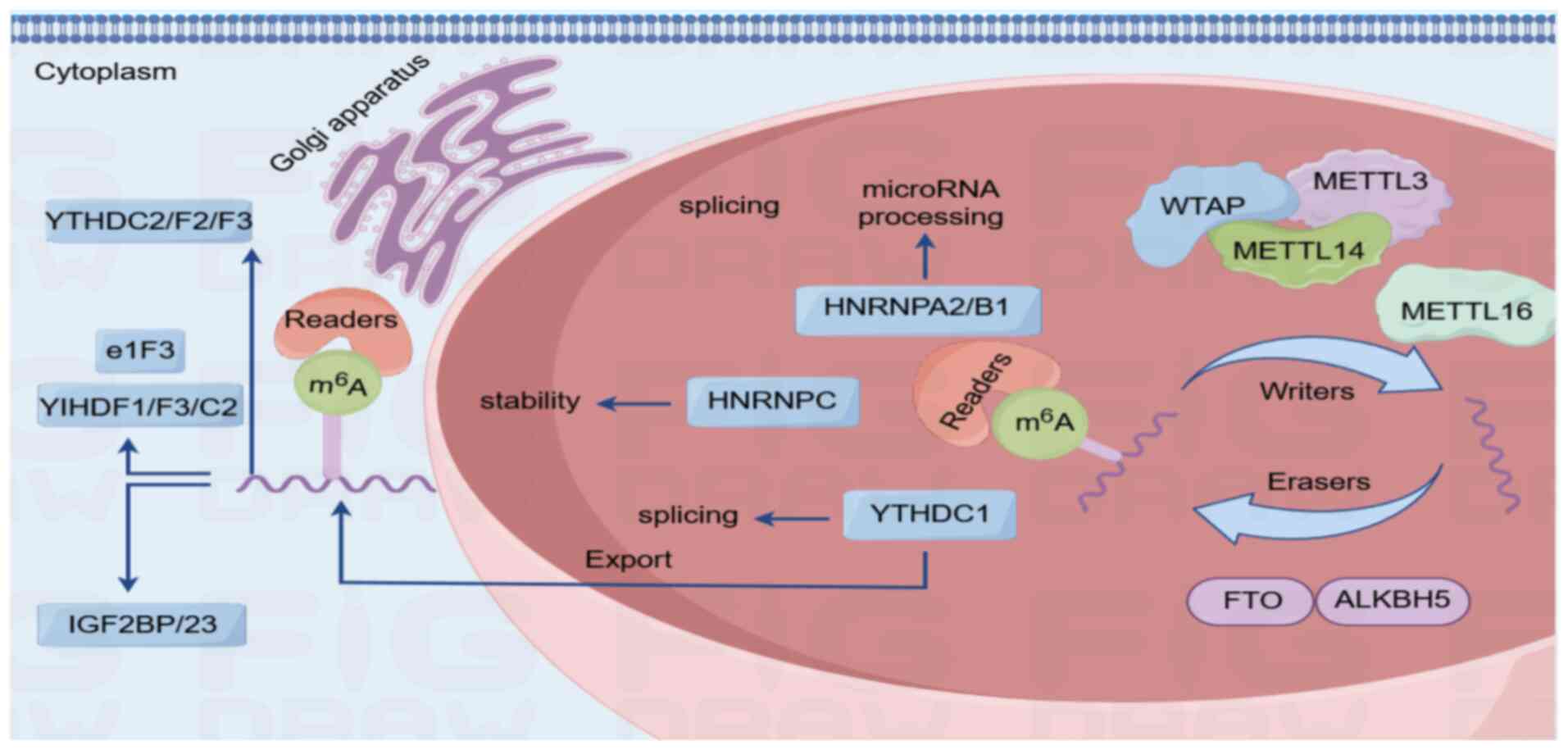
|
1
|
Sakalihasan N, Limet R and Defawe OD:
Abdominal aortic aneurysm. Lancet. 365:1577–1589. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Baman JR and Eskandari MK: What is an
abdominal aortic aneurysm? JAMA. 328:22802022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Haque K and Bhargava P: Abdominal aortic
aneurysm. Am Fam Physician. 106:165–172. 2022.PubMed/NCBI
|
|
4
|
Marcaccio CL and Schermerhorn ML:
Epidemiology of abdominal aortic aneurysms. Semin Vasc Surg.
34:29–37. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schanzer A and Oderich GS: Management of
abdominal aortic aneurysms. N Engl J Med. 385:1690–1698. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lederle FA, Wilson SE, Johnson GR, Reinke
DB, Littooy FN, Acher CW, Ballard DJ, Messina LM, Gordon IL, Chute
EP, et al: Immediate repair compared with surveillance of small
abdominal aortic aneurysms. N Engl J Med. 346:1437–1444. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Memon AA, Zarrouk M, Ågren-Witteschus S,
Sundquist J, Gottsäter A and Sundquist K: Identification of novel
diagnostic and prognostic biomarkers for abdominal aortic aneurysm.
Eur J Prev Cardiol. 27:132–142. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cho IY, Han K, Lee KN, Koo HY, Cho YH, Lee
JH, Park YJ and Shin DW: Risk factors for abdominal aortic aneurysm
in patients with diabetes. J Vasc Surg. 81:128–136.e4. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Silva LF, Vangipurapu J, Oravilahti A,
Lusis AJ and Laakso M: Metabolomics, genetics, and environmental
factors: Intersecting paths in abdominal aortic aneurysm. Int J Mol
Sci. 26:14982025. View Article : Google Scholar
|
|
10
|
Gao J, Cao H, Hu G, Wu Y, Xu Y, Cui H, Lu
HS and Zheng L: The mechanism and therapy of aortic aneurysms.
Signal Transduct Target Ther. 8:552023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li T, Wu Y, Yang J, Jing J, Ma C and Sun
L: N6-methyladenosine-associated genetic variants in NECTIN2 and
HPCAL1 are risk factors for abdominal aortic aneurysm. iScience.
27:1094192024. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mangum KD and Farber MA: Genetic and
epigenetic regulation of abdominal aortic aneurysms. Clin Genet.
97:815–826. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Barkhordarian M, Tran HH, Menon A,
Pulipaka SP, Aguilar IK, Fuertes A, Dey S, Chacko AA, Sethi T,
Bangolo A and Weissman S: Innovation in pathogenesis and management
of aortic aneurysm. World J Exp Med. 14:914082024. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mangum K, Gallagher K and Davis FM: The
role of epigenetic modifications in abdominal aortic aneurysm
pathogenesis. Biomolecules. 12:1722022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tian Z, Li W, Wang J and Li S:
WTAP-mediated m6A modification on BASP1 mRNA contributes to
ferroptosis in AAA. Gen Thorac Cardiovasc Surg. Feb 19–2025.(Epub
ahead of print). doi: 10.1007/s11748-025-02130-5, 2025. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu ZH, Ma P, He Y, Zhang YF, Mou Z, Fang
T, Wang W and Yu KH: The mechanism and latest progress of m6A
methylation in the progression of pancreatic cancer. Int J Biol
Sci. 21:1187–1201. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Qiao Y, Mei Y, Xia M, Luo D and Gao L: The
role of m6A modification in the risk prediction and notch1 pathway
of Alzheimer's disease. iScience. 27:1102352024. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mattei AL, Bailly N and Meissner A: DNA
methylation: A historical perspective. Trends Genet. 38:676–707.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Moore LD, Le T and Fan G: DNA methylation
and its basic function. Neuropsychopharmacology. 38:23–38. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou W, Wang X, Chang J, Cheng C and Miao
C: The molecular structure and biological functions of RNA
methylation, with special emphasis on the roles of RNA methylation
in autoimmune diseases. Crit Rev Clin Lab Sci. 59:203–218. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen Y, Jiang Z, Yang Y, Zhang C, Liu H
and Wan J: The functions and mechanisms of post-translational
modification in protein regulators of RNA methylation. Current
status and future perspectives. Int J Biol Macromol.
253:1267732023. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ru W, Zhang X, Yue B, Qi A, Shen X, Huang
Y, Lan X, Lei C and Chen H: Insight into m6 A methylation from
occurrence to functions. Open Biol. 10:2000912020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang H, Shi X, Huang T, Zhao X, Chen W,
Gu N and Zhang R: Dynamic landscape and evolution of m6A
methylation in human. Nucleic Acids Res. 48:6251–6264. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Maity A and Das B: N6-methyladenosine
modification in mRNA: Machinery, function and implications for
health and diseases. FEBS J. 283:1607–1630. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wu B, Li L, Huang Y, Ma J and Min J:
Readers, writers and erasers of N6-methylated adenosine
modification. Curr Opin Struct Biol. 47:67–76. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shi H, Wei J and He C: Where, when, and
how: Context-dependent functions of RNA methylation writers,
readers, and erasers. Mol Cell. 74:640–650. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xiong X, Hou L, Park YP, Molinie B; GTEx
Consortium, ; Gregory RI and Kellis M: Genetic drivers of
m6A methylation in human brain, lung, heart and muscle.
Nat Genet. 53:1156–1165. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen X, Yuan Y, Zhou F, Li L, Pu J and
Jiang X: m6A RNA methylation: A pivotal regulator of tumor immunity
and a promising target for cancer immunotherapy. J Transl Med.
23:2452025. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen T, Ye W, Gao S, Li Y, Luan J, Lv X
and Wang S: Emerging importance of m6A modification in liver cancer
and its potential therapeutic role. Biochim Biophys Acta Rev
Cancer. 13:1892992025. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen XY, Zhang J and Zhu JS: The role of
m6A RNA methylation in human cancer. Mol Cancer.
18:1032019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jiang X, Liu B, Nie Z, Duan L, Xiong Q,
Jin Z, Yang C and Chen Y: The role of m6A modification in the
biological functions and diseases. Signal Transduct Target Ther.
6:742021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang H, Han J and Zhang XA: Interplay of
m6A RNA methylation and gut microbiota in modulating gut injury.
Gut Microbes. 17:24672132025. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kumari R, Ranjan P, Suleiman ZG, Goswami
SK, Li J, Prasad R and Verma SK: mRNA modifications in
cardiovascular biology and disease: With a focus on m6A
modification. Cardiovasc Res. 118:1680–1692. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zaccara S, Ries RJ and Jaffrey SR:
Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell
Biol. 20:608–624. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang X, Li MJ, Xia L and Zhang H: The
biological function of m6A methyltransferase KIAA1429 and its role
in human disease. PeerJ. 10:e143342022. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang
Z, Cheng T, Gao M, Shu X, Ma H, et al: VIRMA mediates preferential
m6 A mRNA methylation in 3′UTR and near stop codon and associates
with alternative polyadenylation. Cell Discov. 4:102018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
van Tran N, Ernst FGM, Hawley BR, Zorbas
C, Ulryck N, Hackert P, Bohnsack KE, Bohnsack MT, Jaffrey SR,
Graille M and Lafontaine DLJ: The human 18S rRNA m6A
methyltransferase METTL5 is stabilised by TRMT112. Nucleic Acids
Res. 47:7719–7733. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Su R, Dong L, Li Y, Gao M, He PC, Liu W,
Wei J, Zhao Z, Gao L, Han L, et al: METTL16 exerts an
m6A-independent function to facilitate translation and
tumourigenesis. Nat Cell Biol. 24:205–216. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li Y, Su R, Deng X, Chen Y and Chen J: FTO
in cancer: Functions, molecular mechanisms, and therapeutic
implications. Trends Cancer. 8:598–614. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Meyer KD and Jaffrey SR: The dynamic
epitranscriptome: N6-methyladenosine and gene expression control.
Nat Rev Mol Cell Biol. 15:313–326. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yu F, Zhu AC, Liu S, Gao B, Wang Y,
Khudaverdyan N, Yu C, Wu Q, Jiang Y, Song J, et al: RBM33 is a
unique m6A RNA-binding protein that regulates ALKBH5
demethylase activity and substrate selectivity. Mol Cell.
83:2003–2019.e6. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Qu J, Yan H, Hou Y, Cao W, Liu Y, Zhang E,
He J and Cai Z: RNA demethylase ALKBH5 in cancer: From mechanisms
to therapeutic potential. J Hematol Oncol. 15:82022. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhai J, Chen H, Wong CC, Peng Y, Gou H,
Zhang J, Pan Y, Chen D, Lin Y, Wang S, et al: ALKBH5 drives immune
suppression via targeting AXIN2 to promote colorectal cancer and is
a target for boosting immunotherapy. Gastroenterology. 165:445–462.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang X, Zhao BS, Roundtree IA, Lu Z, Han
D, Ma H, Weng X, Chen K, Shi H and He C: N(6)-methyladenosine
modulates messenger RNA translation efficiency. Cell.
161:1388–1399. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Jain S, Koziej L, Poulis P, Kaczmarczyk I,
Gaik M, Rawski M, Ranjan N, Glatt S and Rodnina MV: Modulation of
translational decoding by m6A modification of mRNA. Nat
Commun. 14:47842023. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liao J, Wei Y, Liang J, Wen J, Chen X,
Zhang B and Chu L: Insight into the structure, physiological
function, and role in cancer of m6A readers-YTH domain-containing
proteins. Cell Death Discov. 8:1372022. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xiao W, Adhikari S, Dahal U, Chen YS, Hao
YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, et al: Nuclear m(6)A
reader YTHDC1 regulates mRNA splicing. Mol Cell. 61:507–519. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Mao Y, Dong L, Liu XM, Guo J, Ma H, Shen B
and Qian SB: m6A in mRNA coding regions promotes
translation via the RNA helicase-containing YTHDC2. Nat Commun.
10:53322019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y,
Qi M, Lu Z, Shi H, Wang J, et al: Ythdc2 is an N6-methyladenosine
binding protein that regulates mammalian spermatogenesis. Cell Res.
27:1115–1127. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kretschmer J, Rao H, Hackert P, Sloan KE,
Höbartner C and Bohnsack MT: The m6A reader protein
YTHDC2 interacts with the small ribosomal subunit and the 5′-3′
exoribonuclease XRN1. RNA. 24:1339–1350. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yuan Z, Lu Y, Wei J, Wu J, Yang J and Cai
Z: Abdominal aortic aneurysm: Roles of inflammatory cells. Front
Immunol. 11:6091612021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wagenhäuser MU, Mulorz J, Krott KJ,
Bosbach A, Feige T, Rhee YH, Chatterjee M, Petzold N, Böddeker C,
Ibing W, et al: Crosstalk of platelets with macrophages and
fibroblasts aggravates inflammation, aortic wall stiffening, and
osteopontin release in abdominal aortic aneurysm. Cardiovasc Res.
120:417–432. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Sun W, Zheng J and Gao Y: Targeting
platelet activation in abdominal aortic aneurysm: Current knowledge
and perspectives. Biomolecules. 12:2062022. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Fan YN, Ke X, Yi ZL, Lin YQ, Deng BQ, Shu
XR, Yang DH, Liao ZY and Nie RQ: Plasma D-dimer as a predictor of
intraluminal thrombus burden and progression of abdominal aortic
aneurysm. Life Sci. 240:1170692020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zheng Y, Li Y, Ran X, Wang D, Zheng X,
Zhang M, Yu B, Sun Y and Wu J: Mettl14 mediates the inflammatory
response of macrophages in atherosclerosis through the NF-κB/IL-6
signaling pathway. Cell Mol Life Sci. 79:3112022. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhai Z, Zhang X, Ding Y, Huang Z, Li Q,
Zheng M, Cho K, Dong Z, Fu W, Chen Z and Jiang B: Eugenol restrains
abdominal aortic aneurysm progression with down-regulations on
NF-κB and COX-2. Phytother Res. 36:928–937. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Du J, Liao W, Liu W, Deb DK, He L, Hsu PJ,
Nguyen T, Zhang L, Bissonnette M, He C and Li Y:
N6-Adenosine methylation of Socs1 mRNA is required to
sustain the negative feedback control of macrophage activation. Dev
Cell. 55:737–753.e7. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhong L, He X, Song H, Sun Y, Chen G, Si
X, Sun J, Chen X, Liao W, Liao Y and Bin J: METTL3 induces AAA
development and progression by modulating
N6-Methyladenosine-Dependent primary miR34a processing. Mol Ther
Nucleic Acids. 21:394–411. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
He Y, Xing J, Wang S, Xin S, Han Y and
Zhang J: Increased m6A methylation level is associated with the
progression of human abdominal aortic aneurysm. Ann Transl Med.
7:7972019. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Xu T, Wang S, Li X, Li X, Qu K, Tong H,
Zhang R, Bai S and Fan J: Lithium chloride represses abdominal
aortic aneurysm via regulating GSK3β/SIRT1/NF-κB signaling pathway.
Free Radic Biol Med. 166:1–10. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Mandal P, Berger SB, Pillay S, Moriwaki K,
Huang C, Guo H, Lich JD, Finger J, Kasparcova V, Votta B, et al:
RIP3 induces apoptosis independent of pronecrotic kinase activity.
Mol Cell. 56:481–495. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Li K, Zhang D, Zhai S, Wu H and Liu H:
METTL3-METTL14 complex induces necroptosis and inflammation of
vascular smooth muscle cells via promoting N6 methyladenosine mRNA
methylation of receptor-interacting protein 3 in abdominal aortic
aneurysms. J Cell Commun Signal. 17:897–914. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Lu S, White JV, Nwaneshiudu I, Nwaneshiudu
A, Monos DS, Solomides CC, Oleszak EL and Platsoucas C: Human
abdominal aortic aneurysm (AAA): Evidence for an autoimmune
antigen-driven disease. Autoimmun Rev. 21:1031642022. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Gong W, Tian Y and Li L: T cells in
abdominal aortic aneurysm: Immunomodulation and clinical
application. Front Immunol. 14:12401322023. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Li J, Xia N, Li D, Wen S, Qian S, Lu Y, Gu
M, Tang T, Jiao J, Lv B, et al: Aorta regulatory T cells with a
tissue-specific phenotype and function promote tissue repair
through Tff1 in abdominal aortic aneurysms. Adv Sci (Weinh).
9:e21043382022. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Chan WL, Pejnovic N, Liew TV and Hamilton
H: Predominance of Th2 response in human abdominal aortic aneurysm:
Mistaken identity for IL-4-producing NK and NKT cells? Cell
Immunol. 233:109–114. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Stepien KL, Bajdak-Rusinek K, Fus-Kujawa
A, Kuczmik W and Gawron K: Role of extracellular matrix and
inflammation in abdominal aortic aneurysm. Int J Mol Sci.
23:110782022. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Yang X, Tang H, Sun X and Gui Q: M6A
modification and T cells in adipose tissue inflammation. Cell
Biochem Funct. 42:e40892024. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Swedenborg J, Mäyränpää MI and Kovanen PT:
Mast cells: Important players in the orchestrated pathogenesis of
abdominal aortic aneurysms. Arterioscler Thromb Vasc Biol.
31:734–740. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Leoni C, Bataclan M, Ito-Kureha T,
Heissmeyer V and Monticelli S: The mRNA methyltransferase Mettl3
modulates cytokine mRNA stability and limits functional responses
in mast cells. Nat Commun. 14:38622023. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Qi C, Li H, Yu Y, Hao J, Zhang H, Wang L,
Jin J, Zhou Q, Hu Y, Zhang C and Zhang Q: m6A RNA
methylation decreases atherosclerotic vulnerable plaque through
inducing T cells. Braz J Cardiovasc Surg. 38:124–131. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Chao Y, Li HB and Zhou J: Multiple
functions of RNA methylation in T cells: A review. Front Immunol.
12:6274552021. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Fu C, Feng L, Zhang J and Sun D:
Bioinformatic analyses of the role of m6A RNA methylation
regulators in abdominal aortic aneurysm. Ann Transl Med.
10:5472022. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Li T, Wang T, Jing J and Sun L: Expression
pattern and clinical value of Key m6A RNA modification regulators
in abdominal aortic aneurysm. J Inflamm Res. 14:4245–4258. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Wang K, Kan Q, Ye Y, Qiu J, Huang L, Wu R
and Yao C: Novel insight of N6-methyladenosine modified
subtypes in abdominal aortic aneurysm. Front Genet. 13:10553962022.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Kuivaniemi H, Ryer EJ, Elmore JR and Tromp
G: Understanding the pathogenesis of abdominal aortic aneurysms.
Expert Rev Cardiovasc Ther. 13:975–987. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Domagała D, Data K, Szyller H, Farzaneh M,
Mozdziak P, Woźniak S, Zabel M, Dzięgiel P and Kempisty B:
Cellular, molecular and clinical aspects of aortic
aneurysm-vascular physiology and pathophysiology. Cells.
13:2742024. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Rombouts KB, van Merrienboer TAR, Ket JCF,
Bogunovic N, van der Velden J and Yeung KK: The role of vascular
smooth muscle cells in the development of aortic aneurysms and
dissections. Eur J Clin Invest. 52:e136972022. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Lattanzi S: Abdominal aortic aneurysms:
Pathophysiology and clinical issues. J Intern Med. 288:376–378.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Lei Y, Zhan E, Chen C, Hu Y, Lv Z, He Q,
Wang X, Li X and Zhang F: ALKBH5-mediated m6A
demethylation of Runx2 mRNA promotes extracellular matrix
degradation and intervertebral disc degeneration. Cell Biosci.
14:792024. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Zheng Y, Qi F, Li L, Yu B, Cheng Y, Ge M,
Qin C and Li X: LncNAP1L6 activates MMP pathway by stabilising the
m6A-modified NAP1L2 to promote malignant progression in prostate
cancer. Cancer Gene Ther. 30:209–218. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Zhao Y, Xia A, Li C, Long X, Bai Z, Qiu Z,
Xiong W, Gu N, Shen Y, Zhao R and Shi B: Methyltransferase like
3-mediated N6-methylatidin methylation inhibits vascular smooth
muscle cells phenotype switching via promoting phosphatidylinositol
3-kinase mRNA decay. Front Cardiovasc Med. 9:9130392022. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Fang ZM, Zhang SM, Luo H, Jiang DS, Huo B,
Zhong X, Feng X, Cheng W, Chen Y, Feng G, et al:
Methyltransferase-like 3 suppresses phenotypic switching of
vascular smooth muscle cells by activating autophagosome formation.
Cell Prolif. 56:e133862023. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Liao M, Zou S, Wu J, Bai J, Liu Y, Zhi K
and Qu L: METTL3-mediated m6A modification of NORAD inhibits the
ferroptosis of vascular smooth muscle cells to attenuate the aortic
dissection progression in an YTHDF2-dependent manner. Mol Cell
Biochem. 479:3471–3487. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Chen M, Yang D, Zhou Y, Yang C, Lin W, Li
J, Liu J, Ye J, Huang W, Ma W, et al: Colchicine blocks abdominal
aortic aneurysm development by maintaining vascular smooth muscle
cell homeostasis. Int J Biol Sci. 20:2092–2110. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Griffiths-Jones S, Saini HK, van Dongen S
and Enright AJ: miRBase: Tools for microRNA genomics. Nucleic Acids
Res. 36:D154–D158. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Diener C, Keller A and Meese E: Emerging
concepts of miRNA therapeutics: From cells to clinic. Trends Genet.
38:613–626. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Xu Y, Weng X, Qiu J and Wang S: Biogenesis
of circRBM33 mediated by N6-methyladenosine and its function in
abdominal aortic aneurysm. Epigenetics. 19:23924012024. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Adam M, Raaz U, Spin JM and Tsao PS:
MicroRNAs in abdominal aortic aneurysm. Curr Vasc Pharmacol.
13:280–290. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Carvalho LS: Can microRNAs improve
prediction of abdominal aortic aneurysm growth? Atherosclerosis.
256:131–133. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Thanigaimani S, Iyer V, Bingley J, Browne
D, Phie J, Doolan D and Golledge J: Association between serum
MicroRNAs and abdominal aortic aneurysm diagnosis and growth. Eur J
Vasc Endovasc Surg. 65:573–581. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Milewicz DM: MicroRNAs, fibrotic
remodeling, and aortic aneurysms. J Clin Invest. 122:490–493. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Kumar S, Boon RA, Maegdefessel L, Dimmeler
S and Jo H: Role of noncoding RNAs in the pathogenesis of abdominal
aortic aneurysm. Circ Res. 124:619–630. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Zhang H, Zhang K, Gu Y, Tu Y and Ouyang C:
Roles and mechanisms of miRNAs in abdominal aortic aneurysm:
Signaling pathways and clinical insights. Curr Atheroscler Rep.
26:273–287. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Li X, Yang L and Chen LL: The biogenesis,
functions, and challenges of circular RNAs. Mol Cell. 71:428–442.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Chen LL: The expanding regulatory
mechanisms and cellular functions of circular RNAs. Nat Rev Mol
Cell Biol. 21:475–490. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Mei X and Chen SY: Circular RNAs in
cardiovascular diseases. Pharmacol Ther. 232:1079912022. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Alarcón CR, Lee H, Goodarzi H, Halberg N
and Tavazoie SF: N6-methyladenosine marks primary microRNAs for
processing. Nature. 519:482–485. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Jayasree PJ, Dutta S, Karemore P and
Khandelia P: Crosstalk between m6A RNA methylation and miRNA
biogenesis in cancer: An unholy nexus. Mol Biotechnol.
66:3042–3058. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Qin S, Mao Y, Chen X, Xiao J, Qin Y and
Zhao L: The functional roles, cross-talk and clinical implications
of m6A modification and circRNA in hepatocellular carcinoma. Int J
Biol Sci. 17:3059–3079. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Xu GE, Zhao X, Li G, Gokulnath P, Wang L
and Xiao J: The landscape of epigenetic regulation and therapeutic
application of N6-methyladenosine modifications in
non-coding RNAs. Genes Dis. 11:1010452023. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Zhang BY, Han L, Tang YF, Zhang GX, Fan
XL, Zhang JJ, Xue Q and Xu ZY: METTL14 regulates M6A
methylation-modified primary miR-19a to promote cardiovascular
endothelial cell proliferation and invasion. Eur Rev Med Pharmacol
Sci. 24:7015–7023. 2020.PubMed/NCBI
|
|
103
|
Fang X, Ao X, Xiao D, Wang Y, Jia Y, Wang
P, Li M and Wang J: Circular RNA-circPan3 attenuates cardiac
hypertrophy via miR-320-3p/HSP20 axis. Cell Mol Biol Lett.
29:32024. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Deeg MA, Meijer CA, Chan LS, Shen L and
Lindeman JH: Prognostic and predictive biomarkers of abdominal
aortic aneurysm growth rate. Curr Med Res Opin. 32:509–517. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Khan H, Abu-Raisi M, Feasson M, Shaikh F,
Saposnik G, Mamdani M and Qadura M: Current prognostic biomarkers
for abdominal aortic aneurysm: A comprehensive scoping review of
the literature. Biomolecules. 14:6612024. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Stather PW, Sidloff DA, Dattani N, Gokani
VJ, Choke E, Sayers RD and Bown MJ: Meta-analysis and
meta-regression analysis of biomarkers for abdominal aortic
aneurysm. Br J Surg. 101:1358–1372. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Li T, Yang C, Jing J, Sun L and Yuan Y:
Granzyme K-A novel marker to identify the presence and rupture of
abdominal aortic aneurysm. Int J Cardiol. 338:242–247. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Davis FM, Rateri DL and Daugherty A:
Abdominal aortic aneurysm: Novel mechanisms and therapies. Curr
Opin Cardiol. 30:566–573. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Piechota-Polanczyk A, Demyanets S,
Nykonenko O, Huk I, Mittlboeck M, Domenig CM, Neumayer C, Wojta J,
Nanobachvili J and Klinger M: Decreased tissue levels of
cyclophilin A, a cyclosporine a target and phospho-ERK1/2 in
simvastatin patients with abdominal aortic aneurysm. Eur J Vasc
Endovasc Surg. 45:682–688. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Mata KM, Tefé-Silva C, Floriano EM,
Fernandes CR, Rizzi E, Gerlach RF, Mazzuca MQ and Ramos SG:
Interference of doxycycline pretreatment in a model of abdominal
aortic aneurysms. Cardiovasc Pathol. 24:110–120. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Kristensen KE, Torp-Pedersen C, Gislason
GH, Egfjord M, Rasmussen HB and Hansen PR: Angiotensin-converting
enzyme inhibitors and angiotensin II receptor blockers in patients
with abdominal aortic aneurysms: Nation-wide cohort study.
Arterioscler Thromb Vasc Biol. 35:733–740. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Son BK, Kojima T, Ogawa S and Akishita M:
Testosterone inhibits aneurysm formation and vascular inflammation
in male mice. J Endocrinol. 241:307–317. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Twine CP and Williams IM: Systematic
review and meta-analysis of the effects of statin therapy on
abdominal aortic aneurysms. Br J Surg. 98:346–353. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Liu Y, Yang D, Liu T, Chen J, Yu J and Yi
P: N6-methyladenosine-mediated gene regulation and therapeutic
implications. Trends Mol Med. 29:454–467. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Peng L, Long T, Li F and Xie Q: Emerging
role of m6A modification in cardiovascular diseases.
Cell Biol Int. 46:711–722. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Zhang YS, Liu ZY, Liu ZY, Lin LC, Chen Q,
Zhao JY and Tao H: m6A epitranscriptomic modification of
inflammation in cardiovascular disease. Int Immunopharmacol.
134:1122222024. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Zhang B, Jiang H, Dong Z, Sun A and Ge J:
The critical roles of m6A modification in metabolic abnormality and
cardiovascular diseases. Genes Dis. 8:746–758. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
You Y, Fu Y, Huang M, Shen D, Zhao B, Liu
H, Zheng Y and Huang L: Recent advances of m6a demethylases
inhibitors and their biological functions in human diseases. Int J
Mol Sci. 23:58152022. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Ramesh-Kumar D and Guil S: The IGF2BP
family of RNA binding proteins links epitranscriptomics to cancer.
Semin Cancer Biol. 86:18–31. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Peng Z, Lv SJ, Chen H, Rao H, Guo Z, Wan
Q, Yang J, Zhang Y, Liu DP, Chen HZ and Wang M: Disruption of pcsk9
suppresses inflammation and attenuates abdominal aortic aneurysm
formation. Arterioscler Thromb Vasc Biol. 45:e1–e14. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Chen S, Liu X, Zhou X, Lin W, Liu M, Ma H,
Zhong K, Ma Q and Qin C: Atractylenolide-I prevents abdominal
aortic aneurysm formation through inhibiting inflammation. Front
Immunol. 16:14860722025. View Article : Google Scholar : PubMed/NCBI
|