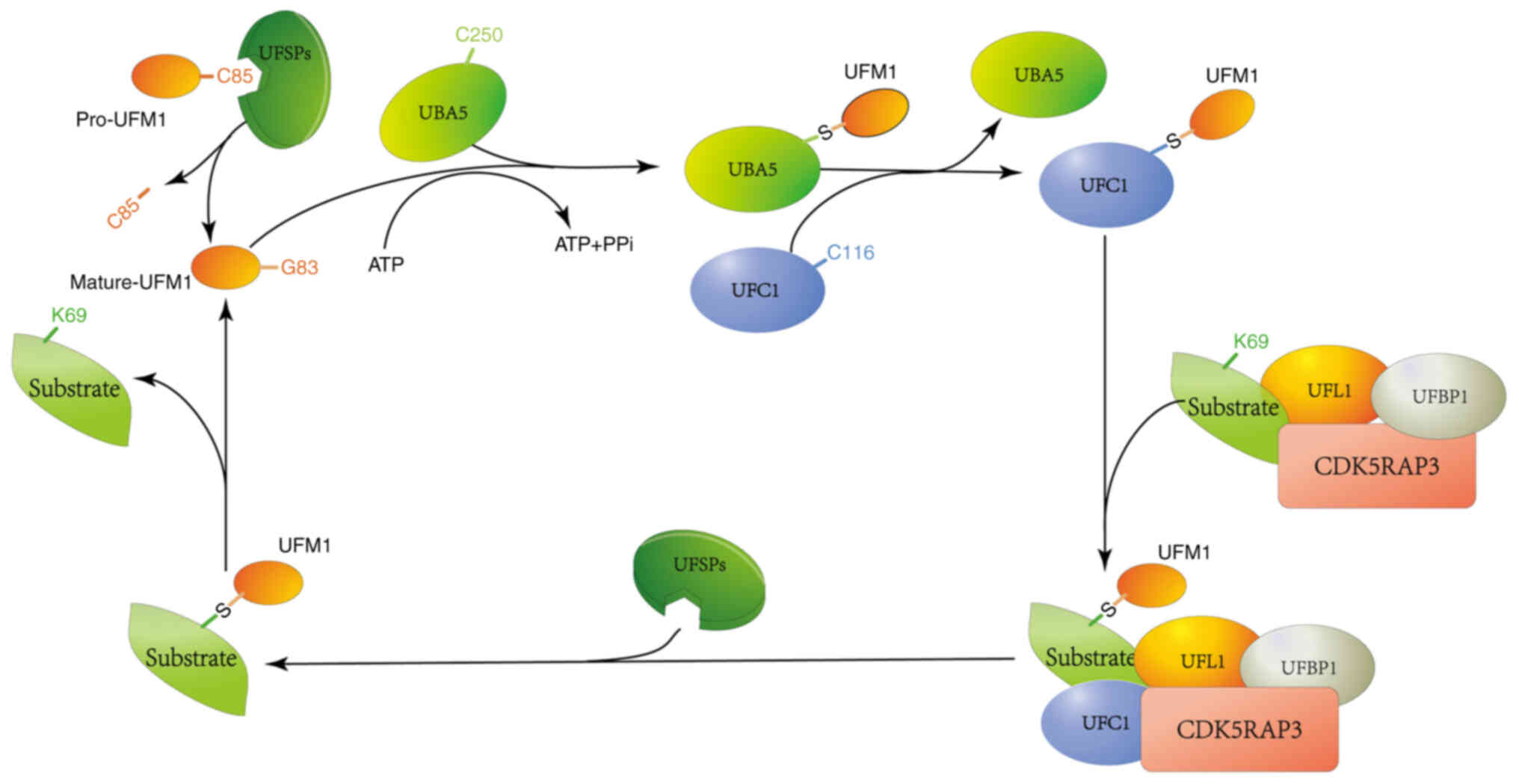
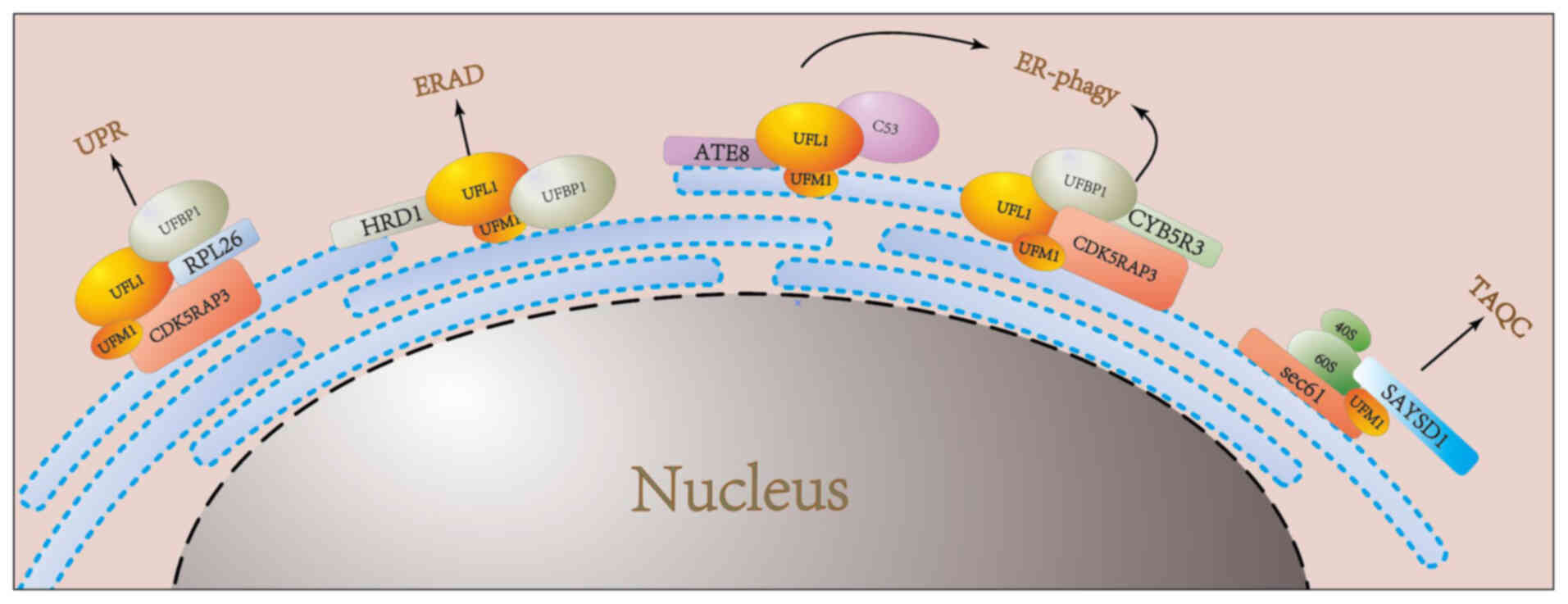
|
1
|
Millar AH, Heazlewood JL, Giglione C,
Holdsworth MJ, Bachmair A and Schulze WX: The scope, functions, and
dynamics of posttranslational protein modifications. Annu Rev Plant
Biol. 70:119–151. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hsu YC, Hsieh YH, Liao CC, Chong LW, Lee
CY, Yu YL and Chou RH: Targeting post-translational modifications
of histones for cancer therapy. Cell Mol Biol (Noisy-le-grand).
30:69–84. 2015.
|
|
3
|
Hitosugi T and Chen J: Post-translational
modifications and the Warburg effect. Oncogene. 33:4279–4285. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Herhaus L and Dikic I: Expanding the
ubiquitin code through post-translational modification. EMBO Rep.
16:1071–1083. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hochstrasser M: Origin and function of
ubiquitin-like proteins. Nature. 458:422–429. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sahtoe DD and Sixma TK: Layers of DUB
regulation. Trends Biochem Sci. 40:456–467. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Karpiyevich M and Artavanis-Tsakonas K:
Ubiquitin-like modifiers: Emerging regulators of protozoan
parasites. Biomolecules. 10:14032020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Komatsu M, Chiba T, Tatsumi K, Iemura S,
Tanida I, Okazaki N, Ueno T, Kominami E, Natsume T and Tanaka K: A
novel protein-conjugating system for Ufm1, a ubiquitin-fold
modifier. EMBO J. 23:1977–1986. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gerakis Y, Quintero M, Li H and Hetz C:
The UFMylation system in proteostasis and beyond. Trends Cell Biol.
29:974–986. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Banerjee S, Kumar M and Wiener R:
Decrypting UFMylation: How proteins are modified with UFM1.
Biomolecules. 10:14422020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Witting KF, van der Heden van Noort GJ,
Kofoed C, Ormeno CT, El Atmioui D, Mulder MPC and Ovaa H:
Generation of the UFM1 toolkit for profiling UFM1-specific
proteases and ligases. Angew Chem Int Ed Engl. 57:14164–14168.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lorenz S, Cantor AJ, Rape M and Kuriyan J:
Macromolecular juggling by ubiquitylation enzymes. BMC Biol.
11:652013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jing Y, Mao Z and Chen F: UFMylation
system: An emerging player in tumorigenesis. Cancers (Basel).
14:35012022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Clague MJ, Urbe S and Komander D: Breaking
the chains: Deubiquitylating enzyme specificity begets function.
Nat Rev Mol Cell Biol. 20:338–352. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lin M, Zheng X and Jin J: Nontraditional
translation is the key to UFMylation and beyond. J Biol Chem.
298:1024312022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang R, Wang H, Kang B, Chen B, Shi Y,
Yang S, Sun L, Liu Y, Xiao W, Zhang T, et al: CDK5RAP3, a UFL1
substrate adaptor, is crucial for liver development. Development.
146:dev1692352019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang S, Moy N and Yang R: The UFM1
conjugation system in mammalian development. Dev Dyn. 252:976–985.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cai Y, Pi W, Sivaprakasam S, Zhu X, Zhang
M, Chen J, Makala L, Lu C, Wu J, Teng Y, et al: UFBP1, a key
component of the ufm1 conjugation system, is essential for
ufmylation-mediated regulation of erythroid development. PLoS
Genet. 11:e10056432015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tandra V, Anderson T, Ayala JD, Weintraub
NL, Singh N, Li H and Li J: Ufmylation of UFBP1 is dispensable for
endoplasmic reticulum stress response, embryonic development, and
cardiac and intestinal homeostasis. Cells. 12:19232023. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang X, Lv X, Ma J and Xu G: UFMylation:
An integral post-translational modification for the regulation of
proteostasis and cellular functions. Pharmacol Ther.
260:1086802024. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ding LJ, Jiang X, Li T and Wang S: Role of
UFMylation in tumorigenesis and cancer immunotherapy. Front
Immunol. 15:14548232024. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang X, Xu X and Wang Z: The
post-translational role of UFMylation in physiology and disease.
Cells. 12:25432023. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang X, Zhou T, Wang X, Xia Y, Cao X,
Cheng X, Cao Y, Ma P, Ma H, Qin A and Zhao J: Loss of DDRGK1
impairs IRE1α UFMylation in spondyloepiphyseal dysplasia. Int J
Biol Sci. 19:4709–4725. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang G, Tang S, Wang H, Pan H, Zhang W,
Huang Y, Kong J and Wang Y, Gu J and Wang Y: UFSP2-related
spondyloepimetaphyseal dysplasia: A confirmatory report. Eur J Med
Genet. 63:1040212020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Colin E, Daniel J, Ziegler A, Wakim J,
Scrivo A, Haack TB, Khiati S, Denommé AS, Amati-Bonneau P, Charif
M, et al: Biallelic variants in UBA5 reveal that disruption of the
UFM1 cascade can result in early-onset encephalopathy. Am J Hum
Genet. 99:695–703. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sasakawa H, Sakata E, Yamaguchi Y, Komatsu
M, Tatsumi K, Kominami E, Tanaka K and Kato K: Solution structure
and dynamics of Ufm1, a ubiquitin-fold modifier 1. Biochem Biophys
Res Commun. 343:21–26. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Millrine D, Cummings T, Matthews SP, Peter
JJ, Magnussen HM, Lange SM, Macartney T, Lamoliatte F, Knebel A and
Kulathu Y: Human UFSP1 is an active protease that regulates UFM1
maturation and UFMylation. Cell Rep. 40:1111682022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kang SH, Kim GR, Seong M, Baek SH, Seol
JH, Bang OS, Ovaa H, Tatsumi K, Komatsu M, Tanaka K and Chung CH:
Two novel ubiquitin-fold modifier 1 (Ufm1)-specific proteases,
UfSP1 and UfSP2. J Biol Chem. 282:5256–5262. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ha BH, Jeon YJ, Shin SC, Tatsumi K,
Komatsu M, Tanaka K, Watson CM, Wallis G, Chung CH and Kim EE:
Structure of ubiquitin-fold modifier 1-specific protease UfSP2. J
Biol Chem. 286:10248–10257. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Millrine D, Peter JJ and Kulathu Y: A
guide to UFMylation, an emerging posttranslational modification.
FEBS J. 290:5040–5056. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liang Q, Jin Y, Xu S, Zhou J, Mao J, Ma X,
Wang M and Cong YS: Human UFSP1 translated from an upstream
near-cognate initiation codon functions as an active UFM1-specific
protease. J Biol Chem. 298:1020162022. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mashahreh B, Hassouna F, Soudah N,
Cohen-Kfir E, Strulovich R, Haitin Y and Wiener R: Trans-binding of
UFM1 to UBA5 stimulates UBA5 homodimerization and ATP binding.
FASEB J. 32:2794–2802. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Soudah N, Padala P, Hassouna F, Kumar M,
Mashahreh B, Lebedev AA, Isupov MN, Cohen-Kfir E and Wiener R: An
N-terminal extension to UBA5 adenylation domain boosts UFM1
activation: isoform-specific differences in ubiquitin-like protein
activation. J Mol Biol. 431:463–478. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kumar M, Padala P, Fahoum J, Hassouna F,
Tsaban T, Zoltsman G, Banerjee S, Cohen-Kfir E, Dessau M,
Rosenzweig R, et al: Structural basis for UFM1 transfer from UBA5
to UFC1. Nat Commun. 12:57082021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tolmachova KA, Farnung J, Liang JR, Corn
JE and Bode JW: Facile preparation of UFMylation activity-based
probes by chemoselective installation of electrophiles at the
C-terminus of recombinant UFM1. ACS Cent Sci. 8:756–762. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kumari S, Banerjee S, Kumar M, Hayashi A,
Solaimuthu B, Cohen-Kfir E, Shaul YD, Rouvinski A and Wiener R:
Overexpression of UBA5 in cells mimics the phenotype of cells
lacking UBA5. Int J Mol Sci. 23:74452022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liess AKL, Kucerova A, Schweimer K, Yu L,
Roumeliotis TI, Diebold M, Dybkov O, Sotriffer C, Urlaub H,
Choudhary JS, et al: Autoinhibition mechanism of the
ubiquitin-conjugating enzyme UBE2S by autoubiquitination.
Structure. 27:1195–1210. e72019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tatsumi K, Sou YS, Tada N, Nakamura E,
Iemura S, Natsume T, Kang SH, Chung CH, Kasahara M, Kominami E, et
al: A novel type of E3 ligase for the Ufm1 conjugation system. J
Biol Chem. 285:5417–5427. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Peter JJ, Magnussen HM, DaRosa PA,
Millrine D, Matthews SP, Lamoliatte F, Sundaramoorthy R, Kopito RR
and Kulathu Y: A non-canonical scaffold-type E3 ligase complex
mediates protein UFMylation. EMBO J. 41:e1110152022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Deshaies RJ and Joazeiro CA: RING domain
E3 ubiquitin ligases. Annu Rev Biochem. 78:399–434. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Deol KK, Lorenz S and Strieter ER:
Enzymatic logic of ubiquitin chain assembly. Front Physiol.
10:8352019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jiang Q, Wang Y, Xiang M, Hua J, Zhou T,
Chen F, Lv X, Huang J and Cai Y: UFL1, a UFMylation E3 ligase,
plays a crucial role in multiple cellular stress responses. Front
Endocrinol (Lausanne). 14:11231242023. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yoo HM, Kang SH, Kim JY, Lee JE, Seong MW,
Lee SW, Ka SH, Sou YS, Komatsu M, Tanaka K, et al: Modification of
ASC1 by UFM1 is crucial for ERα transactivation and breast cancer
development. Mol Cell. 56:261–274. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lee L, Oliva AB, Martinez-Balsalobre E,
Churikov D, Peter J, Rahmouni D, Audoly G, Azzoni V, Audebert S,
Camoin L, et al: UFMylation of MRE11 is essential for telomere
length maintenance and hematopoietic stem cell survival. Sci Adv.
7:eabc73712021. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Qin B, Yu J, Nowsheen S, Wang M, Tu X, Liu
T, Li H, Wang L and Lou Z: UFL1 promotes histone H4 ufmylation and
ATM activation. Nat Commun. 10:12422019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang Z, Gong Y, Peng B, Shi R, Fan D, Zhao
H, Zhu M, Zhang H, Lou Z, Zhou J, et al: MRE11 UFMylation promotes
ATM activation. Nucleic Acids Res. 47:4124–4135. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu J, Guan D, Dong M, Yang J, Wei H,
Liang Q, Song L, Xu L, Bai J, Liu C, et al: UFMylation maintains
tumour suppressor p53 stability by antagonizing its ubiquitination.
Nat Cell Biol. 22:1056–1063. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lee JH and Paull TT: Activation and
regulation of ATM kinase activity in response to DNA double-strand
breaks. Oncogene. 26:7741–7748. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Bakkenist CJ and Kastan MB: DNA damage
activates ATM through intermolecular autophosphorylation and dimer
dissociation. Nature. 421:499–506. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Qin B, Yu J, Nowsheen S, Zhao F, Wang L
and Lou Z: STK38 promotes ATM activation by acting as a reader of
histone H4 ufmylation. Sci Adv. 6:eaax82142020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Fang Z and Pan Z: Essential role of
ubiquitin-fold modifier 1 conjugation in DNA damage response. DNA
Cell Biol. 38:1030–1039. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Sun Y, Jiang X, Chen S, Fernandes N and
Price BD: A role for the Tip60 histone acetyltransferase in the
acetylation and activation of ATM. Proc Natl Acad Sci USA.
102:13182–13187. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Liu J, Wang Y, Song L, Zeng L, Yi W, Liu
T, Chen H, Wang M, Ju Z and Cong YS: A critical role of DDRGK1 in
endoplasmic reticulum homoeostasis via regulation of IRE1α
stability. Nat Commun. 8:141862017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Rusiecki R, Witkowski J and
Jaszczewska-Adamczak J: MDM2-p53 interaction inhibitors: The
current state-of-art and updated patent review (2010-Present).
Recent Pat Anticancer Drug Discov. 14:324–369. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang S, Zhao Y, Aguilar A, Bernard D and
Yang CY: Targeting the MDM2-p53 protein-protein interaction for new
cancer therapy: Progress and challenges. Cold Spring Harb Perspect
Med. 7:a0262452017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Adams KE, Medhurst AL, Dart DA and Lakin
ND: Recruitment of ATR to sites of ionising radiation-induced DNA
damage requires ATM and components of the MRN protein complex.
Oncogene. 25:3894–3904. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Vazifehmand R, Ali DS, Homaie FM,
Jalalvand FM, Othman Z, Deming C, Stanslas J and Sekawi Z: Effects
of HSV-G47Delta oncolytic virus on telomerase and telomere length
alterations in glioblastoma multiforme cancer stem cells under
hypoxia and normoxia conditions. Curr Cancer Drug Targets.
24:1262–1274. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Ufmylation of ASC1 is essential for breast
cancer development. Cancer Discov. 4:OF102014. View Article : Google Scholar
|
|
59
|
Chen Q, Xiao Y, Chai P, Zheng P, Teng J
and Chen J: ATL3 is a tubular ER-Phagy receptor for
GABARAP-mediated selective autophagy. Curr Biol. 29:846–855.
e62019. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Stephani M, Picchianti L, Gajic A,
Beveridge R, Skarwan E, de Medina Hernandez V, Mohseni A, Clavel M,
Zeng Y, Naumann C, et al: A cross-kingdom conserved ER-phagy
receptor maintains endoplasmic reticulum homeostasis during stress.
Elife. 9:e583962020. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Simsek D, Tiu GC, Flynn RA, Byeon GW,
Leppek K, Xu AF, Chang HY and Barna M: The mammalian
Ribo-interactome reveals ribosome functional diversity and
heterogeneity. Cell. 169:1051–1065. e182017. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Komatsu M, Inada T and Noda NN: The UFM1
system: Working principles, cellular functions, and
pathophysiology. Mol Cell. 84:156–169. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Ishimura R, Ito S, Mao G, Komatsu-Hirota
S, Inada T, Noda NN and Komatsu M: Mechanistic insights into the
roles of the UFM1 E3 ligase complex in ufmylation and
ribosome-associated protein quality control. Sci Adv.
9:eadh36352023. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Walczak CP, Leto DE, Zhang L, Riepe C,
Muller RY, DaRosa PA, Ingolia NT, Elias JE and Kopito RR: Ribosomal
protein RPL26 is the principal target of UFMylation. Proc Natl Acad
Sci USA. 116:1299–1308. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Scavone F, Gumbin SC, Da Rosa PA and
Kopito RR: RPL26/uL24 UFMylation is essential for
ribosome-associated quality control at the endoplasmic reticulum.
Proc Natl Acad Sci USA. 120:e22203401202023. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Wang L, Xu Y, Rogers H, Saidi L, Noguchi
CT, Li H, Yewdell JW, Guydosh NR and Ye Y: UFMylation of RPL26
links translocation-associated quality control to endoplasmic
reticulum protein homeostasis. Cell Res. 30:5–20. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
French SW, Masouminia M, Samadzadeh S,
Tillman BC, Mendoza A and French BA: Role of protein quality
control failure in alcoholic hepatitis pathogenesis. Biomolecules.
7:112017. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Luo H, Jiao QB, Shen CB, Gong WY, Yuan JH,
Liu YY, Chen Z, Liu J, Xu XL, Cong YS and Zhang XW: UFMylation of
HRD1 regulates endoplasmic reticulum homeostasis. FASEB J.
37:e232212023. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Wilkinson S: Emerging principles of
selective ER autophagy. J Mol Biol. 432:185–205. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Chino H and Mizushima N: ER-Phagy: Quality
control and turnover of endoplasmic reticulum. Trends Cell Biol.
30:384–398. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Picchianti L, de Medina Hernandez V, Zhan
N, Irwin NA, Groh R, Stephani M, Hornegger H, Beveridge R,
Sawa-Makarska J, Lendl T, et al: Shuffled ATG8 interacting motifs
form an ancestral bridge between UFMylation and autophagy. EMBO J.
42:e1120532023. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Stephani M, Picchianti L and Dagdas Y: C53
is a cross-kingdom conserved reticulophagy receptor that bridges
the gap betweenselective autophagy and ribosome stalling at the
endoplasmic reticulum. Autophagy. 17:586–587. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Klebanovych A, Vinopal S, Draberova E,
Sladkova V, Sulimenko T, Sulimenko V, Vosecká V, Macůrek L, Legido
A and Dráber P: C53 Interacting with UFM1-protein ligase 1
regulates microtubule nucleation in response to ER stress. Cells.
11:5552022. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Reggiori F and Molinari M: ER-phagy:
Mechanisms, regulation, and diseases connected to the lysosomal
clearance of the endoplasmic reticulum. Physiol Rev. 102:1393–1448.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Zhu H, Bhatt B, Sivaprakasam S, Cai Y, Liu
S, Kodeboyina SK, Patel N, Savage NM, Sharma A, Kaufman RJ, et al:
Ufbp1 promotes plasma cell development and ER expansion by
modulating distinct branches of UPR. Nat Commun. 10:10842019.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Ishimura R, El-Gowily AH, Noshiro D,
Komatsu-Hirota S, Ono Y, Shindo M, Hatta T, Abe M, Uemura T,
Lee-Okada HC, et al: The UFM1 system regulates ER-phagy through the
ufmylation of CYB5R3. Nat Commun. 13:78572022. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Liang JR, Lingeman E, Luong T, Ahmed S,
Muhar M, Nguyen T, Olzmann JA and Corn JE: A genome-wide ER-phagy
screen highlights key roles of mitochondrial metabolism and
ER-resident UFMylation. Cell. 180:1160–1177. e202020. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Raymundo DP, Doultsinos D, Guillory X,
Carlesso A, Eriksson LA and Chevet E: Pharmacological targeting of
IRE1 in cancer. Trends Cancer. 6:1018–1030. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Cai Y, Zhu G, Liu S, Pan Z, Quintero M,
Poole CJ, Lu C, Zhu H, Islam B, Riggelen JV, et al: Indispensable
role of the Ubiquitin-fold modifier 1-specific E3 ligase in
maintaining intestinal homeostasis and controlling gut
inflammation. Cell Discov. 5:72019. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Hwang J and Qi L: Quality control in the
endoplasmic reticulum: Crosstalk between ERAD and UPR pathways.
Trends Biochem Sci. 43:593–605. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Lemaire K, Moura RF, Granvik M,
Igoillo-Esteve M, Hohmeier HE, Hendrickx N, Newgard CB, Waelkens E,
Cnop M and Schuit F: Ubiquitin fold modifier 1 (UFM1) and its
target UFBP1 protect pancreatic beta cells from ER stress-induced
apoptosis. PLoS One. 6:e185172011. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
DaRosa PA, Penchev I, Gumbin SC, Scavone
F, Wachalska M, Paulo JA, Ordureau A, Peter JJ, Kulathu Y, Harper
JW, et al: UFM1 E3 ligase promotes recycling of 60S ribosomal
subunits from the ER. Nature. 627:445–452. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Makhlouf L, Peter JJ, Magnussen HM, Thakur
R, Millrine D, Minshull TC, Harrison G, Varghese J, Lamoliatte F,
Foglizzo M, et al: The UFM1 E3 ligase recognizes and releases 60S
ribosomes from ER translocons. Nature. 627:437–444. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
von der Malsburg K, Shao S and Hegde RS:
The ribosome quality control pathway can access nascent
polypeptides stalled at the Sec61 translocon. Mol Biol Cell.
26:2168–2180. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Brandman O and Hegde RS:
Ribosome-associated protein quality control. Nat Struct Mol Biol.
23:7–15. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Wan L, Juszkiewicz S, Blears D, Bajpe PK,
Han Z, Faull P, Mitter R, Stewart A, Snijders AP, Hegde RS and
Svejstrup JQ: Translation stress and collided ribosomes are
co-activators of cGAS. Mol Cell. 81:2808–2822. e102021. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Inada T: Quality controls induced by
aberrant translation. Nucleic Acids Res. 48:1084–1096. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Wang L, Xu Y, Yun S, Yuan Q,
Satpute-Krishnan P and Ye Y: SAYSD1 senses UFMylated ribosome to
safeguard co-translational protein translocation at the endoplasmic
reticulum. Cell Rep. 42:1120282023. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Joazeiro CAP: Mechanisms and functions of
ribosome-associated protein quality control. Nat Rev Mol Cell Biol.
20:368–383. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Cai Y, Singh N and Li H: Essential role of
Ufm1 conjugation in the hematopoietic system. Exp Hematol.
44:442–446. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Hattangadi SM, Wong P, Zhang L, Flygare J
and Lodish HF: From stem cell to red cell: Regulation of
erythropoiesis at multiple levels by multiple proteins, RNAs, and
chromatin modifications. Blood. 118:6258–6268. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Tatsumi K, Yamamoto-Mukai H, Shimizu R,
Waguri S, Sou YS, Sakamoto A, Taya C, Shitara H, Hara T, Chung CH,
et al: The Ufm1-activating enzyme Uba5 is indispensable for
erythroid differentiation in mice. Nat Commun. 2:1812011.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Zhang M, Zhu X, Zhang Y, Cai Y, Chen J,
Sivaprakasam S, Gurav A, Pi W, Makala L, Wu J, et al: RCAD/Ufl1, a
Ufm1 E3 ligase, is essential for hematopoietic stem cell function
and murine hematopoiesis. Cell Death Differ. 22:1922–1934. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Zhang Y, Zhang M, Wu J, Lei G and Li H:
Transcriptional regulation of the Ufm1 conjugation system in
response to disturbance of the endoplasmic reticulum homeostasis
and inhibition of vesicle trafficking. PLoS One. 7:e485872012.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Di Rocco M, Rusmini M, Caroli F, Madeo A,
Bertamino M, Marre-Brunenghi G and Ceccherini I: Novel
spondyloepimetaphyseal dysplasia due to UFSP2 gene mutation. Clin
Genet. 93:671–674. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Zhang G, Tang S, Wang H, Pan H, Zhang W,
Huang Y, Kong J and Wang Y, Gu J and Wang Y: Corrigendum to
UFSP2-related spondyloepimetaphyseal dysplasia: A confirmatory
report. Eur J Med Genet. 63:1040212020. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Franceschi R, Iascone M, Maitz S,
Marchetti D, Mariani M, Selicorni A, Soffiati M and Maines E: A
missense mutation in DDRGK1 gene associated to Shohat-type
spondyloepimetaphyseal dysplasia: Two case reports and a review of
literature. Am J Med Genet A. 188:2434–2437. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Weisz-Hubshman M, Egunsula AT, Dawson B,
Castellon A, Jiang MM, Chen-Evenson Y, Zhiyin Y, Lee B and Bae Y:
DDRGK1 is required for the proper development and maintenance of
the growth plate cartilage. Hum Mol Genet. 31:2820–2830. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Ni M, Afroze B, Xing C, Pan C, Shao Y, Cai
L, Cantarel BL, Pei J, Grishin NV, Hewson S, et al: A pathogenic
UFSP2 variant in an autosomal recessive form of pediatric
neurodevelopmental anomalies and epilepsy. Genet Med. 23:900–908.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Muona M, Ishimura R, Laari A, Ichimura Y,
Linnankivi T, Keski-Filppula R, Herva R, Rantala H, Paetau A,
Pöyhönen M, et al: Biallelic variants in UBA5 link dysfunctional
ufm1 ubiquitin-like modifier pathway to severe infantile-onset
encephalopathy. Am J Hum Genet. 99:683–694. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Zhang J, Zhu H, Liu S, Quintero M, Zhu T,
Xu R, Cai Y, Han Y and Li H: Deficiency of murine UFM1-Specific E3
ligase causes microcephaly and inflammation. Mol Neurobiol.
59:6363–6372. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Arnadottir GA, Jensson BO, Marelsson SE,
Sulem G, Oddsson A, Kristjansson RP, Benonisdottir S, Gudjonsson
SA, Masson G, Thorisson GA, et al: Compound heterozygous mutations
in UBA5 causing early-onset epileptic encephalopathy in two
sisters. BMC Med Genet. 18:1032017. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Nahorski MS, Maddirevula S, Ishimura R,
Alsahli S, Brady AF, Begemann A, Mizushima T, Guzmán-Vega FJ, Obata
M, Ichimura Y, et al: Biallelic UFM1 and UFC1 mutations expand the
essential role of ufmylation in brain development. Brain.
141:1934–1945. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Serrano RJ, Oorschot V, Palipana D,
Calcinotto V, Sonntag C, Ramm G and Bryson-Richardson RJ: Genetic
model of UBA5 deficiency highlights the involvement of both
peripheral and central nervous systems and identifies widespread
mitochondrial abnormalities. Brain Commun. 5:fcad3172023.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Cabrera-Serrano M, Coote DJ, Azmanov D,
Goullee H, Andersen E, McLean C, Davis M, Ishimura R, Stark Z,
Vallat JM, et al: A homozygous UBA5 pathogenic variant causes a
fatal congenital neuropathy. J Med Genet. 57:835–842. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Mignon-Ravix C, Milh M, Kaiser CS, Daniel
J, Riccardi F, Cacciagli P, Nagara M, Busa T, Liebau E and Villard
L: Abnormal function of the UBA5 protein in a case of early
developmental and epileptic encephalopathy with suppression-burst.
Hum Mutat. 39:934–938. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Duan R, Shi Y, Yu L, Zhang G, Li J, Lin Y,
Guo J, Wang J, Shen L, Jiang H, et al: UBA5 mutations cause a new
form of autosomal recessive cerebellar ataxia. PLoS One.
11:e01490392016. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Al-Saady ML, Kaiser CS, Wakasuqui F,
Korenke GC, Waisfisz Q, Polstra A, Pouwels PJW, Bugiani M, van der
Knaap MS, Lunsing RJ, et al: Homozygous UBA5 variant leads to
hypomyelination with thalamic involvement and axonal neuropathy.
Neuropediatrics. 52:489–494. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Yiu SPT, Zerbe C, Vanderwall D, Huttlin
EL, Weekes MP and Gewurz BE: An Epstein-Barr virus protein
interaction map reveals NLRP3 inflammasome evasion via MAVS
UFMylation. Mol Cell. 83:2367–2386. e152023. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Snider DL, Park M, Murphy KA, Beachboard
DC and Horner SM: Signaling from the RNA sensor RIG-I is regulated
by ufmylation. Proc Natl Acad Sci USA. 119:e21195311192022.
View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Xie Z, Fang Z and Pan Z: Ufl1/RCAD, a Ufm1
E3 ligase, has an intricate connection with ER stress. Int J Biol
Macromol. 135:760–767. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
DeSantis CE, Ma J, Gaudet MM, Newman LA,
Miller KD, Sauer A, Jemal A and Siegel RL: Breast cancer
statistics, 2019. CA Cancer J Clin. 69:438–451. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Li Z, Wei H, Li S, Wu P and Mao X: The
role of progesterone receptors in breast cancer. Drug Des Devel
Ther. 16:305–314. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Jozwik KM and Carroll JS: Pioneer factors
in hormone-dependent cancers. Nat Rev Cancer. 12:381–385. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Yoo HM, Park JH, Kim JY and Chung CH:
Modification of ERα by UFM1 increases its stability and
transactivity for breast cancer development. Mol Cells. 45:425–434.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Yoo HM, Park JH, Jeon YJ and Chung CH:
Ubiquitin-fold modifier 1 acts as a positive regulator of breast
cancer. Front Endocrinol (Lausanne). 6:362015. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Le Romancer M, Poulard C, Cohen P, Sentis
S, Renoir JM and Corbo L: Cracking the estrogen receptor's
posttranslational code in breast tumors. Endocr Rev. 32:597–622.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Schulten HJ: Pleiotropic effects of
metformin on cancer. Int J Mol Sci. 19:28502018. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Yang J, Zhou Y, Xie S, Wang J, Li Z, Chen
L, Mao M, Chen C, Huang A, Chen Y, et al: Metformin induces
ferroptosis by inhibiting UFMylation of SLC7A11 in breast cancer. J
Exp Clin Cancer Res. 40:2062021. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Mao M, Chen Y, Yang J, Cheng Y, Xu L, Ji
F, Zhou J, Zhang X, Li Z, Chen C, et al: Modification of PLAC8 by
UFM1 affects tumorous proliferation and immune response by
impacting PD-L1 levels in triple-negative breast cancer. J
Immunother Cancer. 10:e0056682022. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Feng X, Wei Z, Tao X, Du Y, Wu J, Yu Y, Yu
H and Zhao H: PLAC8 promotes the autophagic activity and improves
the growth priority of human trophoblast cells. FASEB J.
35:e213512021. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Hsu JM, Li CW, Lai YJ and Hung MC:
Posttranslational modifications of PD-L1 and their applications in
cancer therapy. Cancer Res. 78:6349–6353. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Lim SO, Li CW, Xia W, Cha JH, Chan LC, Wu
Y, Chang SS, Lin WC, Hsu JM, Hsu YH, et al: Deubiquitination and
stabilization of PD-L1 by CSN5. Cancer Cell. 30:925–939. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Kulsuptrakul J, Wang R, Meyers NL, Ott M
and Puschnik AS: A genome-wide CRISPR screen identifies UFMylation
and TRAMP-like complexes as host factors required for hepatitis A
virus infection. Cell Rep. 34:1088592021. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
French SW, Bardag-Gorce F, Li J, French BA
and Oliva J: Mallory-Denk body pathogenesis revisited. World J
Hepatol. 2:295–301. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Bardag-Gorce F, Oliva J, Villegas J,
Fraley S, Amidi F, Li J, Dedes J, French B and French SW:
Epigenetic mechanisms regulate Mallory Denk body formation in the
livers of drug-primed mice. Exp Mol Pathol. 84:113–121. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Li J, Li XM, Caudill M, Malysheva O,
Bardag-Gorce F, Oliva J, French BA, Gorce E, Morgan K, Kathirvel E,
et al: Betaine feeding prevents the blood alcohol cycle in rats fed
alcohol continuously for 1 month using the rat intragastric tube
feeding model. Exp Mol Pathol. 91:540–547. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Oliva J, Bardag-Gorce F, Li J, French BA,
Nguyen SK, Lu SC and French SW: Betaine prevents Mallory-Denk body
formation in drug-primed mice by epigenetic mechanisms. Exp Mol
Pathol. 86:77–86. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Liu H, Gong M, French BA, Li J, Tillman B
and French SW: Mallory-Denk body (MDB) formation modulates
Ufmylation expression epigenetically in alcoholic hepatitis (AH)
and non-alcoholic steatohepatitis (NASH). Exp Mol Pathol.
97:477–483. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Dong Y and Wang A: Aberrant DNA
methylation in hepatocellular carcinoma tumor suppression (Review).
Oncol Lett. 8:963–968. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Esteller M, Corn PG, Baylin SB and Herman
JG: A gene hypermethylation profile of human cancer. Cancer Res.
61:3225–3229. 2001.PubMed/NCBI
|
|
133
|
Liu H, Li J, Tillman B, French BA and
French SW: Ufmylation and FATylation pathways are downregulated in
human alcoholic and nonalcoholic steatohepatitis, and mice fed DDC,
where Mallory-Denk bodies (MDBs) form. Exp Mol Pathol. 97:81–88.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Li H, Rauch T, Chen ZX, Szabo PE, Riggs AD
and Pfeifer GP: The histone methyltransferase SETDB1 and the DNA
methyltransferase DNMT3A interact directly and localize to
promoters silenced in cancer cells. J Biol Chem. 281:19489–19500.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Fagerberg L, Hallstrom BM, Oksvold P,
Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S,
Danielsson A, Edlund K, et al: Analysis of the human
tissue-specific expression by genome-wide integration of
transcriptomics and antibody-based proteomics. Mol Cell Proteomics.
13:397–406. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Chen E, Zhou B, Bian S, Ni W and Chen Z:
The lncRNA B3GALT5-AS1 functions as an HCC suppressor by regulating
the miR-934/UFM1 axis. J Oncol. 2021:17764322021. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Yang S, Yang R, Wang H, Huang Y and Jia Y:
CDK5RAP3 deficiency restrains liver regeneration after partial
hepatectomy triggering endoplasmic reticulum stress. Am J Pathol.
190:2403–2416. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Sheng L, Li J, Rao S, Yang Z and Huang Y:
Cyclin-dependent kinase 5 regulatory subunit associated protein 3:
Potential functions and implications for development and disease.
Front Oncol. 11:7604292021. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Bade BC and Cruz CS: Lung cancer 2020:
Epidemiology, etiology, and prevention. Clin Chest Med. 41:1–24.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Xia C, Dong X, Li H, Cao M, Sun D, He S,
Yang F, Yan X, Zhang S, Li N and Chen W: Cancer statistics in China
and United States, 2022: Profiles, trends, and determinants. Chin
Med J (Engl). 135:584–590. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Nabet BY, Hamidi H, Lee MC, Banchereau R,
Morris S, Adler L, Gayevskiy V, Elhossiny AM, Srivastava MK, Patil
NS, et al: Immune heterogeneity in small-cell lung cancer and
vulnerability to immune checkpoint blockade. Cancer Cell.
42:429–443. e42024. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Wang X, Wu Y, Gu J and Xu J:
Tumor-associated macrophages in lung carcinoma: From mechanism to
therapy. Pathol Res Pract. 229:1537472022. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Hu J, Zhang L, Xia H, Yan Y, Zhu X, Sun F,
Sun L, Li S, Li D, Wang J, et al: Tumor microenvironment remodeling
after neoadjuvant immunotherapy in non-small cell lung cancer
revealed by single-cell RNA sequencing. Genome Med. 15:142023.
View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Xu D, Zhang D, Wei W and Zhang C: UBA5
inhibition restricts lung adenocarcinoma via blocking macrophage M2
polarization and cisplatin resistance. Exp Cell Res.
440:1141482024. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Kim CH, Nam HS, Lee EH, Han SH, Cho HJ,
Chung HJ, Lee NS, Choi SJ, Kim H, Ryu JS, et al: Overexpression of
a novel regulator of p120 catenin, NLBP, promotes lung
adenocarcinoma proliferation. Cell Cycle. 12:2443–2453. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Zhou J, Ma X, Xu L, Liang Q, Mao J, Liu J,
Wang M, Yuan J and Cong YS: Genomic profiling of the UFMylation
family genes identifies UFSP2 as a potential tumour suppressor in
colon cancer. Clin Transl Med. 11:e6422021. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Smyth EC, Nilsson M, Grabsch HI, van
Grieken NC and Lordick F: Gastric cancer. Lancet. 396:635–648.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Karimi P, Islami F, Anandasabapathy S,
Freedman ND and Kamangar F: Gastric cancer: Descriptive
epidemiology, risk factors, screening, and prevention. Cancer
Epidemiol Biomarkers Prev. 23:700–713. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Lin M, Lian NZ, Cao LL, Huang CM, Zheng
CH, Li P, Xie JW, Wang JB, Lu J, Chen QY, et al: Down-regulated
expression of CDK5RAP3 and UFM1 suggests a poor prognosis in
gastric cancer patients. Front Oncol. 12:9277512022. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Shiwaku H, Yoshimura N, Tamura T, Sone M,
Ogishima S, Watase K, Tagawa K and Okazawa H: Suppression of the
novel ER protein Maxer by mutant ataxin-1 in Bergman glia
contributes to non-cell-autonomous toxicity. EMBO J. 29:2446–2460.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Wu J, Lei G, Mei M, Tang Y and Li H: A
novel C53/LZAP-interacting protein regulates stability of C53/LZAP
and DDRGK domain-containing Protein 1 (DDRGK1) and modulates
NF-kappaB signaling. J Biol Chem. 285:15126–15136. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Lin JX, Xie XS, Weng XF, Zheng CH, Xie JW,
Wang JB, Lu J, Chen QY, Cao LL, Lin M, et al: Low expression of
CDK5RAP3 and DDRGK1 indicates a poor prognosis in patients with
gastric cancer. World J Gastroenterol. 24:3898–3907. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Xi P, Ding D, Zhou J, Wang M and Cong YS:
DDRGK1 regulates NF-κB activity by modulating IκBα stability. PLoS
One. 8:e642312013. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Lin JX, Xie XS, Weng XF, Qiu SL, Yoon C,
Lian NZ, Xie JW, Wang JB, Lu J, Chen QY, et al: UFM1 suppresses
invasive activities of gastric cancer cells by attenuating the
expres7sion of PDK1 through PI3K/AKT signaling. J Exp Clin Cancer
Res. 38:4102019. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Zhou Y, Ye X, Zhang C, Wang J, Guan Z, Yan
J, Xu L, Wang K, Guan D, Liang Q, et al: Ufl1 deficiency causes
kidney atrophy associated with disruption of endoplasmic reticulum
homeostasis. J Genet Genomics. 48:403–410. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Momayyezi P, Bilev E, Ljunggren HG and
Hammer Q: Viral escape from NK-cell-mediated immunosurveillance: A
lesson for cancer immunotherapy? Eur J Immunol. 53:e23504652023.
View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Zhou J, Ma X, He X, Chen B, Yuan J, Jin Z,
Li L, Wang Z, Xiao Q, Cai Y, et al: Dysregulation of PD-L1 by
UFMylation imparts tumor immune evasion and identified as a
potential therapeutic target. Proc Natl Acad Sci USA.
120:e22157321202023. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Ma EH, Poffenberger MC, Wong AH and Jones
RG: The role of AMPK in T cell metabolism and function. Curr Opin
Immunol. 46:45–52. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
He C, Xing X, Chen HY, Gao M, Shi J, Xiang
B, Xiao X, Sun Y, Yu H, Xu G, et al: UFL1 ablation in T cells
suppresses PD-1 UFMylation to enhance anti-tumor immunity. Mol
Cell. 84:1120–1138. e82024. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Zhu MMT, Shenasa E and Nielsen TO:
Sarcomas: Immune biomarker expression and checkpoint inhibitor
trials. Cancer Treat Rev. 91:1021152020. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Brown HK, Schiavone K, Gouin F, Heymann MF
and Heymann D: Biology of bone sarcomas and new therapeutic
developments. Calcif Tissue Int. 102:174–195. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Meng H, Ai H, Li D, Jiang X, Zhang H, Xu J
and Huang S: Bombyx mori UFBP1 regulates apoptosis and promotes
BmNPV proliferation by affecting the expression of ER chaperone
BmBIP. Int J Biol Macromol. 283:1376812024. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Wang K, Chen S, Wu Y, Wang Y, Lu Y, Sun Y
and Chen Y: The ufmylation modification of ribosomal protein L10 in
the development of pancreatic adenocarcinoma. Cell Death Dis.
14:3502023. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
MacLeod G, Bozek DA, Rajakulendran N,
Monteiro V, Ahmadi M, Steinhart Z, Kushida MM, Yu H, Coutinho FJ,
Cavalli FMG, et al: Genome-wide CRISPR-Cas9 screens expose genetic
vulnerabilities and mechanisms of temozolomide sensitivity in
glioblastoma stem cells. Cell Rep. 27:971–986.e9. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Wang S, Jia M, Su M, Hu X, Li J, Xu Y and
Qiu W: Ufmylation is activated in renal cancer and is not
associated with von hippel-lindau mutation. DNA Cell Biol.
39:654–660. 2020. View Article : Google Scholar : PubMed/NCBI
|