Introduction
The majority of the human genome consists of regions
that do not code for proteins and within these regions are the
non-coding RNAs (ncRNAs), which are broadly classified as small
ncRNAs (sncRNAs) and long ncRNAs (lncRNAs). Since their discovery,
several studies have reported roles of ncRNAs as master regulators
of gene expression by acting through innumerable mechanisms
(1–4). Previous studies have shown that
microRNAs (miRNAs; miRs), RNYs-a class of ncRNAs that are
components of the Ro60 ribonucleoprotein particle-lncRNAs and
fragments derived from certain ncRNAs physically interact with
Toll-like receptors (TLRs), which results in oncogenesis (5), the induction and suppression of
inflammatory processes in a neurodegenerative context (6), the induction of nephritis and lung
disease (7), the elimination of
bacterial infections (8) and the
reactivation of the human immunodeficiency virus (HIV) (9). In addition to this, Rodríguez-Corona
et al (10) predicted the
physical interaction of RNYs and their fragments with TLRs and
other cellular receptors, which indicate a range of signaling
pathways and cellular processes that could be regulated by
ncRNAs.
TLRs recognize unique microbial patterns known as
pathogen-associated molecular patterns (PAMPs) (11–15)
and damage-associated molecular patterns (DAMPs) that lead to the
activation of the innate immune system. PAMPs have been associated
with molecules derived from pathogenic bacteria and viruses
(proteins and lipopeptides, LPS, RNA species, DNA and flagellin)
that activate pattern recognition receptor (PRR) families; however,
PRRs also respond to molecules from commensal organisms (16). Meanwhile, DAMPs are molecules
associated with tissue damage (heat shock proteins, transcription
factors, native RNA and DNA, tissue matrix components, such as
hyaluronan and fibronectin and humoral proteins such as
fibrinogen), which activate PRRs, particularly TLRs (17). To date, 10 TLRs have been
identified in humans (TLR1-10) (18) and 12 in mice (TLR1-9, TLR11-13)
(19), which are expressed on the
cell surface or in intracellular vesicles. Due to the importance of
TLR activation in the induction of the innate and adaptive immune
response, several studies have focused on the development of
activators and inhibitors of these receptors for the treatment of
various diseases (20–27). Currently, biomolecules are used for
the treatment of bladder cancer (28,29)
and several other biomolecules are still under study to establish
their use in clinical practice (20–22).
Previous studies reviewed information on the activation of TLRs by
miRNAs (30) and by sncRNAs,
proposing the sequential activation hypothesis in autoimmune
diseases (22,31). The purpose of the present review
was to describe the protective or harmful effects that activation
of TLRs by ncRNAs has, including the activation of these receptors
by lncRNAs and by fragments derived from ncRNAs. The present review
described the involvement of TLR-ncRNAs/fragments interactions in
other diseases such as cancer and viral infections, as well as in
neurodegenerative and neurocognitive diseases, and discussed
advances in the generation of TLR agonists and/or antagonists or
ncRNA mimics or inhibitors for the treatment of these diseases.
Advances in the study of ncRNAs/fragments-TLRs interaction could
lead to the development of clinically useful immunotherapies in the
future.
TLRs activation by ncRNAs
Currently, there is relatively little information
about the physical interaction of ncRNAs with cellular receptors.
Well-studied interactions include those between miRNAs and TLRs 7/8
and ribosomal RNAs (rRNAs) and RNYs with TLRs 13/2 and 3/7/8,
respectively (5,7,32–37)
and recent evidence indicates TLRs interaction with lncRNAs
(38,39). In addition, Yang et al
(40) demonstrated the miRNA-1
interaction with the pore-facing G-loop of Kir2.1 via the core
sequence AAGAAC, which does not include the seed region of this
miRNA. To the best of our knowledge, there is no evidence for the
interaction of other ncRNA with TLRs or other cellular receptors.
However, based on the existing evidence, it is possible that other
ncRNAs can interact with a variety of cellular receptors (10).
miRNAs
miRNAs are sncRNAs with length of ~22 nucleotides
(nt) in mature form (41–43). A study by Hansen et al
(44) demonstrated the presence of
miRNAs, with a size of 80–100 nt, known as Agotrons, which are
bound to and stabilized by Argonaute (Ago) proteins in the
cytoplasm. Although Agotrons also repress gene expression by
interacting with the 3′UTR region of their target mRNA, their main
function could be related to giving specificity to free Ago
proteins by certain RNA species.
miRNAs are produced by diverse biogenic pathways,
which seems to indicate that cells ‘secure’ the control of gene
expression by miRNAs using diverse molecular elements for their
synthesis and processing (45–47).
The best characterized function of miRNAs is their binding to the
3′UTR region of their target mRNAs via the seed region (48); however, there are organisms that do
not express Ago proteins (49),
suggesting different modes of action for these RNAs. Indeed, miRNAs
interact with specific promoters (50,51),
regulate gene expression post-transcriptionally at the nuclear
level (52), and act as ligands
for Kir2.1 (40) and for TLRs 7
and 8.
miRNA-mediated activation of TLRs in
cancer
TLRs recognize both PAMPs and DAMPs, where DAMPs are
secreted by cells in response to tissue damage or cell death and
their excessive release is associated with autoimmune diseases and
cancer (48,49). In relation to this, an increase in
the production of DAMPs (also called ‘alarmins’) has been observed
in different types of cancer as (e.g., pancreatic cancer, breast
cancer, lung cancer, among others) a response to cell death and
chronic inflammation (53,54).
The involvement of TLRs in cancer is a complex issue
as it involves several factors such as: i) Expression of specific
TLRs (55,56); ii) expression of TLRs in specific
cell strains (57); iii)
mutagenesis (58); and iv) TLR
adaptor proteins (59), among
others. In spite of the aforementioned issues, certain ligands of
TLRs 2,4,7 have been used for the treatment of various types of
cancer (28,29,60)
and several ligands have antitumor effects (61–63).
The efficiency of TLR ligands as immunotherapeutic agents lies
predominately in the initiation of T-cell immunity: Antigen uptake,
processing and presentation, dendritic cell maturation and T-cell
activation (64).
Activation of TLRs by miRNAs and their relation to
cancer was first demonstrated by Fabbri et al (5), who demonstrated that activation of
TLRs 7/8 by miRs-21 and −29a induced a prometastatic inflammatory
response and promoted tumor growth and metastasis in a
NF-κB-dependent manner. Similar to the observation of Heil et
al (65), Fabbri et al
(5) identified that GU-rich motifs
were required for the activation of TLRs by miRNAs. Notably,
survival of TLR7−/− mice was markedly longer compared
with that of wild-type mice injected with Lewis lung carcinoma
cells. In addition, depletion of tumor exosomes markedly attenuated
the induction of tumor necrosis factor-α (TNF-α), interleukin
(IL)-6 and early activation antigen CD69. This demonstrated the
impact of activation of TLRs 7/8 by specific miRNAs and suggested
the use of miRNA inhibitors to attenuate the effects in lung
cancer; however, further studies are needed to translate these
experimental results into clinical use for humans.
Studies subsequent to that of Fabbri et al
(5) reported a relationship
between TLRs-miRNAs, but superficially (32–34).
For instance, He et al (32) demonstrated that TLR7 activation, in
a c-Jun N-terminal kinase-dependent manner, by miR-21 induced
myoblasts apoptosis in cancer cachexia. More recently, a feedback
loop was demonstrated between prostaglandin 2 (PGE2) and the
activation of TLRs 7/8 by circulating miR-574-5p (33). These authors suggested that
decreased intracellular expression levels of both miR-574-5p and
PGE2 were associated with the activation-dependent antitumor
effects of TLRs 7 and 8; however, this needs to be validated
experimentally. Finally, miR-16-5p was overexpressed in gastric
cancer and its function as a ligand of TLRs was predicted using
bioinformatics, but its biological impact is unknown. In addition,
it was also postulated that regulatory networks,
lncRNAs-miRNAs-mRNAs, were modified in gastric cancer (34).
Currently, understanding of TLR activation by miRNAs
and their relationship with cancer is limited. Some differentially
expressed miRNAs that function as ligands of TLRs have been
identified in cancer (5,32–34),
which could function as circulating biomarkers for the detection in
various types of cancer. The activation of TLRs by various ligands
is known to have both pro- (66–68)
and anti-tumor effects (69–71)
and this depends on several factors, which should be further
studied in depth in each type of cancer. The identification and
establishment of all the molecular events underlying the activation
of TLRs by miRNAs will likely result in the establishment of
activators and/or inhibitors useful in clinical practice.
miRNAs and TLRs in myocardial
ischemia
Free RNA concentration increases shortly after a
transient myocardial ischemia event and the presence of RNases
attenuates necrotic cell-induced cytokine production in
cardiomyocytes and immune cells and reduces myocardial infarction
after transient ischemia (72,73).
Feng et al (12) reported
that certain circulating miRNAs facilitated the production of
specific cytokines, an effect that was markedly attenuated by: i)
Mutating the uridines of miRs-133a, −146a and −208a by adenosines;
ii) treatment with RNases; iii) eliminating the expression of TLR7
and that of the signal transduction adapter innate immune system
(myeloid differentiation primary response 8, MyD88); or iv) using
TLR7 antagonists. Similarly, the exposure of murine cardiomyocytes
to extracellular vesicles (EVs) containing miR-146a-5p induced the
production of macrophage inflammatory protein 2 (MIP-2), IL-6 and
TNF-α in a TLR7-dependent manner. These effects were observed in
vivo, and cytokine production and activation of the innate
immune response, which was attenuated in mice lacking TLR7
(74).
Although there are still numerous studies that
should be carried out, it is reasonable to consider that the
increased expression of circulating RNA, particularly miRNAs, could
be useful for the diagnosis of a myocardial ischemia event. It
would be potentially beneficial to investigate whether the
expression of other ncRNAs is also increased during these ischemic
events and whether they function as TLR ligands. The search for and
establishment of inhibitors of these miRNAs or TLR7 could be
clinically useful for the treatment of this disease.
Involvement of miRNAs and TLRs in
sepsis
Sepsis is a serious clinical condition that leads to
organ dysfunction in the body caused by an uncontrolled host
response to infection characterized by systemic inflammation, which
is partially mediated by TLRs and certain miRNAs (75,76).
Xu et al (75) demonstrated
that specific circulating miRNAs that are secreted into EVs have a
proinflammatory effect and stimulated the production of IL-6,
TNF-α, interleukin-β and MIP-2. These effects were attenuated by
miR-34a, miR-122 and miR-146a inhibitors, as well as in cells that
did not express TLR7 or MyD88. Notably, injection of EVs from
septic mice into wild-type mice promoted peritoneal neutrophil
migration and this decreased in mice that did not express MyD88.
Similarly, intrathecal administration of exogenous miR-146a
triggered pulmonary inflammation, activated endothelium and
increased endothelial permeability in a TLR7-dependent manner,
indicating the involvement of miR146a-TLR7 in sepsis-associated
acute respiratory distress syndrome (76).
Based on the aforementioned studies, several
therapeutic strategies could be developed for the treatment of this
clinical condition, such as the establishment of miRNA inhibitors,
TLR7 antagonists or MyD88 inhibitors. In particular, some of these
therapeutic strategies may be effective for the treatment of
sepsis-associated acute respiratory distress syndrome.
miRNAs and TLRS in neonatal
morbidity
Several lines of evidence indicate that certain
miRNAs (miR-21a, miR-29a, miR-146a-3p and Let-7b) function as
endogenous danger signals by activating TLRs 7/8, which are viral
single strand RNA (ssRNA) sensors. Exposure to bacterial
lipopolysaccharides (LPS) from human fetal membrane (FM) explants
and wild-type mouse FM increased the expression of miR-146a-3p
(77). In women with preterm birth
and chorioamnionitis, increased expression of this miRNA was also
observed and TLR8 activation by miR-146a-3p induced the IL-8 and
IL-1β production in an LPS-dependent manner (77).
Involvement of miRNAs/TLRs 7/8 in
coronavirus disease 2019 (COVID-19)
COVID-19 is an infectious disease caused by severe
acute respiratory syndrome coronavirus 2 (SARS-CoV-2), producing
symptoms such as fever, cough, dyspnea, myalgia and fatigue, among
others (78). In severe cases, it
is characterized by pneumonia, acute respiratory distress syndrome,
sepsis and circulatory shock (79,80).
In order to improve the understanding of the etiology of the
inflammatory processes triggered by SARS-COV-2, Wallach et
al (81) investigated whether
RNA fragments of SARS-COV-2 could activate TLRs. In fact, several
SARS-COV-2 RNA fragments, the majority of which were 22 nt in
length, activated TLRs 7/8 and induced the release of cytokines
from macrophages and microglia to induce the human immune response
against the virus. Regarding this, Liao et al (82) observed that the SARS-CoV-2 spike
protein activated platelets and induced the expression and
secretion of miR-21 and let-7b in EVs to activate TLRs 7 and 8. The
activation of these receptors induced p47phox phosphorylation and
the NADPH oxidase activation in neutrophils, resulting in reactive
oxygen species (ROS) production and the formation of hyperactive
neutrophil extracellular traps, which was related to disease
severity. Therefore, increased circulating expression of miRs-21
and -let-7b could be used as predisposing factors and the
TLR7/8-miRNA axis as a therapeutic target for severe COVID-19.
TLR activation by miRNAs in the
brain
Several studies indicate the aberrant release of
miRNAs in neurodegenerative diseases, such as Alzheimer's (83–85)
and Parkinson's disease (86–88);
however, the physiological significance of this is currently
unknown. To investigate the possible relationship of miRNAs and the
establishment and/or progression of these diseases, Wallach et
al (6) demonstrated that
miRNAs secreted by cortical neurons induced the secretion of
cytokines and chemokines by microglia and macrophages, which had a
neurotoxic effect mediated by TLRs 7 and 8 (Fig. 1A and B). miRs 100–5p and 298–5p
were internalized within microglia and activated endosomal TLR8 in
a miRNA sequence-dependent manner, but not on its secondary
structure (Fig. 1C). In a similar
manner, these miRNAs also activated neuronal TLR7 and decreased
cell viability in a dose-dependent manner (Fig. 1D). The presence of these miRNAs in
the cerebrospinal fluid induced neurodegeneration and accumulation
of microglia in the mouse cerebral cortex through TLR7 activation
(Fig. 1E) (89). Similarly, in a murine model of
sepsis, it was observed that the activation of TLR7/MyD88 by
specific miRNAs activated the inflammatory processes in the brain
and induced neuronal apoptosis (90).
In addition, TLR7 and miR-let7b co-localize in
hippocampal CA1 neurons, suggesting the TLR7 activation by this
miRNA. Markedly reduced levels of both TLR7 and miR-let7b increased
hippocampal-dependent memory and attenuated inflammatory cytokine
production (91). These results
demonstrated for the first time that specific miRNAs secretion, in
a particular physiological context, can act on specific TLRs in the
brain. It has been postulated that these miRNAs could be important
in the establishment and/or development of neurodegenerative
diseases, as well as in neurocognitive disorders; however, further
studies are needed to ascertain this. An improved understanding of
the mechanisms underlying the activation of TLRs by miRNAs could
help to establish treatments to ameliorate neurodegenerative
diseases or neurocognitive disorders
Sequence characteristics of miRNAs in
TLR activation
Early work that demonstrated the ssRNA detection by
TLRs was carried out by Heil et al (65). The authors reported that
HIV-derived ssRNA, rich in guanosine (G) and uridine (U), induced
the secretion of interferon-α and proinflammatory and regulatory
cytokines in response to murine TLR7 activation and human TLR8.
Similarly, miRs-21 (GUUG) and −29a (GGUU) have a GU motif in the
nucleotide region 18–21, where base number 20 (U) was very
important for the activation of human TLR8 (1). Meanwhile, Wu et al (92) identified the ‘UGUUAU’ motif and
certain uridines and cytosines of miR-20a-5p as determinants for
TLR7 activation and cytokine secretion. The shortest sequence to
induce TLR7 activation was 10 nt in length (ssRNA), including the
‘UGUUAU’ motif. The activation of TLR7 promoted the secretion of
multiple proinflammatory molecules, which was dependent on PI3K,
MAPK and NF-κB1. The administration of miR-20a-5p in wild-type mice
increased leukocyte migration and this was attenuated in both TLR7
knockout mice and in mice administered with miR-20a-5p but with the
‘UGUUAU’ motif mutated.
TLR activation by miRNAs and other types of ncRNAs
is a focus of ongoing research and existing evidence indicates that
the recognition of miRNAs by TLRs is primarily dictated by GU-rich
motifs, where the sequence and position of nucleotides within the
mature miRNA sequence is a determinant for TLR activation (5).
TLR8 is activated by the binding of ssRNA
degradation products at two different sites (93). At the first binding site, uridine
mononucleoside (Kd, 55 µM) binds to TLR8 by stacking interactions
between TLR8 and the aromatic moiety of ligands and hydrogen bonds
(94). The second site of TLR8 is
recognized by the UG dinucleotide, which increases the affinity of
uridine for TLR8 by 50-fold. TLR8 and TLR7 receptors have
functional and sequence similarities, so some structural features
of TLR8 can be applied to those of TLR7. Regarding this, TLR7 has
an activation mode similar to that of TLR8, in which guanosine and
its derivatives, instead of uridine, bind to TLR7 and this binding
is also modulated by oligonucleotide binding at a second site
(95,96).
Similar to TLRs 7/8, TLR13 is activated by ssRNA 13,
derived from the 23S ribosomal subunit, by binding at a site formed
by the interaction of the N- and C-terminal regions of this
receptor (97). The binding of
ssRNA13 to TLR13 depends on the formation of a stem-loop structure
(G2057 and C2064) and the establishment of hydrogen bonds between
specific nucleotides (A2058, A2060 and G2061) and the TLR13 leucine
rich regions LRR2-LRR25. Notably, the interaction of ssRNA13 with
TLR13 was pH-dependent, with a preference for acid pH.
miRNAs retain the viral consensus motifs for TLR
interaction and activation, but this does not rule out other
sequences and mechanisms involved in TLR activation by ssRNA.
Ligand binding to a first site of TLRs 7 and 8 increases the
affinity of another ligand at a second site, which may occur by
allosteric events. More evidence is needed to know whether
activation of TLRs by miRNAs also occurs in this way.
Interactions of RNYs with TLRs
RNYs are highly conserved sncRNAs in vertebrates
(98) and have also been detected
in prokaryotes (99) and viruses
(100). In humans, four RNYs have
been identified: RNY1, RNY3, RNY4 and RNY5, and ~1,000 pseudogenes
(101). The physiological
importance of YRNAs was partially identified by studying their
interaction with proteins and by bioinformatics approaches
(6). Functions regulated by RNYs
include control of the immune system (102,103), DNA replication (99), development (98), cell proliferation (104), enhanced translation efficiency
and virus assembly and retrotransposition control (105).
Circulating ribonucleoprotein complexes associated
with RNYs predominantly induce immune activation and several of
these effects depend on signaling pathways mediated by TLRs. A
previous study reported that each of the TLRs do not respond in the
same way to the RNY activation and have diverse physiological
effects (Table I) (7). For instance, TLR7 activation by RNY1
induced nephritis, but that of TLR3 by RNY3 did not. It has also
been shown that the activation of a particular TLR by a specific
RNY can induce inflammation in a given tissue and repress it in
another. Regarding this, TLR3 or TLR7 activation by U1 RNA induced
lung disease, but that of TLR7 by RNY1 did not have the same effect
(7). In addition, it has been
shown that RNYs that are not bound to Ro60 or La proteins have a
greater effect on TLR activation, because these proteins recognize
the hairpin structure and the 5′-phosphate group of RNYs, which are
important sites for their interaction with TLRs (35).
 | Table I.Activation of specific TLRs by ncRNAs
and their fragments. |
Table I.
Activation of specific TLRs by ncRNAs
and their fragments.
| A, ncRNAs:
miRNAs |
|---|
|
|---|
| First author/s,
year | ncRNA type | Activated TLR | Experimental
model | Biological
function | (Refs.) |
|---|
| Fabbri et
al, 2012 | miR-21 and
miR-29a | 7,8 | Cancer | Induction of
prometastatic inflammatory response: | (5) |
|
|
|
|
| Promotion of tumor
growth and metastasis. |
|
| Feng et al,
2017 | miR-133a, miR-146a
and miR-208a | 7 | Myocardial
ischemia | Production of the
cytokines MIP-2, TNFα and IL-6. Migration of peritoneal neutrophils
and monocytes. | (12) |
| Borchert et
al, 2007 | miR-146a-5p | 7 | Myocardial
ischemia | Production of
MIP-2, IL-6 and TNFα and activation of the innate immune
response. | (46) |
| Chung et al,
2011 | miR-34a, miR-122
and miR-146a | 7 | Sepsis | Proinflammatory and
production of MIP-2, IL-6, IL-β and TNFα: Peritoneal neutrophil
migration. | (47) |
| Wallach et
al, 2020 | miR-340-3p and
miR-132-5p | 7,8 | Neuronal
injury | Release of
cytokines and chemokines from microglia: Neurodegenerative
effect. | (6) |
| Mardente et
al, 2012 | miR-100-5p and
miR-298-5p | 7,8 | Neurodegenerative
disease model | Secretion of
cytokines and chemokines from microglia and macrophages: Increased
phagocytosis. Activation of autonomic apoptosis of cortical
neurons. miRNAs in cerebrospinal fluid: Neuro-degeneration and
accumulation of microglia in mouse cerebral cortex. | (54) |
| Hao et al,
2018 | miR-20a-5p and
miR-148b-3p | 7 | Central nervous
system pathology | Secretion of
proinflammatory molecules dependent on PI3K, MAPKs and NF-κβ.
Migration of leukocytes. | (56) |
|
| B, ncRNAs:
rRNAs |
|
| Oldenburg et
al, 2012; Huang et al, 2023; Chen et al; 2014;
Huang et al, 2022 | 23S rRNA | 13 | Bacterial
infections, action of lactic acid bacteria | Activation by
bacterial infections, regulates the beneficial actions of lactic
acid bacteria. | (8,34,72,76) |
| Shimada et
al, 2020 | 23S (Sa19) and 16S
mt-rRNA | 8 | Bacterial
infections | Activation of PBMCs
in a MyD88-dependent manner. | (74) |
| Chen et al,
2014 | 18S rRNA | 2 | Inflammation | Activation of
differentiated human monocytoid THP-1 cells. | (72) |
| Alvarez-Carbonell
et al, 2017 | Bacterial rRNA | 3 | HIV-associated
neurocognitive disorders | Synergic activation
of TLR2 stimulated transcriptional expression of inflammatory genes
and TNF-α release from macrophages. | (9) |
|
| C, ncRNAs:
RNYs |
|
| Greidinger et
al, | RNY3 | 3,7,8 | Autoimmunity | RNY3 was a strong
activator | (7) |
| 2007 | RNY1 | 7, 8 |
| strong activator of
TLRs 3,7, |
|
|
| RNY4 | 7 |
| and 8. |
|
|
| RNY5 | 7 |
| TLR3 activation by
RNY3 did |
|
|
|
|
|
| not induce
nephritis. |
|
|
|
|
|
| RNY4 was a strong
activator of TLR7. |
|
|
|
|
|
| TLR7 activation by
RNY5 induced nephritis. |
|
| Rodriguez-Corona
et al, 2023 | RNYs 1,3-5 | 5,7,8,10 | Pediatric
astrocytoma | The interaction of
RNYs 1,3-5 with TLRs 5,7,8,10 was predicted. | (10) |
|
|
|
|
| Alzheimer
disease-amyloid secretase, B cell activation, ubiquitin proteasome,
angiogenesis, apoptosis, endothelin and dopamine receptor
mediated-signaling pathways. |
|
|
| D, ncRNA
fragments: tRNAs |
|
| Deng et al,
2024 | 5′-tRNAHisGUG | 7 | Bacterial
infections | TLR7
activation | (91) |
| Wu et al,
2021 | 5′-tRNAVal
CAC/AAC | 7 | Bacterial
infections | Elimination of
Mycobacterium tuberculosis in a GUUU sequence-dependent
manner. | (92) |
|
| E, ncRNA
fragments: s-RNYs |
|
| Onafuwa- | RNY1-5p, | 7 |
Atherosclerosis | Induction of
apoptosis and | (100) |
| Nuga et al,
2005 | RNY3-5p and
RNY4-5p |
|
| NF-κB-mediated
inflammation |
|
| Rodriguez-Corona
et al, 2023 | s-RNYs loop
domains | 3,7,10 | Pediatric
astrocytoma | Bioinformatic
prediction. | (10) |
Based on the aforementioned studies, the TLR-RNY
interactions seem to depend on the type of TLR and RNY involved as
well as on the cellular tissue. In certain cases, this activation
was related to inflammatory processes, but the opposite effect was
observed in others. It is necessary to identify which other
molecular components and signaling pathways are determining these
biological differences.
In pediatric patients with astrocytoma,
Rodríguez-Corona et al (10) demonstrated differential circulating
expression of RNYs (Fig. 2A-C) and
the interaction of RNYs with cellular receptors was predicted.
Bioinformatics analysis [HIPPIE database
(cbdm-01.zdv.uni-mainz.de/~mschaefer/hippie/), by using the
following commands: NETWORK QUERY; direct interaction; GO
(biological process); show KEGG effect; and PANTHER database
(pantherdb.org), by using the following commands: ID List; Homo
sapiens; Functional classification viewed in gene list;
Pathway)] indicated the RNY interaction with receptors other than
TLRs to regulate a myriad of cellular pathways, such as apoptosis,
angiogenesis, cholecystokinin receptor and p53 signaling pathways,
as well as Parkinson's disease (Fig.
3). Identifying the target organs of RNYs and understanding the
biological processes that are being modified by acting on TLRs or
on other types of receptors may be important to understand.
rRNA interactions with TLRs
rRNA is the most abundant RNA species in organisms
and, similar to miRNAs and RNYs, rRNAs also interact with TLRs.
TLR13 is known to recognize a specific sequence of the bacterial
ribosomal 23S subunit, which induces the production of
infection-fighting cytokines (8,36,97,106); however, the 23S ribosomal subunit
derived from erythromycin-resistant Staphylococcus aureus
(S. aureus) strains or methylated oligoribonucleotides,
which mimics MLS antibiotic resistance, did not stimulate TRL13
(36). This observation
demonstrates a mechanism of antibiotic resistance and parallels
host cell ‘detection’ systems, whereby exposure of bacteria to
antibiotics establishes epigenetic marks that render them less or
totally inert to antibiotics and also evade detection by TLR13.
Similarly, fish TLR13a and b receptors were activated by bacterial
23S rRNA and induced the immune response by stimulating specific
effector proteins (Table II)
(106).
 | Table II.Differential activation of
intracellular pathways mediated by TLR13a and b. |
Table II.
Differential activation of
intracellular pathways mediated by TLR13a and b.
| Co-expression in
293T cells | Signaling pathway
activated |
|---|
| TLR13a/MyD88 | Increased
NF-κB-mediated pathway activation. |
|
| Low effect on AP1
pathway activation. |
|
| Significant
activation of the IFN-β-mediated pathway |
| TLR13b | Activation of the
AP1-mediated pathway. |
Li and Chen (36)
demonstrated that the Sa19 sequence (ACGGAAAGACCCC) of the 23S
subunit is a ligand of murine TLR13, which acts mainly against
infection by Gram-positive bacteria. The Sa19 sequence and
mitochondrial 16S rRNA (mt-rRNA) stimulate peripheral blood
mononuclear cells in a MyD88-dependent manner. Sa19 and S.
aureus and Escherichia coli (E. coli) derived
sequences and mitochondrial (mt)-RNA also activate human monocytoid
THP1 cells via TLR8. In addition to the 23S and 16S subunits,
Krüger et al (37) reported
that the 5S subunit from S. aureus and E. coli, as
well as mt-rRNA and P-/DAMPs stimulators, activate human TLR8. In
addition to the UGG motif, the UAA and UGA motifs are required for
TLR8 activation.
In addition to the 23S and 16S ribosomal subunits,
the 18S ribosomal subunit is involved in TLR activation. The
interaction of the 18S rRNA subunit with Pam2 (Pam2 CSK4), ligand
of TLRs 2 and 6, increased the affinity of Pam2 for TLR2 to act
synergistically. TLR2 activation stimulated the transcriptional
expression of inflammatory genes and TNF-α release from
macrophages, which was NF-κB-dependent (107). Usually, the acute cellular
responses to numerous signals are additive and, in certain cases,
the response to simultaneous stimuli is notably greater than the
sum of the responses to each stimulus separately; this is known as
synergism. This has biological significance since the cell will
only respond when two or more signals are present simultaneously.
Therefore, it will be necessary to identify whether all TLRs act
synergistically, and which ligand combinations and which mechanisms
govern the regulation of specific cellular processes.
Although TLRs are a primary detection system that
fight diverse infections, there is also evidence of their
participation in the beneficial action exerted by lactic acid
bacteria in organisms (e.g. anti-inflammatory effects and
activation of the intestinal immune system) (108–110). In mice, these bacteria have
immunomodulatory effects that are mediated, at least in part, by
IL-12 and TLR13. Since humans do not express this receptor, it
could be considered that the effects of this type of bacteria are
mediated by another TLR. Nishibayashi et al (111) demonstrated that the 23S and 16S
rRNA subunits of Enterococcus faecalis Ec-12 strain
activated TLR8 and induced IL-12 production.
Organisms have ‘engineered’ molecular elements that
allow them to fight infectious agents. The bacterial rRNA binding
site to the TLR is also involved in antibiotic resistance, which
ensures evasion of these two defense systems (36). Conversely, the infected cell
detects the infectious agent by means of TLRs and, additionally, it
secretes mtRNA to ensure the innate immune system response to the
infection (37). In short, each
organism fights for its survival by generating innumerable ‘attack’
or ‘defense’ mechanisms to ensure its survival.
i) rRNAs and TLRs in the brain. Alvarez-Carbonell
et al (9) demonstrated the
reactivation of HIV in microglia cultures, an effect that was
mediated by TLR3 activation by poly (I:C) and bacterial rRNA. The
activation of other TLRs revealed a minor effect on the
reactivation of this virus, and the activation of TLRs 2/1, 4–6
induced the NF-κB-mediated pathway, but the activation of TLR3
induced IRF3. Diverse effects were observed in human monocytes and
rat microglia. These results demonstrate that the activation of
TLRs may have functions other than the immune system control. As
aforementioned, both, the biological effect and the potency of this
effect, depends on the type of TLR activated and by the ligand and
the coactivators that may or may not be present in a given
tissue.
lncRNAs control TLR activity
lncRNAs are RNAs with a size of 200–1,000 kb, which
exert a variety of cellular functions by diverse mechanisms
(112,113). It is established that the
activation of TLRs 7 and 9 is associated with immune dysfunction
and severe disease states, and that blockade of these receptors
improved the health status of mice susceptible to develop lupus
(7–9,20).
Similarly, Yang et al (38)
demonstrated that lnc-Atg1611 binds to TLR7 and activates the
MyD88-mediated signaling pathway in immune cells and deletion of
this lncRNA attenuated TLR7-related autoimmune phenotypes in murine
models. The binding of lnc-Atg1611 to TLR7 is complex, as on one
side it binds to TLR7 and on the other to MyD88, thus functioning
as a ‘scaffold’, which induces structural changes in the TLR7
stem-loop and induces signal transduction (113). The authors suggested that
self-RNAs, particularly lnc-Atg1611, may be potential therapeutic
targets to control the development of autoimmune disorders. In this
regard, Mussari et al (20)
identified an antagonist (7f) that has a high affinity for TLR7
and, to a lesser extent, for TLR9, which blocks cytokine production
mediated by TLRs 7 and 9 and markedly decreased proteinuria,
antibody production and IL-10 secretion.
i) lncRNAs and TLRs in cancer. Anaplastic thyroid
carcinoma (ATC) is a rare and aggressive type of thyroid cancer
that spreads rapidly to other parts of the body. In thyroid cancer,
the lncRNA C5AR1 shows increased expression and correlates
positively with short patient survival (39). Molecularly, C5AR1 activates TLRs 1
and 2 receptors and MyD88, which results in tumor growth and lung
metastasis of ATC cells in a murine model and inhibition of the
expression of this lncRNA, considerably decreased the oncogenic
effects regulated by its interaction with TLRs (39). Regarding this, miR-355-5p
negatively regulated C5AR1 expression, which could be used as a
therapeutic tool to downregulate the levels of this lncRNA and
attenuate the oncogenic effects produced by C5AR1-TLRs 1/2-MyD88
signaling.
These results demonstrate that several ncRNAs can
function as ligands for TLRs and exert oncogenic effects. The
identification of all ncRNAs and molecular elements regulating the
TLR-mediated signaling at a specific point in time will allow for
the understanding of the interaction mechanisms underlying TLR
activation and/or inhibition to develop immunotherapies. lncRNAs
have been identified as positive or negative regulators of TLRs,
affecting immune response and drug sensitivity; however, further
studies are needed to understand the mechanisms by which these
ncRNAs control TLR pathways.
Interaction of ncRNA-derived fragments with
TLRs
Since 1971, the production of fragments (~50 nt)
originating from the 16S ribosomal subunit has been observed
(114). Similarly, Borek et
al (115) reported a higher
rate of turnover and excretion of transfer RNA (tRNA) ‘degradation
products’ in patients with cancer relative to individuals without
cancer. Subsequent studies continued to report the presence of
these ‘degradation products’ (116–120). It was not until 2008 that
Thompson et al (121)
reported that these RNA fragments have specific biological
functions in addition to controlling the levels of full-length
sequence transfer RNA (tRNAs). To date, it is acknowledged that all
known ncRNAs are fragmented and that this appears to occur by
specific biogenic pathways; however, the molecular elements
involved in these machineries are not yet known.
Fragments derived from tRNAs
Several types of fragments generated from tRNAs have
been identified and they are classified into two categories: tRNA
halves (tiRNAs; 31–40 nt) and tRNA-derived fragments (14–30 nt)
based on the nuclease cleavage sites in mature or premature tRNAs
(116,117).
Several physiological functions have now been
described for transfer RNA-derived small RNAs (tsRNAs) (122,123), including TLR7 activation. Pawar
et al (118) demonstrated
that mycobacterium infection activates TLRs expressed on the
surface of human monocyte-derived macrophages, which promoted the
production and secretion of 5′-tRNA halves in EVs. Specifically,
the 5′-tRNAHisGUG half was recognized and endocytosed by
recipient cells to subsequently activate endosomal TLR7. Later, the
authors demonstrated TLR7 activation by the
5′-tRNAValCAC/AAC where its GUUU terminal sequence was
determinant for the activation of this receptor and for the
elimination of bacterial infection. The mutation of this tRNA-half
showed that the GUUU sequence was determinant for TLR7 activation.
Notably, this motif is present in other RNAs that also stimulate
this receptor (124).
To date, tsRNAs are the best characterized fragments
and are known to be involved in rRNA maturation, reverse
transcription, adaptation of parasites to their environment and as
ligands for TLRs. The tsRNA-TLR interaction is key for eliminating
Mycobacterium tuberculosis infection and certainly for other
types of bacteria and viruses, so understanding the mechanisms
underlying the tsRNA-TLR regulatory axis may be important for
generating drugs to mitigate these types of infections, which is
particularly important due to the growing resistance to antibiotics
(125,126).
Fragments derived from RNYs and
lncRNAs
RNY fragments (s-RNYs) are generated from stem
terminal regions during apoptosis (119) from miRNAs or Piwi-interacting
RNAs (127) or by RNase L
activity or by activation of the innate immune system mediated by
Poly: I:C (120). Specific
biogenic pathways have been proposed for the generation of s-RNYs,
which retain specific sequences from their parental RNYs to
regulate particular cellular processes (Table I). Currently, limited information
exists regarding TLR activation by s-RNYs. To the best of our
knowledge, there is only one study which revealed that the release
of s-RNYs (sRNYs1-5p, 3–5p and 4–5p)/Ro60 by macrophages resulted
in the activation of TLR7 and in the induction of
caspase-3-dependent apoptosis and NF-κB-mediated inflammation. The
full form of RNYs/Ro60 did not have the same effect, indicating
specific functions in this cellular context for the s-RNYs
(128). Related to this,
Rodriguez-Corona et al (10) predicted the RNYs and their
fragments interactions with TLRs and with other receptors (Table I).
RNYs and their fragments have a central role in
immune system responses and at least part of these responses depend
on RNYs/s-RNYs-TLRs interactions. Studies indicate that the action
of s-RNYs is not necessarily the same as that of parental RNYs
(10,128); therefore, it is necessary to
identify the molecular traits that are determining these
differences.
In relation to lncRNAs, Giraldez et al
(129) demonstrated the presence
of fragments of mRNAs and lncRNAs in blood plasma that are missed
by standard sequencing techniques. The detection of these fragments
in plasma has potential for use as biomarkers and their functions
need to be gradually elucidated.
Conclusion
The discovery of ncRNAs, considerably changed the
understanding and approach to the study of life sciences. Initial
studies demonstrated the interfering function that miRNAs (22,23)
and small-interfering RNAs (siRNAs) (97) have; subsequent studies have shown
the diversity of functions and mechanisms of action by which ncRNAs
may act.
Up to now, several studies have demonstrated that
TLR activity is mediated by ncRNAs and their fragments (5,7,8,32,36,38);
however, several questions arise regarding the molecular mechanisms
underlying this activation and the physiological effect this may
have. The majority of data indicate that activation of TLRs by
ncRNAs or their fragments primarily induces the innate immune
system, protecting organisms from microbial and viral infections
(11–15); however, this activation is also
associated with oncogenic (5,39),
neurodegenerative effects (6,90)
and viral reactivation (9).
Although in the majority of cases the end result is the activation
of the immune system, each ncRNA and its fragments activate
specific intracellular signaling pathways, resulting in the control
of specific biological effects (113,118,128). Uncovering the molecular
mechanisms involved in these differences will allow for improved
understanding of the ncRNAs/fragment-TLR relationship and the
biological functions that are mediated by these interactions.
Regarding this, Wallach et al (6) reported that secondary structure
formation did not determine the binding of miRNAs to TLRs; however,
miRs-21, −93 and 296 can adopt hairpin and/or homoduplex
structures, depending on the miRNA concentration and ionic
conditions, which determined their specificity for target mRNAs
(130,131). Similarly, there is evidence
indicating that secondary structures of mature siRNAs influenced
the efficiency of siRNA-mRNA interaction, where unstructured siRNAs
exert a greater silencing effect than those with secondary
structures. Belter et al (131) observed that miRNA hairpins
resemble those of the anti-Tn-C aptamer, indicating that miRNAs can
directly regulate their targets in an RISC-independent manner,
making them highly specific regulators, more so than previously
considered.
TLRs regulate both the innate and adaptive immune
system and are mainly associated with autoimmune disorders, chronic
inflammation, chronic viral infections and cancer, making these
receptors a target for the development of immunotherapeutic
strategies. Currently, there are several TLR ligands used as
vaccine adjuvants to increase the efficacy of vaccines, and TLR
agonists and antagonists have been identified for the treatment of
chronic viral infections such as hepatitis B virus (HBV), HIV
(23,132), autoimmune diseases (20,21,133) or for the treatment of cancer
(25–27). For instance, TLR7 agonists have
been tested in clinical trials for HBV infection and a TLR9 ligand
as an adjuvant in a vaccine against HBV (132). The activation of specific TLRs
inhibited HIV replication and reduced viral spread; however, TLR
activation also led to the reactivation of latent HIV in cell
cultures. This may be important for the development of anti-HIV
therapies, since reactivation of latently infected reservoir cells
could facilitate the recognition by cytotoxic immune cells, which
has already been observed in clinical studies. Therefore,
immunotherapy against chronic HBV or HIV infection is promising and
antiviral drugs have been approved for the clinical treatment of
HBV and HIV (134).
In previous years, several groups have focused on
designing oligonucleotides that antagonize TLRs 7–9 and have been
proposed as potential treatments for autoimmune and inflammatory
diseases (20–22). Of these antagonists, IMO-8400
reduced moderate-to-severe plaque psoriasis in patients
participating in a phase IIa clinical trial (21). Other research groups have focused
on developing small molecules that function as antagonists of TLRs
7–9. For example, the 7f antagonist of TLRs 7 and 9 has a strong
potency and high selectivity for these receptors, emphasizing the
use of small molecules for the treatment of autoimmune diseases
(20). Meanwhile, TAC5, an
antagonist of endosomal TLRs, reduced inflammation and prevented
the progression of psoriasis and systemic lupus erythematosus in
mice, indicating its therapeutic potential for their treatment
(22). Despite this, there is a
need to evaluate the sensitivity and specificity as well as the
side effects that the application of these molecules may have. For
example, a study conducted by Christensen et al (133) reported that TLR9 knockout mice
developed more aggressive disease pathology; however, dual TLR 7
and 9 knockout demonstrated protection.
For cancer treatment, several TLR3 agonists were
observed to induce cancer regression by restoring immunogenicity to
chemotherapy (25,26). Similarly, intratumoral injection of
the TLR7 and 8 receptor agonist MEDI9197 suppressed tumor growth
and increased survival in murine models by regulating the immune
response and by increasing the expression of associated genes in
innate and adaptive immunity (27,135,136). Notably, the antitumor effects of
MEDI9197 were most potent when this drug was applied in combination
with a programmed death ligand 1 inhibitor (135). Although no tumor response was
observed, the application of MEDI9197 or in combination with
durvalumab and/or palliative radiation induced local and systemic
immune activation in patients with solid tumors (136). In addition, sugar-conjugated TLR7
ligands increased autoimmunostimulatory activity, demonstrating
potential for application as adjuvants in vaccines and cancer
therapies (137).
The activation of TLRs to induce innate and
adaptive immunity and to treat various diseases has great clinical
potential, as there is considerable experimental evidence
indicating their application for the treatment of viral and immune
diseases and different types of cancer. Nevertheless, the
translation of experimental observations into effective clinical
application is a major challenge and further work is still needed
to increase the therapeutic effect and increase the precision of
treatments.
Acknowledgements
The authors thank Mrs. Verónica Vratny for
proofreading and editing.
Funding
Funding: No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
RREG and MAVF contributed to the study conception
and design and conducted the literature review and wrote the
manuscript. All authors read and approved the final version of the
manuscript. Data authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient content for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Costa S, La Rocca G and Cavalieri V:
Epigenetic regulation of chromatin functions by MicroRNAs and Long
Noncoding RNAs and implications in human diseases. Biomedicines.
13:7252025. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Godden AM, Silva WTAF, Kiehl B, Jolly C,
Folkes L, Alavioon G and Immler S: Environmentally induced
variation in sperm sRNAs is linked to gene expression and
transposable elements in zebrafish offspring. Heredity (Edinb). Mar
22–2025.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Oliveira-Rizzo C, Colantuono CL,
Fernández-Alvarez AJ, Boccaccio GL, Garat B, Sotelo-Silveira JR,
Khan S, Ignatchenko V, Lee YS, Kislinger T, et al: Multi-omics
study reveals Nc886/vtRNA2-1 as a positive regulator of prostate
cancer cell immunity. J Proteome Res. 24:433–448. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Estravís M, García-Sánchez A, Martin MJ,
Pérez-Pazos J, Isidoro-García M, Dávila I and Sanz C: RNY3
modulates cell proliferation and IL13 mRNA levels in a T lymphocyte
model: A possible new epigenetic mechanism of IL-13 regulation. J
Physiol Biochem. 79:59–69. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fabbri M, Paone A, Calore F, Galli R,
Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ, et al:
MicroRNAs bind to Toll-like receptors to induce prometastatic
inflam-matory response. Proc Natl Acad Sci USA. 109:E2110–E2116.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wallach T, Wetzel M, Dembny P, Staszewski
O, Krüger C, Buonfiglioli A, Prinz M and Lehnardt S: Identification
of CNS Injury-Related microRNAs as Novel Toll-Like Receptor 7/8
Signaling Activators by Small RNA Sequencing. Cells. 9:1862020.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Greidinger EL, Zang YJ, Martinez L, Jaimes
K, Nassiri M, Bejarano P, Barber GN and Hoffman RW: Differential
tissue targeting of autoimmunity manifestations by
autoantigen-associated Y RNAs. Arthritis Rheum. 56:1589–1597. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Oldenburg M, Krüger A, Ferstl R, Kaufmann
A, Nees G, Sigmund A, Bathke B, Lauterbach H, Suter M, Dreher S, et
al: TLR13 recognizes bacterial 23S rRNA devoid of erythromycin
resistance-forming modification. Science. 337:1111–1115. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Alvarez-Carbonell D, Garcia-Mesa Y, Milne
S, Das B, Dobrowolski C, Rojas R and Karn J: Toll-like receptor 3
activation selectively reverses HIV latency in microglial cells.
Retrovirology. 14:92017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rodríguez-Corona JM, Ruiz Esparza-Garrido
R, Horta-Vega JV and Velázquez-Flores M: Circulating Y-RNAs: A
predicted function mainly in controlling the innate immune system,
cell signaling and DNA replication in pediatric patients with
pilocytic and diffuse astrocytoma. Clin Pediatr. 8:1–9. 2023.
|
|
11
|
Akira S, Takeda K and Kaisho T: Toll-like
receptors: Critical proteins linking innate and acquired immunity.
Nature Immunology. 2:675–680. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Feng Y, Zou L, Yan D, Chen H, Xu G, Jian
W, Ping Cui and Chao W: Extracellular MicroRNAs induce potent
innate immune responses via TLR7/MyD88-dependent mechanisms. J
Immunol. 199:2106–2117. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wiese MD, Manning-Bennett AT and Abuhelwa
AY: Investigational IRAK-4 Inhibitors for the treatment of
rheumatoid arthritis. Expert Opin invest Drugs. 29:475–482. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xiao L, Liu Y and Wang N: New paradigms in
inflammatory signaling in vascular endothelial cells. Am J Physiol
Heart Circ Physiol. 306:H317–H325. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Van Tassell BW, Seropian IM, Toldo S,
Salloum FN, Smithson L, Varma A, Hoke NN, Gelwix C, Chau V and
Abbate A: Pharmacologic inhibition of myeloid differentiation
factor 88 (MyD88) prevents left ventricular dilation and
hypertrophy after experimental acute myocardial infarction in the
mouse. J Cardiovasc Pharmacol. 55:385–390. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rakoff-Nahoum S, Paglino J,
Eslami-Varzaneh F, Edberg S and Medzhitov R: Recognition of
commensal microflora by Toll-like receptors is required for
intestinal homeostasis. Cell. 118:229–241. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tsan MF and Gao B: Endogenous ligands of
Toll-like receptors. J Leukoc Biol. 76:514–519. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rock FL, Hardiman G, Timans JC, Kastelein
RA and Bazan JF: A family of human receptors structurally related
to Drosophila Toll. Proc Natl Acad Sci USA. 95:588–593. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Takeda K and Akira S: Toll-like receptors
in innate immunity. Int Immunol. 17:1–14. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mussari CP, Dodd DS, Sreekantha RK,
Pasunoori L, Wan H, Posy SL, Critton D, Ruepp S, Subramanian M,
Watson A, et al: Discovery of potent and orally bioavailable small
molecule antagonists of toll-like receptors 7/8/9 (TLR7/8/9). ACS
Med Chem Lett. 11:1751–1758. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Balak DM, van Doorn MB, Arbeit RD,
Rijneveld R, Klaassen E, Sullivan T, Brevard J, Thio HB, Prens EP,
Burggraaf J and Rissmann R: IMO-8400, a toll-like receptor 7, 8,
and 9 antagonist, demonstrates clinical activity in a phase 2a,
randomized, placebo-controlled trial in patients with
moderate-to-severe plaque psoriasis. Clin Immunol. 174:63–72. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Patra MC, Achek A, Kim GY, Panneerselvam
S, Shin HJ, Baek WY, Lee WH, Sung J, Jeong U, Cho EY, et al: A
novel small-molecule inhibitor of endosomal TLRs reduces
inflammation and alleviates autoimmune disease symptoms in Murine
models. Cells. 9:16482020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Winckelmann AA, Munk-Petersen LV,
Rasmussen TA, Melchjorsen J, Hjelholt TJ, Montefiori D, Østergaard
L, Søgaard OS and Tolstrup M: Administration of a Toll-like
receptor 9 agonist decreases the pro viral reservoir in
virologically suppressed HIV-infected patients. PLoS One.
8:e620742013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Christensen S, Shupe J, Nickerson K,
Kashgarian M, Flavell R and Shlomchik M: Toll-like receptor 7 and
TLR9 dictate autoantibody specificity and have opposing
inflammatory and regulatory roles in a murine model of lupus.
Immunity. 25:417–428. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Komal A, Noreen M and El-Kott AF: TLR3
agonists: RGC100, ARNAX, and poly-IC: A comparative review. Immunol
Res. 69:312–322. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
LeNaour J, Thierry S, Scuderi SA,
Boucard-Jourdin M, Liu P, Bonnin M, Pan Y, Perret C, Zhao L, Mao M,
et al: A chemically defined TLR3 agonist with anticancer activity.
Oncoimmunology. 12:22275102023. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Singh M, Khong H, Dai Z, Huang XF, Wargo
JA, Cooper ZA, Vasilakos JP, Hwu P and Overwijk WW: Effective
innate and adaptive anti-melanoma immunity through localized
TLR-7/8 activation. J Immunol. 193:4722–4731. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fuge O, Vasdev N, Allchorne P and Green
JS: Immunotherapy for bladder cancer. Res Rep Urol. 7:65–79.
2015.PubMed/NCBI
|
|
29
|
Morales A, Eidinger D and Bruce AW:
Intracavitary Bacillus Calmette-Guerin in the treatment of
superficial bladder tumors. J Urol. 116:180–183. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bayraktar R, Bertilaccio MTS and Calin GA:
The interaction between two worlds: MicroRNAs and Toll-like
receptors. Front Immunol. 14:10532019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yu J, Zhang X, Cai C, Zhou T and Chen Q:
Small RNA and Toll-like receptor interactions: Origins and disease
mechanisms. Trends Biochem Sci. Feb 15–2025.doi:
10.1016/j.tibs.2025.01.004 (Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
32
|
He WA, Calore F, Londhe P, Canella A,
Guttridge DC and Croce CM: Microvesicles containing miRNAs promote
muscle cell death in cancer cachexia via TLR7. Proc Natl Acad Sci
USA. 111:4525–4529. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Donzelli J, Proestler E, Riedel A,
Nevermann S, Hertel B, Guenther A, Gattenlöhner S, Savai R, Larsson
K and Saul MJ: Small extracellular vesicle-derived miR-574-5p
regulates PGE2-biosynthesis via TLR7/8 in lung cancer. J Extracell
Vesicles. 10:e121432021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Huang X, Ma Z and Qin W: Screening and
bioinformatics analyses of key miRNAs associated with Toll-like
receptor activation in gastric cancer cells. Medicina (Kaunas).
59:5112023. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Clancy RM, Alvarez D, Komissarova E,
Barrat FJ, Swartz J and Buyon JP: Ro60-associated single-stranded
RNA links inflammation with fetal cardiac fibrosis via ligation of
TLRs: A novel pathway to autoimmune-associated heart block. J
Immunol. 184:2148–2155. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li XD and Chen ZJ: Sequence specific
detection of bacterial 23S ribosomal RNA by TLR13. Elife.
1:e001022012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Krüger A, Oldenburg M, Chebrolu C, Beisser
D, Kolter J, Sigmund AM, Steinmann J, Schäfer S, Hochrein H,
Rahmann S, et al: Human TLR8 senses UR/URR motifs in bacterial and
mitochondrial RNA. EMBO Rep. 16:1656–1663. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yang Z, Ji S, Liu L, Liu S, Wang B, Ma Y
and Cao X: Promotion of TLR7-MyD88-dependent inflammation and
autoimmunity in mice through stem-loop changes in Lnc-Atg16l1. Nat
Commun. 15:102242024. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu B, Sun Y, Geng T, Wang H, Wu Z, Xu L,
Zhang M, Niu X, Zhao C, Shang J and Shang F: C5AR1-induced TLR1/2
pathway activation drives proliferation and metastasis in
anaplastic thyroid cancer. Mol Carcinog. 63:1938–1952. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yang D, Wan X, Dennis AT, Bektik E, Wang
Z, Costa MGS, Fagnen C, Vénien-Bryan C, Xu X, Gratz DH, et al:
MicroRNA biophysically modulates cardiac action potential by direct
binding to ion channel. Circulation. 143:1597–1613. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lee RC, Feinbaum RL and Ambros V: The
C. elegans heterochronic gene lin-4 encodes small RNAs with
antisense complementarity to lin-14. Cell. 75:843–854. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wightman B, Ha I and Ruvkun G:
Posttranscriptional regulation of the heterochronic gene lin-14 by
lin-4 mediates temporal pattern formation in C. elegans.
Cell. 75:855–862. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Llave C, Kasschau KD, Rector MA and
Carrington JC: Endogenous and silencing-associated small RNAs in
plants. Plant Cell. 14:1605–1619. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hansen TB, Venø MT, Jensen TI, Schaefer A,
Damgaard CK and Kjems J: Argonaute-associated short introns are a
novel class of gene regulators. Nat Commun. 7:115382016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Smalheiser NR and Torvik VI: Mammalian
microRNAs derived from genomic repeats. Trends Genet. 21:318–322.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Borchert GM, Lanier W and Davidson BL: RNA
polymerase III transcribes human microRNAs. Nat Struct Mol Biol.
13:1097–1101. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chung W, Agius P, Westholm JO, Chen M,
Okamura K, Robine N, Leslie CS and Lai EC: Computational and
experimental identification of mirtrons in Drosophila melanogaster
and Caenorhabditis elegans. Genome Res. 21:286–300. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lai EC: MicroRNAs are complementary to
3′UTR sequence motifs that mediate negative post-transcriptional
regulation. Nat Genet. 30:363–364. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Drinnenberg IA, Weinberg DE, Xie KT, Mower
JP, Wolfe KH, Fink GR and Bartel DP: RNAi in budding yeast.
Science. 326:544–550. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zardo G, Ciolfi A, Vian L, Starnes LM,
Billi M, Racanicchi S, Maresca C, Fazi F, Travaglini L, Noguera N,
et al: Polycombs and microRNA-223 regulate human granulopoiesis by
transcriptional control of target gene expression. Blood.
119:4034–4046. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Di Mauro V, Crasto S, Colombo FS, Di
Pasquale E and Catalucci D: Wnt signalling mediates miR-133a
nuclear re-localization for the transcriptional control of Dnmt3b
in cardiac cells. Sci Rep. 9:1–15. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Leucci E, Patella F, Waage J, Holmstrøm K,
Lindow M, Porse B, Kauppinen S and Lund AH: MicroRNA-9 targets the
long non-coding RNA MALAT1 for degradation in the nucleus. Sci Rep.
3:25352013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
van Beijnum JR, Buurman WA and Griffioen
AW: Convergence and amplification of toll-like receptor (TLR) and
receptor for advanced glycation end products (RAGE) signaling
pathways via high mobility group B1 (HMGB1). Angiogenesis.
11:91–99. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Mardente S, Mari E, Consorti F, Gioia CD,
Negri R, Etna M, Zicari A and Antonaci A: HMGB1 induces the
overexpression of miR-222 and miR-221 and increases growth and
motility in papillary thyroid cancer cells. Oncol Rep.
28:2285–2289. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ronkainen H, Hirvikoski P, Kauppila S,
Vuopala KS, Paavonen TK, Selander KS and Vaarala MH: Absent
Toll-like receptor-9 expression predicts poor prognosis in renal
cell carcinoma. J Exp Clin Cancer Res. 30:842011. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Hao B, Chen Z, Bi B, Yu M, Yao S, Feng Y,
Yu Y, Pan L, Di D, Luo G and Zhang X: Role of TLR4 as a prognostic
factor for survival in various cancers: A meta-analysis.
Oncotarget. 9:13088–13099. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Kabelitz D: Expression and function of
toll-like receptors in T lymphocytes. Curr Opin Immunol. 19:39–45.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Elliott DRF, Perner J, Li X, Symmons MF,
Verstak B, Eldridge M, Bower L, O'Donovan M and Gay NJ; OCCAMS
Consortium; Fitzgerald RC, : Impact of mutations in Toll-like
receptor pathway genes on esophageal carcinogenesis. PLoS Genet.
13:e10068082017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Ngo VN, Young RM, Schmitz R, Jhavar S,
Xiao W, Lim KH, Kohlhammer H, Xu W, Yang Y, Zhao H, et al:
Oncogenically active MYD88 mutations in human lymphoma. Nature.
470:115–119. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Li R, Hao Y, Pan W, Wang W and Min Y:
Monophosphoryl lipid A-assembled nanovaccines enhance tumor
immunotherapy. Acta Biomater. 171:482–944. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zhao BG, Vasilakos JP, Tross D, Smirnov D
and Klinman DM: Combination therapy targeting toll like receptors
7, 8 and 9 eliminates large established tumors. J Immunother
Cancer. 2:122014. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Sato-Kaneko F, Yao S, Ahmadi A, Zhang SS,
Hosoya T, Kaneda MM, Varner JA, Pu M, Messer KS, Guiducci C, et al:
Combination immunotherapy with TLR agonists and checkpoint
inhibitors suppresses head and neck cancer. JCI Insight.
2:933972017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Takeda Y, Kataoka K, Yamagishi J, Ogawa S,
Seya T and Matsumoto M: A TLR3-specific adjuvant relieves innate
resistance to PD-L1 blockade without cytokine toxicity in tumor
vaccine immunotherapy. Cell Rep. 19:1874–1887. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Adams S: Toll-like receptor agonists in
cancer therapy. Immunotherapy. 1:949–964. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Heil F, Hemmi H, Hochrein H, Ampeberger F,
Kirchning C, Akira S, Lipford G, Wagner H and Bauer S:
Species-specific recognition of single-stranded RNA via toll-like
receptor 7 and 8. Science. 303:1526–1529. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Yoshida T, Miura T, Matsumiya T, Yoshida
H, Morohashi H, Sakamoto Y, Kurose A, Imaizumi T and Hakamada K:
Toll-like receptor 3 as a recurrence risk factor and a potential
molecular therapeutic target in colorectal cancer. Clin Exp
Gastroenterol. 13:427–438. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Bianchi F, Milione M, Casalini P, Centonze
G, Le Noci VM, Storti C, Alexiadis S, Truini M, Sozzi G, Patorino
U, et al: Toll-like receptor 3 as a new marker to detect high risk
early stage Non-small-cell Lung Cancer patients. Sci Rep.
9:142882019. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Yuan MM, Xu YY, Chen L, Li XY, Qin J and
Shen Y: TLR3 expression correlates with apoptosis, proliferation
and angiogenesis in hepatocellular carcinoma and predicts
prognosis. BMC Cancer. 15:2452015. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Ceccarelli S, Marzolesi VP, Vannucci J,
Bellezza G, Floridi C, Nocentini G, Cari L, Traina G, Petri D, Puma
F and Conte C: Toll-like receptor 4 and 8 are overexpressed in lung
biopsies of human Non-small cell lung carcinoma. Lung. 203:382025.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Gonzalez-Reyes S, Fernandez JM, Gonzalez
LO, Aguirre A, Suarez A, Gonzalez JM, Escaff S and Vizoso FJ: Study
of TLR3, TLR4, and TLR9 in prostate carcinomas and their
association with biochemical recurrence. Cancer Immunol Immunother.
60:217–226. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Chuang HC, Huang CC, Chien CY and Chuang
JH: Toll-like receptor 3-mediated tumor invasion in head and neck
cancer. Oral Oncol. 48:226–232. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Chen C, Feng Y, Zou L, Wang L, Chen HH,
Cai JY, Xu JM, Sosnovik DE and Chao W: Role of extracellular RNA
and TLR3-trif signaling in myocardial Ischemia-reperfusion injury.
J Am Heart Assoc. 3:e0006832014. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Cabrera-Fuentes HA, Ruiz-Meana M,
Simsekyilmaz S, Kostin S, Inserte J, Saffarzadeh M, Galuska SP,
Vijayan V, Barba I, Barreto G, et al: RNase1 prevents the damaging
interplay between extracellular RNA and tumour necrosis factor
alpha in cardiac ischaemia/reperfusion injury. Thromb Haemost.
112:1110–1119. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Shimada BK, Yang Y, Zhu J, Wang S, Suen A,
Kronstadt SM, Jeyaram A, Jay SM, Zou L and Chao W: Extracellular
miR-146a-5p induces cardiac innate immune response and
cardiomyocyte dysfunction. Immunohorizons. 4:561–572. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Xu J, Feng Y, Jeyaram A, Jay SM, Zou L and
Chao W: Circulating plasma extracellular vesicles from septic mice
induce inflammation via MicroRNA- and TLR7-Dependent mechanisms. J
Immunol. 201:3392–3400. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Huang H, Zhu J, Gu L, Hu J, Feng X, Huang
W, Wang S, Yang Y, Cui P, Lin SH, et al: TLR7 Mediates acute
respiratory distress syndrome in sepsis by sensing extracellular
miR-146a. Am J Respir Cell Mol Biol. 67:375–388. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Georges HM, Cassin C, Tong M and Abrahams
VM: TLR8-activating miR-146a-3p is an intermediate signal
contributing to fetal membrane inflammation in response to
bacterial LPS. Immunology. 172:577–587. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Hannestad UlF, Allard A, Nilsson K and
Rosén A: Prevalence of EBV, HHV6, HCMV, HAdV, SARS-CoV-2, and
Autoantibodies to Type I Interferon in Sputum from Myalgic
Encephalomyelitis/Chronic Fatigue Syndrome Patients. Viruses.
17:4222025. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Simpkin AJ, McNicholas BA, Hannon D,
Bartlett R, Chiumello D, Dalton HJ, Gibbons K, White N, Merson L,
Fan E, et al: Correction: Effect of early and later prone
positioning on outcomes in invasively ventilated COVID-19 patients
with acute respiratory distress syndrome: Analysis of the
prospective COVID-19 critical care consortium cohort study. Ann
Intensive Care. 15:442025. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Fakhraei R, Song Y, Kazi DS, Wadhera RK,
Lemos JA, Das SR, Morrow DA, Dahabreh IJ, Rutan CM, Thomas K and
Yeh RW: Social vulnerability and Long-term cardiovascular outcomes
after COVID-19 hospitalization: An analysis of the american heart
association COVID-19 registry linked with medicare claims data. J
Am Heart Assoc. 14:e0380732025. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Wallach T, Raden M, Hinkelmann L, Brehm M,
Rabsch D, Weidling H, Krüger C, Kettenmann H, Backofen R and
Lehnardt S: Distinct SARS-CoV-2 RNA fragments activate Toll-like
receptors 7 and 8 and induce cytokine release from human
macrophages and microglia. Front Immunol. 13:10664562023.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Liao TL, Liu HJ, Chen DY, Tang KT, Chen YM
and Liu PY: SARS-CoV-2 primed platelets-derived microRNAs enhance
NETs formation by extracellular vesicle transmission and TLR7/8
activation. Cell Commun Signal. 21:3042023. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Guedes JR, Santana I, Cunha C, Duro D,
Almeida MR, Cardoso AM, de Lima MCP and Cardoso AL: MicroRNA
deregulation and chemotaxis and phagocytosis impairment in
Alzheimer's disease. Alzheimers Dement (Amst). 3:7–17. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Évora A, Garcia G, Rubi A, De Vitis E,
Matos AT, Vaz AR, Gervaso F, Gigli G, Polini A and Brites D:
Exosomes enriched with miR-124-3p show therapeutic potential in a
new microfluidic triculture model that recapitulates neuron-glia
crosstalk in Alzheimer's disease. Front Pharmacol. 16:14740122025.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Kazemi M, Sanati M, Shekari Khaniani M and
Ghafouri-Fard S: A review on the lncRNA-miRNA-mRNA regulatory
networks involved in inflammatory processes in Alzheimer's disease.
Brain Res. 1856:1495952025. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Ravanidis S, Bougea A, Papagiannakis N,
Koros C, Simitsi AM, Pachi I, Breza M, Stefanis L and Doxakis E:
Validation of differentially expressed brain-enriched microRNAs in
the plasma of PD patients. Ann Clin Transl Neurol. 7:1594–1607.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Currim F, Brown-Leung J, Syeda T, Corson
M, Schumann S, Qi W, Baloni P, Shannahan JH, Rochet JC, Singh R and
Cannon JR: Rotenone induced acute miRNA alterations in
extracellular vesicles produce mitochondrial dysfunction and cell
death. NPJ Parkinsons Dis. 11:592025. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Ishtiaq B, Paracha RZ, Nisar M, Ejaz S and
Hussain Z: Discovering promising drug candidates for Parkinson's
disease: Integrating miRNA and DEG analysis with molecular dynamics
and MMPBSA. J Comput Aided Mol Des. 39:82025. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Wallach T, Mossmann ZJ, Szczepek M, Wetzel
M, Machado R, Raden M, Miladi M, Kleinau G, Krüger C, Dembny P, et
al: MicroRNA-100-5p and microRNA-298-5p released from apoptotic
cortical neurons are endogenous Toll-like receptor 7/8 ligands that
contribute to neurodegeneration. Mol Neurodegener. 16:802021.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Park C, Lei Z, Li Y, Ren B, He J, Huang H,
Chen F, Li H, Brunner K, Zhu J, et al: Extracellular vesicles in
sepsis plasma mediate neuronal inflammation in the brain through
miRNAs and innate immune signaling. J Neuroinflammation.
21:2522024. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Deng L, Gao R, Chen H, Jiao B, Zhang C,
Wei L, Yan C, Ye-Lehmann S, Zhu T and Chen C: Let-7b-TLR7 signaling
axis contributes to the Anesthesia/Surgery-induced cognitive
impairment. Mol Neurobiol. 61:1818–1832. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Wu N, Morsey BM, Emanuel KM and Fox HS:
Sequence-specific extracellular microRNAs activate TLR7 and induce
cytokine secretion and leukocyte migration. Mol Cell Biochem.
476:4139–4151. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Tanji H, Ohto U, Shibata T, Taoka M,
Yamauchi Y, Isobe T, Miyake K and Shimizu T: Toll-like receptor 8
senses degradation products of single-stranded RNA. Nat Struct Mol
Biol. 22:109–115. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Tanji H, Ohto U, Shibata T, Miyake K and
Shimizu T: Structural reorganization of the Toll-like receptor 8
dimer induced by agonistic ligands. Science. 339:1426–1429. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Shibata T, Ohto U, Nomura S, Kibata K,
Motoi Y, Zhang Y, Murakami Y, Fukui R, Ishimoto T, Sano S, et al:
Guanosine and its modified derivatives are endogenous ligands for
TLR7. Int Immunol. 28:211–222. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Davenne T, Bridgeman A, Rigby RE and
Rehwinkel J: Deoxyguanosine is a TLR7 agonist. Eur J Immunol.
50:56–62. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Wang J, Chai J and Wang H: Structure of
the mouse toll-like receptor 13 ectodomain in complex with a
conserved sequence from bacterial 23S ribosomal RNA. FEBS J.
283:1631–1635. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Boccitto M and Wolin SL: Ro60 and Y RNAs:
Structure, functions, and roles in autoimmunity. Crit Rev Biochem
Mol Biol. 54:133–152. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Kowalski MP and Krude T: Functional roles
of non-coding Y RNAs. Int J Biochem Cell Biol. 66:20–29. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Onafuwa-Nuga AA, King SR and Telesnitsky
A: Nonrandom packaging of host RNAs in moloney murine leukemia
virus. J Virol. 79:13528–13537. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Perreault J, Noël JF, Brière F, Cousineau
B, Lucier JF, Perreault JP and Boire G: Retropseudogenes derived
from the human Ro/SS-A autoantigen-associated hY RNAs. Nucleic
Acids Res. 33:2032–2041. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Telesnitsky A and Wolin SL: The host RNAs
in retroviral particles. Viruses. 8:1–15. 2016. View Article : Google Scholar
|
|
103
|
Haderk F, Schulz R, Iskar M, Cid LL, Worst
T, Willmund KV, Schulz A, Warnken U, Seiler J, Benner A, et al:
Tumor-derived exosomes modulate PD-L1 expression in monocytes. Sci
Immunol. 2:eaah55092017. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Irimie AI, Zimta AA, Ciocan C, Mehterov N,
Dudea D, Braicu C and Berindan-Neagoe I: The Unforeseen Non-Coding
RNAs in head and neck cancer. Genes (Basel). 9:1342018. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Apolonia L, Schulz R, Curk T, Rocha P,
Swanson CM, Schaller T, Ule J and Malim MH: Promiscuous RNA binding
ensures effective encapsidation of APOBEC3 proteins by HIV-1. PLoS
Pathog. 11:e10046092015. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Gao FY, Pang JC, Wang M, Lu MX, Liu ZG,
Cao JM, Ke XL and Yi MM: Structurally diverse genes encode TLR13 in
Nile tilapia: The two receptors can recognize Streptococcus 23S RNA
and conduct signal transduction through MyD88. Mol Immunol.
132:60–78. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Grote K, Nicolai M, Schubert U, Schieffer
B, Troidl C, Preissner KT, Bauer S and Fischer S: Extracellular
ribosomal RNA acts synergistically with Toll-like receptor 2
agonists to promote inflammation. Cells. 11:14402022. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Ren C, Cheng L, Sun Y, Zhang Q, de Haan
BJ, Zhang H, Faas MM and de Vos P: Lactic acid bacteria secrete
toll like receptor 2 stimulating and macrophage immunomodulating
bioactive factors. J Functional Foods. 66:1037832020. View Article : Google Scholar
|
|
109
|
Kanmani P and Kim H: Protective effects of
lactic acid bacteria against TLR4 induced inflammatory response in
hepatoma HepG2 cells through modulation of Toll-like receptor
negative regulators of Mitogen-activated protein kinase and NF-κB
signaling. Front Immunol. 9:15372018. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Yamasaki-Yashiki S, Shiraishi T, Gyobu M,
Sasaki H, Kunisawa J, Yokota SI and Katakura Y: Immunostimulatory
activity of lipoteichoic acid with three fatty acid residues
derived from Limosilactobacillus antri JCM
15950T. Appl Environ Microbiol. 90:e01197242024.
View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Nishibayashi R, Inoue R, Harada Y,
Watanabe T, Makioka Y and Ushida K: RNA of enterococcus faecalis
Strain EC-12 Is a major component inducing Interleukin-12
production from human monocytic cells. PLoS One. 10:e01298062015.
View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Mattick JS, Amaral PP, Carninci P,
Carpenter S, Chang HY, Chen LL, Chen R, Dean C, Dinger ME,
Fitzgerald KA, et al: Long non-coding RNAs: Definitions, functions,
challenges and recommendations. Nat Rev Mol Cell Biol. 24:430–447.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Velázquez-Flores MÁ, Rodríguez-Corona JM,
López-Aguilar JE, Siordia-Reyes G, Ramírez-Reyes G,
Sánchez-Rodríguez G and Ruiz Esparza-Garrido R: Noncoding RNAs as
potential biomarkers for DIPG diagnosis and prognosis: XIST and
XIST-210 involvement. Clin Transl Oncol. 23:501–513. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Bowman CM, Dahlberg JE, Ikemura T, Konisky
J and Nomura M: Specific inactivation of 16S ribosomal RNA induced
by colicin E3 in vivo. Proc Natl Acad Sci USA. 8:964–968. 1971.
View Article : Google Scholar
|
|
115
|
Borek E, Baliga BS, Gehrke CW, Kuo CW,
Belman S, Troll W and Waalkes TP: High turnover rate of transfer
RNA in tumor tissue. Cancer Res. 37:3362–3366. 1977.PubMed/NCBI
|
|
116
|
Li S, Xu Z and Sheng J: tRNA-derived small
RNA: A novel regulatory small non-coding RNA. Genes (Basel).
9:e2462018. View Article : Google Scholar
|
|
117
|
Couvillion MT, Sachidanandam R and Collins
K: A growth essential Tetrahymena Piwi protein carries tRNA
fragment cargo. Genes Dev. 24:2742–2747. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Pawar K, Shigematsu M, Sharbati S and
Kirino Y: Infection-induced 5′-half molecules of tRNAHisGUG
activate Toll-like receptor 7. PLoS Biol. 18:e30009822020.
View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Nicolas FE, Hall AE, Csorba T, Turnbull C
and Dalmay T: Biogenesis of YRNA derived small RNAs is independent
of the microRNA pathway. FEBS Lett. 586:1226–1230. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Driedonks TAP and Nolte-'t Hoen ENM:
Circulating Y-RNAs in extracellular vesicles and ribonucleoprotein
complexes; implications for the immune system. Front Immunol.
9:31642018. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Thompson DM, Lu C, Green PJ and Parker R:
tRNA cleavage is a conserved response to oxidative stress in
eukaryotes. RNA. 14:2095–2103. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Di Fazio A, Schlackow M, Pong SK, Alagia A
and Gullerova M: Dicer dependent tRNA derived small RNAs promote
nascent RNA silencing. Nucleic Acids Res. 50:1734–1752. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Yeung ML, Bennasser Y, Watashi K, Le SY,
Houzet L and Jeang KT: Pyrosequencing of small non-coding RNAs in
HIV-1 infected cells: Evidence for the processing of a
viral-cellular double stranded RNA hybrid. Nucleic Acids Res.
37:6575–6586. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Pawar K, Kawamura T and Kirino Y: The
tRNAVal half: A strong endogenous Toll-like receptor 7
ligand with a 5′-terminal universal sequence signature. Proc Natl
Acad Sci USA. 121:e23195691212024. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Ventola CL: The Antibiotic Resistance
Crisis Part 1: Causes and Threats. PT. 40:277–283. 2015.PubMed/NCBI
|
|
126
|
Mori V, Grant G and Hattingh L: Evaluation
of antimicrobial resistance surveillance data sources in primary
care setting: A scoping review. Fam Pract. 42:cmaf0132025.
View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Verhagen AP and Pruijn GJ: Are the Ro
RNP-associated Y RNAs concealing microRNAs? Y RNA-derived miRNAs
may be involved in autoimmunity. Bioessays. 33:674–682. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Hizir Z, Bottini S, Grandjean V, Trabucchi
M and Repetto E: RNY (YRNA)-derived small RNAs regulate cell death
and inflammation in monocytes/macrophages. Cell Death Dis.
8:e25302017. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Giraldez MD, Spengler RM, Etheridge A,
Goicochea AJ, Tuck M, Choi SW, Galas DJ and Tewari M:
Phospho-RNA-seq: A modified small RNA-seq method that reveals
circulating mRNA and lncRNA fragments as potential biomarkers in
human plasma. EMBO J. 38:e1016952019. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Harharan M, Scaria V and Brahmachari SK:
DbSMR: A novel resource of genome-wide SNPs affecting microRNA
mediated regulation. BMC Bioinformatics. 10:1082009. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Belter A, Gudanis D, Rolle K, Piwecka M,
Gdaniec Z, Naskręt-Barciszewska MZ and Barciszewski J: Mature
miRNAs form secondary structure, which suggests their function
beyond RISC. PLoS One. 9:e1138482014. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Du K, Liu J, Broering R, Zhang X, Yang D,
Dittmer U and Lu M: Recent advances in the discovery and
development of TLR ligands as novel therapeutics for chronic HBV
and HIV infections. Expert. Opin. Drug Discov. 13:661–670.
2018.
|
|
133
|
Christensen S, Shupe J, Nickerson K,
Kashgarian M, Flavell R and Shlomchik M: Toll-like receptor 7 and
TLR9 dictate autoantibody specificity and have opposing
inflammatory and regulatory roles in a murine model of lupus.
Immunity. 25:417–428. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Cheng Z, Lin P and Cheng N: HBV/HIV
coinfection: Impact on the development and clinical treatment of
liver diseases. Front Med (Lausanne). 8:7139812021. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Mullins SR, Vasilakos JP, Deschler K,
Grigsby I, Gillis P, John J, Elder MJ, Swales J, Timosenko E,
Cooper Z, et al: Intratumoral immunotherapy with TLR7/8 agonist
MEDI9197 modulates the tumor microenvironment leading to enhanced
activity when combined with other immunotherapies. J Immunother
Cancer. 7:2442019. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Siu L, Brody J, Gupta S, Marabelle A,
Jimeno A, Munster P, Grilley-Olson J, Rook AH, Hollebecque A, Wong
RKS, et al: Safety and clinical activity of intratumoral MEDI9197
alone and in combination with durvalumab and/or palliative
radiation therapy in patients with advanced solid tumors. J
Immunother Cancer. 8:e0010952020. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Baba A, Wakao M, Shinchi H, Chan M,
Hayashi T, Yao S, Cottam HB, Carson DA and Suda Y: Synthesis and
immunostimulatory activity of sugar-conjugated TLR7 ligands. Bioorg
Med Chem Lett. 30:1268402020. View Article : Google Scholar : PubMed/NCBI
|
















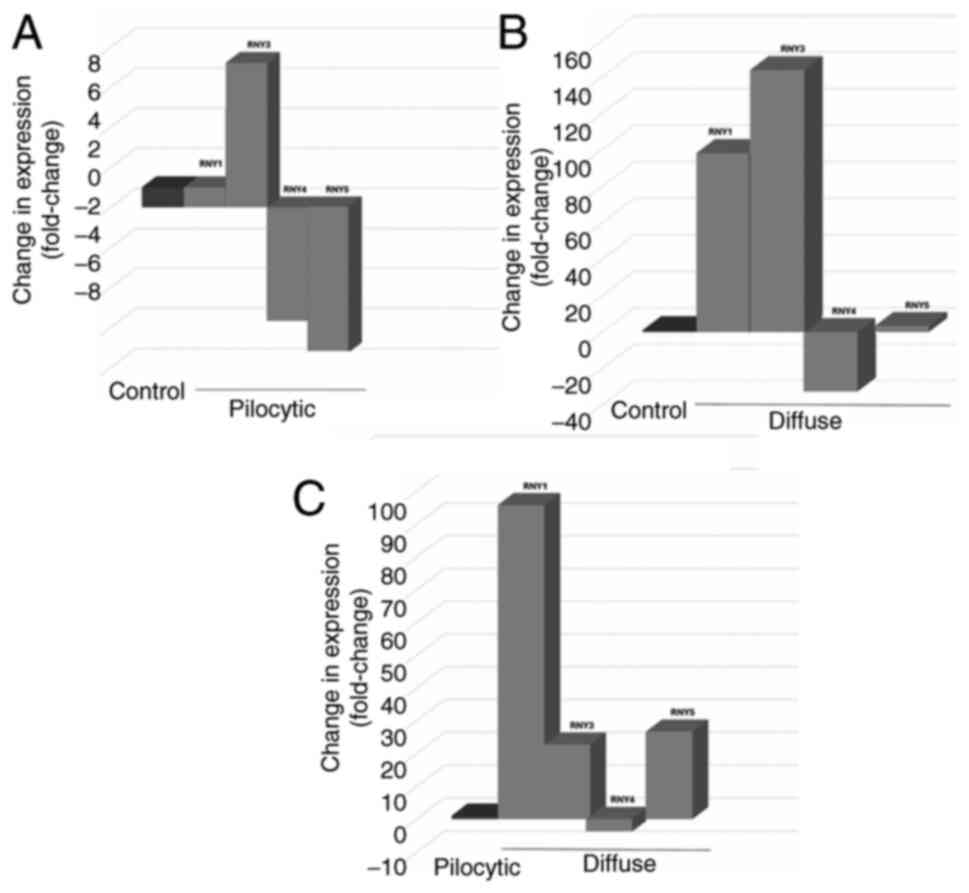
![Predicted functions for circulating
RNYs in pediatric patients with astrocytoma. Differential
expression of the four human RNYs was detected in exosomes of
pediatric patients with astrocytoma. Bioinformatic analyses
indicate that RNYs potentially interact with various protein
receptors. The establishment of protein-protein interaction
networks for these receptors [HIPPIE database
(cbdm-01.zdv.uni-mainz.de/~mschaefer/hippie)] and subsequent
analysis with the PANTHER database (pantherdb.org) showed the
potential involvement of RNYs-receptors in the control of
apoptosis, angiogenesis, cholecystokinin receptor and p53 signaling
pathways, and in Parkinson's disease. RNYs are a class of
non-coding RNAs that are components of the Ro60 ribonucleoprotein
particle. Data obtained from Rodríguez-Corona et al
(10). The data were analyzed with
the Hippie and Panther databases. Interactome images were obtained
from the Hippie database.](/article_images/mmr/32/4/mmr-32-04-13650-g02.jpg)