Introduction
Graves' disease (GD), Hashimoto's thyroiditis (HT)
and other autoimmune thyroid diseases (AITDs) are organ-specific
autoimmune diseases that are more common than type 1 diabetes and
multiple sclerosis (1). The
thyroid-stimulating hormone receptor (TSHR), as a G protein-coupled
receptor (GPCR), serves a key role in regulating thyroid function
(2). Specific autoantibodies such
as thyroid-stimulating hormone receptor antibodies (TRAb) bind to
TSHR, inducing abnormal changes in thyroid function, which are the
fundamental causes of diseases such as GD and HT (3).
TSHR autoantibodies have stimulatory
thyroid-stimulating immunoglobulins (TSAb) that block TSHR-blocking
antibodies (TBAb) or neutral activity, and the different activities
of TRAb are associated with the progression of AITDs, leading to
the occurrence of clinically relevant diseases. TSAb serves a key
role in the development of GD, as it activates the adenylyl cyclase
signaling system upon binding with TSHR, leading to an abnormal
elevation of thyroid hormones (4).
Additionally, TSAb is not controlled by the physiological
hypothalamic-pituitary-thyroid negative feedback loop. After TSAb
binds to TSHR, it causes the activation of the cAMP pathway and the
secondary activation of inositol 1,4,5-triphosphate, which promotes
the proliferation of thyroid follicular cells and the excessive
synthesis and secretion of thyroxine, and ultimately leads to the
pathogenesis of GD (5). However,
TBAb blocks the binding of thyroid-stimulating hormone (TSH) to
TSHR, causing hypothyroidism and subsequently inducing HT (6). The serum TRAb positivity rate reaches
80–90% in patients with newly diagnosed GD, and as treatment
progresses, TRAb titers change accordingly (5). The different activities of TRAb are
associated with the progression of AITDs, leading to the
interconversion between hyper- and hypothyroidism (7). At present, it is difficult to
distinguish and differentiate TSAb from TBAb in clinical tests,
which makes clinical diagnosis and treatment difficult. This also
poses challenges in monitoring the disease progression of a patient
and providing precise treatment (8). Therefore, it is key to differentiate
the active types of TRAb in the diagnosis and treatment of
autoimmune diseases, such as GD, HT and mucinous edema.
Different methods of detecting TRAb are being
investigated. Currently, competitive binding assays and bioassays
are used for TRAb detection. However, these binding assays cannot
distinguish between TSAb and TBAb, while bioassays measure the
functional activity of TRAb rather than simple binding (8). As a result, compared with binding
assays, bioassays can more accurately detect the different
activities of TRAb. However, bioassays are complex and
time-consuming, requiring the use of experimental equipment and
technicians to conduct cell experiments, so cannot be widely used
in clinical testing (9). Since
TSHR is the main autoantigen in the pathogenesis of GD and HT
[particularly the extracellular region of TSHR, which is the
cluster region of antigenic determinants, with the A subunit of
TSHR being the binding site for TSAb (10), and the B subunit of TSHR being the
binding site for TBAb, the interaction between TSHR and
thyroid-specific lymphocytes and TRAb induces the occurrence and
development of AITD (11).
Therefore, analysing the characteristics of AITD induced by the
combination of different domains of TSHR and its autoantibodies may
provide a novel approach for precise diagnosis, treatment and
prevention for patients.
Given the key role of TSHR and its antibody, TRAb,
in the occurrence, development and prognosis of AITDs, studies have
been conducted using cloning, sequence analysis and partial
crystallization to confirm that TSHR possesses unique structural
and functional characteristics (12–14).
TSHR is composed of extracellular domains (ECDs) and transmembrane
domains (TMD) involved in membrane-bound signal transduction
(15,16). The ECD structure contains
leucine-rich repeat domains [LRRDs; amino acid (aa)22-279], which
are rich in leucine (16). The
molecular activation mechanism of TSHR differs from that of other
GPCRs, as its ECD (aa1-418) is larger and primarily responsible for
specific binding with TSH and TRAb. The binding of TRAb or TSH to
TSHR can trigger conformational changes in the ECD, and TSAb or
TBAb can only bind to the ECD when the spatial conformation of TSHR
is correct. This interaction affects thyroid function and induces a
cascade of signal transduction via TMD leading to the development
of AITD (17,18). Therefore, understanding the
antigenic epitopes involved in the binding of TSHR to TSAb and TBAb
helps clarify the pathogenic mechanisms of GD, HT and other related
autoimmune diseases. In the present study, different fragments of
the TSHR ECD were designed to simulate their natural activity
through the preparation of fusion proteins. The pathogenic
antigenic epitopes of TSHR and their immunogenicity were explored
in both in vitro and in vivo experiments, with the
aim of understanding the specific binding sites between TSHR and
its pathogenic antibodies. The present study provided a scientific
basis for the development of detection methods for different types
of TRAb, the construction of disease animal models, and the
diagnosis and treatment of GD, HT and associated autoimmune
diseases.
Materials and methods
Serum samples and study
population
In the present study, a total of 120 patients (aged
18–56 years) and 30 healthy individuals (aged 18–52 years) were
recruited between January 2023 and January 2024 from The Second
Hospital and Clinical Medical School of Lanzhou University
(Lanzhou, China). Thyroid assessments were conducted for all
participants based on the guidelines outlined in ‘Thyroid disease:
assessment and management’ [National Institute for Health and Care
Excellence (NICE) guideline NG10074] released by NICE in 2019
(19). The assessments included
the medical history, clinical symptoms, physical examinations,
thyroid ultrasound and thyroid function tests [TSH, free
triiodothyronine (FT3) and free thyroxine (FT4)]. The exclusion
criteria included acute or chronic infections, other autoimmune or
chronic diseases, malignancies, hepatic or renal dysfunction,
pregnancy, lactation or the use of glucocorticoids, antibiotics,
and interferons or other medications affecting the immune system
and thyroid function.
The 120 patients were divided into four groups based
on their levels of TRAb [serum TRAb levels >1.5 IU/l measured by
chemiluminescence were considered to be positive. By combining TRAb
in serum with the TSHR-biotin-streptavidin-magnetic bead complex
(cat. no. 88817; Thermo Fisher Scientific, Inc.) and then with the
alkaline phosphatase-labeled anti-human IgG secondary antibody
(cat. no. PA1-85606; Thermo Fisher Scientific, Inc.; 1:10,000;
diluted in PBS; 2 h at room temperature)], chemiluminescence is
produced. The intensity of the chemiluminescence signal is
positively associated with the concentration of TRAb): 30 patients
with hyperthyroidism who were TRAb+ (age, 33.91±13.06
years; 10 male and 20 female patients), 30 patients with
hyperthyroidism who were TRAb− (age, 35.10±13.76 years;
9 male and 21 female patients), 30 patients with hypothyroidism who
were TRAb+ (age, 33.17±18.97 years; 11 male and 19
female patients), 30 patients with hypothyroidism who were
TRAb− (age, 37.4±16.86 years; 10 male and 20 female
patients). Furthermore, 30 healthy individuals were included as the
control group (age, 34.9±12.23 years; 12 male and 18 female
patients).
All patients were newly diagnosed and did not
receive any treatment, and all subjects had no thyroid nodules,
thyroid cancer or other thyroid changes. Individuals with other
complicating diseases, extrathyroidal manifestations or who
received medications such as steroids that may affect immune
function or thyroid function were excluded from the present study.
The research protocol was reviewed and approved by the Medical
Ethics Committee of The Second Hospital of Lanzhou University
(approval no. 2023A-802; Lanzhou, China). All participants provided
written informed consent prior to participating in the present
study.
Thyroid function and thyroid-specific
autoantibody testing
Fasting whole blood samples were collected from all
participants after an 8-h fast. Samples were analyzed at the
Department of Nuclear Medicine, Second Hospital of Lanzhou
University (Lanzhou, China), using a chemiluminescence immunoassay
to assess the levels of TSH (cat. no. 06491080; Siemens
Healthineers), FT3 (cat. no. 119781; Siemens Healthineers), FT4
(cat. no. 06490106; Siemens Healthineers), TG (cat. no. 11201760;
Siemens Healthineers), TPOAb (cat. no. 10630887; Siemens
Healthineers) and TRAb (cat. no. 130203009M; Shenzhen New
Industries Biomedical Engineering Co., Ltd.) in the serum. The kits
were all used according to the manufacturer's instructions.
Cloning of the human TSHR (hTSHR)
recombinant gene
The hTSHR gene sequence, protein amino acid sequence
and spatial structural characteristics were determined by querying
the Uniprot (https://www.uniprot.org/) and
National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/) databases. Primers were
designed using Primer 3.0 (https://primer3.ut.ee/) according to the Champion™ pET
vector connection requirements and synthesized by Sangon Biotech
Co., Ltd. (Table I). The hTSHR
plasmid, preserved by the Endocrinology and Metabolism Laboratory
of Lanzhou University Second Hospital, and PrimeSTAR® HS
DNA polymerase [Takara Biomedical Technology (Beijing) Co., Ltd.]
were used for PCR amplification of the target gene. PCR primer
(5′-3′) sequences were as follows: hTSHR410 forward,
CACCGGAATGGGGTGTTCGTCTCC and reverse, CACAATTCTCAGGAACTTGTAGCCCA;
hTSHR290 forward, CACCAAGAAAATCAGAGGAATCCT and reverse,
CACAATTCTCAGGAACTTGTAGCCCA; and hTSHR289 forward,
CACCATGAGGCCGGCGGA and reverse, CTGATTCTTAAAAGCACAGCAGTGGC. The
following thermocycling conditions were used: 98°C for 5 min,
followed by 30 cycles of 98°C for 10 sec, 55°C for 5 sec and 72°C
for 30–90 sec; 72°C for 5 min, followed by a 4°C hold. PCR
experiments were performed using the T100 Thermal Cycler (Bio-Rad
Laboratories, Inc.). Gene size was determined by 2% agarose gel
electrophoresis (visualization method, green fluorescent nucleic
acid dye; cat. no. G8140; Beijing Solarbio Science & Technology
Co., Ltd.) and imaging (BluSight Pro; GD50502; Monad Biotech Co.,
Ltd.), and the purified gene was extracted using the Gene JET Gel
extraction kit (Invitrogen; Thermo Fisher Scientific, Inc.). The
gene concentration was determined using a UV spectrophotometer.
According to the Champion™ pET102 Directional TOPO™ Expression Kit
(Invitrogen; Thermo Fisher Scientific, Inc.) requirements, the
purified and recovered hTSHR target gene was connected to
pET102-D-TOPO and transformed into competent TOP10 cells
[Invitrogen; Thermo Fisher Scientific, Inc.; TOP10 Escherichia
coli cells were cultured in S.O.C. medium (Thermo Fisher
Scientific, Inc.) supplemented with 100 µg/ml ampicillin (Beijing
Solarbio Science & Technology Co., Ltd.) at 37°C] by adding 3
µl reaction mixture to 50 µl TOP10 cells (Invitrogen; Thermo Fisher
Scientific, Inc.). The recovered bacterial liquid was spread onto
LB (Beijing Solarbio Science & Technology Co., Ltd.) solid
plates containing 100 µg/ml ampicillin and incubated overnight at
37°C.
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Gene | Forward primer | Reverse primer |
|---|
| hTSHR410 |
5′-CACCGGAATGGGGTGTTCGTCTCC-3′ |
3′-CACAATTCTCAGGAACTTGTAGCCCA-5′ |
| hTSHR290 |
5′-CACCAAGAAAATCAGAGGAATCCT-3′ |
3′-CACAATTCTCAGGAACTTGTAGCCCA-5′ |
| hTSHR289 |
5′-CACCATGAGGCCGGCGGA-3′ |
3′-CTGATTCTTAAAAGCACAGCAGTGGC-5′ |
Monoclonal colonies were picked out from the petri
dish, the picked colonies were mixed in sterile and enzyme-free
water, and then PCR was performed under the aforementioned PCR
reaction conditions. They were confirmed and identified using 2%
agarose gel electrophoresis (visualization method, green
fluorescent nucleic acid dye; cat. no. G8140; Beijing Solarbio
Science & Technology Co., Ltd.) and imaging (BluSight Pro;
GD50502; Monad Biotech Co., Ltd.). Positive colonies were
inoculated into LB liquid medium (Beijing Solarbio Science &
Technology Co., Ltd.) with 100 µg/ml ampicillin (Beijing Solarbio
Science & Technology Co., Ltd.) and shaken overnight (37°C; 220
rpm), and plasmids were extracted from recombinant TOP10-hTSHR
cells using a plasmid mini-prep kit (TIANprep Mini Plasmid Kit;
cat. no. DP103; Tiangen Biotech Co., Ltd.) according to the
manufacturer's instructions.
After collecting the bacteria, using the plasmid
mini-prep kit, Buffer P1 solution, Buffer P2 solution and Buffer P3
solution containing RNaseA were added sequentially, and the
supernatant was moved into the adsorption column CP3 and
centrifuged for 2 min at room temperature at 13,400 × g, and the
elution buffer EB was added dropwise to the middle part of the
adsorption membrane, and the recombinant plasmid solution
containing the target gene was collected into the centrifuge tube
after centrifugation for 2 min at room temperature at 13,400 × g.
Using a 3730×l DNA Analyzer (cat. no. A41046; Thermo Fisher
Scientific Inc.), the extracted recombinant plasmid was used as a
template, and the primers used were: Forward, TTCCTCGACGCTAACCTG
and reverse, TAGTTATTGCTCAGCGGTGG. In the 3730×l DNA Analyzer, the
plasmid to be tested first replicates DNA from the primer under the
catalysis of DNA polymerase until the reaction stops when it
encounters ddNTP. Then, the sequence of the DNA obtained from the
reaction is read by gel electrophoresis, and ultimately the gene
sequence of the plasmid to be tested is inferred (Sequencing
Analysis v7.1; Thermo Fisher Scientific Inc.).
Transformation of different hTSHR
recombinant plasmids into Escherichia coli for expression
Each TOP10-hTSHR recombinant plasmid was transformed
into Escherichia coli BL21Star™ (DE3) (Invitrogen; Thermo
Fisher Scientific, Inc.). Following expansion culture, induction of
hTSHR fusion protein expression was carried out using 0.5 mmol/l
isopropyl β-D-1-thiogalactopyranoside (IPTG). After 5 h of
induction at 25°C, the bacterial culture was collected. The cells
were resuspended in 1X PBS or binding buffer (20 mM sodium
phosphate; 0.5 M NaCl; 5 mM imidazole; pH 7.4). The cells were
disrupted using a cell disruptor, and the bacterial lysate was
centrifuged at 13,400 × g for 20 min at 4°C to collect the
supernatant and pellet. SDS-PAGE was conducted to determine the
expression of each hTSHR fusion protein, distinguishing between
expression in the supernatant and inclusion bodies. A total of 10
ng of protein was loaded per lane in a 10% gel.
Expression and purification of hTSHR
recombinant protein
Expression and purification of the hTSHR fusion
protein, tagged with histidine (His), were performed using a
Nickel-nitrilotriacetic acid (Ni-NTA) column. The process involved
collecting precipitated bacteria from LB culture following IPTG
induction. The collected cells were sonicated on ice (ultrasonic
power 200 W; 40 min), and the resulting mixture was subjected to
low-temperature centrifugation at 13,400 × g for 20 min at 4°C to
separate the precipitate and supernatant. The inclusion bodies in
the precipitate were dissolved in binding buffer (20 mM sodium
phosphate; 0.5 M NaCl; 5 mM imidazole; 8 M urea; pH 7.4). Following
centrifugation at 13,400 × g for 20 min at 4°C, the supernatant
containing the dissolved inclusion bodies was collected. This
supernatant was then combined with Ni-NTA and incubated for 1 h at
4°C. Similarly, the supernatant from the expression of the fusion
protein was also combined with Ni-NTA and incubated for 1 h at 4°C.
The inclusion bodies in the precipitate were dissolved in binding
buffer (20 mM sodium phosphate; 0.5 M NaCl; 5 mM imidazole; 8 M
urea; pH 7.4). Following centrifugation at 13,400 × g for 20 min at
4°C, the supernatant containing the dissolved inclusion bodies was
collected. This supernatant was then combined with Ni-NTA and
incubated for 1 h at 4°C. Similarly, the supernatant from the
expression of the fusion protein was also combined with Ni-NTA and
incubated for 1 h at 4°C. The inclusion body-expressed protein
underwent gradient renaturation in refolding buffer (20 mM sodium
phosphate; 0.5 M NaCl; 6-0 M urea gradient; pH 7.4). The purified
protein was concentrated using an ultrafiltration tube, and the
concentration of the purified fusion protein was determined using a
BCA assay kit. Subsequently, the complete amino acid sequences of
hTSHR410, hTSHR290 and hTSHR289 were imported into ExpaSy-ProtParam
software (https://web.expasy.org/protparam/), and various
physical and chemical parameters, including the molecular weight,
of the protein were calculated through the software analysis, and
the molecular weight of each fusion protein was estimated. The
predicted molecular weights of hTSHR289, hTSHR410 and hTSHR290 are
~47, 70 and 39 kDa respectively.
Western blot analysis
SDS-PAGE and western blot analysis were used to
characterize the three hTSHR fusion proteins purified in the
previous step. Proteins were extracted using the ultrasonic
disruption method (power 200 w; ultrasound was on for 2 sec and off
for 3 sec), and SDS-PAGE loading buffer (cat. no. P1040, Beijing
Solarbio Science & Technology Co., Ltd.) was added to the
extracted proteins, which were boiled for 5–10 min. The
concentration of the protein was detected using a BCA protein
quantification kit. Each lane was loaded with 5 µg of protein.
Prepared samples (purified hTSHR fusion protein with protein
extraction buffer) were loaded into the sample wells of the
SDS-PAGE gel (10%) and electrophoresed at 120 V. The SDS-PAGE gel
was stained with Coomassie Brilliant Blue (Beijing Solarbio Science
& Technology Co., Ltd.), or the proteins in the SDS-PAGE gel
were electrotransferred to a PVDF membrane (Beijing Solarbio
Science & Technology Co., Ltd.). The membrane was blocked with
5% non-fat powdered milk (cat. no. D8340; Beijing Solarbio Science
& Technology Co., Ltd.) at room temperature for 2 h. The PVDF
membrane was then blocked and incubated with antibodies, including
TSHR 3B12 (1:1,000; sc-53542; Santa Cruz Biotechnology, Inc.; 4°C;
12 h) and His (1:1,000; 1B7G5; Proteintech Group, Inc.; 4°C; 12 h).
The next day, following incubation with secondary antibodies [Goat
Anti-Mouse IgG H&L (HRP); 1:10,000; ab205719; Abcam] for 2 h at
room temperature, visualization was carried out using ECL reagent
(Beijing Solarbio Science & Technology Co., Ltd.) on an
automated chemiluminescence imager.
Direct ELISA using hTSHR fusion
protein as the antigen (hTSHR ELISA) and specificity
The three fusion proteins were coated as antigens on
the plate of enzymes. Following repeated experiments, it was
determined that coating with 1 µg/well of antigen at 4°C overnight
and blocking with 200 µl 1% BSA (Jiangsu Aidisheng Biotechnology
Co., Ltd.) per well at 37°C for 2 h were the optimal conditions.
Different serum samples were diluted in a gradient of 1:1,000
(optimized dilution ratio) and used as the primary antibody, with
100 µl diluted serum added per well. Duplicate test wells and
control wells were set up and then incubated at 37°C for 2 h.
Following the addition of 100 µl HRP-labeled antibody (1:10,000;
cat. no. D110150; Sangon Biotech Co., Ltd.), the plate was
incubated at 37°C for a further 2 h, followed by color development
with tetramethylbenzidine substrate for 15 min at 37°C. The
absorbance value at 450 nm was then determined using an ELISA
reader.
Animal study
A total of 48 female BALB/c mice aged 5–6 weeks and
weighing 20±2 g were obtained from Jiangsu Cavens Experimental
Animals Co., Ltd. The mice were housed under specific pathogen-free
conditions at the Experimental Animal Research Platform of The
Second Hospital of Lanzhou University. The mice were raised in a
specific pathogen-free environment, the temperature was 18–22°C and
the humidity was 50–60%, with a 12/12-h light/dark cycle. The feed
was purchased from Beijing Keao Xieli Feed Co., Ltd. (cat. no.
1016706714625204224). The drinking water came from the experimental
animal research platform. Both the feed and water underwent
high-temperature and high-pressure sterilization treatment. Mice
had ad libitum access to food and water. Food, water, air,
bedding and all other supplies entering the animal center barrier
system were sterilized with high temperature and high pressure, and
were sterilized by irradiation through the transfer window before
entering and exiting the animal research platform. The mice were
randomly divided into 4 groups, with 12 mice in each group, which
were the hTSHR410 fusion protein immune mouse group, hTSHR289
fusion protein immune mouse group, hTSHR290 fusion protein immune
mouse group and normal saline control group. The animal experiments
received approval from the Animal Welfare and Ethics Committee of
The Second Hospital of Lanzhou University (approval no. D2023-156;
Lanzhou, China). After reviewing the literature and combining the
test results of previous studies, mice were immunized with a 1:1
mixture of fusion protein injected at 4 points on the back of the
mice with complete Freund's adjuvant (the first injection) and
incomplete Freund's adjuvant (the second two injections) (20,21).
Each female BALB/c mouse was injected subcutaneously with 50 µg
hTSHR fusion protein at 4 points on the back, and the control group
was injected with normal saline. Three immunizations were given at
weeks 0, 3 and 6. Mice were anesthetized by intraperitoneal
injection of sodium pentobarbital (50 mg/kg) and euthanized by
anesthesia overdose (intraperitoneal injection of sodium
pentobarbital; 100 mg/kg). The Animal Research: Reporting of In
Vivo Experiments guidelines were followed (22).
Mouse serum analysis
Blood was collected via cardiac puncture 2 weeks
after the completion of immunization under deep anesthesia (30
mg/kg sodium pentobarbital; intraperitoneal injection). The
extracted blood was placed in 1.5-ml Eppendorf tubes and left to
stand at room temperature. Next, a centrifuge was set to 3,000 × g
and samples were centrifuged for 15 min at room temperature to
separate the serum, which was then stored at −20°C. The present
study utilized the thyroxine (T4) ELISA kit (cat. no. JL10849;
Shanghai Jonlnbio Industrial Co., Ltd.), the TSH ELISA kit (cat.
no. JM-11776M1; Jingmei Biotechnology Co., Ltd.) and the TRAb ELISA
kit (cat. no. ZY-TRAb-MU; Zeye Biotechnology Co., Ltd.) to assess
the levels of TSH, T4 and TRAb in the serum of each group of mice.
A total of 10 µl of serum was extracted from each mouse in each
group. The serum was diluted by 1:4 using the sample dilution
solution in the aforementioned kits, and then the analysis was
carried out according to the manufacturer's instructions. Readings
were obtained at 450 nm using an ELISA reader. The procedure was
repeated three times for each assessment.
Histological analysis
Mice were euthanized using an overdose anesthetic
method (intraperitoneal injection of sodium pentobarbital; 100
mg/kg), and the thyroid glands were dissected. The thyroid tissue
was soaked in 4% paraformaldehyde for >48 h at room temperature.
After fixation, the tissues were dehydrated, sequentially soaked in
75% alcohol, 95% alcohol and absolute ethanol for gradient
dehydration, sequentially soaked in two xylenes (Tianjin Fuyu Fine
Chemical Co., Ltd.) and paraffin-embedded, and were then sectioned
using a Leica microtome (thickness, 5 µm; Leica Microsystems,
Inc.). Subsequently, the slices were placed in a baking machine and
baked at 75°C for 1 h. After deparaffinization, hematoxylin was
added dropwise to the slices and these were incubated at room
temperature for 5 min. Differentiation was performed by adding
hydrochloric acid ethanol dropwise to the slices and incubating at
room temperature for 3–5 sec. Finally, eosin solution was added
dropwise to the slices and the slices were incubated at room
temperature for 1–2 min. After the neutral gum was completely
dried, observations and scans were performed using a light
microscope.
Flow cytometry of T cell subsets
Following euthanasia, splenic tissues were retrieved
from the immunized mice and placed in 1640 culture medium (Gibco;
Thermo Fisher Scientific, Inc.). The spleen tissues were dissected
into small pieces using a surgical blade. Subsequently, the spleen
tissue was ground on a 100-mesh cell strainer to obtain a
single-cell suspension. Following centrifugation at 350 × g at 4°C
for 10 min, cells were collected, treated with red blood cell lysis
buffer at room temperature for 10 min, washed with PBS and then
centrifuged at 350 × g at 4°C for 10 min to obtain lymphocytes.
Lymphocytes (2×106 cells) were stained using
allophycocyanin (APC)/phycoerythrin (PE)/FITC markers. Cells were
analyzed using the CytoFLEX LX Flow Cytometer (Beckman Coulter,
Inc.). Reported results were analyzed using the CytExpert software
(version 2.5; Beckman Coulter, Inc.) of the CytoFLEX LX flow
cytometer to characterize T cell subsets. The analyte detectors
were as follows: CD3+ antibody (cat. no. 565643; Becton,
Dickinson and Company), CD4+ antibody (cat. no.
F21004A02; Multi Sciences Biotech), CD8+ antibody (cat.
no. F2100801; Multi Sciences Biotech), CD25 antibody (cat. no.
E-AB-F1102C; Wuhan Elabscience Biotechnology Co., Ltd.) and CD122
antibody (cat. no. E-AB-F1029D; Wuhan Elabscience Biotechnology
Co., Ltd.). The analyte reporters were as follows: CD3+,
APC; CD4+, PE; CD8+, FITC; CD25, FITC; and
CD122, PE.
Statistical analysis
Statistical analysis was performed using GraphPad
Prism 8.0 software (Dotmatics) and SPSS 26.0 software (IBM Corp.).
Quantitative data with normal distribution are presented as the
mean ± standard deviation, while data with a skewed distribution
are presented as the median (quartile), and count data are
presented as absolute values or percentages. One-way ANOVA with
Tukey's post hoc test was used for comparisons of multiple groups
of parametric data. Categorical variables were compared using
χ2 tests to compare all groups and Bonferroni correction
was applied to the P-values. The Kruskal-Wallis test with Dunn's
post hoc test was used for non-parametric data. P<0.05 was
considered to indicate a statistically significant difference. All
experiments were repeated in triplicate.
Results
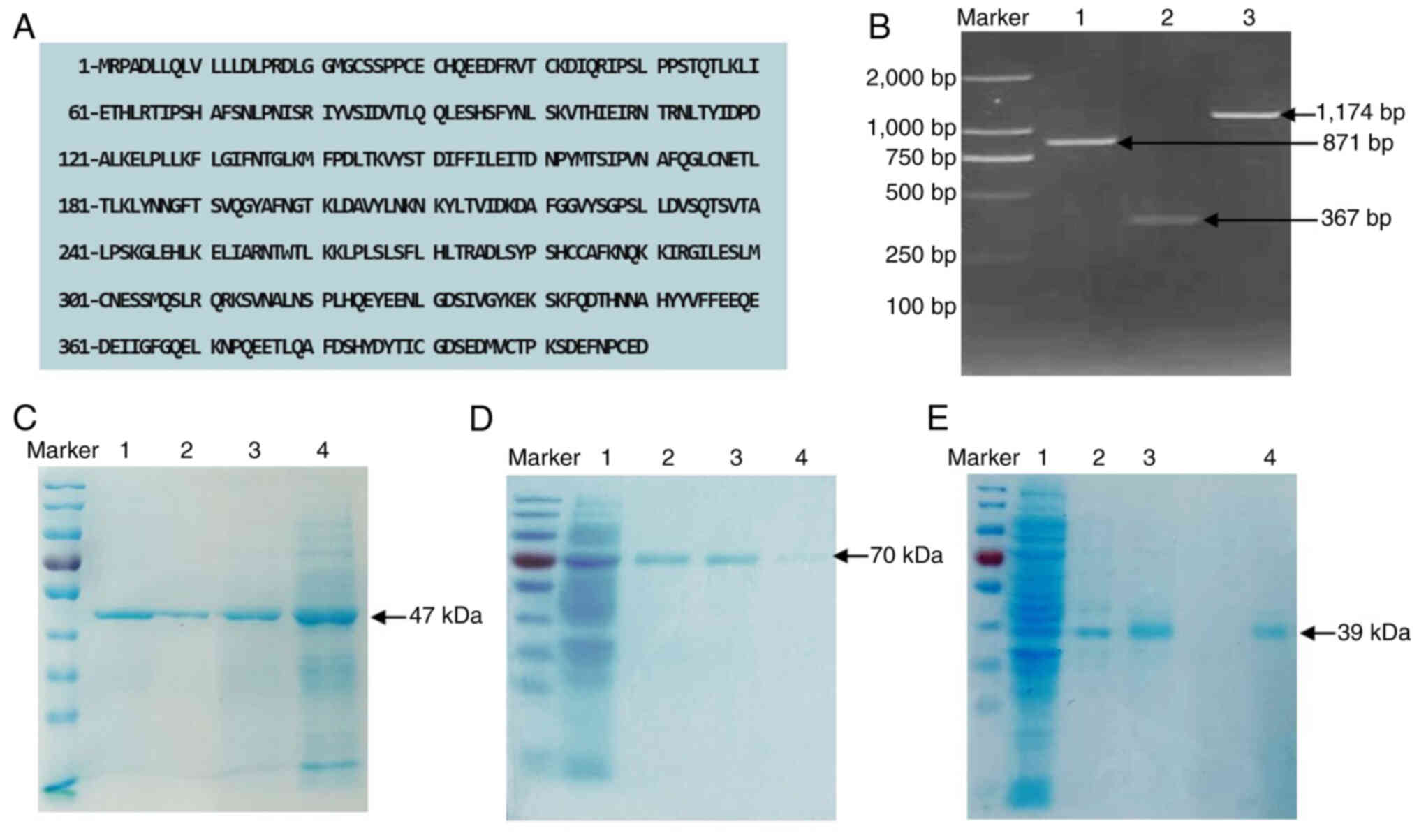
Different fusion proteins of
hTSHR
According to the full-length receptor gene sequence
of human TSHR, different hTSHR protein sequences were extracted:
hTSHR410, hTSHR289 and hTSHR290 (amino acid sequences shown in
Fig. 1A). The recombinant plasmid
was obtained by ligating the gene sequences corresponding to the
aforementioned hTSHR amino acid sequences with the vector. The
results of agarose gel electrophoresis after PCR amplification are
shown in Fig. 1B. Recombinant
plasmids with correctly framed reading sequences were selected for
expression in Escherichia coli, followed by induction with
IPTG, purification and visualization on SDS-PAGE gels, where three
distinct bands were observed (Fig.
1C-E). Based on SDS-PAGE, the molecular weight of the single
band in Fig. 1C was estimated to
be ~47 kDa, that of the single band in Fig. 1D was ~70 kDa and that of the single
band in Fig. 1E was ~39 kDa. These
results indicated that the purified proteins had molecular weights
consistent with the expected values, indicating that they were
hTSHR fusion proteins.
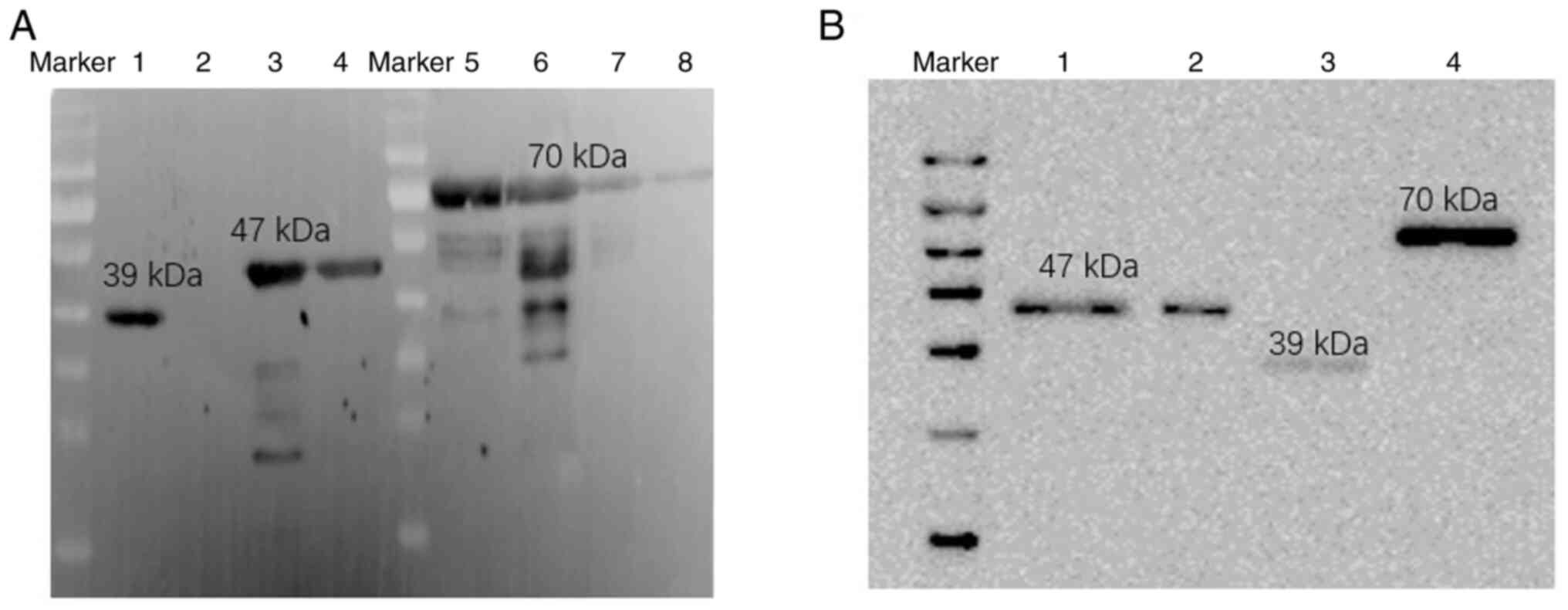
hTSHR fusion protein validation
Fusion proteins were expressed in the prokaryotic
system and single bands were obtained by SDS-PAGE after
purification. In order to accurately identify the three different
hTSHR fusion proteins, validation using western blotting was
performed. TSHR monoclonal antibody (3B12), His antibody and the
three different fusion proteins (hTSHR410, hTSHR289 and hTSHR290)
were assessed using western blotting. The results showed clear
protein bands at 70, 47 and 39 kDa (Fig. 2A and B). This demonstrated that the
purified fusion proteins were hTSHR fusion proteins.
Binding activity of different hTSHR
fusion proteins to an IgG antibody verified using an ELISA
In order to establish a convenient and stable method
for distinguishing the types of TRAb and to validate the biological
activity of the different hTSHR fusion proteins obtained, the age,
sex, and FT3, FT4, TSH and TRAb levels were analyzed in five groups
of recruited patients. There were no statistically significant
differences in the age or sex among the patients in each group.
This is summarized in Table
II.
 | Table II.Clinical characteristics of all
subjects. |
Table II.
Clinical characteristics of all
subjects.
| Variable | Hyperthyroidism
(TRAb+) | Hyperthyroidism
(TRAb−) | Hypothyroidism
(TRAb+) | Hypothyroidism
(TRAb−) | Control | P-value |
|---|
| Age, years | 33.91±13.06 | 35.10±13.76 | 33.17±18.97 | 37.4±16.86 | 34.9±12.23 | 0.631a, 0.346b, 0.593c, 0.275d |
| Sex, n |
|
|
|
|
|
|
|
Male | 10 | 9 | 11 | 10 | 12 | 0.592a, 0.417b, 0.791c, 0.592d |
|
Female | 20 | 21 | 19 | 20 | 18 |
|
| FT3, pmol/l (normal
range, 2.77–6.31 pmol/l) | 15.19±9.23 | 13.67±4.80 | 2.29±1.71 | 1.87±0.99 | 4.13±1.35 | 0.012ae
0.030be, 0.021ce,
0.008de |
| FT4, pmol/l (normal
range, 10.44–24.38 pmol/l) | 41.99±16.21 | 38.28±14.32 | 4.12±3.48 | 3.85±2.52 | 16.61±3.50 | 0.035ae,
0.026be, 0.012ce,
0.007de |
| TSH, µIU/ml (normal
range, 0.38–4.34 µIU/ml) | 0.10 (0.10,
0.30) | 0.08
(0.06,0.40) | 68.09
(33.70,108.76) | 73.77
(41.43,116.29) | 2.64
(1.80,3.41) | 0.006ae,
0.007be, 0.036ce,
0.023de |
| TRAb, IU/l (normal
range, 0.00–60.00 U/ml) | 20.00 (11.00,
30.00) | 0.77
(0.36,1.15) | 16.64 (7.39,
30.00) | 0.54 (0.43,
0.61) | 0.38 (0.14,
1.07) | 0.008ae,
0.558b, 0.021ce,
0.642d |
| TG, ng/ml (normal
range, 1.00–39.00 ng/ml) | 601.56 (28.00,
1300.00) | 548.50 (30.60,
1300.00) | 572.04 (16.30,
1300.00) | 483.41 (21.00,
1300.00) | 15.70 (3.28,
35.40) | 0.016ae,
0.017be, 0.009ce,
0.006de |
| TPOAb, U/ml (normal
range, 0.0–1.5 IU/l) | 54.37 (0.50,
314.10) | 61.43 (0.20,
368.00) | 92.25 (2.00,
600.00) | 85.82 (0.63,
438.50) | 0.53 (0.00,
1.08) | 0.023ae,
0.037be, 0.018ce,
0.025de |
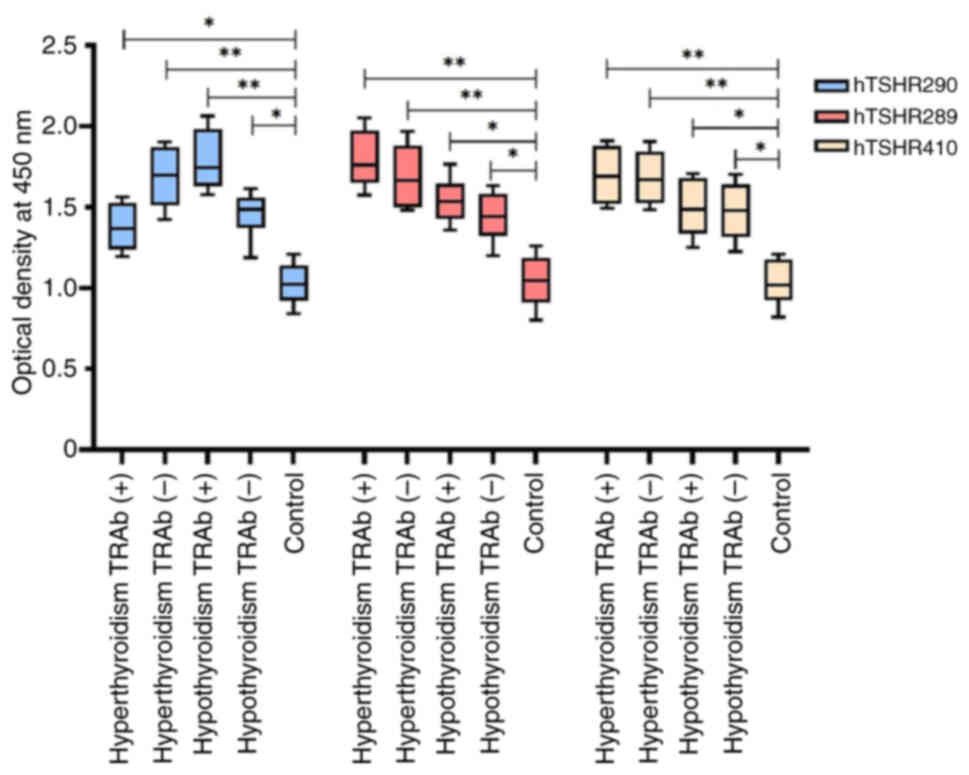
In the present study, an ELISA was carried out to
assess the binding activity of serum IgG antibodies to hTSHR410,
hTSHR289 and hTSHR290 fusion proteins in serum samples from the
aforementioned five groups: Hyperthyroidism (TRAb+ and
TRAb−), hypothyroidism (TRAb+ and
TRAb−) and healthy individuals. The results revealed
that the absorbance [optical density (OD)450 nm] of hTSHR289 fusion
protein with serum from patients in the hyperthyroidism
TRAb+, hyperthyroidism TRAb−, hypothyroidism
TRAb− and hypothyroidism TRAb+ groups was
increased compared with that in healthy individuals (P<0.05).
Among these, the serum from patients with hyperthyroidism
TRAb+ showed the highest binding OD450 value with
hTSHR289 fusion protein. Similarly, the binding OD450 values of
hTSHR290 fusion protein with serum from patients in the
hyperthyroidism TRAb−, hypothyroidism TRAb+,
hypothyroidism TRAb− and hypothyroidism TRAb−
groups were all higher compared with those in healthy individuals
(P<0.05). Among these, the serum from patients with
hypothyroidism TRAb+ exhibited the highest binding OD450
value with the hTSHR290 fusion protein. Furthermore, the binding
OD450 values of the hTSHR410 fusion protein with serum from
patients in the hyperthyroidism TRAb+, hyperthyroidism
TRAb−, hypothyroidism TRAb+ and
hypothyroidism TRAb− groups were all higher compared
with those in healthy individuals (P<0.05; Fig. 3).
Based on Figs. 2
and 3, both the hTSHR290 and
hTSHR289 fusion proteins were revealed to exhibit biological
activity and strong antigenicity. Thus, they could be used as
antigens to establish corresponding serological detection methods
for the determination of TSAb and TBAb.
Changes in thyroid function of
immunized mice
An ELISA was carried out to detect the binding
activity of the three fusion proteins with serum from patients with
hyperthyroidism and hypothyroidism, demonstrating the biological
activity of the prepared fusion proteins. In order to further
validate the immunogenicity of the different fusion proteins,
BALB/c mice were immunized with the fusion proteins.
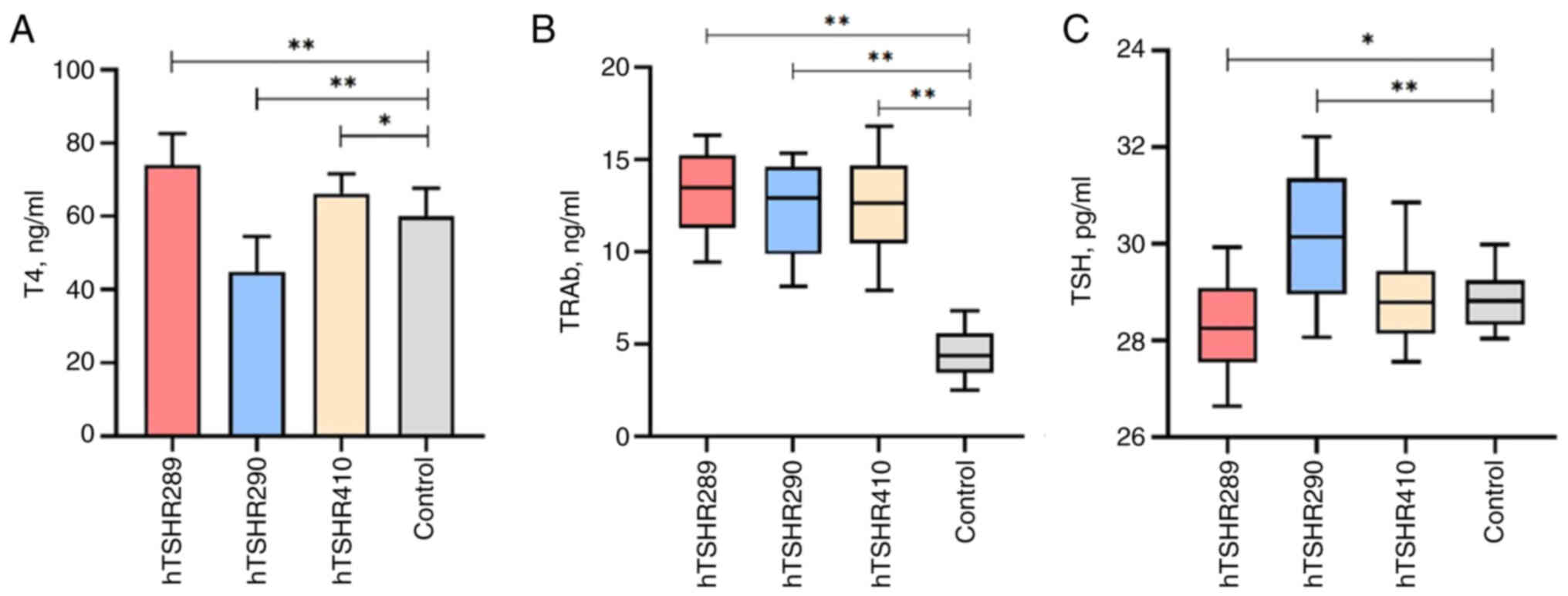
T4, TSH and TRAb may be effective indicators for
evaluating thyroid function. Following the immunization of mice,
serum samples from the four groups of mice were tested using T4,
TSH and TRAb ELISA kits. Analysis revealed that the T4 levels in
mice immunized with hTSHR289 (aa1-289) fusion protein were
74.06±8.55 ng/ml, and were significantly higher than those in the
control group (60.01±7.74 ng/ml) and exceeded the normal range for
T4 levels (52.27–67.75 ng/ml). The T4 levels in mice immunized with
hTSHR410 (aa21-410) were 66.17±5.48 ng/ml, and were higher compared
with those in the control group (60.01±7.74 ng/ml). The T4 levels
in mice immunized with hTSHR290 (aa290-410) were 44.85±9.70 ng/ml,
and were lower compared with those in the control group (60.01±7.74
ng/ml), and these differences were statistically significant
(P<0.05) (Fig. 4A).
The TSH levels in mice in the hTSHR290 (30.13±1.27
pg/ml) group were significantly higher and the TSH levels in the
hTSHR289 (28.34±0.90 pg/ml) group was significantly higher than
those in the control group (28.82±0.55 pg/ml), exhibiting a
statistically significant difference (P<0.05). The TSH levels in
mice in the hTSHR410 group (28.79±0.81 pg/ml) did not differ
significantly from those in mice in the control group (Fig. 4C).
The TRAb levels in mice immunized with hTSHR289
(13.51±2.26 ng/ml), hTSHR410 (12.47±3.48 ng/ml) and hTSHR290
(13.05±2.53 ng/ml) were all increased compared with those in the
control group (4.16±1.36 ng/ml), which was statistically
significant (P<0.05), as shown in Fig. 4B.
Following the determination of thyroid function
indicators, including T4, TSH and TRAb, to further ascertain
alterations in thyroid function in immunized mice, H&E staining
was conducted on the thyroid tissues and various pathological
changes were observed. Thyroid tissues from 7/12 of the hTSHR289
mice exhibited diffuse hyperplasia with increased cellularity.
Epithelial proliferation led to the formation of papillary
projections into the follicular lumen. The colloid within the
follicular lumen was sparse, exhibiting scalloped edges, along with
varying sizes of absorption vacuoles. The epithelium consisted of
tall columnar cells with basal nuclei that were crowded, round or
elliptical and irregular in shape, indicative of pathological
changes in the thyroids of hyperthyroid mice (Fig. 5A and B). In the thyroid tissues of
8/12 of the hTSHR290 mice, the thyroid follicular epithelium was
composed of tall columnar cells with loosely arranged basal nuclei,
exhibiting round or elliptical nuclei that were irregular in shape.
Some follicles exhibited destruction and atrophy, accompanied by
extruded colloid, representing pathological changes in hypothyroid
mouse thyroids (Fig. 5C and D).
The thyroid lesions in 7/12 of the hTSHR410 mice manifested as
focal distribution, with some follicles displaying destruction,
atrophy and extruded colloid. Regeneration of some follicular
epithelium and increased fibrous tissue between the follicles were
observed (Fig. 5E and F). The
thyroid tissue of mice in the control group revealed that the
follicular structure of the thyroid gland was normal, the follicle
was round or oval, the epithelial cells were arranged in a single
flat shape, and the follicle was filled with glia (Fig. 5G and H). No animals were
prematurely euthanized in the present study, and all animals
completed the predetermined protocol. The results reflected the
respective mean values of all measurements for all randomized
animals.
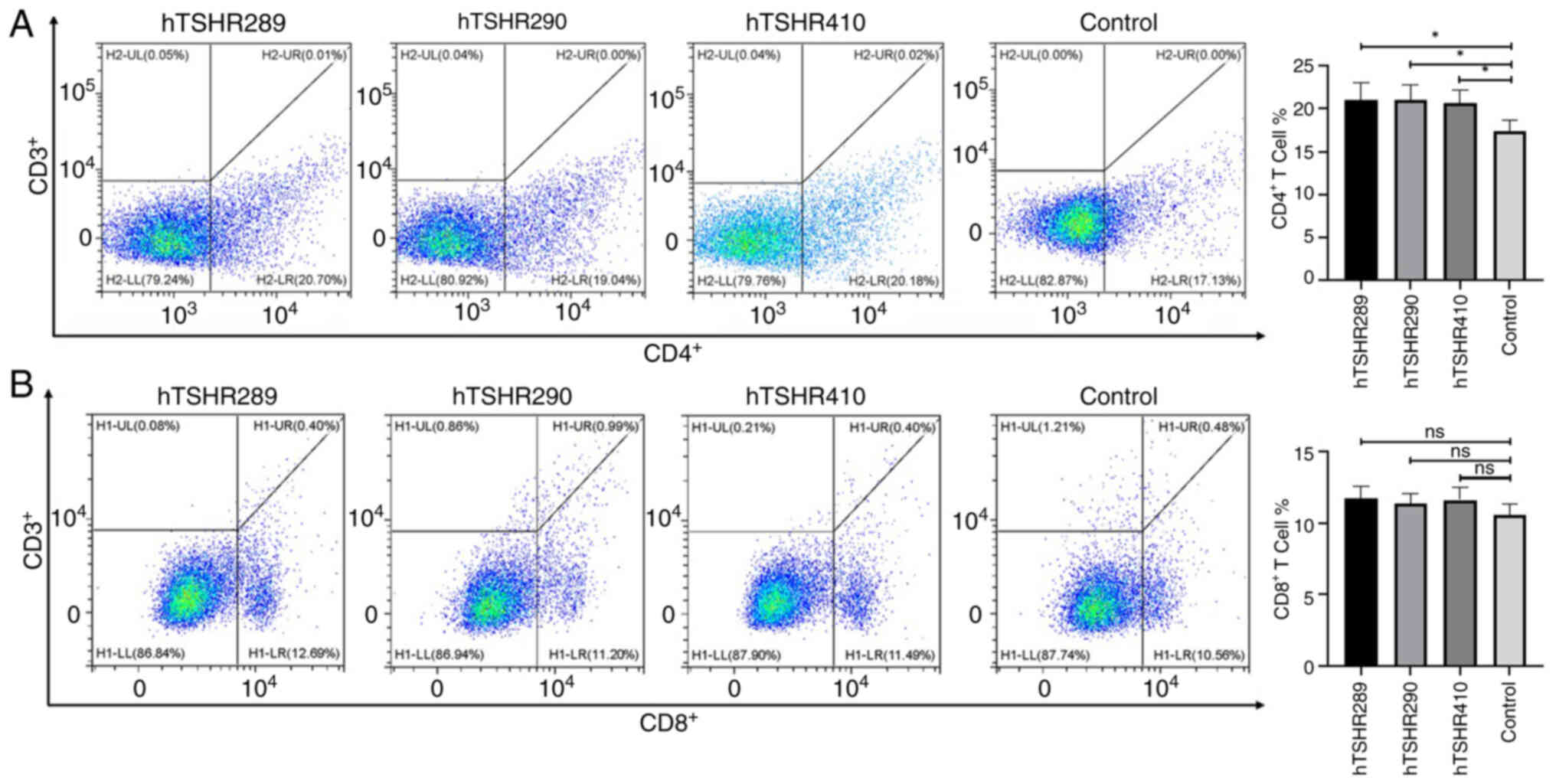
CD4+ T cells and
CD8+ T cells participate in immune responses in
mice
The predominant causes of thyroid cell damage and
TSHR antibody production in AITD include the activation of
CD4+ T cells. Simultaneously, cytokines produced
following the activation of CD4+ T cells stimulate
CD8+ T cells to destroy thyroid cells, particularly
thyroid cells in patients with HT (23,24).
Therefore, the overall balance, proliferation and activation of
CD4+ T cells and CD8+ T cells are of
importance in the pathogenesis of AITD.
In the present study, the immune response was
further investigated following the immunization of mice with three
types of hTSHR fusion proteins as antigens. Flow cytometry was
utilized to determine the proportions of CD4+ and
CD8+ T cells in splenocytes of each group of mice. The
proportion of CD4+ T cells in the three immunization
groups was increased compared with that in the control group, and
the difference was statistically significant. The proportion of
CD8+ T cells in the three immunization groups was
increased compared with that in the control group; however, the
difference was not statistically significant (Fig. 6).
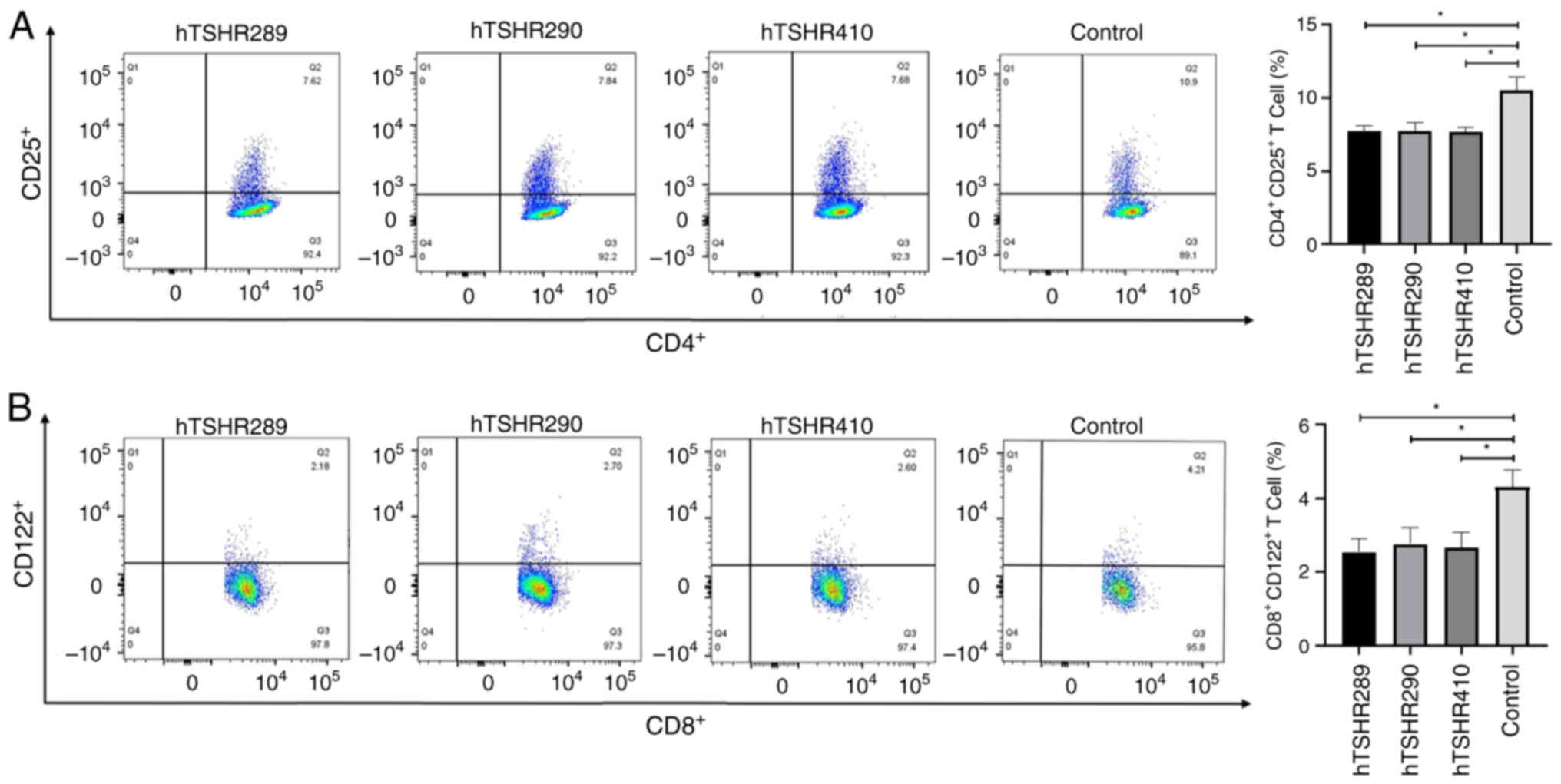
CD4+CD25+ T
cells and CD8+CD122+ T cells participate in
the immune response in mice
CD4+CD25+ T cells and
CD8+CD122+ T cells are associated with the
occurrence and severity of AITD (25,26).
Flow cytometry was carried out to detect the numbers of
CD4+CD25+ T cells and
CD8+CD122+ T cells in spleen cells from each
group of mice. Analysis revealed that the numbers of
CD4+CD25+ T cells and
CD8+CD122+ T cells in spleen cells of the
hTSHR289, hTSHR290 and hTSHR410 groups were decreased compared with
those in the control group (Fig.
7).
Discussion
The occurrence of AITD affects hundreds of millions
of individuals worldwide, with the main types being GD and HT,
characterized by hyperthyroidism and hypothyroidism, respectively
(27). GD is a diffuse goiter and
hyperthyroidism condition associated with the presence of TSHR and
its autoantibodies (28). TSHR
contains three structural domains: The LRRD, hinge region and TMD
(29–31). The full-length TSHR can be
proteolytically cleaved into an A subunit (aa1-289) and a
transmembrane B subunit (aa370-764) (32,33).
The A subunit can be shed from the full-length TSHR, becoming an
inducer of TRAb autoantibodies (34). Studies have shown that TSAb, TSH
and TBAb can bind to the leucine-rich repeat region, and some TBAb
and neutral antibodies can also bind to linear epitopes in other
structural domains of TSHR (35–37).
Furthermore, the epitopes recognized by TSH, TSAb and TBAb on TSHR
are spatially close and exhibit some overlap (36,37).
Therefore, based on the structural characteristics of TSHR and its
binding properties with autoantibodies, active hTSHR fusion
proteins were produced that mimic the natural protein structure and
function of hTSHR. These include the hTSHR289 fusion protein
containing the active LRRD of hTSHR (TSHR A subunit), the hTSHR410
(aa1-410) fusion protein containing the entire ECD of TSHR, and the
hTSHR290 fusion protein containing the carboxyl-terminal region of
the ECD including the hinge region of TSHR (TSHR aa290-410).
The present study further investigated the
immunogenicity and clinical significance of different fusion
proteins of TSHR using serological and in vivo experiments.
In a study by Zulkarnain et al (38), the authors used a prepared fusion
protein of the TSHR169 fragment to bind with the sera of 20
patients with GD, exploring methods for the diagnosis of GD. In the
present study, serum-based validation was conducted using the ELISA
method. Strong specific binding reactions were observed between the
hTSHR289 fusion protein and the sera of patients with clinically
diagnosed hyperthyroidism with TRAb+. The sera of
patients diagnosed with hyperthyroidism TRAb+ primarily
contain TSAb, indicating that TSAb largely binds specifically to
the TSHRA subunit (4). The
hTSHR290 fusion protein exhibited the strongest binding reaction
with the sera of patients with hypothyroidism who were
TRAb+, and even the sera of patients clinically
diagnosed with hypothyroidism who were TRAb− displayed a
binding reaction with the hTSHR290 fusion protein. Furthermore, the
results of the present study indicated that the carboxyl-terminal
end of TSHR (aa290-410) may be an antigenic determinant for
recognizing TBAb.
In previous years, numerous studies have used
eukaryotic or adenoviral vectors or plasmids as the basis for
investigating the role of TSHR. Researchers have expressed concerns
that prokaryotically prepared TSHR proteins may lack
immunogenicity, which was also a focus of the present study. For
studies of TSHR-induced thyroid disease models, current research
often employs adenoviral vectors carrying the TSHR A subunit or
plasmid-induced GD models. This method requires technical skills
and specialized equipment support, and is expensive (16,39–41).
The present study prepared different fusion proteins of TSHR in a
prokaryotic system, which is simple to operate, has high production
yields (higher than eukaryotic systems) (42) and is cost-effective. Subcutaneous
injection of fusion proteins to establish disease models has been
utilized in various disease models and has been demonstrated to be
effective (20,21). Previous investigations have noted
that immunization of BALB/c mice with the ECD protein of human
thyroid-stimulating hormone receptor induced thyrotropin-binding
inhibitory immunoglobulins (TBII) in the serum, subsequently
leading to thyroid pathology (11,39,43).
In addition, a study has demonstrated that repetitive intravenous
injections of 0.1 mg/kg of cyclic peptide 19, a derivative of the
first cytoplasmic loop of TSHR rich in LRRDs, could reduce thyroid
hyperplasia in a long-term mouse model of GD. This intervention
also led to elevated T4 levels and a reduction in sinus tachycardia
(11). Therefore, the important
role of LRRDs needs to be carefully considered. In the present
study, three fusion proteins with different biological activities
were identified through patient serum analysis. To gain a deeper
understanding of the role of TSHR as a primary antigen in the
pathogenesis of AITD, a series of experiments were conducted using
BALB/c mice to investigate the immunogenicity and clinical
significance of TSHR fusion proteins.
In the present investigation, using various fusion
proteins of hTSHR as immunogens, immune responses were induced in
mice that mimicked those triggered by native hTSHR protein
antigens. Analysis revealed that mice immunized with the hTSHR289
fusion protein exhibited significantly elevated levels of TSHR
antibodies and T4 compared with the control group.
Histopathological examination of thyroid tissue sections revealed
pathological changes consistent with hyperthyroidism. These
findings underscore the potential utility of hTSHR fusion proteins
in developing experimental models of thyroid disorders. Compared
with the control group, the hTSHR290 group of mice exhibited a
significant decrease in T4 levels, along with increased levels of
TRAb and TSH. Pathological examination of thyroid tissue sections
revealed pathological changes consistent with hypothyroidism, such
as the presence of tall columnar cells comprising the thyroid
follicular epithelium with nuclei located basally, partial follicle
destruction and atrophy, as well as extracellular colloid leakage.
When comparing the hTSHR410 group with the control group,
significant increases in TRAb and T4 levels were observed, while
TSH values did not significantly differ between the groups. The
pathological presentation of thyroid tissue indicated focal
distribution of fibrotic changes. Collectively, the fusion protein
hTSHR289 (TSHRA subunit; aa1-289) induced primarily stimulatory
antibodies in mice, leading to the development of GD. The fusion
protein hTSHR290 (carboxyl-terminal region of TSHR; aa290-410)
acted as an immunogenic antigen, resulting in the production of
TBAb, and thus, inducing hypothyroidism in mice. hTSHR410 fusion
protein (entire ECD of TSHR) triggered a cross-reactive immune
response in mice, among which the pathological changes associated
with GD were more prominent. However, as a single model for
research into AITD, the hTSHR410 fusion protein as an immunogen may
not be an ideal choice. Therefore, the utilization of hTSHR289 and
hTSHR290 fusion proteins as immunogens to induce hyperthyroidism
and hypothyroidism animal models, respectively, would be beneficial
for in-depth exploration of the occurrence, progression and
treatment of AITD. Furthermore, inducing disease occurrence through
fusion proteins is close to the natural process of autoimmune
disease development. The animal models induced and constructed in
the present study will be more conducive to the research of
AITD.
The activation of CD4+ T cells serves as
the primary mechanism in AITDs leading to thyroid cell damage and
the production of TRAb (31,44).
In HT, thyroid cells are destructed by inflammatory infiltration
composed of CD8+ T cells, CD4+ T cells,
macrophages and plasma cells (24). Both CD4+ and
CD8+ T cells infiltrate the thyroid gland in varying
proportions, resulting in altered thyroid function and the onset of
HT and GD. Furthermore, they are closely associated with TRAb
levels, serving a key role in predicting treatment outcomes
(44,45). The animal experiments in the
present study indicated that the hTSHR fusion protein induced
alterations in mice, leading to hyperthyroidism, hypothyroidism and
inflammatory responses. Further confirming the immunogenicity of
hTSHR fusion proteins and their role in inducing pathogenic TRAb in
mice, the present results indicated that the ratio of
CD4+ T cells and CD8+ T cells was altered in
immunized mice.
The specific expression of CD25 on the surface of
CD4+ T cells accounts for ~10% of peripheral blood
CD4+ T cells. Inhibiting abnormal or excessive immune
responses against self, microbial and environmental antigens is
associated with a wide range of autoimmune diseases (autoimmune
gastritis, thyroiditis and type 1 diabetes) (46). Deficiency of
CD4+CD25+ T cells leads to various autoimmune
and inflammatory states in mice (47). Studies have found that the
frequency of CD4+CD25+ T cells in lupus-prone
mice was lower than that in normal mice (48,49).
A decrease in the number of CD4+CD25+ T cells
may be one of the mechanisms leading to immune dysregulation in
immune thrombocytopenia (ITP), and the changes in the number of
CD4+CD25+ T cells are considered to be
associated with the severity of ITP (25). The lack of
CD4+CD25+ T cells can lead to severe
autoimmune/inflammatory diseases, while normal regulatory T cells
(Tregs) can inhibit disease development (50). CD122 is the β subunit of the IL-2
receptor, not only regulating Tregs but also influencing Treg
function. CD8+CD122+ T cells may regulate
immune responses by producing IL-10, TGFβ1 and IFNγ; however, the
exact mechanism of their inhibitory effect remains largely unknown
(51,52). Previous studies have indicated that
CD8+CD122+ T cells could suppress T cell
responses, modulated autoimmunity and alloimmune responses, and
served an important role in autoimmune diseases (52–54).
The development of systemic lupus erythematosus-like disease in
B6-Yaa mutant mice is associated with a defect in
CD8+CD122+ T cells (55). Disruption of the inhibitory
interaction between CD8+CD122+ T cells and
follicular helper T cells leads to the development of a lethal
systemic lupus erythematosus-like autoimmune disease dependent on
autoantibodies (56). The
depletion of CD8+CD122+ T cells increases the
incidence of AITD (57). Research
data show that CD8+CD122+ T cells in
secondary lymphoid organs of prediabetic non-obese diabetic mice
are reduced by ~50%, highlighting the role of
CD8+CD122+ T cells in suppressing autoimmune
diseases (26). Several studies
have shown that CD4+CD25+ and
CD8+CD122+ T cells serve a key role in the
incidence and severity of GD, HT and hypothyroidism (57,58).
The present study revealed that immunization with hTSHR fusion
protein led to a decrease in the number of
CD4+CD25+ and
CD8+CD122+ T cells in the secondary lymphoid
organs of mice. Collectively, the data in the present study
suggested that the three types of TSHR fusion proteins that were
prepared could induce immune dysregulation in BALB/c mice, leading
to the development of AITD.
The present study offers insights into the
significance of different structural domains of TSHR in the
pathogenesis of AITD. Additionally, the TRAb assay kit may also be
used to monitor changes in the condition of patients, providing
evidence for the early diagnosis and individualized treatment of
patients. The primary concern now is that the ELISA may face issues
when used in clinical testing, such as false positives, and low
sensitivity and specificity, which will affect the diagnosis and
treatment of patients. Therefore, multi-center and large-sample
research and testing are required, which should include a larger
group of patients. A more stable and accurate ELISA detection
system also needs to be created to improve reliability and clinical
applicability. In the present study, three fusion proteins were
prepared according to the characteristics of the TSHR domain, and
the regions that TSHR may bind with TSAb and TBAb were initially
identified. In the subsequent experiment, the extracellular region
of TSHR was segmented into smaller structural units according to
the structural characteristics of the internal functional
characteristics of a single domain. Further refining and clarifying
the binding sites of TSAb, TBAb and TSHR is also an effective way
of reducing false positives and increasing sensitivity and
specificity. In addition, combining the fusion protein with TRAb
detection methods such as the current third-generation TBII method
and biological assays is also one of the methods for more accurate
diagnosis and treatment of patients, which needs further
research.
Therefore, a comprehensive understanding of the
biological effects of the TSHR fusion protein and in-depth
exploration of the specific mechanism of TSHR fusion protein
inducing the immune response can help increase the understanding of
the pathogenesis of AITD, provides a scientific basis for research
of TSHR and the application of fusion proteins, and provides a
basis for the accurate diagnosis, and individualized diagnosis and
treatment of patients with AITD.
In view of the important role of TSHR in AITDs, the
present study expressed fusion proteins of different domains of
TSHR, and the fusion proteins of different domains of TSHR
exhibited different immunogenicity. The TSHR A subunit fusion
protein and TSHR carboxyl-terminal fusion protein prepared in the
present study could serve as a basis for the development of ELISA
kits for the detection of TSAb and TBAb. Fusion proteins of
different structural domains of TSHR induced clinical symptoms of
hyperthyroidism and hypothyroidism in mice. This research provides
a scientific basis for future studies on the etiology and
mechanisms of AITD, as well as the invention of novel methods for
the detection of TRAb.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Science and Technology
Plan Project of Gansu Province (Basic Research Plan; grant no.
25JRRA597) and the Lanzhou University Second Hospital ‘Cuiying
Technological Innovation’ Program (grant no. CY2018-ZD02).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
XC contributed to conceptualization,
methodology,-visualization, writing of the original draft and
editing. HC contributed to conceptualization, acquired funding,
provided resources, supervised the study and revised the
manuscript. Both authors have read and approved the final version
of the manuscript. XC and HC confirm the authenticity of all the
raw data.
Ethics approval and consent to
participate
The animal experiments received approval from the
Animal Welfare and Ethics Committee of The Second Hospital of
Lanzhou University (approval no. D2023-156; Lanzhou, China). The
research protocol was reviewed and approved by the Medical Ethics
Committee of The Second Hospital of Lanzhou University (approval
no. 2023A-802; Lanzhou, China). All participants provided written
informed consent prior to participating in the present study. All
experiments have been approved by the appropriate ethics committee
and have therefore been performed in accordance with the ethical
standards laid down in the 1964 Declaration of Helsinki and its
later amendments.
Patient consent for publication
Informed consent from patients was obtained for
publication of the present study, and relevant written consent has
been provided.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
McLachlan SM and Rapoport B: Thyroid
autoantibodies display both ‘Original antigenic sin’ and epitope
spreading. Front Immunol. 8:18452017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Furmaniak J, Sanders J, Núñez Miguel R and
Rees Smith B: Mechanisms of action of TSHR autoantibodies. Horm
Metab Res. 47:735–752. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Furmaniak J, Sanders J, Sanders P, Li Y
and Rees Smith B: TSH receptor specific monoclonal autoantibody
K1-70TM targeting of the TSH receptor in subjects with
Graves' disease and Graves' orbitopathy-Results from a phase I
clinical trial. Clin Endocrinol (Oxf). 96:878–887. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gupta AK and Kumar S: Utility of
antibodies in the diagnoses of thyroid diseases: A review article.
Cureus. 14:e312332022.PubMed/NCBI
|
|
5
|
Smith TJ and Hegedüs L: Graves' disease. N
Engl J Med. 375:1552–1565. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Latif R, Mezei M, Morshed SA, Ma R,
Ehrlich R and Davies TF: A Modifying autoantigen in Graves'
disease. Endocrinology. 160:1008–1020. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Núñez Miguel R, Sanders P, Allen L, Evans
M, Holly M, Johnson W, Sullivan A, Sanders J, Furmaniak J and Rees
Smith B: Structure of full-length TSH receptor in complex with
antibody K1-70™. J Mol Endocrinol. 70:e2201202022.PubMed/NCBI
|
|
8
|
George A, Diana T, Längericht J and Kahaly
GJ: Stimulatory thyrotropin receptor antibodies are a biomarker for
graves' orbitopathy. Front Endocrinol (Lausanne). 11:6299252021.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lytton SD, Schluter A and Banga PJ:
Functional diagnostics for thyrotropin hormone receptor
autoantibodies: Bioassays prevail over binding assays. Front Biosci
(Landmark Ed). 23:2028–2043. 2018. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bahn RS: Current Insights into the
Pathogenesis of Graves' Ophthalmopathy. Horm Metab Res. 47:773–778.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Faßbender J, Holthoff HP, Li Z and Ungerer
M: Therapeutic effects of short cyclic and combined epitope
peptides in a Long-term model of Graves' disease and orbitopathy.
Thyroid. 29:258–267. 2019. View Article : Google Scholar
|
|
12
|
Rapoport B, Aliesky HA, Chen CR and
McLachlan SM: Evidence that TSH receptor A-subunit multimers, not
monomers, drive antibody affinity maturation in Graves' disease. J
Clin Endocrinol Metab. 100:E871–E875. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li CW, Osman R, Menconi F, Concepcion E
and Tomer Y: Cepharanthine blocks TSH receptor peptide presentation
by HLA-DR3: Therapeutic implications to Graves' disease. J
Autoimmun. 108:1024022020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Holthoff HP, Goebel S, Li Z, Faßbender J,
Reimann A, Zeibig S, Lohse MJ, Münch G and Ungerer M: Prolonged TSH
receptor A subunit immunization of female mice leads to a long-term
model of Graves' disease, tachycardia, and cardiac hypertrophy.
Endocrinology. 156:1577–1589. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rapoport B, Chazenbalk GD, Jaume JC and
McLachlan SM: The thyrotropin (TSH) receptor: Interaction with TSH
and autoantibodies. Endocr Rev. 19:673–716. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Davies TF, Ando T, Lin RY, Tomer Y and
Latif R: Thyrotropin receptor-associated diseases: From adenomata
to Graves disease. J Clin Invest. 115:1972–1983. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Latif R, Teixeira A, Michalek K, Ali MR,
Schlesinger M, Baliram R, Morshed SA and Davies TF: Antibody
protection reveals extended epitopes on the human TSH receptor.
PLoS One. 7:e446692012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Marcinkowski P, Kreuchwig A, Mendieta S,
Hoyer I, Witte F, Furkert J, Rutz C, Lentz D, Krause G and Schülein
R: Thyrotropin receptor: Allosteric modulators illuminate
intramolecular signaling mechanisms at the interface of Ecto- and
transmembrane domain. Mol Pharmacol. 96:452–462. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Thyroid disease, . Assessment and
management. London: National Institute for Health and Care
Excellence (NICE); November 20–2019
|
|
20
|
Holthoff HP, Uhland K, Kovacs GL, Reimann
A, Adler K, Wenhart C and Ungerer M: Thyroid-stimulating hormone
receptor (TSHR) fusion proteins in Graves' disease. J Endocrinol.
246:135–147. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang Y, Xia Q, Zhou L, Zhang Y, Guan Z,
Zhang J, Li Z, Liu K, Li B, Shao D, et al: B602L-Fc fusion protein
enhances the immunogenicity of the B602L protein of the African
swine fever virus. Front Immunol. 14:11862992023. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Percie du Sert N, Hurst V, Ahluwalia A,
Alam S, Avey MT, Baker M, Browne WJ, Clark A, Cuthill IC, Dirnagl
U, et al: The ARRIVE guidelines 2.0: Updated guidelines for
reporting animal research. PLoS Biol. 18:e30004102020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zheng H, Xu J, Chu Y, Jiang W, Yao W, Mo
S, Song X and Zhou J: A global regulatory network for dysregulated
gene expression and abnormal metabolic signaling in immune cells in
the microenvironment of Graves' disease and Hashimoto's
thyroiditis. Front Immunol. 13:8798242022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kristensen B: Regulatory B and T cell
responses in patients with autoimmune thyroid disease and healthy
controls. Dan Med J. 63:B51772016.PubMed/NCBI
|
|
25
|
Hsu WT, Suen JL and Chiang BL: The role of
CD4CD25 T cells in autoantibody production in murine lupus. Clin
Exp Immunol. 145:513–519. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Arndt B, Witkowski L, Ellwart J and
Seissler J: CD8+ CD122+ PD-1-effector cells promote the development
of diabetes in NOD mice. J Leukoc Biol. 97:111–120. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Benvenga S, Elia G, Ragusa F, Paparo SR,
Sturniolo MM, Ferrari SM, Antonelli A and Fallahi P: Endocrine
disruptors and thyroid autoimmunity. Best Pract Res Clin Endocrinol
Metab. 34:1013772020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Taylor PN, Albrecht D, Scholz A,
Gutierrez-Buey G, Lazarus JH, Dayan CM and Okosieme OE: Global
epidemiology of hyperthyroidism and hypothyroidism. Nat Rev
Endocrinol. 14:301–316. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Duan J, Xu P, Cheng X, Mao C, Croll T, He
X, Shi J, Luan X, Yin W, You E, et al: Structures of full-length
glycoprotein hormone receptor signalling complexes. Nature.
598:688–692. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jiang X, Liu H, Chen X, Chen PH, Fischer
D, Sriraman V, Yu HN, Arkinstall S and He X: Structure of
follicle-stimulating hormone in complex with the entire ectodomain
of its receptor. Proc Natl Acad Sci USA. 109:12491–12496. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kleinau G, Mueller S, Jaeschke H, Grzesik
P, Neumann S, Diehl A, Paschke R and Krause G: Defining structural
and functional dimensions of the extracellular thyrotropin receptor
region. J Biol Chem. 286:22622–22631. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rapoport B and McLachlan SM: TSH receptor
cleavage into subunits and shedding of the A-subunit; a molecular
and clinical perspective. Endocr Rev. 37:114–134. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Couët J, de Bernard S, Loosfelt H, Saunier
B, Milgrom E and Misrahi M: Cell surface protein
disulfide-isomerase is involved in the shedding of human
thyrotropin receptor ectodomain. Biochemistry. 35:14800–14805.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Duan J, Xu P, Luan X, Ji Y, He X, Song N,
Yuan Q, Jin Y, Cheng X, Jiang H, et al: Hormone- and
antibody-mediated activation of the thyrotropin receptor. Nature.
609:854–859. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Faust B, Billesbølle CB, Suomivuori CM,
Singh I, Zhang K, Hoppe N, Pinto AFM, Diedrich JK, Muftuoglu Y,
Szkudlinski MW, et al: Autoantibody mimicry of hormone action at
the thyrotropin receptor. Nature. 609:846–853. 2022.PubMed/NCBI
|
|
36
|
Morshed SA and Davies TF: Graves' disease
mechanisms: The role of stimulating, blocking, and cleavage region
TSH receptor antibodies. Horm Metab Res. 47:727–734. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Akamizu T, Kosugi S, Kohn LD and Mori T:
Anti-thyrotropin (TSH) receptor antibody binding epitopes of TSH
receptor: Site-directed mutagenesis approach. Nihon Rinsho.
52:1024–1030. 1994.(In Japanese). PubMed/NCBI
|
|
38
|
Zulkarnain Z, Ulhaq ZS, Sujuti H,
Soeatmadji DW, Zufry H, Wuragil DK, Marhendra APW, Riawan W,
Kurniawati S, Oktanella Y and Aulanni'am A: Comparative performance
of ELISA and dot blot assay for TSH-receptor antibody detection in
Graves' disease. J Clin Lab Anal. 36:e242882022. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
McLachlan SM, Aliesky HA, Garcia P,
Banuelos B and Rapoport B: Thyroid hemiagenesis in a thyroiditis
prone mouse strain. Eur Thyroid J. 7:187–192. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Rapoport B, Aliesky HA, Banuelos B, Chen
CR and McLachlan SM: A unique mouse strain that develops
spontaneous, Iodine-accelerated, pathogenic antibodies to the human
thyrotrophin receptor. J Immunol. 194:4154–4161. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Schlüter A, Horstmann M, Diaz-Cano S,
Plöhn S, Stähr K, Mattheis S, Oeverhaus M, Lang S, Flögel U,
Berchner-Pfannschmidt U, et al: Genetic immunization with mouse
thyrotrophin hormone receptor plasmid breaks Self-tolerance for a
murine model of autoimmune thyroid disease and Graves' orbitopathy.
Clin Exp Immunol. 191:255–267. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Porowińska D, Wujak M, Roszek K and
Komoszyński M: Prokaryotic expression systems. Postepy Hig Med Dosw
(Online). 67:119–129. 2013.(In Polish). View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Costagliola S, Alcalde L, Tonacchera M,
Ruf J, Vassart G and Ludgate M: Induction of thyrotropin receptor
(TSH-R) autoantibodies and thyroiditis in mice immunised with the
recombinant TSH-R. Biochem Biophys Res Commun. 199:1027–1034. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Janyga S, Marek B, Kajdaniuk D,
Ogrodowczyk-Bobik M, Urbanek A and Bułdak Ł: CD4+ cells in
autoimmune thyroid disease. Endokrynol Pol. 72:572–583. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yin L, Zeng C, Yao J and Shen J: Emerging
roles for noncoding RNAs in autoimmune thyroid disease.
Endocrinology. 161:bqaa0532020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sakaguchi S, Mikami N, Wing JB, Tanaka A,
Ichiyama K and Ohkura N: Regulatory T cells and human disease. Annu
Rev Immunol. 38:541–566. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Grindebacke H, Wing K, Andersson AC,
Suri-Payer E, Rak S and Rudin A: Defective suppression of Th2
cytokines by CD4CD25 regulatory T cells in birch allergics during
birch pollen season. Clin Exp Allergy. 34:1364–1372. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Rosenberger S, Undeutsch R, Akbarzadeh R,
Ohmes J, Enghard P, Riemekasten G and Humrich JY: Regulatory T
cells inhibit autoantigen-specific CD4+ T cell responses in
lupus-prone NZB/W F1 mice. Front Immunol. 14:12541762023.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yan JJ, Lee JG, Jang JY, Koo TY, Ahn C and
Yang J: IL-2/anti-IL-2 complexes ameliorate lupus nephritis by
expansion of CD4+CD25+Foxp3+ regulatory T cells. Kidney Int.
91:603–615. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Malek TR and Castro I: Interleukin-2
receptor signaling: At the interface between tolerance and
immunity. Immunity. 33:153–165. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Rifa'i M, Kawamoto Y, Nakashima I and
Suzuki H: Essential roles of CD8+CD122+ regulatory T cells in the
maintenance of T cell homeostasis. J Exp Med. 200:1123–1134. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yuan X, Dong Y, Tsurushita N, Tso JY and
Fu W: CD122 blockade restores immunological tolerance in autoimmune
type 1 diabetes via multiple mechanisms. JCI Insight. 3:e966002018.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Liu J, Chen D, Nie GD and Dai Z:
CD8(+)CD122(+) T-cells: A newly emerging regulator with central
memory cell phenotypes. Front Immunol. 6:4942015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Mishra S, Srinivasan S, Ma C and Zhang N:
CD8+ regulatory T cell-a mystery to be revealed. Front Immunol.
12:7088742021. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kim HJ, Wang X, Radfar S, Sproule TJ,
Roopenian DC and Cantor H: CD8+ T regulatory cells express the Ly49
Class I MHC receptor and are defective in autoimmune prone B6-Yaa
mice. Proc Natl Acad Sci USA. 108:2010–2015. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Kim HJ, Verbinnen B, Tang X, Lu L and
Cantor H: Inhibition of follicular T-helper cells by CD8(+)
regulatory T cells is essential for self tolerance. Nature.
467:328–332. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Saitoh O, Abiru N, Nakahara M and Nagayama
Y: CD8+CD122+ T cells, a newly identified regulatory T subset,
negatively regulate Graves' hyperthyroidism in a murine model.
Endocrinology. 148:6040–6046. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
McLachlan SM, Nagayama Y, Pichurin PN,
Mizutori Y, Chen CR, Misharin A, Aliesky HA and Rapoport B: The
link between Graves' disease and Hashimoto's thyroiditis: A role
for regulatory T cells. Endocrinology. 148:5724–5733. 2007.
View Article : Google Scholar : PubMed/NCBI
|