Introduction
An aneurysm is defined as a permanent, irreversible,
localized dilatation of a blood vessel (1). The term abdominal aortic aneurysm
(AAA) is defined as an anomalous dilation of the infrarenal
abdominal aorta measuring ≥3.0 cm. A reduction in the thickness of
the abdominal aorta wall can cause it to bulge, resulting in a
permanent and progressively expanding focal dilation (2,3). The
prevalence of AAA in developed countries is estimated to be between
2 and 8%. If left untreated, it can progress to rupture and become
life-threatening, with a mortality rate of ≤80% (4). Treatment of AAA is dependent on a
number of factors, including the size, location and rate of
expansion of the aneurysm. In cases of small AAAs, the
recommendation is to conduct long-term imaging surveillance, such
as ultrasound or computed tomography (CT) scans, to confirm that
the aneurysm does not enlarge. An AAA with a diameter >5.5 cm in
men and 5.0 cm in women, or one that expands swiftly (0.5 cm in 6
months or >1.0 cm in 1 year), should be monitored, as it is at
an increased risk of rupture. Repair is usually recommended in such
cases. Treatment options include open surgical repair or
endovascular aortic aneurysm repair (5). Randomized trials have demonstrated
that for AAA ≤5.5 cm, surgical treatment has no advantage when
compared with close monitoring in terms of survival (6). Surveillance programs have been shown
to be efficacious, but the associated costs can place a marked
financial burden on healthcare systems (7), making it particularly important to
investigate alternatives. The identification of biomarkers
associated with aortic diameter and AAA growth may prove beneficial
in identifying patients who require additional monitoring.
Furthermore, biomarkers associated with AAA may represent potential
targets for future therapeutic interventions (7). Nevertheless, there are insufficient
efficacious medical therapies to either prevent the formation of
AAA or to constrain aneurysm growth subsequent to diagnosis. This
is due to the incomplete understanding of the mechanisms underlying
the formation of AAA. To the best of our knowledge, the majority of
previous studies have identified advanced age, male sex, tobacco
smoking, hypertension and family history of AAA as the most notable
risk factors for AAA (8,9). However, as the smooth muscle cells of
the abdominal aorta originate from the splenic mesoderm, the
pathogenesis of AAA is distinct from other forms of aortic
aneurysms (10). Genetic
regulation of gene expression has been well described in various
types of human tissue, and several studies have emphasized the
important role of genetic factors in the development and
progression of AAA (9,11). Genome-wide association studies
(GWAS) have pinpointed numerous single nucleotide polymorphisms
(SNPs) linked to AAA in both coding and non-coding regions of the
genome. These SNPs specific to AAA and their associated genes have
been associated with pathological factors, such as extracellular
matrix (ECM) organization, inflammation, lipid metabolism,
oxidative stress and smooth muscle cell function during AAA
development, suggesting that some of the pathological features of
AAA may originate from genetic abnormalities (11,12).
DNA methylation is an inheritable epigenetic change that takes
place at the CpG islands of gene promoters, leading to
transcriptional silencing and the modification of cytosine by DNA
methyltransferase. The most notable effect of DNA methylation on
AAA risk is the disruption of gene transcription related to
aneurysms, and it may be a sensitive epigenetic modification
(13). Furthermore, epigenetic
mechanisms, including DNA methylation, post-translational histone
modifications and non-coding RNAs, result in dysregulated aneurysm
gene expression levels. Nonetheless, these mechanisms do not lead
to substantial sequence variation, unlike genetic causes. Research
involving human tissue samples and animal models has revealed that
mRNA transcriptional changes are key in AAA development (14). Among the numerous RNA
modifications, m6A methylation is the most prevalent reversible and
dynamic eukaryotic mRNA transcriptional modification, and it is
also a key epigenetic mechanism in AAA (15).
m6A methylation
Methylation modification represents a considerable
alteration to nucleic acids and proteins. It is associated with a
number of pathological conditions, including cancer, the ageing
process and Alzheimer's disease (16,17).
DNA and histone methylation are among the most frequent methylation
modifications. DNA methylation, which is the addition of methyl
groups to DNA, is the primary mechanism by which gene transcription
is repressed or silenced. By contrast, histone methylation refers
to the process of methylation that occurs on the N-terminal
arginine or lysine residues of H3 and H4 histones (18,19).
This process is catalyzed by histone methyltransferases, which
facilitate the transfer of a methyl group to the substrate, and
mainly serves a role in forming heterochromatin, imprinting genes,
inactivating the X-chromosome and regulating transcription
(20). RNA methylation has been
identified as a key factor in RNA metabolism and a multitude of
cellular biological functions (21). The m6A modification is the most
prevalent methylation modification observed in mRNAs with ~25% of
mRNAs carrying ≥1 m6A site. These sites are enriched in proximity
to the termination codon and 30 untranslated regions (UTRs) of
human mRNAs and are present in a substantial number of mRNAs and
long non-coding RNAs (lncRNAs) (22).
m6A RNA methylation is facilitated by a
methyltransferase complex composed of methyltransferase 3-like
(METTL3), METTL14 and Wilms' tumor 1-associated protein (WTAP)
(23), and this complex has been
observed to accelerate the processing time of mRNA precursors and
their subsequent export from the nucleus. The methyltransferase
complex is capable of regulating the eukaryotic transcriptome,
influencing processes such as mRNA splicing, export, localization,
translation and mRNA stability (24). The function of m6A is dependent on
the presence of m6A readers, writers and erasers (25). In the nucleus, m6A may affect
alternative splicing of pre-mRNAs, as well as the storage and
export of mature mRNAs. The complexity of gene regulation mediated
by RNA is influenced by the reversible and dynamic nature of
distinct internal RNA modifications (26). m6A methylation modifications can
affect the expression of target genes, thereby regulating a wide
range of physiological processes, including self-renewal, invasion
and proliferation (27). In
addition to its involvement in neurodevelopmental regulation,
several studies indicate that m6A carries out a pivotal role in
tumor metabolism and growth (28,29).
The expression and activity of writers and erasers mainly determine
the abnormal m6A levels in cancer. The regulation of target mRNAs
subsequent to modification is predominantly determined by readers
(30). Additionally, m6A carries
out a pivotal role in spermatogenesis, stem cell differentiation,
immune response and other processes, e.g., regulating the stability
of the internal environment (31,32).
Previous research has identified m6A modifications as serving an
important role in the development of cardiovascular diseases
(33). Further studies are
required to determine the role of m6A modifications in AAA
pathophysiological processes, as this will provide new mechanistic
and therapeutic insights (Fig.
1).
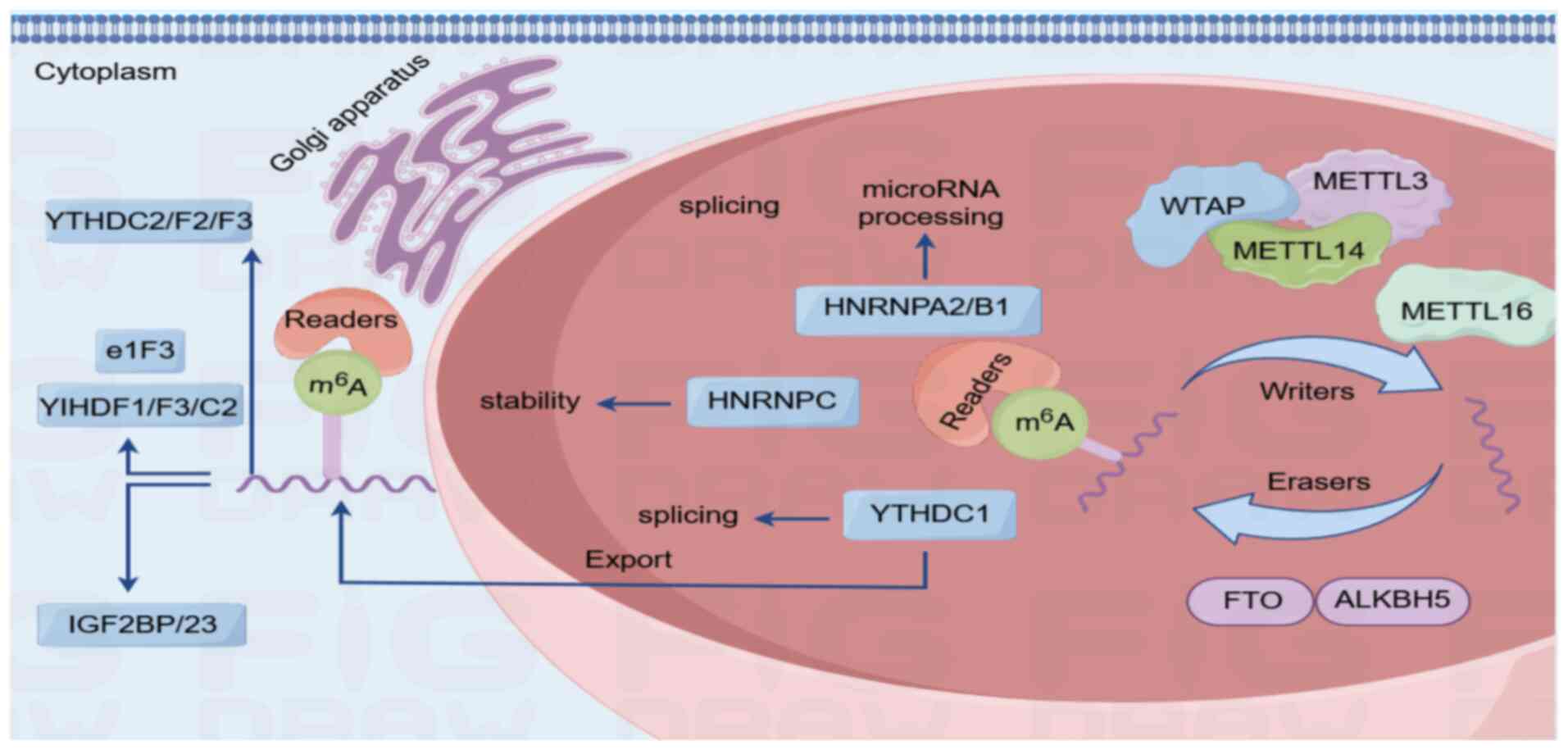 | Figure 1.Process of m6A methylation. m6A
methylation represents the most prevalent internal chemical
modification in eukaryotic mRNA and is dynamically regulated by a
tripartite enzymatic system (writers, erasers and readers) to
fine-tune RNA metabolism and cellular responses. Mechanistic
scheme: i) Methyltransferase complex (writers)-core components:
METTL3 (catalytic subunit), METTL14 (structural scaffold) and WTAP
(co-factor for localization). This complex catalyzes adenosine
methylation at conserved RRACH motifs (R, purine; H, non-guanine
base); ii) demethylases (erasers)-FTO and ALKBH5: Iron-dependent
oxidases that remove m6A marks to ensure reversibility for adaptive
gene regulation; and iii) m6A-binding proteins (readers), including
the YTHDF family (YTHDF1/2/3): Direct mRNA fate. YTHDF1 enhances
translation (ribosome recruitment), while YTHDF2 targets
transcripts for decay (localization to processing bodies) and
IGF2BP proteins stabilize mRNAs in an m6A-dependent manner. FTO,
fat mass and obesity-associated protein; ALKBH5, ALKB homologue 5;
METTL3, methyltransferase 3-like; IGF2BP, recombinant insulin like
growth factor 2 mRNA binding protein 2k; YTHDF1, YTH domain family
protein 1; YTHDC, YTH domain-containing protein 1; e1F3, eukaryotic
translation initiation factor 3; HNRNPC, heterogeneous nuclear
ribonucleoprotein C; m6A, N6-methyladenosine; WTAP, Wilms tumor 1
associated protein; HNRNPA2/B1, heterogeneous nuclear
ribonucleoprotein A2/B1. |
m6A writers
The m6A methyltransferase complex comprises METTL3,
METTL14 and WTAP as its principal constituents. Catalytic activity
resides in the METTL3-METTL14 dimeric unit, where METTL3 provides
enzymatic function, while METTL14 stabilizes the structure and
guides RNA binding. Together, they coordinate site-specific m6A
deposition across RNA polymerase II transcripts. The METTL3-METTL14
heterodimer catalyzes the majority of m6A modifications in
eukaryotic mRNAs (34). WTAP, a
regulatory partner of this methyltransferase complex, recruits RNA
substrates and directs methyl group deposition (26). In addition, KIAA1429 governs m6A
installation at 3′UTRs through dual mechanisms (both dependent on
and independent of m6A modification) to modulate RNA metabolism
spanning splicing, maturation, translation and decay. These
regulatory effects extend beyond mRNAs to include lncRNAs and
circular RNAs (circRNAs) (35). In
parallel, ZC3H13 promotes the nuclear translocation of the
methyltransferase complex, functioning as an essential assembly
cofactor (36). Vir-like m6A
methyltransferase associated exhibits substrate specificity,
preferentially marking mRNA regions near 3′UTRs and termination
codons for methylation (37).
Meanwhile, the METTL5-tRNA methyltransferase subunit 11-2 (TRMT112)
complex generates unique m6A signatures in 18S rRNA, with METTL5
providing catalytic activity and TRMT112 serving as a structural
scaffold (38). Beyond canonical
methylation roles, METTL16 engages eukaryotic translation
initiation factor 3 subunit a/b and ribosomal RNAs to coordinate
translation initiation and mRNA-ribosome interactions (38).
m6A erasers
Fat mass and obesity-associated protein (FTO) and
ALKB homologue 5 (ALKBH5) are the principal m6A demethylases. FTO
was the first enzyme to be identified as an m6A demethylase. It is
primarily located in nucleosomes (nuclear speckles), which are rich
in RNA-binding proteins. It carries out an important role in the
regulation of energy homeostasis, adipogenesis and cellular
autophagy (39). The mechanism of
FTO demethylation involves the catalysis of m6A oxidation, which
produces intermediates that gradually demethylate and eventually
form adenosine (40). The second
m6A demethylase to be identified was ALKBH5, which has the capacity
to interact directly with RNA in the nucleus, thereby demethylating
m6A to adenosine without the production of intermediates (40,41).
In addition to its role in mRNA stability, splicing and
translation, ALKBH5 also carries out a key role in regulating the
tumor immune microenvironment and mediating the effects of
immunotherapy (42,43).
m6A readers
Methylated reading proteins are the executors of
specific regulatory functions of m6A methylation modification. They
are capable of recognizing and binding RNAs with the m6A
modification, thereby mediating various processes related to RNA
metabolic homeostasis (44).
Different methylated reading proteins exercise different biological
functions in m6A modification. Methylated reading proteins are
primarily comprised of the YTH domain family (YTH) structural
domain family [encompassing YTH N6-methyladenosine RNA binding
protein F (YTHDF) 1, 2 and 3] and YTH domain containing 1–2
(including YTHDC1 and 2) (45).
The YTH structural domains fulfill different functions through a
family of YTH proteins that are expressed in different regions of
the cell. YTHDC1 is widely expressed and mainly localized in the
nucleus, and YTHDF1-3 are mainly distributed in the cytoplasm and
can facilitate translation of mRNAs containing m6A modifications
(46). YTHDF1 has the capacity to
regulate the translation initiation process through its interaction
with initiation factors, thereby enhancing the translation
efficiency of target RNAs. YTHDF2 is capable of selectively binding
to RNAs bearing m6A modifications, thereby mediating a range of
processes pertaining to RNA metabolic homeostasis. YTHDF2
selectively binds to RNAs with m6A modifications and transports
them to the decay site to promote their degradation (40). YTHDC1 regulates the binding region
of target RNAs by recruiting and regulating pre-mRNA splicing
factors into the mRNA binding region of the target mRNAs. The mRNA
binding regions regulate mRNA splicing (47). YTHDC2 regulates the stability of
m6A-containing mRNAs by recruiting RNA degradation machinery
(48). Furthermore, YTHDC2
enhances translational efficiency by directly recognizing mRNAs
with m6A modifications (49).
Additionally, it can accelerate mRNA decay by modifying the head
region of the ribosomal 40S subunit, which promotes disassembly of
the ribosomal mRNA complex (50)
(Table I).
 | Table I.Common types of m6A methylases and
their functions. |
Table I.
Common types of m6A methylases and
their functions.
| Typology | Common
genes/proteins | Functionality |
|---|
| m6A Writer | METTL3 | An m6A
methyltransferase that acts as an enzyme facilitator to form a
METTL3-METTL14 heterodimer with METTL14 to catalyze m6A methylation
modification of RNA in vitro and in vivo. |
|
| METTL14 | An m6A
methyltransferase that acts as a heterologous activator to form a
METTL3-METTL14 heterodimer with METTL3 to catalyze m6A methylation
modification of RNA in vitro and in vivo. |
|
| WTAP | Targets RNA and
recruits the methyltransferase complex to RNA. |
|
| KIAA1429 | The ability to
promote m6A modification of the 3′-UTR terminus not only affects
the function of mRNAs, but also by affecting the function of
lncRNAs and circRNAs. |
|
| ZC3H13 | Induction of
methylase complex translocation into the nucleus as a novel
cofactor for the m6A methyltransferase complex. |
|
| VIRMA | A
methyltransferase-associated protein that mediates preferential
mRNA methylation near the 3′UTR and termination codon. |
|
| METTL5 | An m6A
methyltransferase that acts as the catalytic subunit to form a
METTL5-TRMT112 complex with TRMT112, which may be associated with
ribosome generation and function. |
|
| TRMT112 | As a heterologous
aptamer can form a METTL5-TRMT112 complex with METTL5, which may be
related to ribosome production and function. |
|
| METTL16 | An m6A
methyltransferase that promotes the assembly of translation
initiation complexes and facilitates the translation of multiple
mRNA transcripts. |
| m6A Eraser | FTO | An m6A demethylase,
which catalyzes the oxidation of m6A to generate intermediates that
are progressively demethylated and ultimately form adenosine, plays
important functions in the regulation of energy homeostasis,
adipogenesis and cellular autophagy. |
|
| ALKBH5 | An m6A demethylase
that carries out a role in mRNA stability, splicing and
translation; involved in nuclear export and processing of mRNAs and
plays an important role in regulating the tumor immune
microenvironment and mediating the effects of immunotherapy. |
| m6A Reader | YTHDF1 | Contains the YTH
structural domain, which regulates the translation initiation
process by interacting with initiation factors, thereby improving
the translation efficiency of the target RNA. |
|
| YTHDF2 | Contains a YTH
structural domain that selectively binds RNA with m6A modifications
and transports it to the decay site, promoting its
degradation. |
|
| YTHDF3 | Contains the YTH
structural domain, taking into account both YTHDF1 and YTHDF2
functions. It can collaborate with YTHDF1 and improve the
translation efficiency of mRNA; it can also promote the degradation
of m6A-containing mRNA by affecting YTHDF2-mediated RNA decay. |
|
| YTHDC1 | Contains a YTH
structural domain that regulates mRNA splicing by over-recruiting
and regulating pre-mRNA splicing factors into the binding region of
the target mRNA. |
|
| YTHDC2 | Contains a YTH
structural domain that recruits the RNA degradation machinery to
regulate the stability of m6A-containing mRNAs; improves
translational efficiency by directly recognizing mRNAs with m6A
modifications; also promotes the disassembly of ribosomal mRNA
complexes, thereby accelerating mRNA decay. |
Mechanism of m6A methylation in AAA
Inflammation and the immune
response
AAA pathogenesis involves a multifaceted interplay
of inflammatory and immune mechanisms. Central to this process is
ECM destabilization, driven by inflammatory cell-derived proteases
that degrade vascular structural components. The compromised ECM
integrity allows inflammatory cell migration through the vascular
adventitia into the media, amplifying local inflammation (51). Platelet-mediated upregulation of
osteopontin in macrophages and aortic tissues further exacerbates
inflammation, vascular remodeling and leukocyte adhesion to the
aneurysmal wall and intraluminal thrombus (ILT) (52). ILT perpetuates inflammation by
generating a microenvironment enriched with neutrophils, proteases
and reactive oxygen species (ROS), collectively weakening the
aortic wall (53,54). Emerging insights highlight
epigenetic regulation via m6A methylation in AAA progression. In
atherosclerotic models, METTL14 silencing promotes
anti-inflammatory M2 macrophage polarization, suppresses foam cell
formation and attenuates migration by stabilizing Myd88 mRNA
through m6A-dependent mechanisms (55). Meanwhile, activated macrophages are
a major source of NF-κB, and the NF-κB signaling pathway is an
important mechanism of action in AAA for the generation of
inflammatory responses (56).
Notably, m6A modification is an upstream regulation
of the NF-κB pathway, while it remains elusive whether m6A is able
to influence the inflammatory response in AAA by regulating the
NF-κB pathway (57). However,
previous research on m6A methylation modification in AAA indicates
that m6A methylation is capable of influencing the inflammatory
response process. A notable correlation between the levels of
macrophage infiltration and YTHDF3 expression was demonstrated by
Zhong et al (58).
Furthermore, METTL14 was also revealed to correlate with
inflammatory infiltration and neovascularization in AAA (58). Cellular senescence and microRNA
(miRNA or miR) dynamics further interconnect m6A with AAA
pathology. Sirtuin 1 (SIRT1) mitigates aneurysm formation by
inhibiting vascular senescence and pro-inflammatory factor
secretion (59). Furthermore,
miR34a has been demonstrated to increase aortic macrophage
infiltration and the expression of age-associated pro-inflammatory
secretory factors. Additionally, miR34a-mediated angiotensin (Ang)
II-induced inflammation in the abdominal aorta has been observed
following a direct reduction in SIRT1 expression (60). From a mechanistic perspective,
elevated levels of AAA-related miR-34a are contingent upon
increased expression of METTL3 in vascular smooth muscle cells
(VSMCs). This is due to the capacity of METTL3 to enhance miR-34a
maturation by recognizing DiGeorge syndrome critical region genes,
thereby promoting m6A methylation (60). Receptor-interacting
serine/threonine-protein kinase 3 (RIP3) is a kinase that carries
out a role in the process of cellular necrosis and can initiate the
cell necrosis process (61).
In the presence of the m6A reader YTHDF3, RIP3 has
been shown to induce vascular endothelial cell necrosis.
Furthermore, the debris produced by VSMC necrosis induces
inflammatory factors in neighboring VSMCS and recruits monocytes or
macrophages to the lesion site, which ultimately leads to the
deterioration of AAA. In addition, mRNA expression of inflammatory
factors [interleukin (IL)-6, TNF, C-C chemokine motif ligand 2 and
IFN] was reported to be markedly elevated; however, it remains
elusive whether m6A modifications regulate these cells (62). The three most prominent
inflammatory cell types involved in the AAA aortic wall are
immune-related lymphocytes, mast cells and macrophages (63). The pertinent lymphocyte populations
implicated in the inflammatory process within the aortic wall are T
and B lymphocytes. The most notable and prevalent cells within the
aneurysmal wall are CD4+ T cells. These cells secrete a range of
cytokines that control the dynamic metabolism of the ECM through
macrophage incorporation, ECM regulation and protein hydrolase
synthesis. Furthermore, T helper (Th) 1 is activated by IL-12 and
secretes IFN-γ, TNF-α and TNF-β through the STAT4 and T-bet
signaling pathways, thereby influencing macrophage activation and
markedly contributing to AAA formation (64,65).
Th2 represents the second important group of cells involved in the
inflammatory process of AAA. These cells are regarded as
anti-inflammatory cells (66).
IL-4 stimulates Th2 cells, inducing the differentiation of CD4+ T
cells into cells with a Th2 cell phenotype (67). m6A modification carries out an
important role in the regulation of T-cell homeostasis and
function. m6A not only has the potential to direct the
differentiation of Th cells, but the abrogation of METTL3 in T
cells also disrupts T-cell homeostasis (68). Mast cells can be activated by
degranulation, which results in the release of proteases and
inflammatory factors that promote AAA (69).
Furthermore, the key enzyme for m6A methylation,
METTL3, has been demonstrated to regulate mast cell proliferation
and effector functions by modulating the stability of IL-13 mRNA
(70). In vascular Parkinson's
disease, Qi et al (71)
observed that Knockdown of the FTO gene mediated by m6A RNA
methylation resulted in a reduction in the proportion of Th-cell
subsets in lymphocytes. It should be noted that the different
functional outcomes of m6A modifications depend largely on the cell
type and cellular environment associated with different m6A
‘readers’ (72). Similarly, Fu
et al (73) identified
notable differences in the expression of natural killer (NK)
CD56-bright cells and immature dendritic cells between different
m6A clusters in Gene Expression Omnibus (GEO) datasets. In a study
conducted by Li et al (74)
in GEO datasets, correlation analyses between the expression of
AAA-associated m6A regulators and infiltrating immune cell scores
in AAA tissues indicated that downregulation of METTL14 or
heterogeneous nuclear ribonucleoproteins C1/C2 and upregulation of
RNA binding motif protein 15B may inhibit central memory T cell,
macrophage and mast cell infiltration, and promote the aggregation
of various immune cells such as Tγδ and NK CD56-bright cells.
However, in the study by Wang et al (75), m6A methylation typing of patients
with AAA based on m6A score was conducted and a positive
correlation was identified between m6A scores and the majority of
Th and effector T cells. Considerable differences were observed in
various T cells across different m6A clusters, indicating a
potential association between m6A methylation regulators and the
effects of different T-cell profiles. This also indicates the
potential for cellular heterogeneity and imbalanced m6A methylation
expression levels in AAA, both at the pathological and molecular
levels. It is evident that inflammation and immunity carry out a
pivotal role in AAA development, and m6A may indirectly influence
this process through its impact on inflammatory and immune
cells.
Cell proliferation and apoptosis
From a physiological perspective, the abdominal
aorta is an elastic artery comprising an intimal, a middle membrane
and an outer membrane layers. The intima is a single layer of
endothelial cells on connective tissue. The mesentery consists of
ECM embedded with structural proteins (elastin and collagen). By
contrast, the ectoderm consists of fibroblasts and collagen fibers
(76). VSMCs are the major
cellular component of the aortic mesentery. Their interactions with
the mesenteric ECM help to maintain the structural and functional
integrity of the aortic wall. The elastin of the mesentery is also
synthesized primarily by VSMCs; thus, apoptosis of VSMCs and
disruption of the ECM affect the structure of the entire aortic
wall (77). Indeed, apoptosis of
VSMCs in the inner layers of the aortic wall is an early marker of
AAA pathogenesis. A reduction in VSMC density weakens the ability
of the aortic wall to maintain its integrity and limits the ability
of the stroma to be repaired, as VSMCs are essential for ECM
regeneration. A reduction in VSMC density and ECM regenerative
capacity make the aortic wall more susceptible to dilatation, which
leads to the onset and progression of AAA (78).
Furthermore, following neovascularization,
inflammatory cells such as macrophages migrate into the vessel
wall, releasing pro-inflammatory cytokines and matrix
metalloproteinases (MMPs), which stimulate smooth muscle cells to
produce MMPs and other proteases. This results in the breakdown of
collagen I and elastin, which in turn stimulates the migration of
further inflammatory cells, thereby initiating a cascade of aortic
wall breakdown (79). The
degradation of the ECM and apoptosis of VSMCs are pivotal factors
in the development of AAA from a histological perspective. m6A
methylation affects the degradation of ECM and the apoptosis of
VSMCs. In the context of intervertebral disc degeneration, Lei
et al (80) demonstrated
that m6A demethylation of Runx2 mRNA (a key regulator of skeletal
development and homeostasis) promoted ECM degradation by
upregulating MMPs. Zheng et al (81) also identified a role for the
METTL14/METTL3 complex in mediating nucleosome assembly protein 1
like (NAP1L)2 apoptosis through m6A methylation of NAP1L2 mRNA in
prostate cancer. m6A methylation of mRNA prompts NAP1L6 to interact
with YinYang-1 (YY1), which promotes the transcription of MMP2 and
MMP9, and ultimately activates the MMP signaling pathway. However,
there is currently a lack of experimental evidence as to whether
m6A methylation influences the degradation of ECM in cardiovascular
diseases.
At least, the impact of m6A methylation on VSMCs has
been established in the context of cardiovascular diseases. Zhao
et al (82) demonstrated
that METTL3 knockdown resulted in the attenuation of
phosphatidylinositol 3-kinase (PI3K) mRNA, which inactivates
PI3K/AKT signaling and inhibits VSMC phenotypic transition.
Furthermore, Fang et al (83) reported that knockdown of METTL3
facilitated the proliferation of human aortic smooth muscle cells
(HASMCs), whereas overexpression of METTL3 inhibited their
proliferation. This phenomenon may be associated with the fact that
METTL3 arrests HASMCs at the G2/M checkpoint and inactivates the
phosphorylation of cell division cycle 2 (CDC2). Conversely, METTL3
knockdown was observed to enhance the migratory and synthetic
phenotypes of HASMCs, whereas METTL3 gene overexpression inhibited
the migratory and synthetic phenotypes of HASMCs. It is noteworthy
that, in HASMCs overexpressing METTL3, the protein levels of MMP2,
MMP7 and MMP9 were reduced, whereas the expression levels of tissue
MMP inhibitor 3 were elevated.
Furthermore, Liao et al (84) observed that METTL3 promoted m6A
methylation of lncRNA activated by DNA damage, an essential lncRNA
for genomic stability, in a YTHDF2-dependent manner, thereby
affecting VSMC proliferation in aortic coarctation. The current
findings on m6A methylation modification in AAA similarly indicate
that m6A methylation can influence vascular cell biological
processes (85). RIP3 has been
demonstrated to promote AAA progression by promoting smooth muscle
cell necrosis. Furthermore, SMAD2/3-mediated increase in
METTL3-METTL14 complex levels was revealed to enhance the m6A
modification of RIP3 mRNA by promoting the binding between YTHDF3
and RIP3 mRNA, which in turn induced the necrosis of VSMCs and
promoted AAA progression (62).
This evidence suggests that m6A modification indirectly affects the
ECM and thus has effects on AAA. In a previous study on
colchicine-mediated retardation of AAA, colchicine increased global
mRNA stability through the inhibition of METTL14/YTHDC1-mediated
m6A modification. This ultimately prevented VSMC phenotypic
transition and apoptosis, thereby slowing AAA progression (85). Furthermore, Xu et al
(60) demonstrated that
METTL3/YTHDC1-mediated m6A modification carried out a regulatory
role in the biogenesis of circRBM33, which is derived from the exon
of the RBM33 gene. Furthermore, knockdown of circRBM33 has been
demonstrated to alleviate AAA by reducing ECM degradation.
Consequently, the regulatory role of m6A modification on the
proliferation and apoptosis of key VSMC cell types, as well as its
effect on ECM degradation, may represent an important contributing
factor in the development of AAA.
Regulation of miRNAs and circRNAs
miRNAs are evolutionarily conserved ncRNA molecules
(~22 nucleotides) that post-transcriptionally modulate gene
expression, targeting >1/3 of human genes (86,87).
Biogenesis begins with RNA polymerase II transcribing primary
miRNAs (pri-miRNAs), which are subsequently processed into
precursor hairpins (pre-miRNAs) by the Drosha-DiGeorge critical
region-8 (DGCR8) complex. Notably, a single pri-miRNA transcript
may encode multiple distinct miRNAs (88). Notably, extracellular miRNAs
exhibit remarkable stability in biofluids such as plasma,
positioning them as promising biomarkers for detecting pathological
states (89). In cardiovascular
pathologies, including hypertension, heart failure and
acute/chronic coronary syndromes, these small RNAs demonstrate
diagnostic potential and therapeutic applicability through targeted
gene regulation (90).
Therapeutic modulation of mRNA expression can be
precision-engineered through synthetic miRNA mimics (upregulating
targets) or anti-miR oligonucleotides (attenuating miRNA activity
via sequence-specific binding) (91). In AAA, miRNAs emerge as master
regulators coordinating ECM homeostasis, cellular reprogramming and
tissue repair across vascular compartments (92). Genome-wide studies have revealed
vascular-bed-specific miRNA signatures targeting endothelial cells,
immune populations and, notably, VSMCs, whose dysregulation is
directly associated with AAA pathomechanics (93). Mechanistically, miRNAs orchestrate
aneurysm initiation/progression through three interlinked axes: i)
VSMC plasticity control: Modulating proliferation/apoptosis balance
and contractile-synthetic phenotype switching; ii)
vasculo-inflammatory crosstalk: Regulating leukocyte infiltration
and endothelial activation; and iii) MMP dysregulation: Driving
collagen/elastin degradation thresholds (94). Emerging evidence indicates that
miRNAs functionally crosstalk with pivotal disease-driving axes in
AAA pathophysiology, particularly NF-κB (inflammatory activation),
PI3K/AKT (cellular survival)/MAPK (proliferation-apoptosis
balance), TGF-β/Wnt (matrix homeostasis) and p53/p21 (senescence
regulation). This multi-axis regulatory network underpins aneurysm
initiation and progression (95).
The regulatory landscape extends to circRNAs
(covalently closed non-coding transcripts generated via
exon-derived backsplicing of pre-mRNAs) (95). Distinguished by their notable
stability (resistant to exonucleases) and evolutionary
conservation, circRNAs exhibit tissue-enriched expression, yet
remain detectable in the circulation. Functioning as miRNA sponges,
they sequester miRNAs via complementary binding to derepress
downstream targets, a competing endogenous RNA mechanism with
therapeutic relevance (96).
Notably, AAA-associated circRNAs (such as circCCDC66, circCBFB and
hsa-circ000595) display altered expression profiles correlating
with disease stages (97). m6A
modification has been identified as a key factor contributing to
miRNA biogenesis. A previous study by Alarcón et al
(98) revealed that m6A was
prevalent in pri-miRNA transcripts and that m6A levels were not
markedly elevated by the presence of m6A.
Furthermore, the enrichment of m6A and the reduction
of m6A levels resulted in the overall downregulation of the
expression of most mature miRNAs (99). Downregulation of METTL3 also
diminished the association between DGCR8 and target pri-miRNAs,
which was accompanied by the nuclear accumulation of unprocessed
pri-miRNAs. Furthermore, mutual regulation between m6A and circRNAs
contributes to the biological properties and functions of circRNAs.
It is notable that m6A modifications are not only widespread in
circRNAs, but also that circRNAs exhibit different m6A patterns
compared with mRNAs (100).
During protein synthesis, circRNAs follow a non-canonical
translation pathway that is positively regulated by m6A levels.
Specifically, this m6A-driven translation is initiated through the
binding of YTHDF3 to the translation initiation factors eIF4G2 and
eIF3A (101). The impact of m6A
modifications on miRNAs and circRNAs has been established in the
context of cardiovascular disease. A study by Zhang et al
(102) revealed that a reduction
in the expression of METTL14 resulted in the inhibition of the
binding of methylated RNAs to the RNA splicing-associated protein
DGCR8. Furthermore, silencing of METTL14 was observed to markedly
inhibit the expression of miR-19a, while promoting the expression
of pre-miR-19a. Increased expression of METTL14 was observed to
considerably increase the expression of DGCR8 and methylated m6A.
Furthermore, silencing of miR-19a was observed to inhibit the
proliferation and invasion of atherosclerotic vascular endothelial
cells.
In a study on cardiac hypertrophy, Fang et al
(103) identified circPan3 as a
key inhibitor of cardiac hypertrophy through its targeting of the
miR-320-3p/heat shock protein 20 axis. This regulatory mechanism
was found to be mediated by the alkylated DNA repair protein ALKBH5
and to be associated with m6A methylation regulation. Previous
findings on m6A methylation modification in AAA similarly revealed
the effect of m6A on the regulatory role of miRNAs vs. circRNAs.
Zhong et al (58) revealed
that knockdown of METTL3 inhibited AAA formation by identifying
that treatment of apolipoprotein E-deficient mice with Ang II,
while METTL3 overexpression played the opposite role. This may be
attributed to the METTL3-dependent methylation of m6A, which
facilitates the maturation of pri-miR34a via DGCR8. In addition,
overexpression of miR-34a was observed to notably reduce SIRT1
expression, thereby exacerbating the formation of AAA. Conversely,
miR-34a deficiency was revealed to have the opposite effect.
Furthermore, the protective effect of METTL3 deficiency on AAA
formation was partially affected by knockdown of miR-34a or forced
expression of SIRT1, respectively.
In a previous study, Xu et al (60) observed elevated m6A levels of
circRBM33 in VSMCs with Ang II-induced AAA, which was indicative of
its potential involvement in the disease process. METTL3 was
observed to positively regulate circRBM33 expression, whereas
YTHDC1 deficiency was revealed to decrease circRBM33 expression.
Analysis revealed that METTL3/YTHDC1 regulated circRBM33 biogenesis
in an m6A-dependent manner. This may be associated with the fact
that METTL3/YTHDC1-mediated m6A modification regulates the
biogenesis of circRBM33 from the exons of the RBM33 gene, thereby
alleviating AAA development by reducing ECM degradation.
Consequently, the effects of m6A methylation modifications on miRNA
precursors and circRNAs are equally important for AAA development
(Table II).
 | Table II.Identification and role of m6A
methylation in abdominal aortic aneurysms. |
Table II.
Identification and role of m6A
methylation in abdominal aortic aneurysms.
| First author/s,
year | AAA-related m6A
methylases | Main
mechanisms | (Refs.) |
|---|
| He et al,
2019 | YTHDF1, YTHDF3,
FTO, METTL14 | METTL14 is
associated with inflammatory infiltration; FTO is associated with
VSMC apoptosis; YTHDF3 is associated with macrophage
infiltration | (59) |
| Zhong et al,
2020 | METTL3 | Promoting
maturation of primary microRNAs | (58) |
| Li et al,
2021 | METTL14, HNRNPC,
RBM15B | Correlates with
degree of immune infiltration | (74) |
| Fu et al,
2022 | RBM15, WTAP,
ALKBH5, IGFBP3 | Immune
infiltration; VSMC apoptosis | (73) |
| Wang et al,
2022 | ALKBH5, HNRNPC,
METTL14, YTHDF1, YTHDF2 | Immune
infiltration; inflammatory response | (75) |
| Li et al,
2023 | METTL3-METTL14
complexes, YTHDF3 | VSMC apoptosis;
inflammatory response | (62) |
| Chen et al,
2024 | METTL14/YTHDC1 | VSMC phenotype
switching and apoptosis; vascular inflammation | (85) |
| Xu et al,
2024 | METTL3/YTHDC | Regulation of
biogenesis from exon circRBM33 of the RBM33 gene | (88) |
m6A methylation in AAA and potential
clinical applications
The determination of medical management for AAA is
based exclusively on the assessment of the aneurysm size, which is
conducted through ultrasound or CT imaging. However, imaging
techniques are retrospective and continuous AAA diameter changes
require lengthy time intervals due to the slow progression of the
disease. Consequently, biomarkers or combinations of biomarkers
that predict the aneurysm growth rate offer numerous advantages in
the clinical setting, both by facilitating clinical decision-making
and evaluation of clinical trials of pharmacological interventions
aimed at reducing aneurysm progression (104). Given the complexity and
multifactorial nature of AAA, these biomarkers are involved in
multiple pathways, including cardiovascular health, hemostasis,
transport proteins, inflammation and immunity, renal function,
cellular structure, and hormones and growth factors (105). MMP-9, IL-6, C-reactive protein
(CRP), α1-antitrypsin, triglycerides, lipoprotein(a),
apolipoprotein A and high-density lipoprotein have been identified
as potential biomarkers associated with AAA progression (106). Furthermore, previous research has
indicated that granzyme K, a proinflammatory factor within the
granzyme family, may offer a promising avenue for diagnosing AAA
and detecting rupture. Its combination with CRP or leukocytes
represents a potential strategy for monitoring AAA (107). Consequently, a comprehensive
understanding of the mechanisms underlying AAA onset and
progression is key to facilitating the development of effective
therapies.
Since it has become known that inflammatory cells
and inflammatory mediators (including IL-1, IL-17, TGF-β and Ang
II) are the predominant factors in the development of AAA (108), novel therapeutic approaches have
emerged, including the immunosuppressive effect of
cyclosporine-specific inhibitors on AAA progression, which has been
previously demonstrated (109).
In addition, the use of doxycycline, a broad-spectrum MMP
inhibitor, has been revealed to slow the progression of AAAs by
inhibiting the permanent dilation of the aorta caused by ECM
degradation (110). Other
approaches include the following: i) The reduction of inflammation
in AAA tissues by Ang-converting enzyme inhibition (111); ii) the use of testosterone to
inhibit AAA by modulating macrophages (112); iii) the use of statins to limit
AAA progression through their anti-inflammatory and antioxidant
properties; and iv) the use of metformin to mitigate AAA
progression by attenuating vascular inflammation, ROS production
and neovascularization (113). It
is therefore important to investigate several pathways in order to
improve the prognosis of this disease.
The structure and function of m6A methylation
modifications affect not only chromatin regulation and
transcriptional regulation, but also post-transcriptional
regulation, which ultimately affects RNA metabolism and cellular
function through different regulatory mechanisms (114). The current literature indicates
that m6A methylation modifications are involved in the onset and
progression of a variety of diseases, including the pathogenesis of
cardiovascular diseases. It has been confirmed that m6A methylation
influences the pathophysiology of cardiovascular disease by
regulating cellular processes such as differentiation,
proliferation, inflammation, autophagy and apoptosis (115), including the potential
pathological mechanisms of METTL14-dependent m6A in vascular
calcification (116), the
existence of differential expression of genes in m6A-SNPs in
coronary heart disease and the potential role of m6A in blood
pressure regulation (117).
Furthermore, m6A methylation plays a pivotal role in regulating the
inflammatory response of macrophages, monocytes and endothelial
cells. It also carries out a key part in the interplay between
transcriptome modification regulation and inflammation in
cardiovascular disease, offering potential targets for disease
diagnosis and treatment (116,117).
Aberrant expression of m6A methylation carries out a
pivotal role in the interconnection between epigenetic
modifications and immune infiltration in the pathogenesis of AAA.
There are notable associations between AAA-related m6A regulators
and key biological processes such as immunity, metabolism and
autophagy. There is a close association between m6A methylation
regulators and AAA immune cell infiltration, which may be a
potential therapeutic target for the diagnosis and inhibition of
AAA rupture (74,75). Emerging therapeutic strategies for
AAA increasingly focus on m6A methylation regulators due to their
dual roles in gene expression regulation and immune modulation.
Pharmacological inhibition of m6A methyltransferases (such as
METTL3) suppresses m6A deposition, thereby attenuating
pro-inflammatory gene signatures and dampening immune-driven
pathological cascades to impede AAA progression (62). Conversely, targeting m6A erasers
(FTO and ALKBH5) stabilizes m6A-modified transcripts, which
modifies immunomodulatory gene expression and alters immune
signaling dynamics to exert therapeutic benefits (118). Additionally, intervening with m6A
reader proteins (such as YTHDF2 and IGF2BP) provides a
complementary approach, since modulating their binding activity
reprograms m6A-mediated transcript fate, thus fine-tuning
immune-related transcriptional programs and downstream effector
responses (119).
Furthermore, miRNAs have emerged as a promising
pharmacological tool for restoring cellular homeostasis and
directing entire cellular pathways through interactions with a wide
range of target genes. In light of the pivotal function of
m6A-mediated miR-34a maturation in AAA development, miRNAs may
offer a novel therapeutic target and diagnostic biomarker for AAA
therapy (57). The inflammatory
process in AAA is one of the key pathways, as this pathway
possesses the most prognostic biomarkers (120,121). An understanding of the role of
inflammation in AAA can facilitate the early detection and
monitoring of the disease, which in turn can lead to an improvement
in patient prognosis. However, the multiplicity of AAA mechanisms
suggests that controlling pathways such as the genetic phenotype is
equally important. By further elucidating the role of m6A
methylation in AAA mechanisms, potential therapeutic targets can be
identified through further investigation of the pathways involved
in this disease, ultimately leading to the development of more
effective treatments.
Although previous studies have found that m6A
methylation has a considerable impact on the genetic risk of
developing AAA, and a number of targets and regulators of m6A
modification have been identified, the understanding of their
functions and interaction mechanisms is still incomplete, and
further functional studies are needed to clarify their exact roles
in the relevant mechanisms. In addition, the current understanding
of the dynamic regulatory mechanisms of m6A modification is still
insufficient and further in-depth studies are needed to reveal the
dynamic changes and regulation of m6A modification during AAA
development. Finally, the application of the findings on m6A
methylation regulators to clinical diagnosis and treatment is still
challenging and further studies are needed to validate their
feasibility and efficacy in patients with AAA.
Summary
AAA is a progressive dilatation of the abdominal
aorta that is caused by a variety of lifestyle and genetic factors
and has a high mortality rate. Despite considerable advances in
imaging technology as well as open and endovascular repair
techniques, pharmacological treatments that are capable of
preventing the onset, dilatation and rupture of AAA and of treating
AAA effectively in the perioperative period remain unavailable.
This is due to the incomplete understanding of the biological
mechanisms that underpin the disease. Genomic research has
transformed the current understanding of the mechanisms that govern
the development of multifactorial diseases such as AAA. This has
been demonstrated through GWAS, which have identified AAA as a
multifactorial, polygenic disease with epigenetic associations.
Methylation represents the most extensively researched epigenetic
mechanism. The methylation pattern constitutes a long-term genetic
trait that induces alterations in gene transcription and is subject
to influence from both genetic and environmental factors. The
present review highlights the potential involvement of m6A
methylation in the etiology of AAA from various perspectives.
m6A methylation, as a key form of RNA modification,
carries out a pivotal role in the pathogenesis of AAA. The m6A
modification affects the expression levels of AAA-related genes and
participates in the regulation of various biological functions by
regulating RNA transcription, splicing, degradation and translation
processes. Previous studies have identified several key mediators
and signaling pathways involved in m6A methylation during AAA
pathogenesis, which may represent potential molecular targets for
the control of AAA.
In light of the pivotal function of m6A modification
in AAA, future research should investigate therapeutic strategies
targeting m6A modification. This could entail regulating the
expression and biological functions of AAA-related genes by
inhibiting or promoting the activity of specific m6A
modification-related proteins. The development of AAA is a
multifactorial and complex process and it is important to develop
standard protocols for early screening of AAA, treatment of lesion
progression and prevention of AAA rupture, from the
characterization and etiology of AAA to the in-depth molecular
mechanisms of the AAA formation process. First, the regulatory
mechanisms of m6A methyltransferases and m6A demethylases should be
thoroughly investigated to understand their expression changes in
AAA and their association with disease progression. Second, the
functions of m6A-modified recognition proteins should be further
explored to understand how they regulate m6A-modified target genes
and immune signaling pathways. Finally, the interactions and
synergistic regulation among m6A methylation regulators should be
considered comprehensively to reveal the integrated effects of the
whole regulatory network in AAA development. Furthermore, the
association between the level of m6A modification and the clinical
prognosis of AAA should be investigated to provide new insights and
methodologies for the early diagnosis and treatment of AAA.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
KW contributed to the conception, wrote the
manuscript and revised the manuscript. ZS contributed to the
conception and revised the manuscript. Both authors contributed to
the review and read and approved the final manuscript. Data
authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
AAA
|
abdominal aortic aneurysm
|
|
ALKBH5
|
alkylated DNA repair protein alkB
homologue 5
|
|
CCD2
|
cell division cycle 2
|
|
CCL2
|
C-C motif chemokine ligand 2
|
|
circRNA
|
circular RNA
|
|
CRP
|
C-reactive protein
|
|
CT
|
computed tomography
|
|
ECM
|
extracellular matrix
|
|
FTO
|
fat mass and obesity-associated
protein
|
|
GEO
|
gene expression omnibus
|
|
HASMC
|
human aortic smooth muscle cell
|
|
IL
|
interleukin
|
|
ILT
|
intraluminal thrombus
|
|
lncRNA
|
long non-coding RNA
|
|
METTL3
|
methyltransferase 3
|
|
m6A
|
N6-methyladenosine
|
|
PI3K
|
phosphatidylinositol 3-kinase
|
|
ROS
|
reactive oxygen species
|
|
Th
|
T helper
|
|
UTR
|
untranslated region
|
|
VSMC
|
vascular smooth muscle cell
|
|
VIRMA
|
Vir-like m6A methyltransferase-related
protein
|
|
WTAP
|
Wilms tumor 1-associating protein
|
|
GWAS
|
genome-wide association study
|
|
SNP
|
single nucleotide polymorphism
|
|
TRMT112
|
tRNA methyltransferase subunit
11-2
|
|
YTH
|
YTH domain family
|
|
YTHDF
|
YTH N6-methyladenosine RNA binding
protein F
|
|
YY1
|
YinYang-1
|
|
miR
|
microRNA
|
|
SIRT1
|
sirtuin 1
|
|
RIP3
|
receptor-interacting
serine/threonine-protein kinase 3
|
|
NK
|
natural killer
|
|
MMP
|
matrix metalloproteinase
|
|
NAP1L
|
nucleosome assembly protein 1-like
|
|
pri-mRNA
|
primary miRNA
|
|
pre-miRNA
|
precursor hairpin
|
|
DGCR8
|
Drosha-DiGeorge critical region-8
|
References
|
1
|
Sakalihasan N, Limet R and Defawe OD:
Abdominal aortic aneurysm. Lancet. 365:1577–1589. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Baman JR and Eskandari MK: What is an
abdominal aortic aneurysm? JAMA. 328:22802022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Haque K and Bhargava P: Abdominal aortic
aneurysm. Am Fam Physician. 106:165–172. 2022.PubMed/NCBI
|
|
4
|
Marcaccio CL and Schermerhorn ML:
Epidemiology of abdominal aortic aneurysms. Semin Vasc Surg.
34:29–37. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schanzer A and Oderich GS: Management of
abdominal aortic aneurysms. N Engl J Med. 385:1690–1698. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lederle FA, Wilson SE, Johnson GR, Reinke
DB, Littooy FN, Acher CW, Ballard DJ, Messina LM, Gordon IL, Chute
EP, et al: Immediate repair compared with surveillance of small
abdominal aortic aneurysms. N Engl J Med. 346:1437–1444. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Memon AA, Zarrouk M, Ågren-Witteschus S,
Sundquist J, Gottsäter A and Sundquist K: Identification of novel
diagnostic and prognostic biomarkers for abdominal aortic aneurysm.
Eur J Prev Cardiol. 27:132–142. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cho IY, Han K, Lee KN, Koo HY, Cho YH, Lee
JH, Park YJ and Shin DW: Risk factors for abdominal aortic aneurysm
in patients with diabetes. J Vasc Surg. 81:128–136.e4. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Silva LF, Vangipurapu J, Oravilahti A,
Lusis AJ and Laakso M: Metabolomics, genetics, and environmental
factors: Intersecting paths in abdominal aortic aneurysm. Int J Mol
Sci. 26:14982025. View Article : Google Scholar
|
|
10
|
Gao J, Cao H, Hu G, Wu Y, Xu Y, Cui H, Lu
HS and Zheng L: The mechanism and therapy of aortic aneurysms.
Signal Transduct Target Ther. 8:552023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li T, Wu Y, Yang J, Jing J, Ma C and Sun
L: N6-methyladenosine-associated genetic variants in NECTIN2 and
HPCAL1 are risk factors for abdominal aortic aneurysm. iScience.
27:1094192024. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mangum KD and Farber MA: Genetic and
epigenetic regulation of abdominal aortic aneurysms. Clin Genet.
97:815–826. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Barkhordarian M, Tran HH, Menon A,
Pulipaka SP, Aguilar IK, Fuertes A, Dey S, Chacko AA, Sethi T,
Bangolo A and Weissman S: Innovation in pathogenesis and management
of aortic aneurysm. World J Exp Med. 14:914082024. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mangum K, Gallagher K and Davis FM: The
role of epigenetic modifications in abdominal aortic aneurysm
pathogenesis. Biomolecules. 12:1722022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tian Z, Li W, Wang J and Li S:
WTAP-mediated m6A modification on BASP1 mRNA contributes to
ferroptosis in AAA. Gen Thorac Cardiovasc Surg. Feb 19–2025.(Epub
ahead of print). doi: 10.1007/s11748-025-02130-5, 2025. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu ZH, Ma P, He Y, Zhang YF, Mou Z, Fang
T, Wang W and Yu KH: The mechanism and latest progress of m6A
methylation in the progression of pancreatic cancer. Int J Biol
Sci. 21:1187–1201. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Qiao Y, Mei Y, Xia M, Luo D and Gao L: The
role of m6A modification in the risk prediction and notch1 pathway
of Alzheimer's disease. iScience. 27:1102352024. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mattei AL, Bailly N and Meissner A: DNA
methylation: A historical perspective. Trends Genet. 38:676–707.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Moore LD, Le T and Fan G: DNA methylation
and its basic function. Neuropsychopharmacology. 38:23–38. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou W, Wang X, Chang J, Cheng C and Miao
C: The molecular structure and biological functions of RNA
methylation, with special emphasis on the roles of RNA methylation
in autoimmune diseases. Crit Rev Clin Lab Sci. 59:203–218. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen Y, Jiang Z, Yang Y, Zhang C, Liu H
and Wan J: The functions and mechanisms of post-translational
modification in protein regulators of RNA methylation. Current
status and future perspectives. Int J Biol Macromol.
253:1267732023. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ru W, Zhang X, Yue B, Qi A, Shen X, Huang
Y, Lan X, Lei C and Chen H: Insight into m6 A methylation from
occurrence to functions. Open Biol. 10:2000912020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang H, Shi X, Huang T, Zhao X, Chen W,
Gu N and Zhang R: Dynamic landscape and evolution of m6A
methylation in human. Nucleic Acids Res. 48:6251–6264. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Maity A and Das B: N6-methyladenosine
modification in mRNA: Machinery, function and implications for
health and diseases. FEBS J. 283:1607–1630. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wu B, Li L, Huang Y, Ma J and Min J:
Readers, writers and erasers of N6-methylated adenosine
modification. Curr Opin Struct Biol. 47:67–76. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shi H, Wei J and He C: Where, when, and
how: Context-dependent functions of RNA methylation writers,
readers, and erasers. Mol Cell. 74:640–650. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xiong X, Hou L, Park YP, Molinie B; GTEx
Consortium, ; Gregory RI and Kellis M: Genetic drivers of
m6A methylation in human brain, lung, heart and muscle.
Nat Genet. 53:1156–1165. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen X, Yuan Y, Zhou F, Li L, Pu J and
Jiang X: m6A RNA methylation: A pivotal regulator of tumor immunity
and a promising target for cancer immunotherapy. J Transl Med.
23:2452025. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen T, Ye W, Gao S, Li Y, Luan J, Lv X
and Wang S: Emerging importance of m6A modification in liver cancer
and its potential therapeutic role. Biochim Biophys Acta Rev
Cancer. 13:1892992025. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen XY, Zhang J and Zhu JS: The role of
m6A RNA methylation in human cancer. Mol Cancer.
18:1032019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jiang X, Liu B, Nie Z, Duan L, Xiong Q,
Jin Z, Yang C and Chen Y: The role of m6A modification in the
biological functions and diseases. Signal Transduct Target Ther.
6:742021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang H, Han J and Zhang XA: Interplay of
m6A RNA methylation and gut microbiota in modulating gut injury.
Gut Microbes. 17:24672132025. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kumari R, Ranjan P, Suleiman ZG, Goswami
SK, Li J, Prasad R and Verma SK: mRNA modifications in
cardiovascular biology and disease: With a focus on m6A
modification. Cardiovasc Res. 118:1680–1692. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zaccara S, Ries RJ and Jaffrey SR:
Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell
Biol. 20:608–624. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang X, Li MJ, Xia L and Zhang H: The
biological function of m6A methyltransferase KIAA1429 and its role
in human disease. PeerJ. 10:e143342022. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang
Z, Cheng T, Gao M, Shu X, Ma H, et al: VIRMA mediates preferential
m6 A mRNA methylation in 3′UTR and near stop codon and associates
with alternative polyadenylation. Cell Discov. 4:102018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
van Tran N, Ernst FGM, Hawley BR, Zorbas
C, Ulryck N, Hackert P, Bohnsack KE, Bohnsack MT, Jaffrey SR,
Graille M and Lafontaine DLJ: The human 18S rRNA m6A
methyltransferase METTL5 is stabilised by TRMT112. Nucleic Acids
Res. 47:7719–7733. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Su R, Dong L, Li Y, Gao M, He PC, Liu W,
Wei J, Zhao Z, Gao L, Han L, et al: METTL16 exerts an
m6A-independent function to facilitate translation and
tumourigenesis. Nat Cell Biol. 24:205–216. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li Y, Su R, Deng X, Chen Y and Chen J: FTO
in cancer: Functions, molecular mechanisms, and therapeutic
implications. Trends Cancer. 8:598–614. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Meyer KD and Jaffrey SR: The dynamic
epitranscriptome: N6-methyladenosine and gene expression control.
Nat Rev Mol Cell Biol. 15:313–326. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yu F, Zhu AC, Liu S, Gao B, Wang Y,
Khudaverdyan N, Yu C, Wu Q, Jiang Y, Song J, et al: RBM33 is a
unique m6A RNA-binding protein that regulates ALKBH5
demethylase activity and substrate selectivity. Mol Cell.
83:2003–2019.e6. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Qu J, Yan H, Hou Y, Cao W, Liu Y, Zhang E,
He J and Cai Z: RNA demethylase ALKBH5 in cancer: From mechanisms
to therapeutic potential. J Hematol Oncol. 15:82022. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhai J, Chen H, Wong CC, Peng Y, Gou H,
Zhang J, Pan Y, Chen D, Lin Y, Wang S, et al: ALKBH5 drives immune
suppression via targeting AXIN2 to promote colorectal cancer and is
a target for boosting immunotherapy. Gastroenterology. 165:445–462.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang X, Zhao BS, Roundtree IA, Lu Z, Han
D, Ma H, Weng X, Chen K, Shi H and He C: N(6)-methyladenosine
modulates messenger RNA translation efficiency. Cell.
161:1388–1399. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Jain S, Koziej L, Poulis P, Kaczmarczyk I,
Gaik M, Rawski M, Ranjan N, Glatt S and Rodnina MV: Modulation of
translational decoding by m6A modification of mRNA. Nat
Commun. 14:47842023. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liao J, Wei Y, Liang J, Wen J, Chen X,
Zhang B and Chu L: Insight into the structure, physiological
function, and role in cancer of m6A readers-YTH domain-containing
proteins. Cell Death Discov. 8:1372022. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xiao W, Adhikari S, Dahal U, Chen YS, Hao
YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, et al: Nuclear m(6)A
reader YTHDC1 regulates mRNA splicing. Mol Cell. 61:507–519. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Mao Y, Dong L, Liu XM, Guo J, Ma H, Shen B
and Qian SB: m6A in mRNA coding regions promotes
translation via the RNA helicase-containing YTHDC2. Nat Commun.
10:53322019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y,
Qi M, Lu Z, Shi H, Wang J, et al: Ythdc2 is an N6-methyladenosine
binding protein that regulates mammalian spermatogenesis. Cell Res.
27:1115–1127. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kretschmer J, Rao H, Hackert P, Sloan KE,
Höbartner C and Bohnsack MT: The m6A reader protein
YTHDC2 interacts with the small ribosomal subunit and the 5′-3′
exoribonuclease XRN1. RNA. 24:1339–1350. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yuan Z, Lu Y, Wei J, Wu J, Yang J and Cai
Z: Abdominal aortic aneurysm: Roles of inflammatory cells. Front
Immunol. 11:6091612021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wagenhäuser MU, Mulorz J, Krott KJ,
Bosbach A, Feige T, Rhee YH, Chatterjee M, Petzold N, Böddeker C,
Ibing W, et al: Crosstalk of platelets with macrophages and
fibroblasts aggravates inflammation, aortic wall stiffening, and
osteopontin release in abdominal aortic aneurysm. Cardiovasc Res.
120:417–432. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Sun W, Zheng J and Gao Y: Targeting
platelet activation in abdominal aortic aneurysm: Current knowledge
and perspectives. Biomolecules. 12:2062022. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Fan YN, Ke X, Yi ZL, Lin YQ, Deng BQ, Shu
XR, Yang DH, Liao ZY and Nie RQ: Plasma D-dimer as a predictor of
intraluminal thrombus burden and progression of abdominal aortic
aneurysm. Life Sci. 240:1170692020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zheng Y, Li Y, Ran X, Wang D, Zheng X,
Zhang M, Yu B, Sun Y and Wu J: Mettl14 mediates the inflammatory
response of macrophages in atherosclerosis through the NF-κB/IL-6
signaling pathway. Cell Mol Life Sci. 79:3112022. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhai Z, Zhang X, Ding Y, Huang Z, Li Q,
Zheng M, Cho K, Dong Z, Fu W, Chen Z and Jiang B: Eugenol restrains
abdominal aortic aneurysm progression with down-regulations on
NF-κB and COX-2. Phytother Res. 36:928–937. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Du J, Liao W, Liu W, Deb DK, He L, Hsu PJ,
Nguyen T, Zhang L, Bissonnette M, He C and Li Y:
N6-Adenosine methylation of Socs1 mRNA is required to
sustain the negative feedback control of macrophage activation. Dev
Cell. 55:737–753.e7. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhong L, He X, Song H, Sun Y, Chen G, Si
X, Sun J, Chen X, Liao W, Liao Y and Bin J: METTL3 induces AAA
development and progression by modulating
N6-Methyladenosine-Dependent primary miR34a processing. Mol Ther
Nucleic Acids. 21:394–411. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
He Y, Xing J, Wang S, Xin S, Han Y and
Zhang J: Increased m6A methylation level is associated with the
progression of human abdominal aortic aneurysm. Ann Transl Med.
7:7972019. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Xu T, Wang S, Li X, Li X, Qu K, Tong H,
Zhang R, Bai S and Fan J: Lithium chloride represses abdominal
aortic aneurysm via regulating GSK3β/SIRT1/NF-κB signaling pathway.
Free Radic Biol Med. 166:1–10. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Mandal P, Berger SB, Pillay S, Moriwaki K,
Huang C, Guo H, Lich JD, Finger J, Kasparcova V, Votta B, et al:
RIP3 induces apoptosis independent of pronecrotic kinase activity.
Mol Cell. 56:481–495. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Li K, Zhang D, Zhai S, Wu H and Liu H:
METTL3-METTL14 complex induces necroptosis and inflammation of
vascular smooth muscle cells via promoting N6 methyladenosine mRNA
methylation of receptor-interacting protein 3 in abdominal aortic
aneurysms. J Cell Commun Signal. 17:897–914. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Lu S, White JV, Nwaneshiudu I, Nwaneshiudu
A, Monos DS, Solomides CC, Oleszak EL and Platsoucas C: Human
abdominal aortic aneurysm (AAA): Evidence for an autoimmune
antigen-driven disease. Autoimmun Rev. 21:1031642022. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Gong W, Tian Y and Li L: T cells in
abdominal aortic aneurysm: Immunomodulation and clinical
application. Front Immunol. 14:12401322023. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Li J, Xia N, Li D, Wen S, Qian S, Lu Y, Gu
M, Tang T, Jiao J, Lv B, et al: Aorta regulatory T cells with a
tissue-specific phenotype and function promote tissue repair
through Tff1 in abdominal aortic aneurysms. Adv Sci (Weinh).
9:e21043382022. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Chan WL, Pejnovic N, Liew TV and Hamilton
H: Predominance of Th2 response in human abdominal aortic aneurysm:
Mistaken identity for IL-4-producing NK and NKT cells? Cell
Immunol. 233:109–114. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Stepien KL, Bajdak-Rusinek K, Fus-Kujawa
A, Kuczmik W and Gawron K: Role of extracellular matrix and
inflammation in abdominal aortic aneurysm. Int J Mol Sci.
23:110782022. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Yang X, Tang H, Sun X and Gui Q: M6A
modification and T cells in adipose tissue inflammation. Cell
Biochem Funct. 42:e40892024. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Swedenborg J, Mäyränpää MI and Kovanen PT:
Mast cells: Important players in the orchestrated pathogenesis of
abdominal aortic aneurysms. Arterioscler Thromb Vasc Biol.
31:734–740. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Leoni C, Bataclan M, Ito-Kureha T,
Heissmeyer V and Monticelli S: The mRNA methyltransferase Mettl3
modulates cytokine mRNA stability and limits functional responses
in mast cells. Nat Commun. 14:38622023. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Qi C, Li H, Yu Y, Hao J, Zhang H, Wang L,
Jin J, Zhou Q, Hu Y, Zhang C and Zhang Q: m6A RNA
methylation decreases atherosclerotic vulnerable plaque through
inducing T cells. Braz J Cardiovasc Surg. 38:124–131. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Chao Y, Li HB and Zhou J: Multiple
functions of RNA methylation in T cells: A review. Front Immunol.
12:6274552021. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Fu C, Feng L, Zhang J and Sun D:
Bioinformatic analyses of the role of m6A RNA methylation
regulators in abdominal aortic aneurysm. Ann Transl Med.
10:5472022. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Li T, Wang T, Jing J and Sun L: Expression
pattern and clinical value of Key m6A RNA modification regulators
in abdominal aortic aneurysm. J Inflamm Res. 14:4245–4258. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Wang K, Kan Q, Ye Y, Qiu J, Huang L, Wu R
and Yao C: Novel insight of N6-methyladenosine modified
subtypes in abdominal aortic aneurysm. Front Genet. 13:10553962022.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Kuivaniemi H, Ryer EJ, Elmore JR and Tromp
G: Understanding the pathogenesis of abdominal aortic aneurysms.
Expert Rev Cardiovasc Ther. 13:975–987. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Domagała D, Data K, Szyller H, Farzaneh M,
Mozdziak P, Woźniak S, Zabel M, Dzięgiel P and Kempisty B:
Cellular, molecular and clinical aspects of aortic
aneurysm-vascular physiology and pathophysiology. Cells.
13:2742024. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Rombouts KB, van Merrienboer TAR, Ket JCF,
Bogunovic N, van der Velden J and Yeung KK: The role of vascular
smooth muscle cells in the development of aortic aneurysms and
dissections. Eur J Clin Invest. 52:e136972022. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Lattanzi S: Abdominal aortic aneurysms:
Pathophysiology and clinical issues. J Intern Med. 288:376–378.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Lei Y, Zhan E, Chen C, Hu Y, Lv Z, He Q,
Wang X, Li X and Zhang F: ALKBH5-mediated m6A
demethylation of Runx2 mRNA promotes extracellular matrix
degradation and intervertebral disc degeneration. Cell Biosci.
14:792024. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Zheng Y, Qi F, Li L, Yu B, Cheng Y, Ge M,
Qin C and Li X: LncNAP1L6 activates MMP pathway by stabilising the
m6A-modified NAP1L2 to promote malignant progression in prostate
cancer. Cancer Gene Ther. 30:209–218. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Zhao Y, Xia A, Li C, Long X, Bai Z, Qiu Z,
Xiong W, Gu N, Shen Y, Zhao R and Shi B: Methyltransferase like
3-mediated N6-methylatidin methylation inhibits vascular smooth
muscle cells phenotype switching via promoting phosphatidylinositol
3-kinase mRNA decay. Front Cardiovasc Med. 9:9130392022. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Fang ZM, Zhang SM, Luo H, Jiang DS, Huo B,
Zhong X, Feng X, Cheng W, Chen Y, Feng G, et al:
Methyltransferase-like 3 suppresses phenotypic switching of
vascular smooth muscle cells by activating autophagosome formation.
Cell Prolif. 56:e133862023. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Liao M, Zou S, Wu J, Bai J, Liu Y, Zhi K
and Qu L: METTL3-mediated m6A modification of NORAD inhibits the
ferroptosis of vascular smooth muscle cells to attenuate the aortic
dissection progression in an YTHDF2-dependent manner. Mol Cell
Biochem. 479:3471–3487. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Chen M, Yang D, Zhou Y, Yang C, Lin W, Li
J, Liu J, Ye J, Huang W, Ma W, et al: Colchicine blocks abdominal
aortic aneurysm development by maintaining vascular smooth muscle
cell homeostasis. Int J Biol Sci. 20:2092–2110. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Griffiths-Jones S, Saini HK, van Dongen S
and Enright AJ: miRBase: Tools for microRNA genomics. Nucleic Acids
Res. 36:D154–D158. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Diener C, Keller A and Meese E: Emerging
concepts of miRNA therapeutics: From cells to clinic. Trends Genet.
38:613–626. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Xu Y, Weng X, Qiu J and Wang S: Biogenesis
of circRBM33 mediated by N6-methyladenosine and its function in
abdominal aortic aneurysm. Epigenetics. 19:23924012024. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Adam M, Raaz U, Spin JM and Tsao PS:
MicroRNAs in abdominal aortic aneurysm. Curr Vasc Pharmacol.
13:280–290. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Carvalho LS: Can microRNAs improve
prediction of abdominal aortic aneurysm growth? Atherosclerosis.
256:131–133. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Thanigaimani S, Iyer V, Bingley J, Browne
D, Phie J, Doolan D and Golledge J: Association between serum
MicroRNAs and abdominal aortic aneurysm diagnosis and growth. Eur J
Vasc Endovasc Surg. 65:573–581. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Milewicz DM: MicroRNAs, fibrotic
remodeling, and aortic aneurysms. J Clin Invest. 122:490–493. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Kumar S, Boon RA, Maegdefessel L, Dimmeler
S and Jo H: Role of noncoding RNAs in the pathogenesis of abdominal
aortic aneurysm. Circ Res. 124:619–630. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Zhang H, Zhang K, Gu Y, Tu Y and Ouyang C:
Roles and mechanisms of miRNAs in abdominal aortic aneurysm:
Signaling pathways and clinical insights. Curr Atheroscler Rep.
26:273–287. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Li X, Yang L and Chen LL: The biogenesis,
functions, and challenges of circular RNAs. Mol Cell. 71:428–442.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Chen LL: The expanding regulatory
mechanisms and cellular functions of circular RNAs. Nat Rev Mol
Cell Biol. 21:475–490. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Mei X and Chen SY: Circular RNAs in
cardiovascular diseases. Pharmacol Ther. 232:1079912022. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Alarcón CR, Lee H, Goodarzi H, Halberg N
and Tavazoie SF: N6-methyladenosine marks primary microRNAs for
processing. Nature. 519:482–485. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Jayasree PJ, Dutta S, Karemore P and
Khandelia P: Crosstalk between m6A RNA methylation and miRNA
biogenesis in cancer: An unholy nexus. Mol Biotechnol.
66:3042–3058. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Qin S, Mao Y, Chen X, Xiao J, Qin Y and
Zhao L: The functional roles, cross-talk and clinical implications
of m6A modification and circRNA in hepatocellular carcinoma. Int J
Biol Sci. 17:3059–3079. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Xu GE, Zhao X, Li G, Gokulnath P, Wang L
and Xiao J: The landscape of epigenetic regulation and therapeutic
application of N6-methyladenosine modifications in
non-coding RNAs. Genes Dis. 11:1010452023. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Zhang BY, Han L, Tang YF, Zhang GX, Fan
XL, Zhang JJ, Xue Q and Xu ZY: METTL14 regulates M6A
methylation-modified primary miR-19a to promote cardiovascular
endothelial cell proliferation and invasion. Eur Rev Med Pharmacol
Sci. 24:7015–7023. 2020.PubMed/NCBI
|
|
103
|
Fang X, Ao X, Xiao D, Wang Y, Jia Y, Wang
P, Li M and Wang J: Circular RNA-circPan3 attenuates cardiac
hypertrophy via miR-320-3p/HSP20 axis. Cell Mol Biol Lett.
29:32024. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Deeg MA, Meijer CA, Chan LS, Shen L and
Lindeman JH: Prognostic and predictive biomarkers of abdominal
aortic aneurysm growth rate. Curr Med Res Opin. 32:509–517. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Khan H, Abu-Raisi M, Feasson M, Shaikh F,
Saposnik G, Mamdani M and Qadura M: Current prognostic biomarkers
for abdominal aortic aneurysm: A comprehensive scoping review of
the literature. Biomolecules. 14:6612024. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Stather PW, Sidloff DA, Dattani N, Gokani
VJ, Choke E, Sayers RD and Bown MJ: Meta-analysis and
meta-regression analysis of biomarkers for abdominal aortic
aneurysm. Br J Surg. 101:1358–1372. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Li T, Yang C, Jing J, Sun L and Yuan Y:
Granzyme K-A novel marker to identify the presence and rupture of
abdominal aortic aneurysm. Int J Cardiol. 338:242–247. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Davis FM, Rateri DL and Daugherty A:
Abdominal aortic aneurysm: Novel mechanisms and therapies. Curr
Opin Cardiol. 30:566–573. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Piechota-Polanczyk A, Demyanets S,
Nykonenko O, Huk I, Mittlboeck M, Domenig CM, Neumayer C, Wojta J,
Nanobachvili J and Klinger M: Decreased tissue levels of
cyclophilin A, a cyclosporine a target and phospho-ERK1/2 in
simvastatin patients with abdominal aortic aneurysm. Eur J Vasc
Endovasc Surg. 45:682–688. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Mata KM, Tefé-Silva C, Floriano EM,
Fernandes CR, Rizzi E, Gerlach RF, Mazzuca MQ and Ramos SG:
Interference of doxycycline pretreatment in a model of abdominal
aortic aneurysms. Cardiovasc Pathol. 24:110–120. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Kristensen KE, Torp-Pedersen C, Gislason
GH, Egfjord M, Rasmussen HB and Hansen PR: Angiotensin-converting
enzyme inhibitors and angiotensin II receptor blockers in patients
with abdominal aortic aneurysms: Nation-wide cohort study.
Arterioscler Thromb Vasc Biol. 35:733–740. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Son BK, Kojima T, Ogawa S and Akishita M:
Testosterone inhibits aneurysm formation and vascular inflammation
in male mice. J Endocrinol. 241:307–317. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Twine CP and Williams IM: Systematic
review and meta-analysis of the effects of statin therapy on
abdominal aortic aneurysms. Br J Surg. 98:346–353. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Liu Y, Yang D, Liu T, Chen J, Yu J and Yi
P: N6-methyladenosine-mediated gene regulation and therapeutic
implications. Trends Mol Med. 29:454–467. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Peng L, Long T, Li F and Xie Q: Emerging
role of m6A modification in cardiovascular diseases.
Cell Biol Int. 46:711–722. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Zhang YS, Liu ZY, Liu ZY, Lin LC, Chen Q,
Zhao JY and Tao H: m6A epitranscriptomic modification of
inflammation in cardiovascular disease. Int Immunopharmacol.
134:1122222024. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Zhang B, Jiang H, Dong Z, Sun A and Ge J:
The critical roles of m6A modification in metabolic abnormality and
cardiovascular diseases. Genes Dis. 8:746–758. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
You Y, Fu Y, Huang M, Shen D, Zhao B, Liu
H, Zheng Y and Huang L: Recent advances of m6a demethylases
inhibitors and their biological functions in human diseases. Int J
Mol Sci. 23:58152022. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Ramesh-Kumar D and Guil S: The IGF2BP
family of RNA binding proteins links epitranscriptomics to cancer.
Semin Cancer Biol. 86:18–31. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Peng Z, Lv SJ, Chen H, Rao H, Guo Z, Wan
Q, Yang J, Zhang Y, Liu DP, Chen HZ and Wang M: Disruption of pcsk9
suppresses inflammation and attenuates abdominal aortic aneurysm
formation. Arterioscler Thromb Vasc Biol. 45:e1–e14. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Chen S, Liu X, Zhou X, Lin W, Liu M, Ma H,
Zhong K, Ma Q and Qin C: Atractylenolide-I prevents abdominal
aortic aneurysm formation through inhibiting inflammation. Front
Immunol. 16:14860722025. View Article : Google Scholar : PubMed/NCBI
|















