Introduction
The human placenta is a transient foetal organ
during pregnancy which is responsible for multiple vital functions,
including the transfer of nutrients/factors/gases between the
mother and the foetus, the removal of waste products from the
foetus, and immunoprotection for the foetus (1,2).
Among these functions, the placenta also acts as an endocrine
organ, secreting a number of key hormones (e.g., steroids) for the
maintenance of pregnancy and foetal development (1,2).
Asprosin was identified by Romere et al
(3) during investigation of
Neonatal Progeroid Syndrome (NPS), showing that its pathogenesis is
due to premature ablation of profibrillin-1 (pro-FBN1) by furin
(3). Patients with NPS have a
unique phenotype characterised by extreme leanness, low appetite,
lipodystrophy and insulin sensitivity. Asprosin is implicated in a
number of physiologic processes and is perhaps most known for its
glucogenic function, namely stimulating hepatic glucose secretion
via the G-protein-cAMP-PKA pathway (4). The receptor by which asprosin carries
out its glucogenic function has been established to be the mouse
olfactory receptor OLFR743, whilst in humans asprosin is thought to
act via the ortholog OR4M1 (4,5).
Asprosin has also been found to induce pro-inflammatory effects in
both skeletal muscle and pancreatic β cells, as well as in
macrophages, inducing the expression and secretion of
pro-inflammatory mediators, such as tumour necrosis factor α
(TNFα), and interleukins IL-1β, IL-8 and IL-12 (4,6,7).
Asprosin exerts these pro-inflammatory effects via Toll-like
receptor 4 (TLR4), Jun N-terminal kinase (JNK) and nuclear
factor-kappa B (NFκB) pathways (4,6,7).
Moreover, asprosin exerts orexigenic effects in
relation to its role in regulating appetite (4). In terms of its orexigenic function,
asprosin is able to cross the blood brain barrier activating
agouti-related protein neurons in the hypothalamus. This role is
modulated via the same G-protein-cAMP-PKA pathway, although here it
is thought to be through a different cell surface receptor, namely
the protein tyrosine phosphatase receptor type D (PTPRD) (8,9).
Mishra et al (8) showed
that genetic ablation of this ligand in a mouse model resulted in
strong loss of appetite, leanness and lack of response to
asprosin's orexigenic effects (8).
Notably, emerging studies point towards a role for asprosin in
reproduction. For example, asprosin levels are elevated in women
with polycystic ovary syndrome (PCOS) (10), gestational diabetes mellitus (GDM)
(11), and preeclampsia (12). Asprosin has also been shown to
induce markers for ovarian folliculogenesis and steroidogenesis in
a mouse model (13). Given the
increasing evidence on the pleiotropic effects/roles of asprosin,
in the present study, we investigated the potential role of
asprosin in relation to placental cells in vitro, by
assessing its effects on the transcriptome of BeWo and JEG-3
placental cell lines. We have also expanded on our observations by
measuring the expression of FBN1/Furin and asprosin's candidate
receptors in normal placentas and comparing these to placentas from
GDM pregnancies.
Materials and methods
Tissue culture
To study placental function in vitro, we used
two established cell lines, namely the BeWo cell line which
secretes hormones and undergoes syncytialisation after treatment
with forskolin, and the JEG-3 cell line which does not undergo
substantial fusion (14). BeWo
cells were cultured using Dulbecco's Modified Eagle's Medium (DMEM)
Ham's F12 (Sigma Aldrich D8437) supplemented with 10% foetal Bovine
Serum (Sigma Aldrich F6765) and 1% penicillin-streptomycin (Gibco
15140122) at 37°C with 5% CO2. JEG-3 cells were cultured
using Eagle's minimum essential medium (EBSS) with 2 mM Glutamine,
1% Non-Essential Amino Acids (NEAAs), 1 mM Sodium Pyruvate (NaP),
10% Foetal Bovine Serum (Sigma Aldrich F6765), and 1%
penicillin-streptomycin (Gibco 15140122) at 37°C with 5%
CO2. Both cell lines were treated with 10 nM Recombinant
Human Asprosin (BioLegend, 761904) for 4 h. A wound healing assay
was also performed, as previously described (15).
RNA isolation and RNA sequencing
Total RNA was extracted from cell lysates using a
Trizol/phenol-chlorophorm based method. Sample purity was assessed
using Nano Drop 2000C (Thermo Fisher Scientific, Inc.). Only
samples with a ratio of absorbance A260/A280 between 1.8 and 2.1
were used. Triplicate samples were sequenced using the Illumina
NextSeq 500/550 Mid Output kit V2.5 (in-house sequencing unit).
Data were de-multiplexed and aligned to the human genome.
Expression data was analysed using R (v.4.2.3, The R Foundation for
Statistical Computinga), with the R studio desktop application
(RStudio) and with the use of specific packages; DSeq2 (v1.44),
pheatmap and ggplot2. For visualisation, volcano plots were
generated using R package ggplot2 (v.3.5.1). Differentially
expressed genes (DEGs) were identified for subsequent enrichment
analysis.
Gene expression/gene ontology
analysis
The identified DEGs, were then subjected to
functional enrichment analysis. Funrich (v3.1.3) (16) was accessed to provide a functional
annotation, including biological processes, pathways and molecular
functions. Enrichment analysis was also performed using Omics
playground (v3.44, BigOmics Analytics) (17) for the function comparison of the
genes in asprosin treated versus untreated BeWo and JEG-3
cells.
Human placental samples
Human placental tissue samples from patients with
GDM (n=4; four experimental replicates per sample, i.e., n=16), as
well as normal healthy placenta control samples (n=4; three
experimental replicates per sample, i.e., n=12) were obtained from
the Arden Tissue Bank at the University Hospital Coventry and
Warwickshire (UHCW) NHS Trust (ethical approval obtained by the
Arden Tissue Bank management committee and by the UHCW ethics
committee; NRES 18/SC/0180). Patient consent to participate in the
study and use their tissues was obtained, as specified in the
Declaration of Helsinki.
Fresh human placental tissue samples were collected
on the same day as the delivery/surgery on wet ice in 50 ml Falcon™
tubes full of RNAlater™ stabilization solution (Thermo Fisher
Scientific, Inc.). Samples were stored at 4°C until further
processing. Paraffin embedded human placental tissue slides were
also obtained from the Arden Tissue Bank at UHCW which were
collected on the same day as the delivery/surgery and were stored
short-term at room temperature until processing.
Isolation of mRNA, cDNA synthesis and
RT-qPCR
Total RNA was extracted from human placental tissue
using the Qiagen RNEasy Plus Mini Kit®. Sample purity
was assessed using Nano Drop 2000C (Thermo Fisher Scientific,
Inc.). Only samples with a ratio of A260/A280 between 1.8 and 2.1
were used. cDNA was synthesized using the High-Capacity cDNA
Reverse-Transcription Kit (Applied Biosystems™) according to the
manufacturer's protocol. The cDNA samples were diluted to 10 ng/µl
RNA with nuclease free water and stored at −20°C until further
use.
SYBR Green-based assays
Exploring the expression of a number of genes in
both normal healthy and GDM human placental cDNA samples. Primers
were obtained from Harvard Primer Bank and RT-qPCR was run using
PowerUp SYBR® Green Master Mix. RT-qPCR was performed in
triplicate using the primers included in Table I.
 | Table I.List of primers used for reverse
transcription-quantitative PCR in human placenta samples. |
Table I.
List of primers used for reverse
transcription-quantitative PCR in human placenta samples.
| Name | Amplicon Size,
bp | Strand | Size (bases) | Sequence
(5′-3′) |
|---|
| Furin | 108 | Forward | 22 |
TCGGGGACTATTACCACTTCTG |
|
|
| Reverse | 20 |
CCAGCCACTGTACTTGAGGC |
| PTPRDδ | 105 | Forward | 21 |
CAGGCGGAAGCGTTAATATCA |
|
|
| Reverse | 23 |
TTGGCATATCATCTTCAGGTGTC |
| OR4M1 | 100 | Forward | 23 |
TCTGTTAATGTCCTATGCCTTCC |
|
|
| Reverse | 20 |
AATGTGGGAATAGCAGGTGG |
| TLR4 | 94 | Forward | 23 |
AGTTGATCTACCAAGCCTTGAGT |
|
|
| Reverse | 23 |
GCTGGTTGTCCCAAAATCACTTT |
| FBN1 | 166 | Forward | 20 |
TTTAGCGTCCTACACGAGCC |
|
|
| Reverse | 21 |
CCATCCAGGGCAACAGTAAGC |
| Beta Actin | 140 | Forward | 20 |
CTGGAACGGTGAAGGTGACA |
|
|
| Reverse | 23 |
AAGGGACTTCCTGTAACAATGCA |
Taqman based-assays
For the purpose of validating the expression of the
top genes identified from RNA sequencing, RT-qPCR was performed in
triplicate using TaqMan™ Gene Expression Assays (Applied
Biosystems) and TaqMan™ Fast Advanced MasterMix (Applied
Biosystems). Details on the genes and assay IDs are presented in
Table II. All RT-qPCR experiments
were carried out on a QuantStudio™ 5 Real-Time PCR System, 96-well
(Thermo Fisher Scientific, Inc.). Amplicon load was measured by
relative quantification using a ΔΔCq method.
 | Table II.List of primers used for RNA
sequencing validation. |
Table II.
List of primers used for RNA
sequencing validation.
| Gene ID | Gene name | Amplicon Size,
bp | Assay ID |
|---|
| DDIT4 | DNA damage
inducible transcript 4 | 68 | Hs01111686_g1 |
| HK2 | Hexokinase II | 149 | Hs00606086_m1 |
| STC2 | Stanniocalcin
2 | 93 | Hs01063215_m1 |
| SLC2A1 | Solute carrier
family 2 member 1/glucose transporter 1 | 76 | Hs00892681_m1 |
| ZNF395 | Zinc finger protein
395 | 97 | Hs00608626_m1 |
| GAPDH |
Glyceraldehyde-3-phosphate
dehydrogenase | 157 | Hs02786624_g1 |
In silico analysis
ThemiRDB, TargetScan, and ENCORI databases were
utilized to identify functional miRNA:TLR4 interactions
(18–20). Following this, the results from
these databases were plotted in a Venn diagram using FunRich
(21) to identify the more
efficacious mRNA target interactions. The database STRING (22) was used to identify the top
co-expressed genes with TLR4.
Statistical analysis
Differences identified in experiments were assessed
for statistical significance using the unpaired Student's t-test.
An assessment for homoscedasticity of data for each data set was
made using the F-test. If homoscedasticity was proven, an unpaired
Student's t-test was performed to assess significance. If the data
were not determined to be homoscedastic, an unpaired Student's
t-test with Welch's correction was performed to account for the
variance. All statistical tests were performed using GraphPad
Prism® software (GraphPad Software, Inc.). P-values
<0.05 were considered significant.
Results
Effect of asprosin on BeWo and JEG-3
cells
To gain a better insight into its role in the human
placental transcriptome, BeWo and JEG-3 cells were treated with
asprosin (10 nM for 4 h). RNA sequencing revealed that asprosin
induced cell specific differential expression for 51 genes (DEGs)
in BeWo cells, and 204 in JEG-3 cells, with nine common DEGs in
both in vitro models (Fig.
1A; Tables SI and SII). A Uniform Manifold Approximation
and Projection (UMAP) was constructed to display the up (red) and
down-regulated (blue) genes for BeWo and JEG-3 asprosin-treated and
control (i.e. untreated) samples (Fig. 1B,C). The results show a contrast
between the two cell lines in terms of treated and control samples
(see volcano plots, Fig. 1D). We
have used Taqman probes for validation of RNAseq data, for two up-
and down-regulated genes in BeWo (Fig.
1E), and JEG-3 (Fig. 1F)
cells, respectively. Despite the lack of significance in BeWo
asprosin-treated cells, a trend for upregulation of ZNF395
and DDIT4 was noted. In JEG-3 cells, both SLC2A1 and DDIT4
were significantly downregulated following treatment with asprosin
(P<0.001 and P<0.05 respectively).
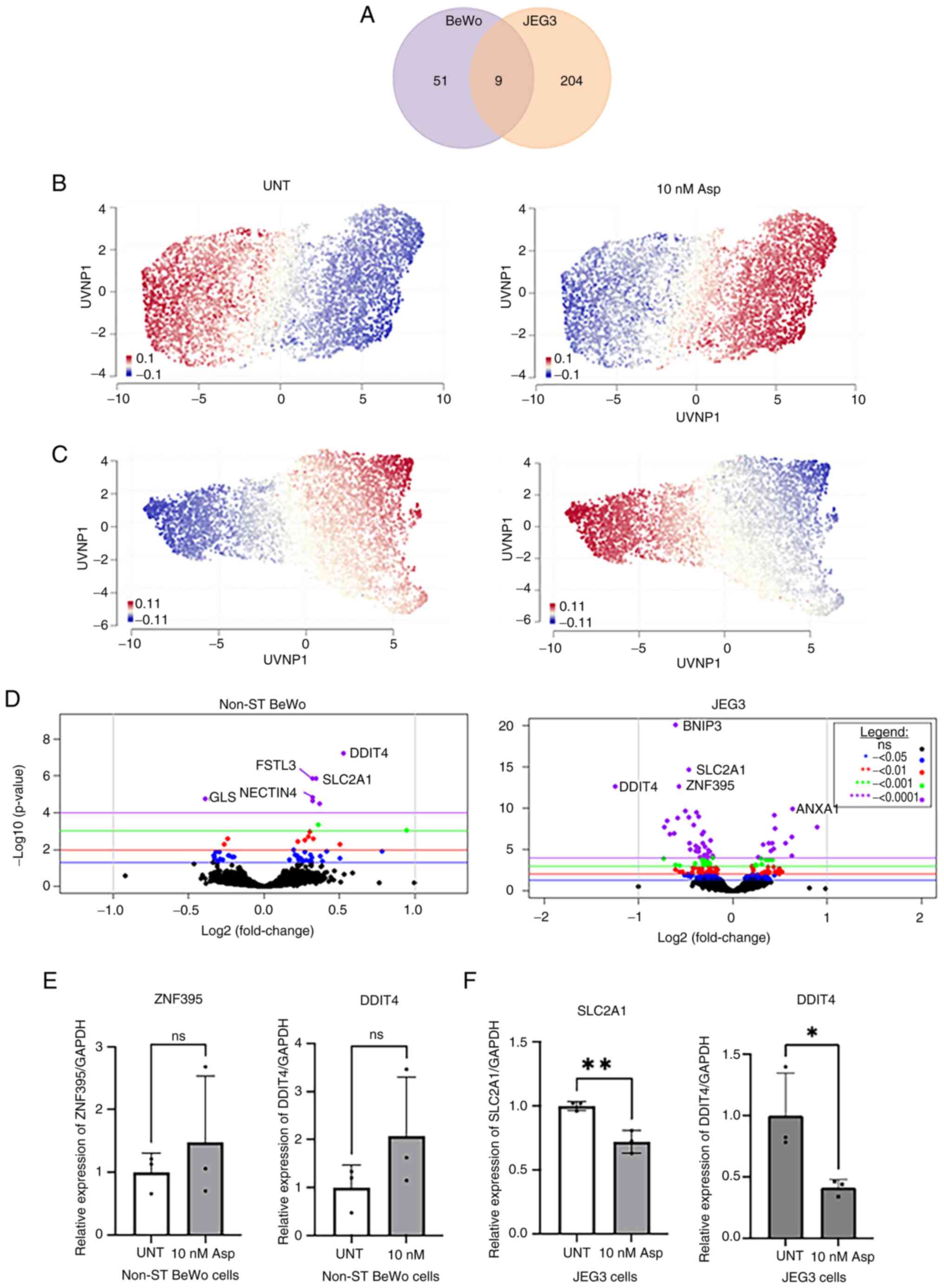 | Figure 1.DEGs in BeWo and JEG-3 cells treated
with Asp. (A) A Venn diagram displaying the DEGs for Asp-treated
BeWo and JEG-3 cells when compared with their respective UNT
controls. (B and C) Geneset signature UMAP displaying up- and
down-regulated genes in different samples. UMAPs display genes
clustered by relative log-expression which were up- (red) or down-
(blue) regulated in (B) BeWo and (C) JEG-3 cells, UNT control and
10 nM Asp-treated samples. (D) Volcano plots for BeWo and JEG-3
cells, indicating the most up- and down-regulated DEGs. RNA
sequencing validation for (E) BeWo and (F) JEG-3 cells, using
reverse transcription-quantitative PCR. *P<0.05, **P<0.001.
DEGs, differentially expressed genes; UMAP, uniform manifold
approximation and projection; UNT, untreated; Asp, asprosin;
ZNF395, Zinc Finger Protein 395; DDIT4, DNA Damage Inducible
Transcript 4; SLC2A1, solute carrier family 2 member 1; Non-ST,
non-syncytialised; ns, not significant. |
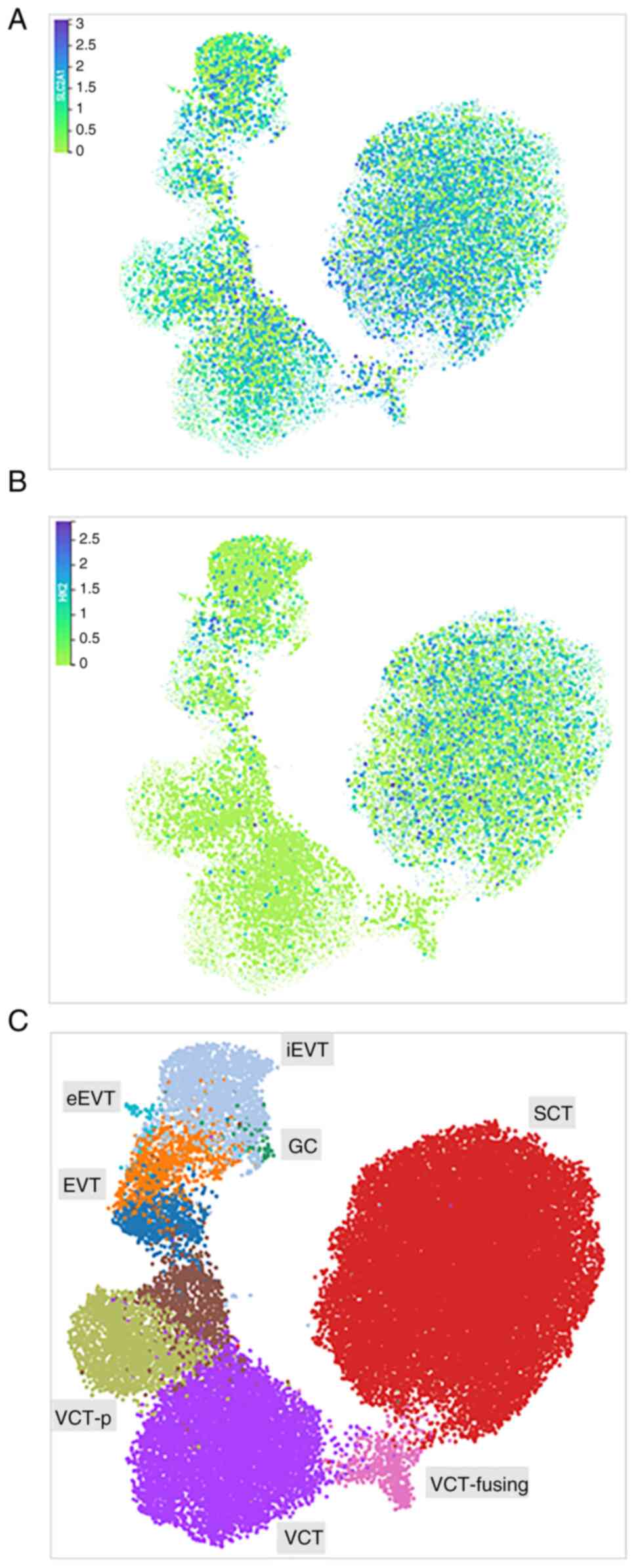
We have used a spatially resolved single-cell
multiomic characterization of the maternal-foetal interface
(reproductivecellatlas.org) (23)
to map the expression of SLCA1 and HK2 in a diverse trophoblast
population (Fig. 2). SLCA1 is
abundantly expressed in villous syncytiotrophoblasts (SCTs),
extravillous trophoblast cells (EVTs), and villous cytotrophoblast
cells (VCTs), as well as in placenta giant cells (GCs) (Fig. 2A and C). HK2 expression was
primarily confound in SCTs (Fig. 2B
and C).
Heat map of top 50 DEGs identifies
four different clusters in both asprosin-treated cell lines
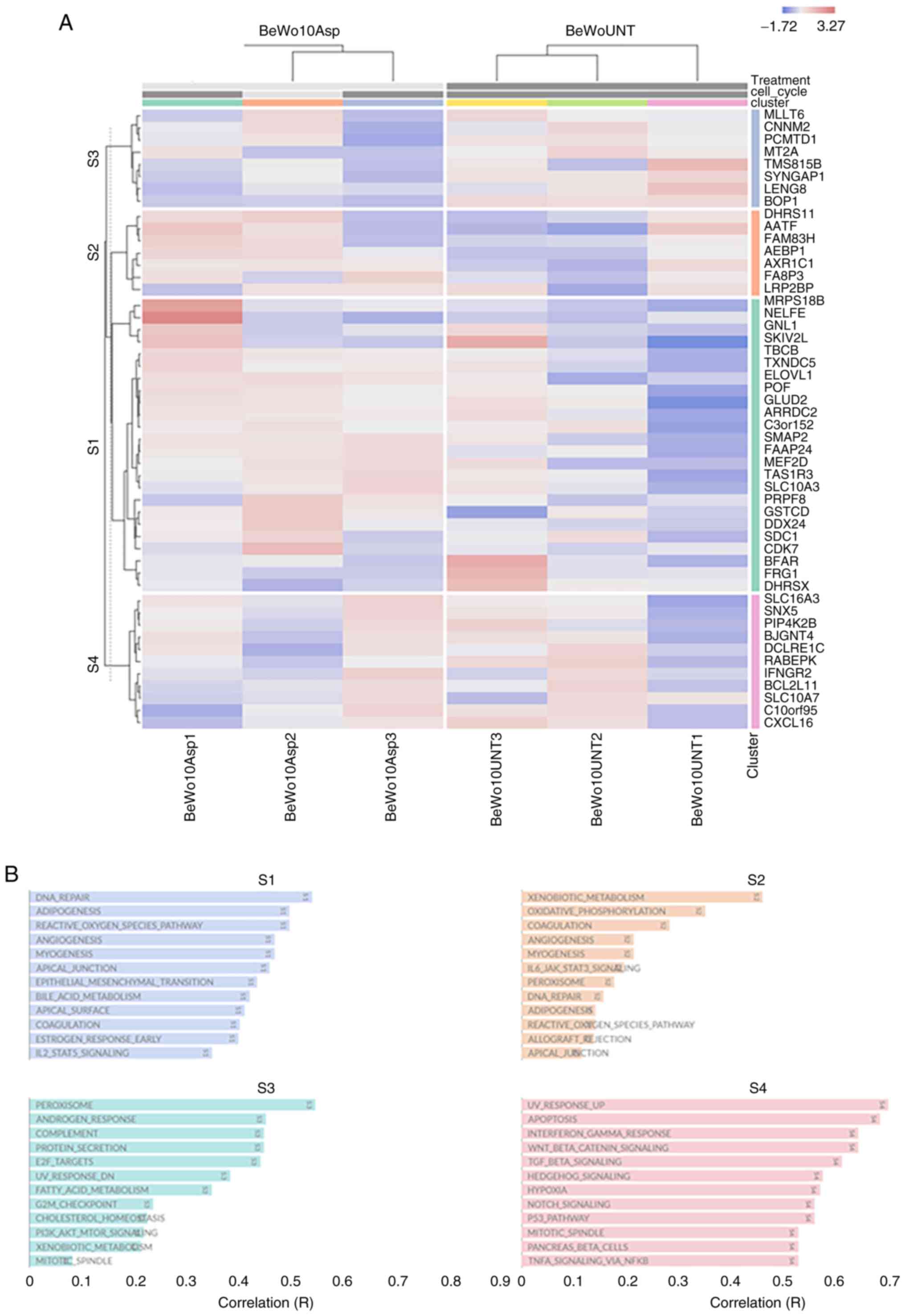
Fig. 3A presents
the functional heat map of the top 50 DEGs with highest standard
deviation across all samples (BeWo-treated vs. control). The
hierarchical clustering was performed at the gene level and
showcased four clusters S1-S4 (Fig.
3B). Many of these functional annotations are related to DNA
Repair, angiogenesis, fatty acid metabolism, mTOR/NOTCH/WNT/p53
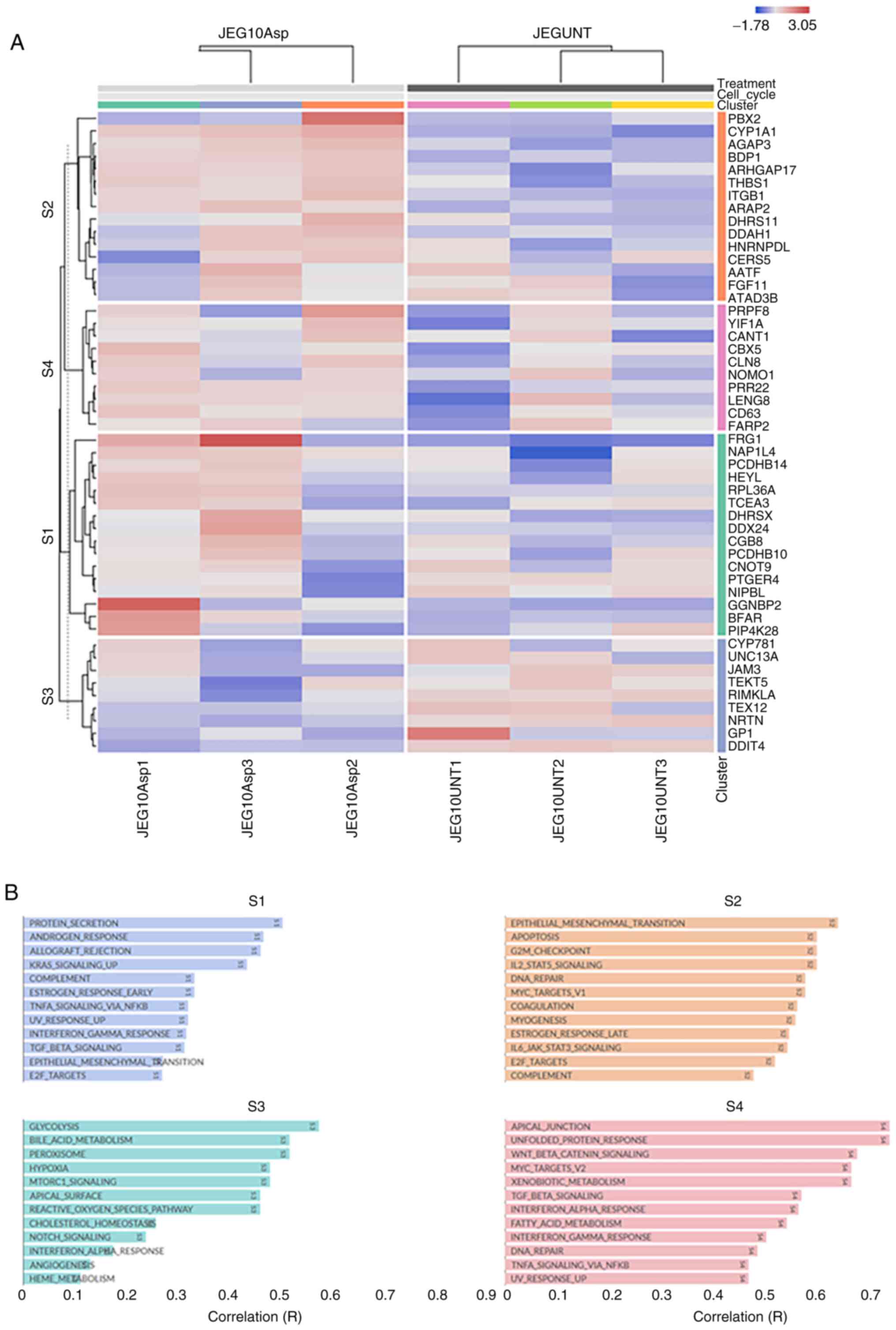
signalling. Similarly, four distinct clusters were identified in
JEG-3 treated samples (Fig. 4A),
including changes in protein secretion, glycolysis,
TGFβ/TNFα/KRAS/L2 signalling, hypoxia and steroid response
(Fig. 4B).
Gene enrichment analysis
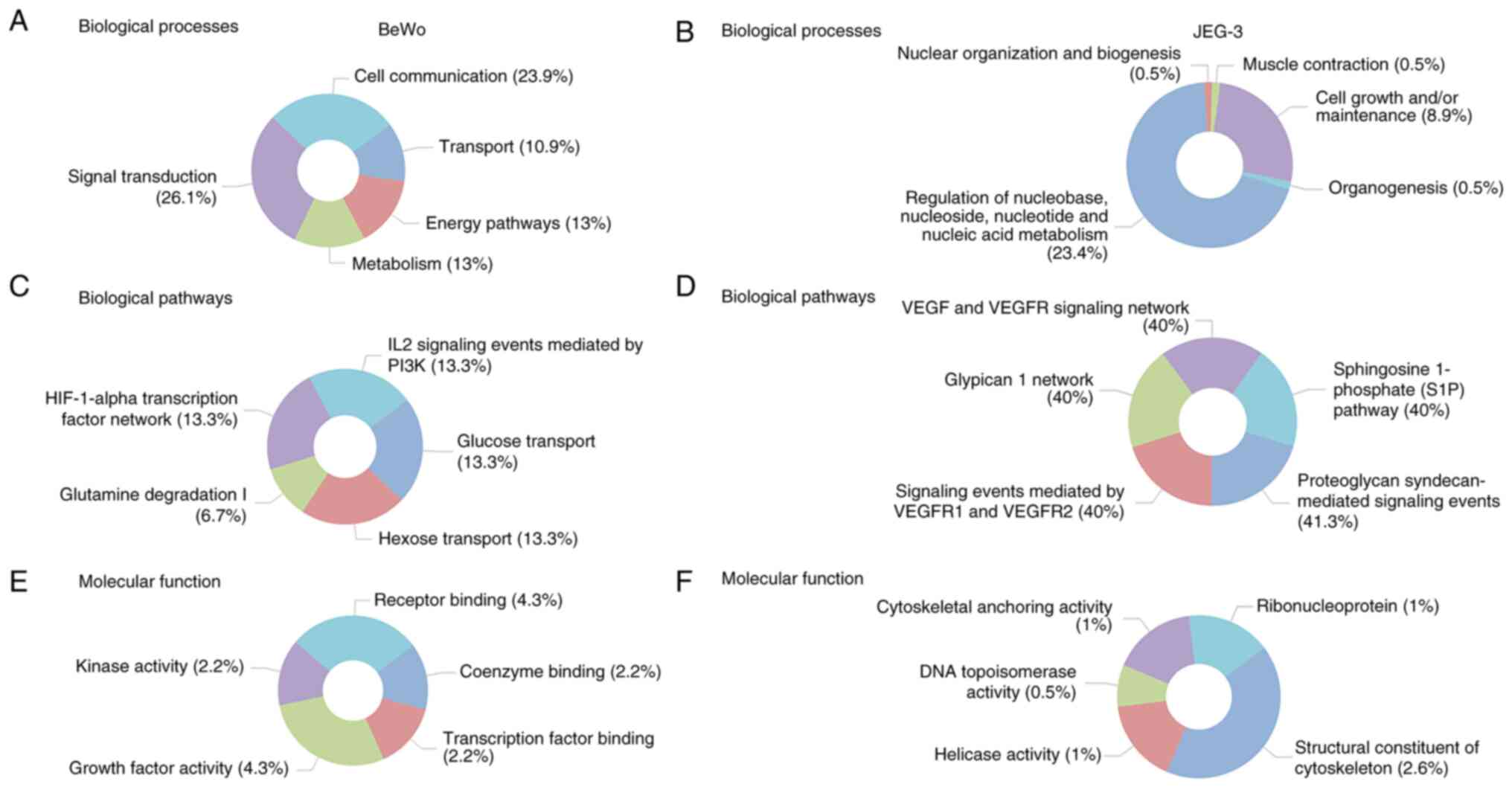
Funrich was used to determine the top five enriched
biological processes (Fig. 5A),
biological pathways (Fig. 5B), and
molecular functions (Fig. 5C) in
BeWo cells treated with asprosin. The main biological processes
identified as: signal transduction, cell communication, energy
pathways/metabolism, and transport. Glucose/Hexose transport were
two of the five biological pathways identified, underpinning the
role of asprosin in cell homeostasis. Enriched molecular functions
included receptor binding, growth factor and kinase activity.
Similar analyses were performed for the JEG-3
related DEGs, identifying the top five enriched biological
processes (Fig. 5D), biological
pathways (Fig. 5E), and molecular
functions (Fig. 5F). Most enriched
biological processes included regulation of nucleic acid
metabolism, and cell maintenance, whereas vascular endothelial
growth factor receptor (VEGFR) signalling predominated under
biological pathways. Contrary to BeWo cells, structural constituent
of the cytoskeleton was the most enriched molecular function in
JEG-3 cells. All p-values for the top five enriched biological
pathways, biological processes, and molecular pathways (depicted in
Fig. 5) are presented in Fig. S1.
Furthermore, we have assessed asprosin's role in
cell proliferation and migration in vitro, using BeWo cells.
When cells were treated with asprosin, no apparent differences were
observed at 24 or 48 h post asprosin treatment (Fig. S2A). The performed wound healing
assay at the same time-points indicated a potential cytostatic
effect for asprosin (Fig. S2B and
S2C).
Expression of FBN1, Furin, OR4M1,
PTPRD and TLR4 in placentas from normal and GDM pregnancies
Expression of FBN1, Furin and putative asprosin
receptors was assessed in normal and GDM placentas at term. There
was no difference in the gene expression of either FBN1 (Fig. 6A) or Furin (Fig. 6B) between these two groups. Similar
expression of OR4M1 (Fig. 6C), and
PTPRD (Fig. 6D) was also noted
between normal and GDM samples, whereas TLR4 expression (Fig. 6E) was significantly downregulated
in GDM placentas compared to the controls (P<0.0001).
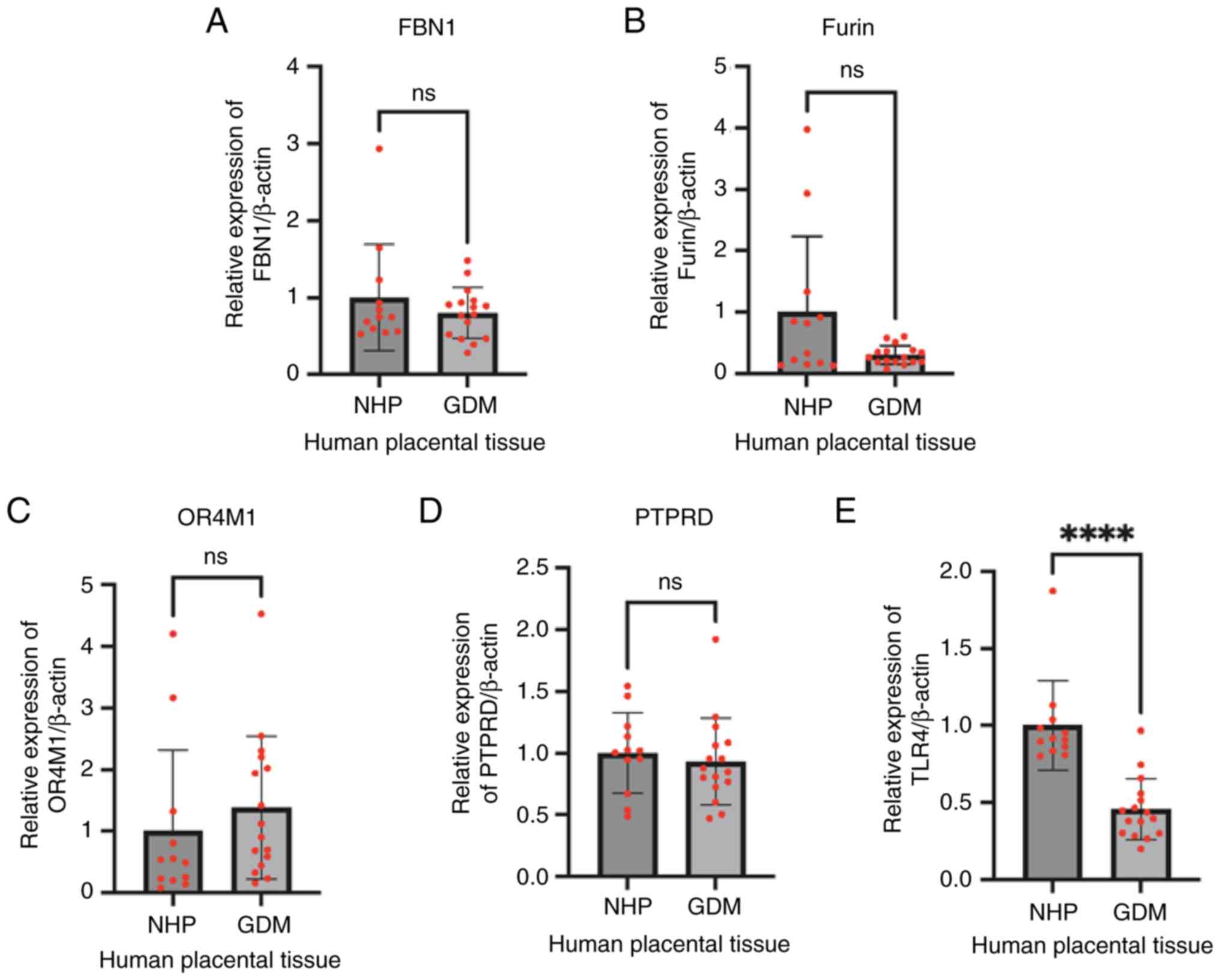 | Figure 6.Gene expression of (A) FBN1, (B)
Furin, (C) OR4M1, (D) PTPRD and (E) TLR4 in NHP (control) and GDM
placenta samples, as assessed by reverse transcription-quantitative
PCR. ****P<0.0001. FBN1, fibrillin-1; OR4M1, olfactory receptor
4M1; PTPRD, protein tyrosine phosphatase receptor type D; GDM,
gestational diabetes mellitus; NHP, normal human placenta; ns, not
significant. |
Identification of miRNA:mRNA
interactions and investigation of TLR4 effects in signalling
pathways
Investigation of three predictive miRNA databases
was undertaken, namely miRDB, TargetScan, and ENCORI, which
identified 95, 1, and 91 interactions, respectively (Fig. S3A). The results showed that there
were 10 common miRNAs between ENCORI and miRDB targeting TLR4, and
only one common miRNA between miRDB and TargetScan. The 10 common
miRNAs are: hsa-miR-448, hsa-miR-642a-5p, hsa-miR-7-5p,
hsa-miR-25-3p, hsa-miR-367-3p, hsa-miR-363-3p, hsa-miR-92a-3p,
hsa-miR-92b-3p, hsa-miR-32-5p, and hsa-miR-655-3p. The one common
miRNA is hsa-miR-140-5p.
Given the most notable changes in receptor
expression rest with TLR4, we have performed STRING analysis, and
we have identified candidate proteins which have a cross-talk with
TLR4. The proteins are: TICAM1, TICAM2, IRAK4, TRAF6, TIRAP, TLR2,
LY96, TLR6, HSPD1, and HMGB1. With the exemption of TLR2, all other
interactions are experimentally determined (Fig. S3B).
Discussion
In the present study, we provide evidence of how
asprosin can change the placental transcriptome using two
well-characterised in vitro models. We have also measured
the expression of FBN1, the proteolytic enzyme furin, as well as
asprosin's putative receptors (OR4M1, PTPRD and TLR4) (9) in healthy (normal pregnancy) and GDM
placentas.
Asprosin treatment altered almost 4-fold more genes
in JEG-3 cells compared to BeWo cells, indicating their inherent
transcriptomic differences, as previously described (24). Of note, nine genes were similarly
affected in both these cell lines, namely SLC2A1, ZNF395, DDIT4,
HK2, STC2, RGS16, SH3PDXD2B, XYLT1 and CENPF. SLC2A1 encodes a
major placental glucose transporter (GLUT1), increases in
expression with gestation, and facilitates glucose uptake (25). Similarly, asprosin also affected
the expression of Hexokinase 2 (HK2), an enzyme which
phosphorylates glucose to glucose-6-phosphate, the first step in
most glucose metabolism pathways (26). Both GLUT1 and HK2 appear to be
upregulated in patients with GDM (27). Another link between asprosin and
glucose is suggested by the differential regulation of
stanniocalcin-2 (STC2), a gene which is also upregulated in GDM
placentas and inhibits trophoblast invasion under high-glucose
conditions (28). DNA Damage
Inducible Transcript 4 (DDIT4) has also been affected by asprosin.
This is also a crucial signalling pathway in the human placenta
which regulates cell proliferation by inhibiting the activity of
the mechanistic target of rapamycin (mTOR) (29,30).
Notably, it has been suggested that DDIT4 is critical for normal
decidualization and possibly involved in the development of
preeclampsia (31). For the
remaining genes (ZNF395, RGS16, SH3PDXD2B, XYLT1, CENPF) no data
are available for specific functions in the human placenta.
However, Xylosyltransferase 1 (XYLT1) has been described as an
insulin sensitizer (32), whilst
depletion of CENPF disrupts GLUT4 trafficking in murine cells
(33), and RGS16 induces insulin
secretion (34). Collectively,
this is the first time that a direct effect of asprosin as a
glucose sensor on key components, such as SLC2A1, HK2, and STC2,
has been shown at the placental level.
In accordance with their genetic/phenotypic
differences as trophoblastic cell lines, BigOmics analytics
generated four distinct pathway-related clusters for BeWo and JEG-3
cells based on RNAseq data. Despite some overlap, notable pathways
for JEG-3 cells include glycolysis, mTORC1, NOTCH, and KRAS
signalling. Common pathways include steroidal responses,
cholesterol homeostasis, xenobiotic and fatty acid metabolism, as
well as cytokine responses and hypoxia. Glycolysis is an important
process for the maintenance of the homeostasis of the
maternal-foetal interface, as well as ensuring normal gestation
(35). Dysregulation of glycolysis
has attracted interest for its role in pregnancy disorders,
including miscarriage, GDM and preeclampsia. Indeed, Lu et
al (36) have shown that NOTCH
signalling, glycolysis, and hypoxia were the main enriched area for
six preeclampsia-related genes, using the Gene Expression Omnibus
public database (36).
We have also explored DEG enrichment for both cell
lines in terms of the role of the DEGs in biological processes and
pathways, as well as molecular function using FunRich, where a
non-overlapping enrichment emerged when the two cell lines were
compared. In BeWo cells, the most enriched biological process with
mapped genes included GLS, HK2, TMX1, SGMS1, XYLT1 and ILVBL.
Corroborating previous data, glucose transport was the most
enriched biological pathway (SLC2A1, HK2); whereas the most
enriched molecular function was that of growth factor activity,
involving GDF15 and PGRN. Interestingly, low expression of growth
differentiation factor 15 (GDF15) has been associated with impaired
invasion of extravillous trophoblasts and predisposition to
pregnancy loss (37), whereas
progranulin (PGRN) deficient mice developed abnormal placental
angiogenesis (38). In JEG-3
cells, nucleic acid metabolism and VEGF signalling were amongst the
most enriched biological processes and pathways. VEGF is a key
angiogenic factor that affects not only endothelial cells, but also
trophoblasts (39). Impaired
glucose tolerance appears to affect the expression of placental
VEGFRs (40). Contrary to BeWo
cells, ‘structural constituents of the cytoskeleton’ was the most
enriched molecular function in JEG-3 cells. For this function,
mapped genes included DSP, KRT19, ERRFI1, ACTB and ACTR1A; none of
which have any known placental-specific functions assigned.
Of note, HK2 and SLC2A1 expression was noted during
embryo morphogenesis (in the period between implantation and
gastrulation) (41); particularly
in cytotrophoblasts and syncytiotrophoblasts, but not in epiblasts
or hypoblasts (see Fig. S4). The
presence of these genes as early as nine days post-fertilisation,
suggests an important role for embryonic development. Moreover,
when cytotrophoblast cells (BeWo cells) were treated with asprosin
over 48 h, no apparent changes were noted in cell proliferation,
corroborating previous human and animal studies (supplementary
data) (42,43). Future studies should use ex
vivo or in vivo models, or even more comprehensive
clinical studies to confirm the role(s) and mechanism(s) of
asprosin in the overall pregnancy environment.
Following RNA sequencing analysis, we investigated
the expression of FBN1, Furin, OR4M1, PTPRD and TLR4 in healthy and
GDM placentas. Early studies have shown increased plasma asprosin
levels in pregnant women with GDM as early as 18–20 weeks of
gestation (44). More recently,
Boz et al (11) showed that
asprosin levels were elevated in pregnant women with normal glucose
tolerance or with GDM when compared to healthy non-pregnant
controls (11). In the present
study, no difference in the expression of FBN1 and Furin was noted
between GDM and healthy placentas. This suggests that the source of
elevated asprosin in GDM pregnancies is not likely the placenta.
Indeed, GDM is associated with elevated maternal body mass index
(BMI), so it is possible that increased in adiposity drives higher
release of asprosin in circulation. For example, a positive
correlation between placental asprosin immunoreactivity and BMI has
been shown (45). In terms of the
putative asprosin receptors, only TLR4 was significantly
downregulated in GDM placentas in our study. Numerous studies have
shown that TLR4-mediated signalling plays a pivotal role in immune
and inflammatory processes (46).
Moreover, a TLR4/NF-κB/PFKFB3 signalling cascade might provide a
link between glycometabolism and trophoblastic pyroptosis (47). However, a previous study has
reported elevated levels of TLR4 in patients with GDM (48). Thus, more studies are needed to
determine the potential differences in the expression of asprosin
receptors at both gene and protein level in GDM and other pregnancy
complications.
Of note, relating to the identified potential miRNA
interactions with TLR4, there is good evidence for involvement of
hsa-miR-7-5p, hsa-miR-92a-3p, and hsa-miR-32-5p in GDM/obesity. For
example, miRNA 32 and 92a-3p are upregulated in GDM (49,50),
whereas hsa-miR-7-5p expression was reduced at 21 days post
bariatric surgery (51). STRING
motif also revealed interactions implicated in GDM. For example, it
has been suggested that the miR-146a-3p/TRAF6 interaction might
play a key role in the pathogenesis of GDM (52), whereas HMGB1 expression is
increased as a result of tissue damage due to inflammation and
oxidative stress related to GDM (53). Following analyses of cord blood
samples from diabetic and normal pregnancies, it was shown that
maternal diabetes drives a profound inflammatory activation in
neonates that involves TLR1/2 or TRL5 (54). It should be noted that there is a
further interplay between TLR2 and TLR4, since it has been shown
that, under foetal exposure to GDM conditions, TLR4 and TLR2 can
activate IL-1β responses in rat offspring spleen cells (55). This corroborates a previous in
vivo study, where C57BL/6 mice lacking TLR4 (TLR4-knockout,
TLR4−/− mice) were partially protected from high-fat
diet-induced insulin resistance, suggesting that TLR4 acts as
molecular link among pro-inflammatory responses, nutrition, and
lipids (56).
To conclude, the present study provides a novel
insight into the actions of asprosin in two well-established in
vitro placental (trophoblast) models, identifying key genes and
signalling pathways. Based on the present findings, a common theme
that emerged is that of glucose homeostasis, in accordance with the
physiologic role of this adipokine. Future work is needed to
understand the exact role of asprosin in health and disease (e.g.
in GDM) expanding on in vitro models (e.g.,
syncytialised BeWo cells), using primary placental cells, as well
as trying to recapitulate better the placental microenvironment
using 3D cultures.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The General Charities of the City of Coventry (grant no.
RMV1169CSA).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
JC, HSR, IK and EK conceptualised the study. SO, SG
and EK acquired and analysed data. SO, VP, SG, JK, SS, EK, IK and
HSR performed the formal analysis. HSR and IK were responsible for
funding. HSR, IK and EK were involved in the investigation. SO, SG,
SS, VP and EK were involved in the methodology. HSR, IK, EK
administered the project. HSR, EK and IK supervised the study. EK,
SS and SO visualised the data. EK, SO, JC and IK wrote the original
draft. JC, EK, SO, IK, SS and HSR wrote and edited the final draft.
EK and HSR confirm the authenticity of the data. All authors have
read and approved the final manuscript.
Ethics approval and consent to
participate
Human placental tissue samples were obtained from
the Arden Tissue Bank at the University Hospital Coventry and
Warwickshire NHS Trust (ethical approval obtained by the Arden
Tissue Bank management committee and by the UHCW ethics committee;
NRES 18/SC/0180). Patients provided written informed consent to
participate.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Costa J, Mackay R, De Aguiar Greca SC,
Corti A, Silva E, Karteris E and Ahluwalia A: The role of the 3Rs
for understanding and modeling the human placenta. J Clin Med.
10:34442021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Herrick EJ and Bordoni B: Embryology,
placenta. StatPearls (Internet). StatPearls Publishing; Treasure
Island, FL: 2023, https://www.ncbi.nlm.nih.gov/books/NBK551634/November
11–2024
|
|
3
|
Romere C, Duerrschmid C, Bournat J,
Constable P, Jain M, Xia F, Saha PK, Del Solar M, Zhu B, York B, et
al: Asprosin, a fasting-induced glucogenic protein hormone. Cell.
165:566–579. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shabir K, Brown JE, Afzal I, Gharanei S,
Weickert MO, Barber TM, Kyrou I and Randeva HS: Asprosin, a novel
pleiotropic adipokine implicated in fasting and obesity-related
cardio-metabolic disease: Comprehensive review of preclinical and
clinical evidence. Cytokine Growth Factor Rev. 60:120–132. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kerslake R, Hall M, Vagnarelli P,
Jeyaneethi J, Randeva H, Pados G, Kyrou I and Karteris E: A
pancancer overview of FBN1, asprosin and its cognate receptor OR4M1
with detailed expression profiling in ovarian cancer. Oncol Lett.
22:6502021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shabir K, Gharanei S, Orton S, Patel V,
Chauhan P, Karteris E, Randeva HS, Brown JE and Kyrou I: Asprosin
exerts pro-inflammatory effects in THP-1 macrophages mediated via
the Toll-like Receptor 4 (TLR4) pathway. Int J Mol Sci. 24:2272022.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lee T, Yun S, Jeong JH and Jung TW:
Asprosin impairs insulin secretion in response to glucose and
viability through TLR4/JNK-mediated inflammation. Mol Cell
Endocrinol. 486:96–104. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mishra I, Xie WR, Bournat JC, He Y, Wang
C, Silva ES, Liu H, Ku Z, Chen Y, Erokwu BO, et al: Protein
tyrosine phosphatase receptor δ serves as the orexigenic asprosin
receptor. Cell Metab. 34:549–563.e8. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Orton S, Karkia R, Mustafov D, Gharanei S,
Braoudaki M, Filipe A, Panfilov S, Saravi S, Khan N, Kyrou I, et
al: In silico and in vitro mapping of receptor-type protein
tyrosine phosphatase receptor type D in health and disease:
Implications for asprosin signalling in endometrial cancer and
neuroblastoma. Cancers (Basel). 16:5822024. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ozturk HA and Arici FN: Achilles tendon
thickness and serum asprosin level significantly increases in
patients with polycystic ovary syndrome. PeerJ. 12:e179052024.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Boz İB, Aytürk Salt S, Salt Ö, Sayın NC
and Dibirdik İ: Association between plasma asprosin levels and
gestational diabetes mellitus. Diabetes Metab Syndr Obes.
16:2515–2521. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shafi N, Bano F and Uraneb S: The role of
novel hormone asprosin in insulin resistance during preeclampsia.
Pak J Pharm Sci. 34 (Suppl 3):1039–1043. 2021.PubMed/NCBI
|
|
13
|
Banerjee A, Vishesh C, Anamika Tripathy M
and Umesh R: Asprosin-mediated regulated of ovarian functions in
mice: An age-dependent study. Peptides. 181:1712932024. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Msheik H, El Hayek S, Bari MF, Azar J,
Abou-Kheir W, Kobeissy F, Vatish M and Daoud G: Transcriptomic
profiling of trophoblast fusion using BeWo and JEG-3 cell lines.
Mol HumReprod. 25:811–824. 2019.
|
|
15
|
Morea A, Saravi S, Sisu C, Hall M, Tosi S,
Karteris E and Storlazzi CT: Effect of MYC and PARP inhibitors in
ovarian cancer using an In-Vitro model. Anticancer Res.
44:1817–1827. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
FunRich, . Functional Enrichment Analysis
Tool. http://www.funrich.org/December
28–2024
|
|
17
|
BigOmics Analytics, . Omics Analysis
Software. BigOmics Analytics SA; Lugano: 2019, https://bigomics.ch/December 28–2024
|
|
18
|
Chen Y and Wang X: miRDB: An online
database for prediction of functional microRNA targets. Nucleic
Acids Res. 48:D127–D131. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42:D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pathan M, Keerthikumar S, Ang CS, Gangoda
L, Quek CY, Williamson NA, Mouradov D, Sieber OM, Simpson RJ, Salim
A, et al: FunRich: An open access standalone functional enrichment
and interaction network analysis tool. Proteomics. 15:2597–2601.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Szklarczyk D, Kirsch R, Koutrouli M,
Nastou K, Mehryary F, Hachilif R, Gable AL, Fang T, Doncheva NT,
Pyysalo S, et al: The STRING database in 2023: Protein-protein
association networks and functional enrichment analyses for any
sequenced genome of interest. Nucleic Acids Res. 51:D638–D646.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Arutyunyan A, Roberts K, Troulé K, Wong
FCK, Sheridan MA, Kats I, Garcia-Alonso L, Velten B, Hoo R,
Ruiz-Morales ER, et al: Spatial multiomics map of trophoblast
development in early pregnancy. Nature. 616:143–151. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Burleigh DW, Kendziorski CM, Choi YJ,
Grindle KM, Grendell RL, Magness RR and Golos TG: Microarray
analysis of BeWo and JEG3 trophoblast cell lines: Identification of
differentially expressed transctripts. Placenta. 28:383–389. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lynch CS, Kennedy VC, Tanner AR, Ali A,
Winger QA, Rozance PJ and Anthony RV: Impact of placental SLC2A3
deficiency during the first-half of gestation. Int J Mol Sci.
23:125302022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tan VP and Miyamoto S: HK2/hexokinase-II
integrates glycolysis and autophagy to confer cellular protection.
Autophagy. 11:963–964. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Song TR, Su GD, Chi YL, Wu T, Xu Y and
Chen CC: Dysregu-lated miRNAs contribute to altered placental
glucose metabolism in patients with gestational diabetes via
targeting GLUT1 and HK2. Placenta. 105:14–22. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lai R, Ji L, Zhang X, Xu Y, Zhong Y, Chen
L, Hu H and Wang L: Stanniocalcin2 inhibits the
epithelial-mesenchymal transition and invasion of trophoblasts via
activation of autophagy under high-glucose conditions. Mol Cell
Endocrinol. 547:1115982022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mparmpakas D, Zachariades E, Goumenou A,
Gidron Y and Karteris E: Placental DEPTOR as a stress sensor during
pregnancy. Clin Sci (Lond). 122:349–359. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mparmpakas D, Zachariades E, Foster H,
Kara A, Harvey A, Goumenou A and Karteris E: Expression of mTOR and
downstream signalling components in the JEG-3 and BeWo human
placental choriocarcinoma cell lines. Int J Mol Med. 25:65–69.
2010.PubMed/NCBI
|
|
31
|
Yang J, Zhang Y, Tong J, Lv H, Zhang C and
Chen ZJ: Dysfunction of DNA damage-inducible transcript 4 in the
decidua is relevant to the pathogenesis of preeclampsia. Biol
Reprod. 98:821–833. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Orioli L, Canouil M, Sawadogo K, Ning L,
Deldicque L, Lause P, de Barsy M, Froguel P, Loumaye A, Deswysen Y,
et al: Identification of myokines susceptible to improve glucose
homeostasis after bariatric surgery. Eur J Endocrinol. 189:409–421.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pooley RD, Moynihan KL, Soukoulis V, Reddy
S, Francis R, Lo C, Ma LJ and Bader DM: Murine CENPF interacts with
syntaxin 4 in the regulation of vesicular transport. J Cell Sci.
121:3413–3421. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Vivot K, Moullé VS, Zarrouki B, Tremblay
C, Mancini AD, Maachi H, Ghislain J and Poitout V: The regulator of
G-protein signaling RGS16 promotes insulin secretion and β-cell
proliferation in rodent and human islets. Mol Metab. 26:988–996.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gou R and Zhang X: Glycolysis: A fork in
the path of normal and pathological pregnancy. FASEB J.
37:e232632023. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lu X, Lan X, Fu X, Li J, Wu M, Xiao L and
Zeng Y: Screening preeclampsia genes and the effects of CITED2 on
trophoblastic function. Int J Gen Med. 17:3493–3509. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lyu C, Ni T, Guo Y, Zhou T, Chen ZJ, Yan J
and Li Y: Insufficient GDF15 expression predisposes women to
unexplained recurrent pregnancy loss by impairing extravillous
trophoblast invasion. Cell Prolif. 56:e135142023. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xu B, Chen X, Ding Y, Chen C, Liu T and
Zhang H: Abnormal angiogenesis of placenta in progranulin-deficient
mice. Mol Med Rep. 22:3482–3492. 2020.PubMed/NCBI
|
|
39
|
Shibuya M: Vascular endothelial growth
factor (VEGF) and its receptor (VEGFR) signaling in angiogenesis: A
crucial target for anti- and pro-angiogenic therapies. Genes
Cancer. 2:1097–1105. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Marini M, Vichi D, Toscano A, Thyrion GD,
Bonaccini L, Parretti E, Gheri G, Pacini A and Sgambati E: Effect
of impaired glucose tolerance during pregnancy on the expression of
VEGF receptors in human placenta. Reprod Fertil Dev. 20:789–801.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Molè MA, Coorens THH, Shahbazi MN,
Weberling A, Weatherbee BAT, Gantner CW, Sancho-Serra C, Richardson
L, Drinkwater A, Syed N, et al: A single cell characterisation of
human embryogenesis identifies pluripotency transitions and
putative anterior hypoblast centre. Nat Commun. 12:36792021.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang Z, Tan Y, Zhu L, Zhang B, Feng P,
Gao E, Xu C, Wang X, Yi W and Sun Y: Asprosin improves the survival
of mesenchymal stromal cells in myocardial infarction by inhibiting
apoptosis via the activated ERK1/2-SOD2 pathway. Life Sci.
231:1165542019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lu Y, Liu C, Pang X, Chen X, Wang C and
Huang H: Bioinformatic identification of signature miRNAs
associated with fetoplacental vascular dysfunction in gestational
diabetes mellitus. Biochem Biophys Reps. 41:1018882024.PubMed/NCBI
|
|
44
|
Zhong L, Long Y, Wang S, Lian R, Deng L,
Ye Z, Wang Z and Liu B: Continuous elevation of plasma asprosin in
pregnant women complicated with gestational diabetes mellitus: A
nested case-control study. Placenta. 93:17–22. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hoffmann T, Morcos YAT, Janoschek R,
Turnwald EM, Gerken A, Müller A, Sengle G, Dötsh J, Appel S and
Hucklenbruch-Rother E: Correlation of metabolic characteristics
with maternal, fetal and placental asprosin in human pregnancy.
Endocr Connect. 11:e2200692022. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Barboza R, Lima FA, Reis AS, Murillo OJ,
Peixoto EPM, Bandeira CL, Fotoran WL, Sardinha LR, Wunderlich G,
Bevilacqua E, et al: TLR4-mediated placental pathology and
pregnancy outcome in experimental malaria. Sci Rep. 7:86232017.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang Y, Liu W, Zhong Y, Li Q, Wu M, Yang
L, Liu X and Zou L: Metformin corrects glucose metabolism
reprogramming and NLRP3 inflammasome-induced pyroptosis via
inhibiting the TLR4/NF-κβ/PFKFB3 signaling in trophoblasts:
Implication for a potential therapy of preeclampsia. Oxid Med Cell
Longev. 2021:18063442021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhou J, Bai J, Guo Y, Fu L and Xing J:
Higher levels of triglyceride, fatty acid translocase, and
toll-like receptor 4 and lower level of HDL-C in pregnant women
with GDM and their close correlation with neonatal weight. Gynecol
Obstet Invest. 86:48–54. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Batalha IM, Maylem ERS, Spicer LJ, Pena
Bello CA, Archilia EC and Shütz LF: Effects of asprosin on
estradiol and progesterone secretion and proliferation of bovine
granulosa cells. Mol Cell Endocrinol. 565:1118902023. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Vasu S, Kumano K, Darden CM, Rahman I,
Lawrence MC and Naziruddin B: MicroRNA signatures as future
biomarkers for diagnosis of diabetes states. Cells. 8:15332019.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Atkin SL, Ramachandran V, Yousri NA,
Benurwar M, Simper SC, McKinlay R, Adams TD, Najafi-Shoushtari SH
and Hunt SC: Changes in blood microRNA expression and early
metabolic responsiveness 21 days following bariatric surgery. Front
Endocrinol (Lausanne). 9:7732019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chen M and Yan J: A preliminary integrated
analysis of miRNA-mRNA expression profiles reveals a role of
miR-146a-3p/TRAF6 in plasma from gestational diabetes mellitus
patients. Ginekol Pol. 95:627–635. 2024.PubMed/NCBI
|
|
53
|
Oğlak SC, Aşir F, Yılmaz EZ, Bolluk G,
Korak T and Ağaçayak E: The immunohistochemical and bioinformatics
analysis of the placental expressions of vascular cell adhesion
protein 1 (VCAM-1) and high mobility group box 1 (HMGB1) proteins
in gestational diabetic mothers. Z Geburtshilfe Neonatol.
229:90–98. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Yanai S, Tokuhara D, Tachibana D, Saito M,
Sakashita Y, Shintaku H and Koyama M: Diabetic pregnancy activates
the innate immune response through TLR5 or TLR1/2 on neonatal
monocyte. J Reprod Immunol. 117:17–23. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Li QP, Pereira TJ, Moyce BL, Mahood TH,
Doucette CA, Rempel J and Dolinsky VW: In utero exposure to
gestational diabetes mellitus conditions TLR4 and TLR2 activated
IL-1beta responses in spleen cells from rat offspring. Biochim
Biophys Acta. 1862:2137–2146. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Shi H, Kokoeva MV, Inouye K, Tzameli I,
Yin H and Flier JS: TLR4 links innate immunity and fatty
acid-induced insulin resistance. J Clin Invest. 116:3015–3025.
2006. View Article : Google Scholar : PubMed/NCBI
|