Introduction
Esophageal carcinoma is a prevalent malignant tumor
originating from the esophageal mucosa, with varying global
incidence in different regions. Increased incidences of esophageal
carcinoma were reported in developing countries, particularly in
the low-middle Social Demographic Index (SDI) region, where
incident cases surged by 105.73% (1), which may be attributed to regional
cultures, dietary patterns and other factors (2,3). In
China, the main histologic type of esophageal carcinoma is
esophageal squamous cell carcinoma (ESCC). Contributors to ESCC
include smoking, alcohol intake (4,5),
poor diet, chronic esophagitis, genetic mutations (6) and viral infections (7). Treatment strategies for ESCC include
surgical resection, endoscopic therapy, radiotherapy, chemotherapy,
targeted therapy and immunotherapy (8–10).
Despite improvements in treatment strategies, the 5-year survival
rate of patients with ESCC is 18% (11), warranting exploration of the
molecular pathogenesis of ESCC to find new therapeutic
strategies.
Endoplasmic reticulum stress (ERS) is a state of
cellular stress caused by ER dysfunction and abnormal protein
accumulation, which triggers an adaptive program response known as
the unfolded protein response (UPR). In response to UPR, three
transmembrane sensors, inositol-requiring enzyme 1α [IRE1α; also
known as endoplasmic reticulum to nucleus signaling 1 (ERN1)],
protein kinase R (PKR)-like endoplasmic reticulum kinase [PERK;
also known as eukaryotic translation initiation factor 2 alpha
kinase 3 (EIF2AK3)] and activating transcription factor (ATF) 6 are
activated by dissociating with the molecular chaperone
glucose-regulated protein 78. Among them, IRE1α catalyzes the
shearing of X-box binding protein 1 (XBP1) mRNA, thus resulting in
translation of active transcription factors called XBP1s. PERK
phosphorylates its catalytic substrate, eIF2α through
autophosphorylation, and then stimulates the production of ATF4,
which subsequently activates CHOP to initiate cell death, including
apoptosis. ATF6 translocates to the golgi apparatus, where it is
cleaved by sphingosine-1-phosphate and site-2 protease to release
amino acid fragments with transcriptional activity called ATF6f
(12). ATF4, XBP1s and ATF6f
regulate transcription of a large range of target genes that
participate in the UPR. In tumors, the biological effects of ERS
are two-fold, the intensity of stress and different cell types will
induce mRNA transcription and promote tumor cell proliferation
(13), migration (14), autophagy (15–17)
and apoptosis (18–20). However, the mechanism of ERS in
ESCC is still not well clarified.
To explore the occurrence and mechanism of ERS in
ESCC, RNA sequencing was carried out on the in vitro ESCC
cell model of ERS and focused on the role of C-X-C motif chemokine
ligand (CXCL) 8, also known as IL-8. CXCL8, is a cytokine and
chemokine that belongs to the CXC chemokine family and is a small
molecule protein produced by a variety of cells. CXCL8 carries out
an important role in inflammation, tumor and immune response
(21). CXCL8 exerts biological
effects by binding to its chemokine receptors (CXCRs) 1 and 2,
among which CXCR1 mainly binds to CXCL8, while CXCR2 also binds to
CXCL1, 2, 3, 5 and 7 (22).
Increased CXCL8 expression is associated with poor prognosis in a
variety of tumors, including breast, colon and lung cancer
(23–25). Furthermore, elevated CXCL8
expression has been associated with resistance to chemotherapy and
immunotherapy in breast cancer, gastric cancer, lung adenocarcinoma
and hepatocellular carcinoma (26–29).
The CXCL8-CXCR1/2 axis promotes migration, proliferation, invasion
and survival of tumor cells (30).
CXCL8 and CXCR1/2 are reported to be highly expressed in ESCC
tissues and are associated with factors and prognosis (31,32).
In particular, CXCL8 promotes migration and invasion of ESCC cells
through phosphorylation of AKT and ERK1/2 (32). Nevertheless, the role of CXCL8 in
ERS and the potential downstream mechanisms promoting ESCC
progression need to be further investigated.
The present study investigated the mechanisms
underlying CXCL8 upregulation during ERS and its tumor-promoting
effects in ESCC, in order to provide a new strategy for ESCC
treatment.
Materials and methods
Cell culture and treatment
Human esophageal cancer cell lines (KYSE-150,
KYSE-170 and TE-1) and the human normal esophageal epithelial cell
line (HEEC) were purchased from the China Center for Type Culture
Collection and cultured in RPMI1640 medium (cat. no. 11879020;
Gibco; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine
serum (FBS; cat. no. A5670201; Gibco; Thermo Fisher Scientific,
Inc.). Cells were maintained under standard culture conditions
(37°C and 5% CO2) in culture medium. For thapsigargin
(TG; cat. no. T9033; MilliporeSigma) treatment, KYSE-150 and TE-1
cells were cultured at 37°C and treated with 100 nM TG for 0 to 24
h. For recombinant human (rh-)CXCL8 (cat no. HY-P7379;
MedChemExpress) treatment, KYSE-150 and TE-1 cells were cultured at
37°C and treated with rh-CXCL8 at varying concentrations (0, 10,
20, 40, 80 or 160 ng/ml) for 0 to 48 h. For TGF-β1 (cat. no.
HY-P7118; MedChemExpress) treatment, KYSE-150 and TE-1 cells were
cultured at 37°C and treated with 10 ng/ml TGF-β1 for 48 h.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR) assays
Total RNA was extracted from KYSE-150, KYSE-170,
TE-1 and HEEC cells (4×106 cells per sample) using
Triquick reagent (cat. no. R1100; Beijing Solarbio Science &
Technology Co., Ltd.) and reverse transcribed into cDNA with the
Transcriptor First Strand cDNA Synthesis Kit (cat. no. 04896866001;
Roche Diagnostics) according to the manufacturer's protocol. SYBR
Green real-time fluorescent quantitative PCR premix (cat. no.
B110031; Sangon Biotech Co., Ltd.) was carried out in StepOnePlus
real-time fluorescent quantitative PCR system (Applied Biosystems;
Thermo Fisher Scientific, Inc.) with the following cycling
conditions: Initial denaturation at 95°C for 10 min; 40 cycles of
95°C for 15 sec, 56°C for 30 sec and 72°C for 30 sec, and a final
extension at 72°C for 7 min. The expression levels of genes were
analyzed with the 2−ΔΔCq method (33) and normalized to endogenous GAPDH.
Primer sequences are listed in Table
SI.
RNA sequencing analysis
TE-1 cells were treated with 100 nM TG for 12 h
under standard culture conditions (37°C; 5% CO2), after
which total RNA was extracted using TRIzol™ Reagent
(cat. no. 15596026CN; Thermo Fisher Scientific, Inc.). The quality
and integrity of the RNA samples were assessed using an Agilent
2100 Bioanalyzer (Agilent Technologies, Inc.). Library
construction, sequencing and data analysis were carried out by
Sangon Biotech Co., Ltd. Sequencing was carried out on an Illumina
platform using paired-end sequencing with a read length of 150 bp.
The sequencing was performed using the Illumina NovaSeq 6000 system
with the NovaSeq 6000 S4 Reagent Kit (300 cycles; cat. no.
20027466; Illumina, Inc.). The final library was quantified using a
Qubit 4.0 Fluorometer (Thermo Fisher Scientific, Inc.) and the
concentration was adjusted to 10–20 pM prior to sequencing, based
on molar concentration calculations. Bioinformatics analysis was
performed using the programs FastQC (v0.11.9; http://www.bioinformatics.babraham.ac.uk/projects/fastqc/),
Cutadapt (v2.10; http://cutadapt.readthedocs.io/), HISAT2 (v2.2.1;
http://daehwankimlab.github.io/hisat2/) and
featureCounts (v2.0.1; http://subread.sourceforge.net/featureCounts.html).
Statistical analysis and visualization were conducted in R (v4.3.2;
https://www.r-project.org/; The R
Foundation). Differential expression analysis was conducted in R
(v4.3.2; http://www.r-project.org) using
DESeq2 (v1.30.1; http://bioconductor.org/packages/release/bioc/html/DESeq2.html).
Differentially expressed genes were screened based on the following
criteria: |log2foldchange|>1, adjusted P<0.05.
Cell transfection
ERN1, EIF2AK3 and ATF6 small interfering (si)RNA,
alongside si-negative control were purchased from Shanghai
GenePharma Co., Ltd., sequences are listed in Table SII. pcDNA3.1-XBP1 and
pcDNA3.1-ATF6 were purchased from GenScript Biotech Corporation.
The cDNA encoding ATF4 was amplified and inserted into the pcDNA3.1
vector (cat. no. V79520; Invitrogen; Thermo Fisher Scientific
Inc.), named pcDNA3.1-ATF4, sequences are listed in Table SIII. KYSE-150 and TE-1 cells were
seeded in 6-well plates at a density of 1×106 cells/well
and cultured for 24 h to allow adherence. Upon reaching 70–80%
confluency, transfection was carried out according to the
manufacturer's protocol for Lipofectamine® 2000 (cat.
no. 11668500; Invitrogen; Thermo Fisher Scientific, Inc.).
Specifically, 3 µg of target overexpression plasmid and empty
vector control plasmid or 100 pmol of siRNA and negative control
siRNA was mixed with transfection reagent at room temperature for
20 min and added to the cell culture system. Post-transfection,
cells were maintained in a 37°C, 5% CO2 incubator. The
culture medium was replaced with fresh medium at 6 h. After 48 h of
total incubation, cells were harvested for transfection efficiency
analysis by RT-qPCR and subsequent experiments.
Lentiviral infection
Full-length human CXCL8 cDNA was obtained via
RT-qPCR and inserted into the lentiviral expression vector
pCDH-CMV-MCS-EF1-Puro. The recombinant plasmid (10 µg), along with
the packaging plasmid psPAX2 (7.5 µg) and the envelope plasmid
pMD2.G (5 µg), was mixed and incubated at room temperature for 20
min to form the transfection complex. The complex was then
co-transfected into 293T cells using Lipofectamine 2000®
to produce lentiviral particles at 37°C for 6 h, after which the
medium was replaced with fresh medium. After 48 h, viral
supernatants were harvested and subsequently used to transduce
KYSE-150 cells. Infected cells were cultured in medium containing
1.0 µg/ml puromycin for 14 days to generate a stable KYSE-150 cell
line with CXCL8 overexpression.
Western blot analysis
KYSE-150 and TE-1 cells were washed in ice-cold PBS
and RIPA lysis buffer (cat. no. R0010; Beijing Solarbio Science
& Technology Co., Ltd.). PMSF (cat. no. P0100; Beijing Solarbio
Science & Technology Co., Ltd.) was used to extract proteins
from cells. Protein concentrations were measured using the BCA
Protein Assay Kit (cat. no. PC0020; Beijing Solarbio Science &
Technology Co., Ltd.) per the manufacturer's instructions. Briefly,
samples were mixed with working reagent (1:50 ratio) and incubated
at 37°C for 30 min before absorbance measurement at 562 nm. Protein
(50 µg/lane) was then separated by 10% SDS-PAGE and transferred to
PVDF membranes (cat. no. 1.15546; MilliporeSigma). Membranes were
blocked with 5% non-fat milk in TBS with 0.05% Tween-20 for 1 h at
room temperature, followed by incubation with primary antibodies
overnight at 4°C and corresponding HRP-conjugated secondary
antibodies for 1 h at room temperature. The enhanced
chemiluminescence detection reagent (cat. no. WBULS0100;
MilliporeSigma) was used to detect the protein bands by Chemi XT 4
(Syngene). The antibodies are listed in Table SIV.
Gene expression profiling interactive
analysis (GEPIA) database analysis
Expression profile analysis of CXCL8 across various
types of cancer and normal tissues was carried out using the GEPIA
web server (http://gepia.cancer-pku.cn/), which integrates RNA
sequencing data from The Cancer Genome Atlas and Genotype-Tissue
Expression projects. The ‘Expression DIY’ module was used to assess
CXCL8 mRNA expression in pan-cancer types, and to compare its
expression levels between ESCC and normal esophageal tissues. The
thresholds were set at |log2 fold change (FC)|>1 and
P<0.01 to define significant differential expression. Box plots
were generated directly through GEPIA for visualization.
PROMO database analysis
Prediction of transcription factor binding sites was
performed using the PROMO database (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3),
which is based on the TRANSFAC transcription factor binding site
database. The promoter region (−2,000 bp upstream of the
transcription start site) of the CXCL8 gene was analyzed, with the
species set to Homo sapiens and the maximum matrix
dissimilarity rate set to 15%. Candidate transcription factors were
selected based on their known relevance to ERS for further
validation.
Cell proliferation assay
Cellular proliferation capability was detected using
an MTS assay. Briefly, 1×103 cells pretreated with
rh-CXCL8 for 24 h under standard culture conditions (37°C, 5%
CO2) were seeded per well in 96-well plates. At 0, 24,
48, 72 and 96 h post-seeding, CellTiter 96® AQueous One
Solution Reagent (cat. no. G3582; Promega Corporation) was added,
followed by a 2-h incubation at 37°C before measuring absorbance at
490 nm.
Cell migration and invasion
assays
Wound healing and Transwell assays were carried out
to detect the migration and invasion ability of cells. For the
wound healing assay, treated cells (KYSE-150 and TE-1 cells
subjected to 24 h treatment with NC or 10 ng/ml rh-CXCL8, 12 h
treatment with 100 nM TG, 48 h transfection with 100 pmol siCXCR1
and combined treatment with 100 nM TG and 100 pmol siCXCR1) were
inoculated in 6-well plates and cultured in medium containing 10%
FBS. A straight scratch was made in each well when the cell
monolayer reached near complete confluence, and cells were then
maintained in serum-free medium. Images were taken at the same
position at 0 and 24 h using an inverted optical microscope (Nikon
Eclipse 50i; Nikon Corporation). Subsequently, ImageJ (National
Institutes of Health) software was employed to analyze the acquired
images. The calculation of scratch area percentage was defined as
the ratio of scratch area at 24 h to the scratch area at 0 h for
each group.
For the Transwell invasion assay, 1×105
indicated cells in 200 µl serum-free medium were placed in the
upper chamber (Corning, Inc.) and cultured at 37°C for 24 h. The
chamber had been previously coated with 200 µl Matrigel (incubated
at 37°C for 6 h), and 0.7 ml medium containing 10% FBS was placed
in the lower chamber. Cells on the upper surface were wiped off
after 24 h of incubation and 4% paraformaldehyde was used to fix
the invasive cells on the lower surface of the chamber which was
then stained with 0.1% crystal violet solution for 20 min at room
temperature. Subsequently, the number of invading cells was counted
in five random fields selected by an inverted microscope, and the
average was calculated. The migration assay was carried out
following the same protocol as the invasion assay, except that
Transwell membranes were not coated with Matrigel.
Dual-luciferase reporter assay
In KYSE-150 cells, the promoter-reporter gene
plasmids (containing promoter regions derived from CXCL8, SNAI2 and
ZEB1) were separately co-transfected with pcDNA3.1-XBP1,
pcDNA3.1-ATF4, pcDNA3.1 empty plasmid or treated with TGF-β1.
Luciferase activity was measured 48 h after transfection and
treatment under standard culture conditions (37°C; 5%
CO2), using the Dual-Luciferase® Reporter
Assay System (cat. no. E1910; Promega Corporation) according to the
manufacturer's instructions and normalized to Renilla luciferase
activity. The primer sequences for the dual-luciferase reporter
plasmids are detailed in Table
SV.
Animal experiments
A total of 10 BALB/c-nude mice (6-week-old male and
body weight of 18–22 g) were purchased from the Beijing Huafukang
Biotechnology Co., Ltd. Mice were housed under standard conditions
and had unlimited access to sterilized food and distilled water.
Room temperature was maintained at 24±2°C, relative humidity was
maintained at 60±10% and a 12-h light/dark cycle was used. After 7
days of acclimation, the mice were randomly divided into two
experimental groups (n=5 per group): i) Control group, ii)
CXCL8-overexpressed group. Group i) mice served as the normal
control group and were subcutaneously injected with
5×106 control (KYSE-150) cells into the dorsal flanks.
Group ii) was the CXCL8-overexpressed group and the mice were
injected with a total of 5×106 CXCL8-overexpressing
KYSE-150 cells into the dorsal flanks. The total duration of the
animal study was 30 days. The mice were monitored daily for food
and water intake, weight, body posture, behavior, distress and
response to external stimuli.
Humane endpoints of the present study were loss of
>20% body weight, severe dehydration, refusal of food, severe
pain or distress or a moribund state, no animals reached these
endpoints or were found dead during the present study. All mice
were anesthetized with an intraperitoneal injection of 50 mg/kg
pentobarbital sodium and 0.5–1 ml blood was collected by cardiac
puncture followed by sacrifice by exsanguination. Indexes such as
breathing, heartbeat, pupils and nerve reflexes were assessed to
confirm death. Mice were euthanized after 30 days, and terminal
volume and weight of tumor tissues were measured. The present study
was approved by the Institutional Animal Care and Use Committee
(IACUC) of the Fourth Hospital of Hebei Medical University
(approval no. IACUC-4th Hos Hebmu-2023001).
Immunohistochemical staining
assay
In the CXCL8-overexpressing mouse model, both
esophageal carcinoma and adjacent normal tissues were collected and
fixed in 4% formaldehyde at 4°C for 24–48 h. After washing with
PBS, the specimens were dehydrated through a graded ethanol series,
embedded in paraffin and sectioned at a thickness of 4-µm. The
sections were then blocked with 3% hydrogen peroxide (15 min at
room temperature) to inhibit endogenous peroxidase activity and
subjected to antigen retrieval in citrate buffer (pH 6.0) at
>95°C for 5 min. Primary antibodies against CXCR1/2, p-SMAD2/3,
SNAI2 and ZEB1 (all at 1:100 dilution) were applied and incubated
overnight at 4°C, followed by a 2 h room temperature incubation
with secondary antibodies. The stained sections were visualized
using an inverted optical microscope. The antibody information is
identical to that used for western blotting (Table SIV).
Prediction of SMAD3-regulated miRNAs
targeting ZEB1
To identify candidate miRNAs that may be
transcriptionally regulated by SMAD3 and simultaneously target
ZEB1, bioinformatics screening was conducted using the
TransmiR v3.0 (http://www.cuilab.cn/transmir) and TargetScan Release
7.1 (http://www.targetscan.org/vert_71/). First, TransmiR
v3.0 was queried to identify miRNAs either known or predicted to be
regulated by SMAD3. Subsequently, TargetScan was used to predict
miRNAs with conserved binding sites in the 3′-untranslated region
(3′-UTR) of ZEB1. The predicted miRNAs from both TransmiR v3.0 and
TargetScan databases were compared, and the overlapping miRNAs were
identified as potential candidates linking SMAD3 and ZEB1.
Statistical analysis
GraphPad Prism 8.0 (Dotmatics) was used to carry out
data analysis and graphing. Statistical significance was determined
using an unpaired Student's t-test or one-way ANOVA with Tukey's
post hoc test. All in vitro experiment data came from three
independent experiments carried out in triplicate and presented as
mean ± SD. P<0.05 was considered to indicate a statistically
significant difference.
Results
CXCL8 is induced by ERS in ESCC
ESCC cells were treated with 100 nM TG (an ERS
inducer) for 0–24 h (34), after
which expression of effectors of the UPR were assessed. The mRNA
and protein expression levels of XBP1, ATF4 and ATF6 peaked at 12 h
after TG treatment (Fig. 1A and
B). To explore the potential mechanism of ERS in ESCC
progression, RNA sequencing analysis was employed to compare the
gene expression profiles between TE-1 cells treated with either TG
or DMSO. CXCL8 was one of the top ranked differentially altered
genes in response to ERS (Fig.
1C). The elevated mRNA and protein expression levels of CXCL8
were further verified at 12 h in TG-treated KYSE-150 and TE-1 cells
by RT-qPCR and western blot analysis (Fig. 1D and E). CXCL8 was significantly
upregulated in a variety of types of cancer, including esophageal
cancer by searching the GEPIA database (Fig. 1F and G). The mRNA expression level
of CXCL8, as well as its receptors, CXCR1 and CXCR2, was increased
in ESCC cells compared with that in HEEC cells (Fig. 1H).
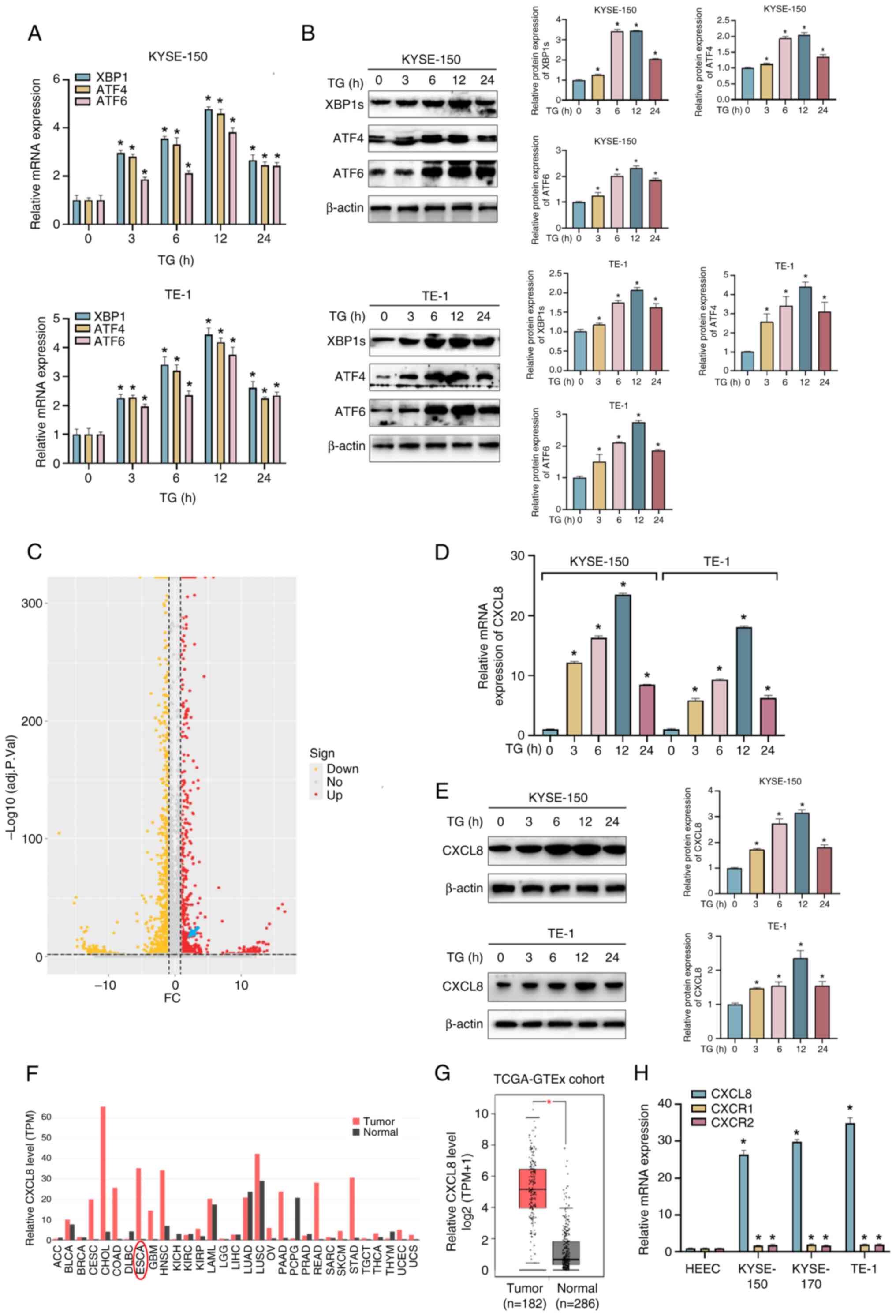 | Figure 1.Endoplasmic reticulum stress induces
CXCL8 expression in ESCC cells. KYSE-150 and TE-1 cells were
treated with 100 nM TG for 0, 3, 6, 12 and 24 h, and expression
levels of effectors in the UPR were examined by (A) RT-qPCR and (B)
western blot analysis. (C) The volcano plot shows differentially
expressed genes in TE-1 cells treated with 100 nM TG for 12 h. The
x-axis represents the log2-transformed of FC ratios and the y-axis
is the log10-transformed adjusted P-value. Red/yellow dots
represent significantly up/downregulated genes (|log2FC|≥1,
adj.P<0.05). The blue arrow indicates the possible location of
CXCL8. The (D) mRNA and (E) protein expression levels of CXCL8 in
ESCC cells treated with 100 nM TG for 24 h. (F) The CXCL8
expression profile across all tumor samples and corresponding
normal tissues was obtained from the GEPIA. (G) The mRNA expression
levels of CXCL8 in ESCC tissues (n=182) and normal tissues (n=286)
were obtained from the GEPIA database. (H) Expression levels of
CXCL8, the receptors CXCR1 and CXCR2 in the normal esophageal
epithelial cell line and ESCC cell lines were determined by RT-qPCR
method. All data are expressed as mean ± SD, n=3/group. *P<0.05
vs. TG-treated 0 h group. CXCL8, C-X-C motif chemokine ligand;
ESCC, esophageal squamous cell carcinoma; TG, thapsigargin; FC,
fold change; GEPIA, Gene Expression Profiling Interactive
£Analysis, RT-qPCR, reverse transcription-quantitative PCR; HEEC,
human normal esophageal epithelial cell; XBP1, X-box binding
protein 1; ATF, activating transcription factor; TCGA-GTEx; The
Cancer Genome Atlas-Genotype-Tissue Expression; sign, significance;
up, upregulated; no, no significance; down, downregulated; TPM,
transcript per million. |
Both the IRE1α and PERK pathways
regulate the transcription of CXCL8
To explore the signaling pathways which may be
responsible for induction of CXCL8, three transmembrane sensors
(IRE1α, PERK and ATF6) were respectively knocked down in KYSE-150
and TE-1 cells using siRNAs, and the knockdown efficiency was
validated by RT-qPCR assay (Fig.
S1A). As shown in Fig. 2A and
B, under the DMSO-treated condition, knockdown of the three
sensors resulted in reduced expression of their downstream
transcription factors (XBP1, ATF4, and ATF6). A decrease in CXCL8
expression was observed only in the siIRE1α + DMSO and siPERK +
DMSO groups under the DMSO-treated condition. Under the TG-treated
condition, the siNS + TG group exhibited significantly increased
mRNA and protein levels of the three sensors, their downstream
transcription factors, and CXCL8, compared with the siNS + DMSO
group. In the siRNA + TG groups, the mRNA and protein levels of the
three sensors, their downstream transcription factors, were
decreased compared with those in the siNS + TG group; however, a
reduction in CXCL8 mRNA and protein expression was observed only in
the siIRE1α + TG and siPERK + TG groups. Elevated levels of CXCL8
mRNA and protein in KYSE-150 and TE-1 cells overexpressing XBP1 and
ATF4, the downstream transcription factors of IRE1α and PERK,
respectively were detected (Fig. 2C
and D). In contrast, the expression level of CXCL8 was not
influenced by the overexpression of ATF6 (Fig. S1B and C), indicating that
upregulation of CXCL8 in ERS may be regulated by the IRE1α/XBP1 and
PERK/ATF4 signaling pathways. As transcription factors, XBP1 and
ATF4 carry out key roles by regulating the transcription of various
genes which are involved in ERS. A possible binding site of XBP1 on
the promoter region of CXCL8 (−181 bp/-175 bp) was predicted by the
PROMO database, and the binding effect was further verified by
dual-luciferase reporter assay in KYSE-150 cells (Fig. 2E). A possible binding site of ATF4
on the promoter region of CXCL8 (−40 bp/-28 bp) was also predicted
and the transcriptional regulatory effect of ATF4 on CXCL8 was also
verified (Fig. 2F).
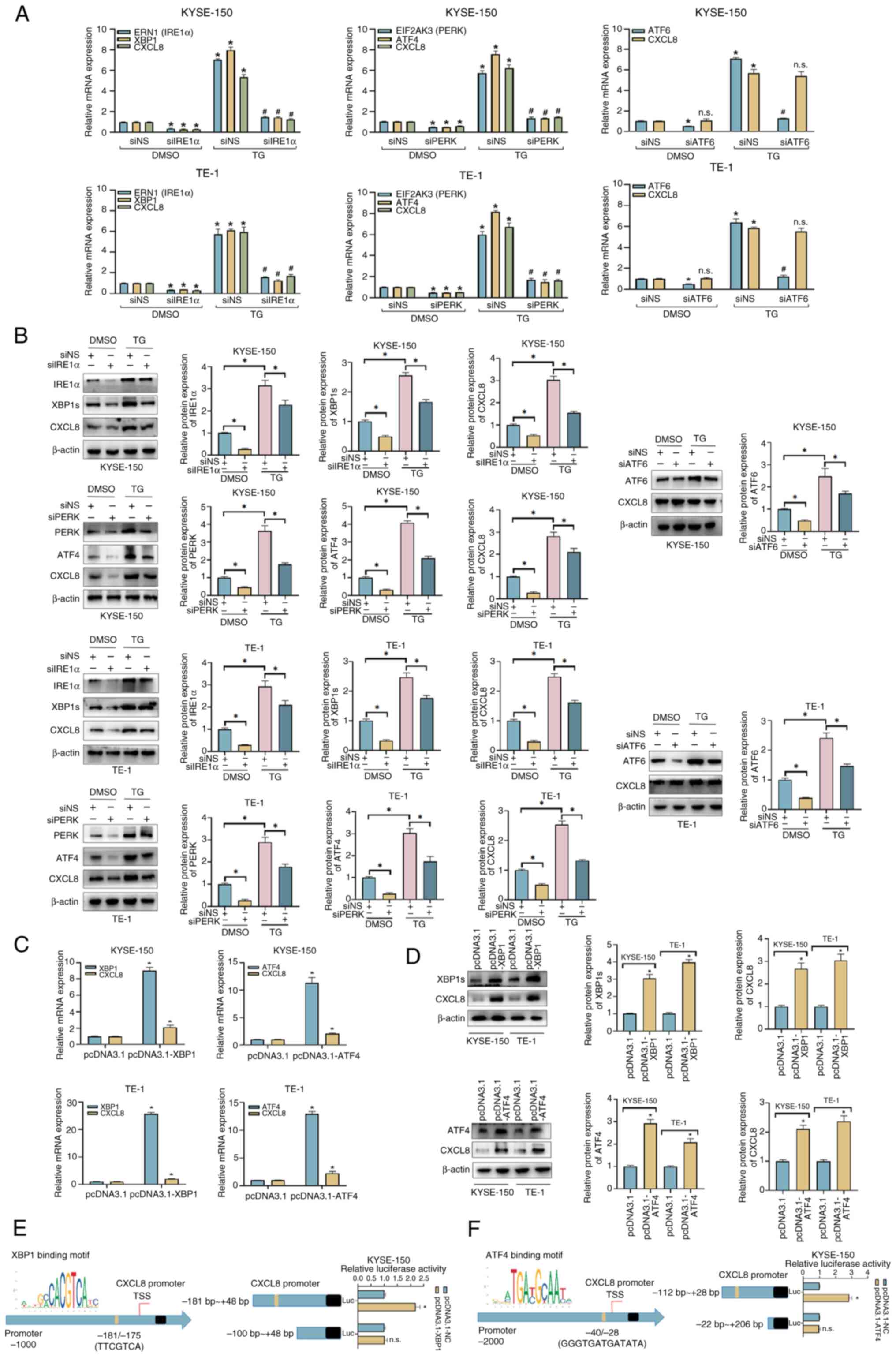 | Figure 2.CXCL8 is induced by the PERK and
IRE1α pathways of the unfolded protein response in esophageal
squamous cell carcinoma cells. Evaluation of the indicated (A) mRNA
and (B) protein expression levels in siRNA-transfected KYSE-150 and
TE-1 cells, which were treated with 100 nM TG or DMSO for 24 h.
Evaluation of (C) mRNA and (D) protein expression levels of CXCL8
in transfected KYSE-150 and TE-1 cells by reverse
transcription-quantitative PCR and western blot analysis.
Dual-luciferase reporter assays were conducted in KYSE-150 cells to
verify the direct binding effects of (E) XBP1 and (F) ATF4 on the
CXCL8 promoter. All data are expressed as mean ± SD, n=3/group.
*P<0.05, vs. siNS + DMSO group, siNS + TG group or pcDNA3.1
group; #P<0.05 vs. siNS + TG group. n.s., not
significant; IRE1α, inositol-requiring enzyme 1α; PERK, protein
kinase R (PKR)-like endoplasmic reticulum kinase; si, small
interfering; ERN, endoplasmic reticulum to nucleus signaling;
EIF2AK3, ATF, eukaryotic translation initiation factor 2 alpha
kinase 3; TG, thapsigargin; NS, non-silencing; CXCL, C-X-C motif
chemokine ligand. |
CXCL8 promotes ESCC cells migration
and invasion in vitro
In order to study the biological function of CXCL8
in ESCC, KYSE-150 and TE-1 cells were treated with rh-CXCL8, and
the expression levels of CXCR1 and CXCR2 were assessed. As shown in
Fig. 3A-D, the mRNA and protein
expression levels of both receptors peaked at 24 h with 10 ng/ml
rh-CXCL8 treatment. ESCC cells were then treated with 10 ng/ml
rh-CXCL8 for 24 h to detect the variation of cell function. CXCL8
had no effect on the viability of ESCC cells (Fig. 3E) but significantly promoted their
migration and invasion (Fig. 3F and
G).
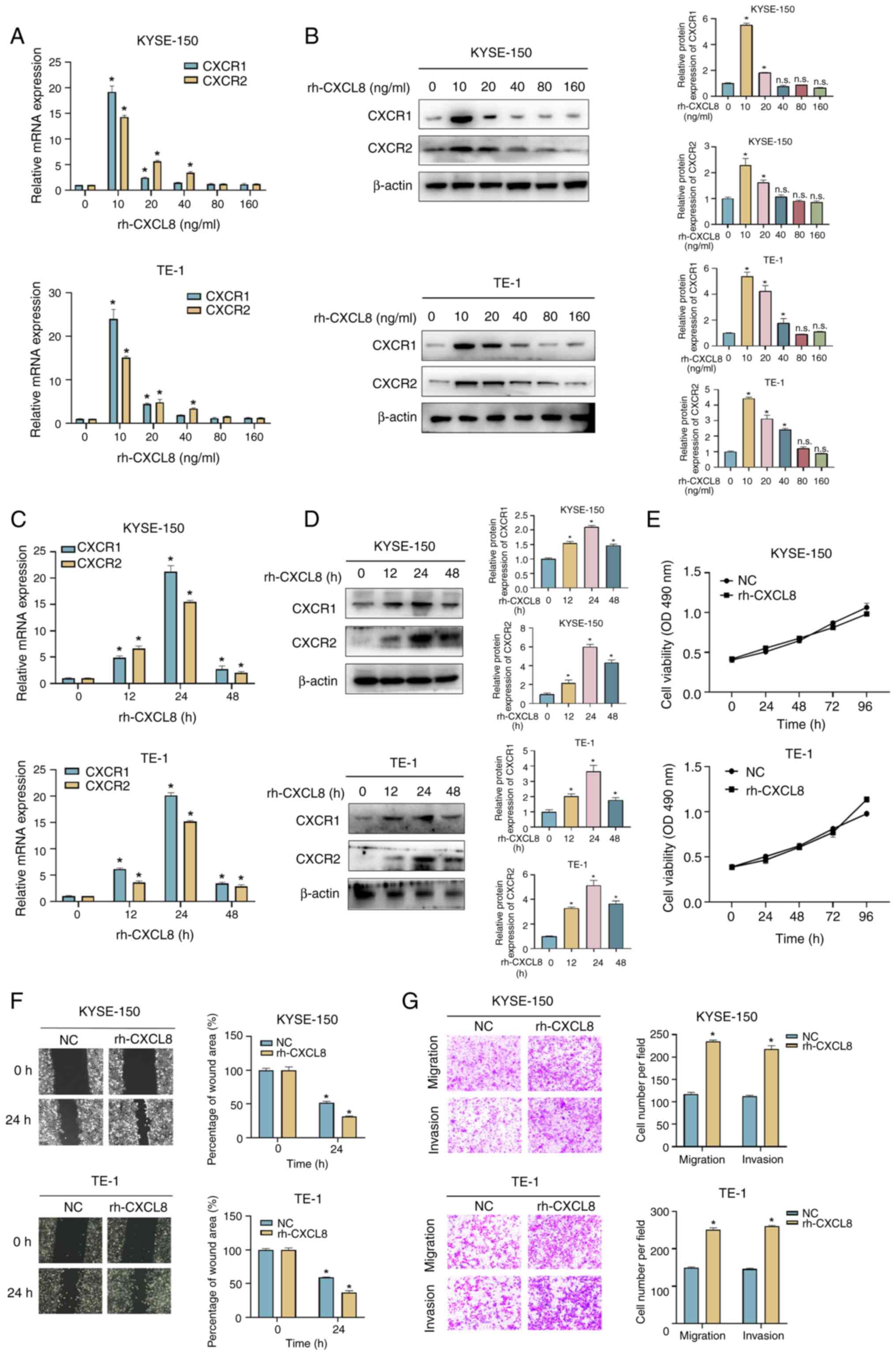 | Figure 3.CXCL8 facilitates migration and
invasion of ESCC cells. Evaluation of (A) mRNA and (B) protein
expression levels of CXCR1 and CXCR2 in KYSE-150 and TE-1 cells
treated with 0, 10, 20, 49, 80 and 160 mg/ml rh-CXCL8 for 24 h.
Evaluation of (C) mRNA and (D) protein expression levels of CXCR1
and CXCR2 in KYSE-150 and TE-1 cells treated with 10 ng/ml rh-CXCL8
for 0, 12, 24 and 48 h. (E) Cell proliferation was analyzed with an
MTS assay in KYSE-150 and TE-1 cells treated with 10 ng/ml rh-CXCL8
for 24 h. (F) Wound healing and (G) Transwell assays were conducted
to assess the migration and invasion abilities in KYSE-150 and TE-1
cells treated with 10 ng/ml rh-CXCL8 for 24 h. All data are
expressed as mean ± SD, n=3/group. *P<0.05 vs. 0 ng/ml
rh-CXCL8-treated group, rh-CXCL8-treated 0 h group, NC-treated 0 h
group or NC group. n.s., not significant; rh-CXCL, recombinant
human-C-X-C motif chemokine ligand; CXCR, chemokine receptor; NC,
negative control. |
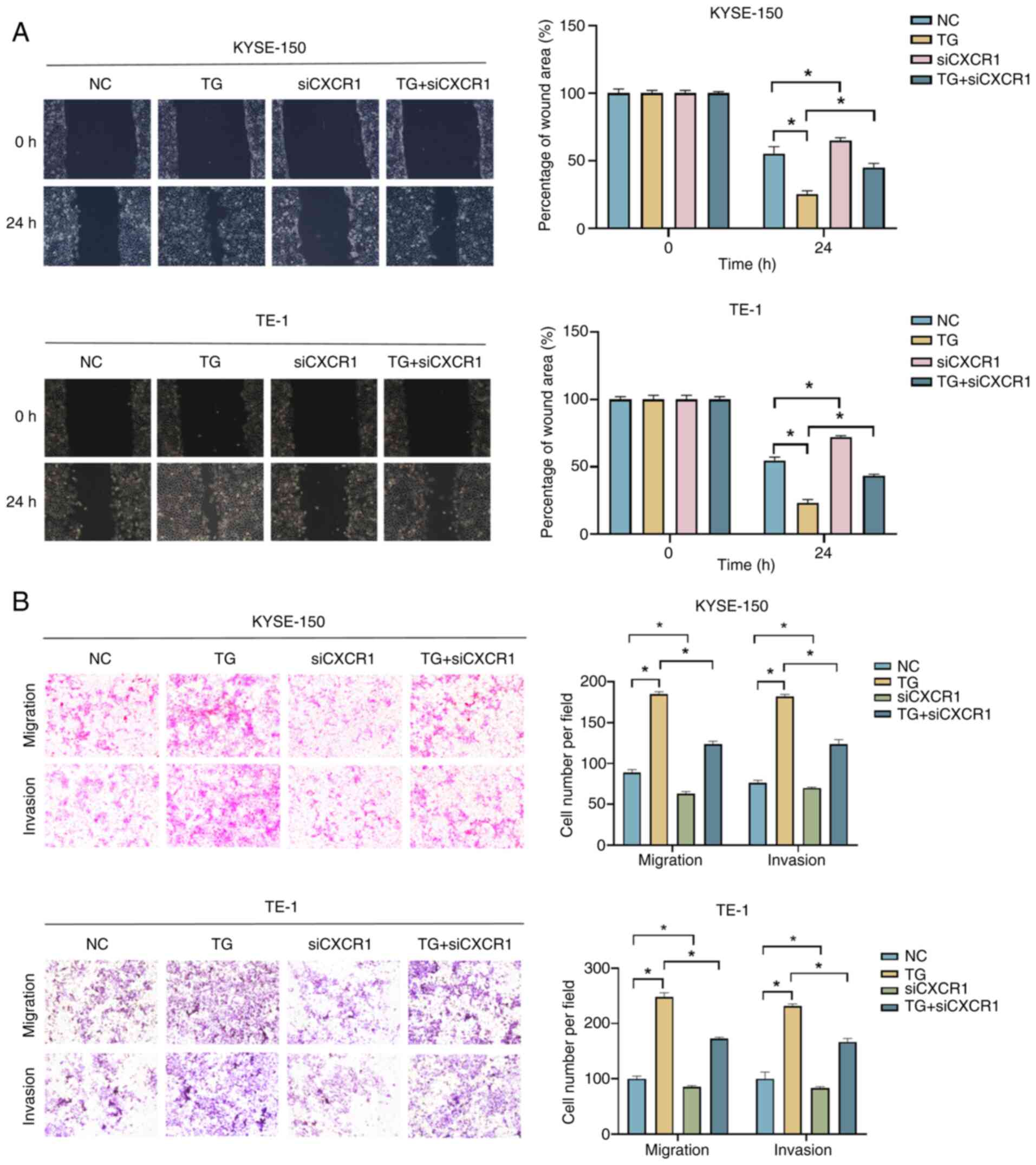
CXCL8/CXCR1 promotes migration and
invasion of ESCC cells under an ERS state
Roles of CXCL8/CXCR1 in the progression of ESCC
cells under ERS state were further examined. The knockdown
efficiency of CXCR1 was validated by an RT-qPCR assay (Fig. S1D). When ESCC cells were treated
with 100 nM TG for 24 h, cell migration and invasion were
significantly enhanced, while knockdown of CXCR1 partially
prevented this effect (Fig. 4A and
B).
CXCL8 increases the expression of
EMT-related genes, SNAI2 and ZEB1
Considering that both ERS and CXCL8 promoted the
migration and invasion of ESCC cells, the mRNA expression levels of
a set of EMT-related genes were screened in rh-CXCL8-treated ESCC
cells, including SNAI2, ZEB1, TWIST1, SNAIL, FN1, TWIST2, CDH2,
VIMENTIN and ZEB2. Among these genes, SNAI2 and ZEB1 demonstrated
the most significant trend of increased expression in KYSE-150
(Fig. 5A) and TE-1 cells (Fig. S2) when treated with 10 ng/ml
rh-CXCL8. Nevertheless, further increases in the rh-CXCL8
concentration led to a progressive decline in their expression
levels. The protein expression levels of SNAI2 and ZEB1, which were
similar to their mRNA expression levels, were further verified in
rh-CXCL8-treated ESCC cells by western blot analysis (Fig. 5B). Given that SNAI2 and ZEB1 are
important genes involved in EMT process, we hypothesized whether
CXCL8 promoted the expression of SNAI2 and ZEB1 by regulating the
activation of EMT-related pathways. The PI3K/AKT, JAK2/STAT3,
Wnt/β-catenin and TGF-β/SMAD pathways are important pathways that
regulate the EMT process, therefore whether CXCL8 promoted the
activation of these pathways was further investigated. As shown in
Fig. 5C and D, rh-CXCL8 treatment
promoted phosphorylation of AKT, STAT3 and SMAD2/3, whereas its
effect on β-catenin appeared to be minimal although this was not
quantitatively assessed, suggesting that CXCL8 may be involved in
the progression of EMT by regulating the activation of AKT, STAT3
and SMAD2/3. The present study revealed several signaling pathways
that may be involved in the action of CXCL8. Since activation of
AKT and STAT3 by CXCL8 has been reported (35–37),
which is consistent with the findings of the present study, the
TGF-β/SMAD signaling pathway was the focus of the present
study.
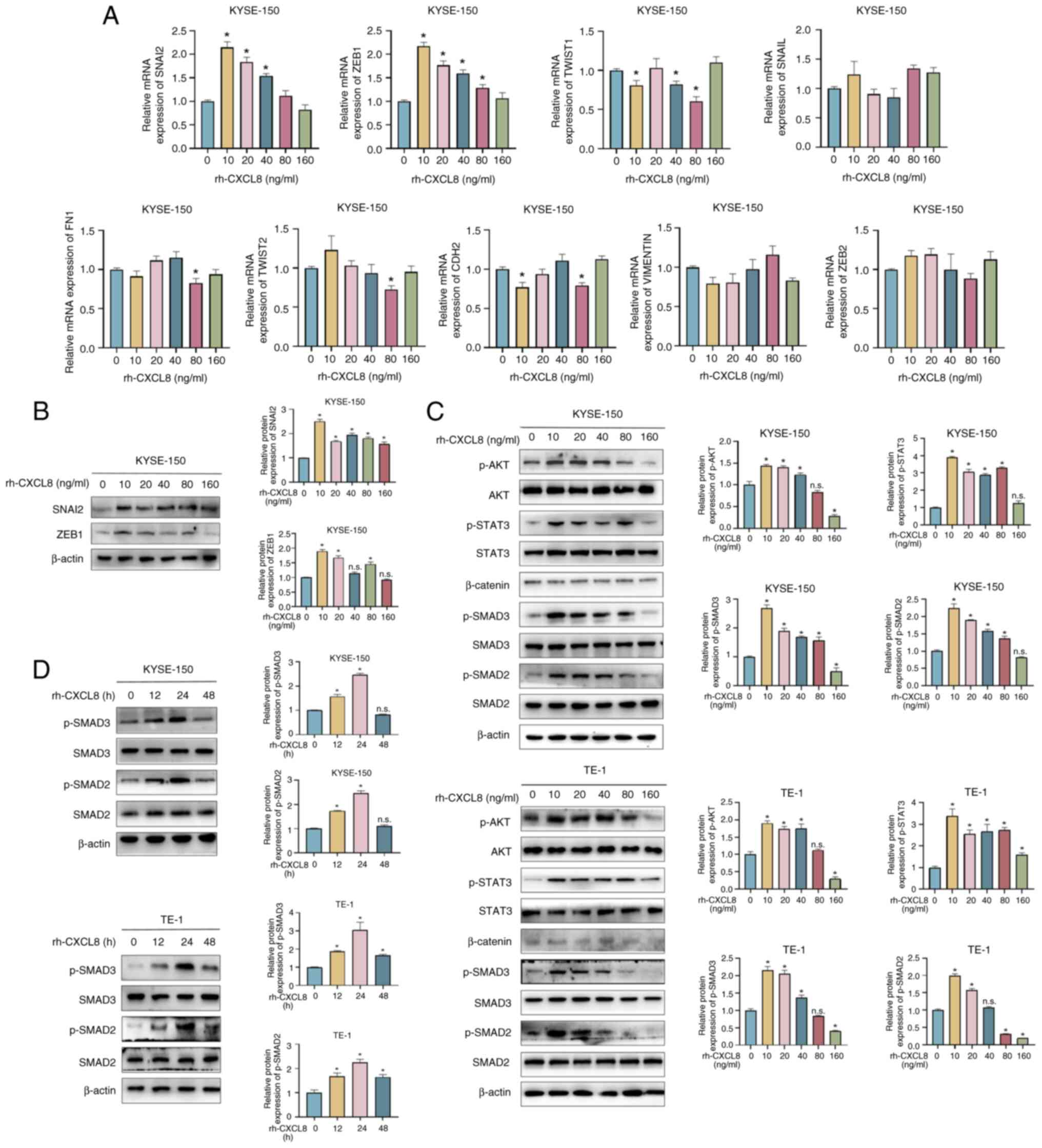 | Figure 5.CXCL8 participates in the EMT process
by regulating the expression of SNAI2 and ZEB1. KYSE-150 cells were
treated with specified concentrations of rh-CXCL8 for 24 h, and
then (A) mRNA expression of SNAI2, ZEB1, TWIST1, SNAIL, FN1,
TWIST2, CDH2, VIMENTIN and ZEB2 was detected by RT-qPCR, and (B)
protein expression of SNAI2 and ZEB1 was detected by western
blotting. (C) KYSE-150 and TE-1 cells were treated with specified
concentrations of rh-CXCL8 for 24 h and the protein expression
levels of EMT-related pathway genes were examined by Western blot
assay. (D) KYSE-150 and TE-1 cells were treated with 10 ng/ml
rh-CXCL8 for indicated times, and the protein expression of SMAD2/3
and p-SMAD2/3 was detected by Western blot assay. All data are
expressed as mean ± SD, n=3/group. *P<0.05 vs. 0 ng/ml
rh-CXCL8-treated group or rh-CXCL8-treated 0 h group. n.s., not
significant; rh-CXCL, recombinant human C-X-C motif chemokine
ligand; EMT, epithelial-mesenchymal transition. |
CXCL8/CXCR1 promotes the EMT process
by activating SMAD2/3
CXCR1 was knocked down to study the activating
effect of CXCL8 on SMAD2/3. As shown in Fig. 6A, the protein expression levels of
p-SMAD2/3 were significantly reduced when CXCR1 was knocked down,
suggesting that CXCL8 may activate SMAD2/3 by binding to CXCR1. The
regulatory role of CXCL8/CXCR1 in the ERS state was further
analyzed. As shown in Fig. 6B, the
expression levels of p-SMAD2/3 were significantly increased in
TG-treated ESCC cells, whereas knockdown of CXCR1 partially
alleviated this effect.
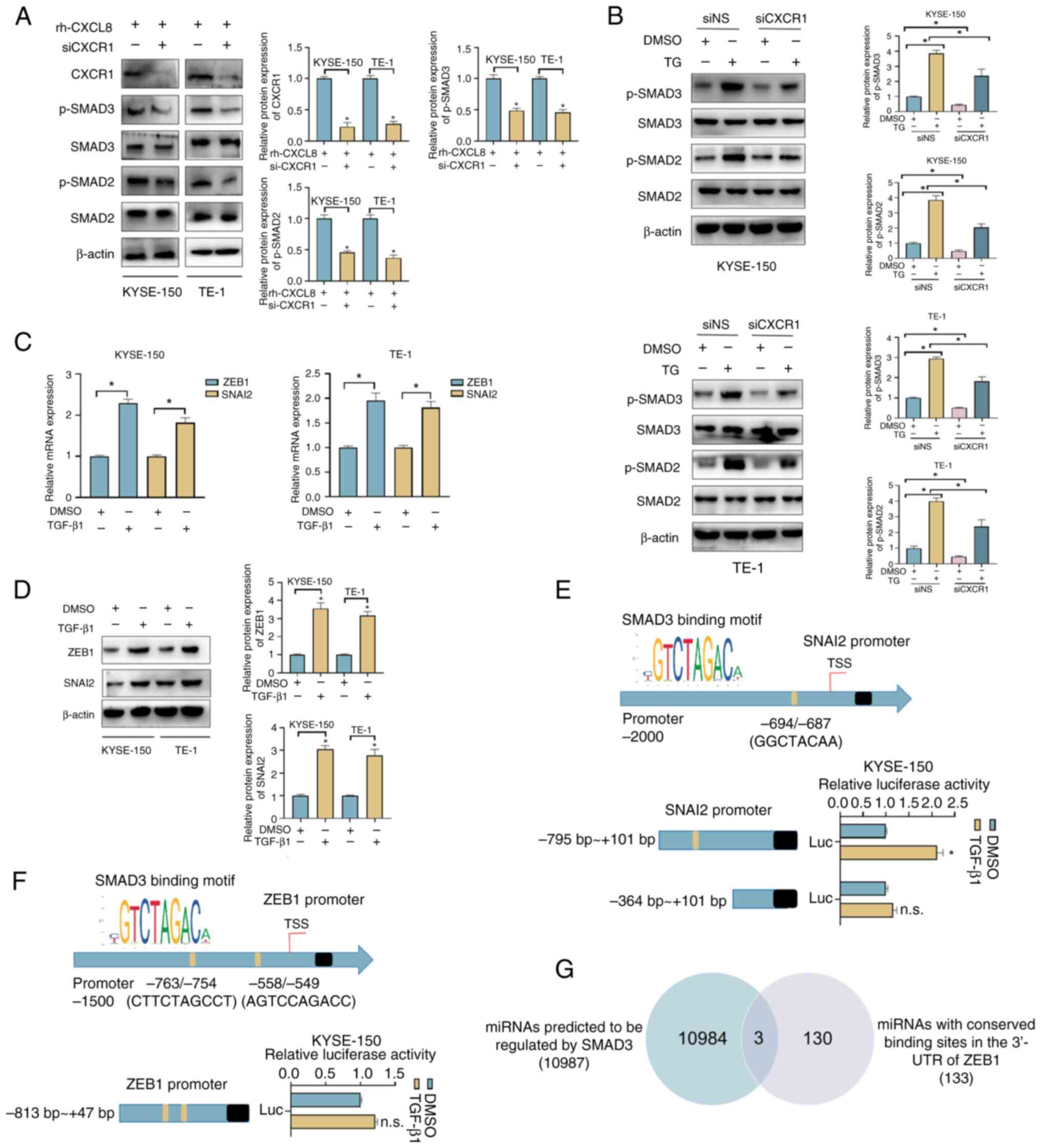 | Figure 6.CXCL8 regulates the expression of
SNAI2 and ZEB1 through phosphorylated activation of SMAD2/3. (A)
Influence of knocking down CXCR1 on the protein expression of
SMAD2/3 and p-SMAD2/3 in KYSE-150 and TE-1 cells, treated with 10
ng/ml rh-CXCL8 for 24 h. (B) Influence of knocking down CXCR1 on
protein expression of SMAD2/3 and p-SMAD2/3 in TG (100 nM; 24 h)
treated KYSE-150 and TE-1 cells. (C) mRNA and (D) protein
expression levels of ZEB1 and SNAI2 were examined in KYSE-150 and
TE-1 cells treated with TGF-β1 (10 ng/ml; 48 h) using reverse
transcription-quantitative PCR and western blot analysis.
Dual-luciferase reporter assays were conducted in KYSE-150 cells to
detect the transcriptional regulatory effect of SMAD3 on (E) SNAI2
and (F) ZEB1. (G) Venn diagram showing the bioinformatics
identification of SMAD3-regulated miRNAs that target ZEB1. All data
are expressed as mean ± SD, n=3/group. *P<0.05 vs. rh-CXCL8
group, DMSO + siNS group, TG + siNS group or DMSO group. n.s., not
significant; rh-CXCL, recombinant human-C-X-C motif chemokine
ligand; CXCR, chemokine receptor; p-, phosphorylated; TG,
thapsigargin; si, small interfering; NS, non-silencing; Luc,
luciferase; miRNA, microRNA; UTR, untranslated region. |
Analysis revealed that 10 ng/ml CXCL8 significantly
promoted the expression of SNAI2 and ZEB1 and whether this effect
was achieved through its activation of SMAD2/3 was further
investigated. KYSE-150 and TE-1 cells were treated with 10 ng/ml
TGF-β1 for 48 h (38). As shown in
Fig. 6C and D, increased mRNA and
protein expression levels of SNAI2 and ZEB1 were detected in
TGF-β1-treated cells, potentially indicating the regulatory effect
of TGF-β/SMAD signaling pathway on the expression of SNAI2 and
ZEB1. A binding site for SMAD3 was identified in the promoter
region of SNAI2, and dual-luciferase reporter assay confirmed that
exogenous addition of TGF-β1 promoted transcription of SNAI2
(Fig. 6E). Two binding sites for
SMAD3 were also found in the promoter region of ZEB1, but ZEB1
transcription was not affected by exogenous incorporation of TGF-β1
(Fig. 6F). miR-3146, miR-3163 and
miR-3171 were predicted to be transcriptionally regulated by SMAD3
and further regulated the expression of ZEB1 by binding to its
3′-UTR (Fig. 6G).
CXCL8 facilitates ESCC progression in
vivo
To evaluate the oncogenic role of CXCL8 in
vivo, KYSE-150 cells stably overexpressing CXCL8 or control
cells were Xeno transplanted subcutaneously into the flanks of
mice. Quantitative analysis at 30 days post-inoculation revealed a
statistically significant augmentation in both tumor volume and
weight in the CXCL8-overexpressing group compared with control
group (Fig. 7A-C), suggesting a
pro-tumorigenic function of CXCL8. To further validate the role of
the CXCL8-CXCR1/2-SMAD2/3-SNAI2/ZEB1 axis in tumor metastasis, the
protein expression levels of CXCR1/2, p-SMAD2/3, SNAI2 and ZEB1
were examined in CXCL8-overexpressing ESCC tissues.
Immunohistochemical analyses revealed enhanced positive staining
for CXCR1/2, p-SMAD2/3, SNAI2 and ZEB1 in the CXCL8-overexpression
group compared with control group (Fig. 7D).
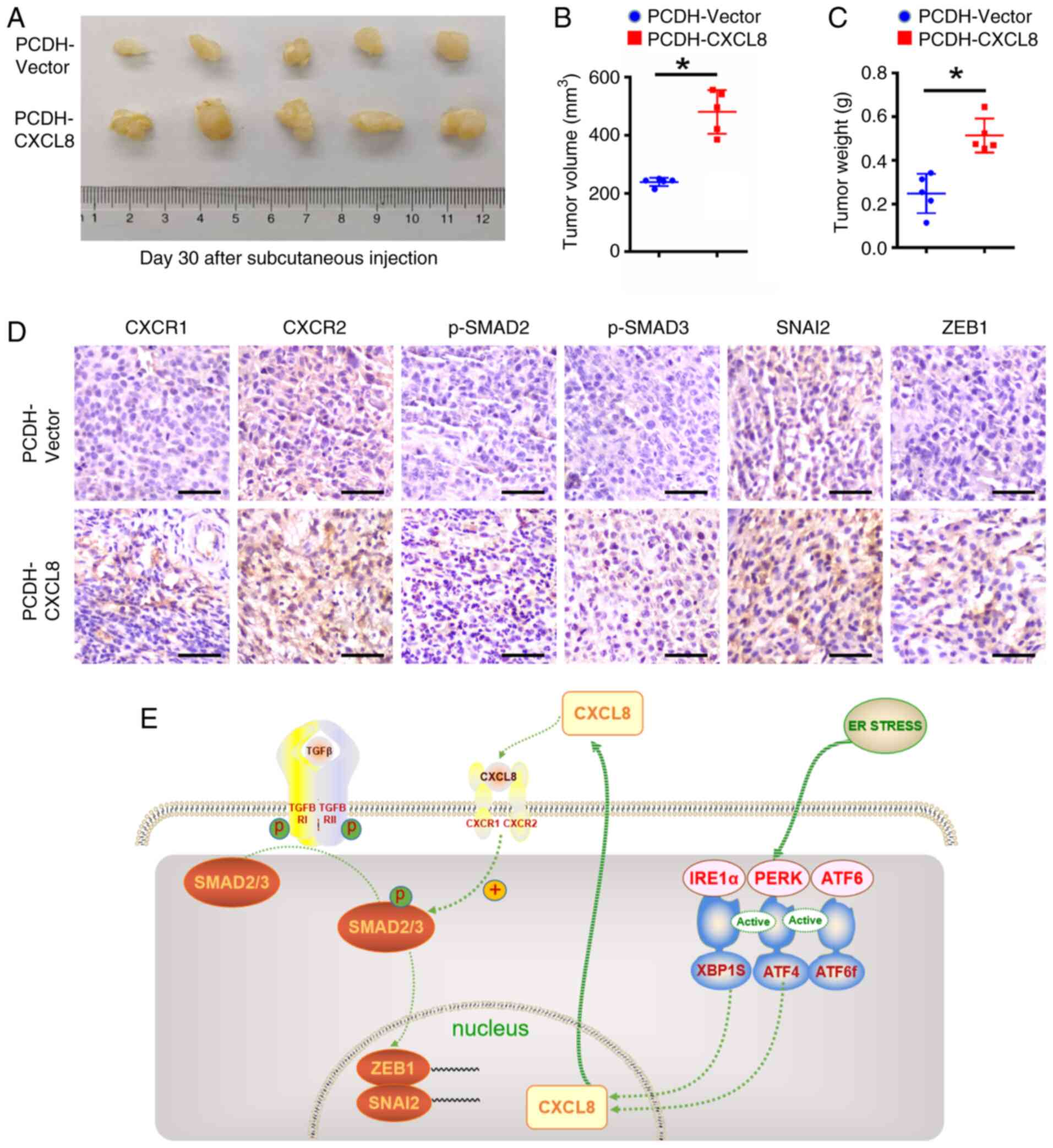 | Figure 7.Animal experiments validate the role
of the CXCL8-CXCR1/2-SMAD2/3-SNAI2/ZEB1 axis in tumor growth and
metastasis. (A) Image of subcutaneous xenograft tumors following
injection of CXCL8-overexpressing KYSE-150 cells (n=5) or control
cells (n=5) in BALB/c-nude mice. The volume (B) and weight (C) of
harvested xenograft tumors were measured (n=5). (D) Representative
immunohistochemical images of CXCR1/2, p-SMAD2/3, SNAI2 and ZEB1
expression in CXCL8-overexpressing ESCC tissues and corresponding
control tissues (n=5). Scale bar, 50 µm. (E) Mechanistic diagram of
CXCL8 in ESCC cells under an ER stress state. All data are
expressed as mean ± SD. *P < 0.05. vs. PCDH-Vector group. CXCL,
C-X-C motif chemokine ligand; IRE1α, inositol-requiring enzyme 1α;
PERK, protein kinase R (PKR)-like endoplasmic reticulum kinase;
ATF, activating transcription factor; XBP, X-box binding protein;
ER, endoplasmic reticulum; p-, phosphorylated; ESCC, esophageal
squamous cell carcinoma. |
The aforementioned results suggested that CXCL8,
which was highly secreted by ESCC cells in the ERS state, promoted
the EMT process by activating SMAD2/3 after binding to its
receptors. Activated SMAD2/3 directly or indirectly regulated the
transcription of SNAI2 and ZEB1 (Fig.
7E).
Discussion
The significance of ERS in tumor progression is well
recognized, where the activation of the UPR intricately governs the
fate of tumor cells (39,40). In different types of human cancer,
the activation of distinct elements within the UPR cascade was
poised to yield diverse cellular outcomes across nearly all stages
of tumor cell development (14,17,41–43).
In addition, a previous study proposed that the UPR may influence
key characteristics defining cancer, encompassing angiogenesis,
metastasis, genome stability, inflammatory responses and resistance
to drugs (39). As for the role of
ERS in the emergence and advancement of ESCC, studies have revealed
that the ERS-induced PERK/eIF2α/CHOP pathway might impede
proliferation and induce apoptosis (44,45).
The activation of the IRE1/JNK pathway in ESCC is implicated in
regulating apoptosis and autophagy (46). Notably, the IRE1α/AKT/mTOR
signaling axis has been identified as an inducer of apoptosis in
ESCC cells (47). Nevertheless, to
the best of our knowledge, the involvement of CXCL8 in the ERS of
ESCC and its specific functional role have not been reported. The
present study revealed that CXCL8 was one of the most significantly
upregulated genes in ESCC cells with ERS, and may be induced by the
IRE1α-XBP1 and PERK-ATF4 pathways. CXCL8-induced activation of
SMAD2/3 and SMAD2/3 subsequently exerted direct or indirect
regulation of SNAI2 and ZEB1 and promoted the progression of EMT in
ESCC cells.
CXCL8, recognized as a multifaceted chemokine,
encompasses diverse functions in biological processes and has been
implicated in numerous diseases including HIV (48), inflammatory bowel disease (49), reperfusion injury (50) and chronic obstructive pulmonary
disease (51,52). The increased expression of CXCL8
has also been reported in several types of cancer. For example,
elevated CXCL8 enhances the tumorigenic properties of human colon
cancer LoVo cells by activating specific signaling pathways
(23); CXCL8 serves as a pivotal
mediator in the tumor-stroma-inflammation network of
triple-negative breast cancer (24) and CXCL8 functions as an autocrine
growth regulator in human lung cancer pathogenesis (25).
Mechanistically, the present study uncovered a
previously unrecognized role of CXCL8 in driving ESCC progression
by engaging in ERS-associated cascades, challenging the
conventional perspective on its pro-metastatic functions.
Functional analyses demonstrated that tumor-derived CXCL8 served in
a dual capacity. It operated in a paracrine manner, modifying the
immune cell composition within the tumor microenvironment.
Simultaneously, CXCL8 acted in an autocrine manner, promoting
oncogenic signaling, angiogenesis and fostering pro-metastatic
traits such as invasion and resistance (53). However, the functional role and
underlying mechanisms of CXCL8 in the ERS state tumors remain
poorly investigated, with limited reports in only a few types of
cancer. In breast cancer cells, CXCL8 has been identified as one of
the secretory factors displaying the highest fold change after
treatment with TG and it might be induced via the PERK-CEBPδ
pathway (54). In thyroid tumors,
TG treatment markedly increased the expression of CXCL8 at both the
mRNA and protein levels (55). In
non-small cell lung adenocarcinoma A549 cells, the transcription
factors ATF4 and P65 downstream of the UPR directly orchestrated
the transcription of CXCL8 (56).
The present study revealed that the upregulation of CXCL8 in the
ERS state ESCC cells was not only regulated by the PERK/ATF4
signaling pathway as observed in non-small cell lung adenocarcinoma
(56), but also regulated by the
IRE1α/XBP1 pathway. The evident upregulation of CXCL8 by the two
pathways promoted ESCC migration and invasion through participating
in the EMT process.
The augmentation of CXCL8-CXCR1/2 expression has
been observed in several tumors. CXCL8/CXCR1/2 promoted
androgen-independent prostate cancer cell growth and proliferation
ability by activating cyclin D1 expression (57). In pancreatic cancer (PaCa),
CXCL8-CXCR2 notably increased cancer cell angiogenesis,
proliferation and invasion ability, and CXCR2 was an
anti-angiogenic target in PaCa (58). CXCR1 also promotes proliferation,
migration and invasion of gastric cancer in vitro and in
vivo (59). CXCL8 enhances
angiogenesis of glioblastoma endothelial cells via CXCR2 signaling
(60). The present study revealed
that CXCL8-CXCR1/2 could facilitate the migration and invasion of
ESCC cells in vitro. The promotion of tumor progression by
CXCL8 occurs due to its binding to CXCR1 and CXCR2 receptors,
indicating that targeting this interaction could be a promising
therapeutic strategy for ESCC.
The CXCL8-CXCR1/2 axis has been reported to be
involved in tumor progression through activating several signaling
pathways, including PI3K/AKT (36,61),
JAK2/STAT3 (37), MAPK (35,62),
phospholipase C (37) and
Rho-GTPase (37). In the present
study, aside from the aforementioned pathways, the phosphorylated
activation of SMAD2/3 by CXCL8/CXCR1 was observed. The activated
SMAD2/3 directly or indirectly modulated the transcription of EMT
markers SNAI2 and ZEB1 to further participate in the EMT process.
The direct transcriptional regulatory effect of p-SMAD2/3 on SNAI2
observed in the present study was consistent with the study by
Brandl et al (63) in panc1
cells, as for the indirect regulation of p-SMAD2/3 on ZEB1, there
may be intermediate genes or mechanisms involved. For example, in
the process of renal fibrosis, the miR-200 family regulates
TGF-β1-induced renal tubular EMT through the SMAD pathway by
targeting ZEB1 (64). Based on
bioinformatics analysis using the TransmiR v3.0 database and
TargetScan Release 7.1, miR-3146, miR-3163 and miR-3171 were
predicted to be regulated by SMAD3 and target the 3′-UTR of ZEB1 to
modulate its expression. However, further studies need to be
carried out to verify this prediction.
In summary, the findings of the present study
highlighted that CXCL8, originating from the IRE1α-XBP1 and
PERK-ATF4 pathways in ERS, facilitated the EMT process in ESCC
cells through the CXCL8-CXCR1/2-SMAD2/3-SNAI2/ZEB1 axis. This
underscored the potential therapeutic strategy of targeting the
CXCL8-CXCR1/2 axis to intervene in ERS signaling, hopefully
improving therapeutic efficacy in ESCC treatment.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by Grants from the National Natural
Science Foundation of China (grant nos. 82372863 and 82203622),
Hebei Natural Science Foundation (grant nos. H2022206598 and
H2024206106), 2024 Government Funded Clinical Medicine Excellent
Talent Training Project (grant nos. ZF2024099), Innovative Research
Team Support Program of the Fourth Hospital of Hebei Medical
University (grant nos. 2023B09 and 2023C15) and S&T Program of
Hebei (grant no. 20377766D).
Availability of data and materials
The sequencing data generated in the present study
may be found in the NCBI SRA database under accession number
PRJNA1256794 or at the following URL: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1256794.
The other data generated in the present study may be requested from
the corresponding author.
Authors' contributions
WG and LL contributed to the conception and design
of the study, data acquisition, analysis and interpretation,
supervision of the experiments, and revision of the manuscript. JW
carried out the experiments, analyzed the data and drafted the
paper. FS and JL collected the data and prepared the tables and
figures. HX, XY and FL carried out the statistical analysis. All
authors read and approved the final manuscript. WG and LL confirm
the authenticity of all raw data.
Ethics approval and consent to
participate
All animal experiments in the present study were
conducted in accordance with the Regulations for the Administration
of Affairs Concerning Experimental Animals and relevant
institutional guidelines. The experiments were approved by the
Institutional Animal Care and Use Committee (IACUC) of the Fourth
Hospital of Hebei Medical University (approval no. IACUC-4th Hos
Hebmu-2023001).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jin W, Huang K, Ding Z, Zhang M, Li C,
Yuan Z, Ma K and Ye X: Global, regional, and national burden of
esophageal cancer: A systematic analysis of the Global Burden of
Disease Study 2021. Biomark Res. 13:32025. View Article : Google Scholar
|
|
2
|
Ilic I, Zivanovic Macuzic I, Ravic-Nikolic
A, Ilic M and Milicic V: Global burden of esophageal cancer and its
risk factors: A systematic analysis of the global burden of disease
study 2019. Life (Basel). 15:242024.
|
|
3
|
Qi L, Sun M, Liu W, Zhang X, Yu Y, Tian Z,
Ni Z, Zheng R and Li Y: Global esophageal cancer epidemiology in
2022 and predictions for 2050: A comprehensive analysis and
projections based on GLOBOCAN data. Chin Med J (Engl).
137:3108–3116. 2024. View Article : Google Scholar
|
|
4
|
Yokoyama A, Tsutsumi E, Imazeki H, Suwa Y,
Nakamura C, Mizukami T and Yokoyama T: Salivary acetaldehyde
concentration according to alcoholic beverage consumed and aldehyde
dehydrogenase-2 genotype. Alcohol Clin Exp Res. 32:1607–1614. 2008.
View Article : Google Scholar
|
|
5
|
Prabhu A, Obi KO and Rubenstein JH: The
synergistic effects of alcohol and tobacco consumption on the risk
of esophageal squamous cell carcinoma: a meta-analysis. Am J
Gastroenterol. 109:822–827. 2014. View Article : Google Scholar
|
|
6
|
Blaydon DC, Etheridge SL, Risk JM, Hennies
HC, Gay LJ, Carroll R, Plagnol V, McRonald FE, Stevens HP, Spurr
NK, et al: RHBDF2 mutations are associated with tylosis, a familial
esophageal cancer syndrome. Am J Hum Genet. 90:340–346. 2012.
View Article : Google Scholar
|
|
7
|
Ludmir EB, Stephens SJ, Palta M, Willett
CG and Czito BG: Human papillomavirus tumor infection in esophageal
squamous cell carcinoma. J Gastrointest Oncol. 6:287–295. 2015.
|
|
8
|
Kato K, Ito Y, Nozaki I, Daiko H, Kojima
T, Yano M, Ueno M, Nakagawa S, Takagi M, Tsunoda S, et al:
Parallel-group controlled trial of surgery versus chemoradiotherapy
in patients with stage I esophageal squamous cell carcinoma.
Gastroenterology. 161:1878–1886.e2. 2021. View Article : Google Scholar
|
|
9
|
Sun JM, Shen L, Shah MA, Enzinger P,
Adenis A, Doi T, Kojima T, Metges JP, Li Z, Kim SB, et al:
Pembrolizumab plus chemotherapy versus chemotherapy alone for
first-line treatment of advanced oesophageal cancer (KEYNOTE-590):
A randomised, placebo-controlled, phase 3 study. Lancet.
398:759–771. 2021. View Article : Google Scholar
|
|
10
|
Fan X, Wang J, Xia L, Qiu H, Tian Y,
Zhangcai Y, Luo X, Gao Y, Li C, Wu Y, et al: Efficacy of endoscopic
therapy for T1b esophageal cancer and construction of prognosis
prediction model: A retrospective cohort study. Int J Surg.
109:1708–1719. 2023. View Article : Google Scholar
|
|
11
|
Li Y, Li Y and Chen X: NOTCH and
esophageal squamous cell carcinoma. Adv Exp Med Biol. 1287:59–68.
2021. View Article : Google Scholar
|
|
12
|
Ron D and Walter P: Signal integration in
the endoplasmic reticulum unfolded protein response. Nat Rev Mol
Cell Biol. 8:519–529. 2007. View
Article : Google Scholar
|
|
13
|
Bobrovnikova-Marjon E, Grigoriadou C,
Pytel D, Zhang F, Ye J, Koumenis C, Cavener D and Diehl JA: PERK
promotes cancer cell proliferation and tumor growth by limiting
oxidative DNA damage. Oncogene. 29:3881–3895. 2010. View Article : Google Scholar
|
|
14
|
Urra H, Henriquez DR, Cánovas J,
Villarroel-Campos D, Carreras-Sureda A, Pulgar E, Molina E, Hazari
YM, Limia CM, Alvarez-Rojas S, et al: IRE1α governs cytoskeleton
remodelling and cell migration through a direct interaction with
filamin A. Nat Cell Biol. 20:942–953. 2018. View Article : Google Scholar
|
|
15
|
Hart LS, Cunningham JT, Datta T, Dey S,
Tameire F, Lehman SL, Qiu B, Zhang H, Cerniglia G, Bi M, et al: ER
stress-mediated autophagy promotes Myc-dependent transformation and
tumor growth. J Clin Invest. 122:4621–4634. 2012. View Article : Google Scholar
|
|
16
|
Avivar-Valderas A, Salas E,
Bobrovnikova-Marjon E, Diehl JA, Nagi C, Debnath J and
Aguirre-Ghiso JA: PERK integrates autophagy and oxidative stress
responses to promote survival during extracellular matrix
detachment. Mol Cell Biol. 31:3616–3629. 2011. View Article : Google Scholar
|
|
17
|
Notte A, Rebucci M, Fransolet M, Roegiers
E, Genin M, Tellier C, Watillon K, Fattaccioli A, Arnould T and
Michiels C: Taxol-induced unfolded protein response activation in
breast cancer cells exposed to hypoxia: ATF4 activation regulates
autophagy and inhibits apoptosis. Int J Biochem Cell Biol. 62:1–14.
2015. View Article : Google Scholar
|
|
18
|
Lu M, Lawrence DA, Marsters S,
Acosta-Alvear D, Kimmig P, Mendez AS, Paton AW, Paton JC, Walter P
and Ashkenazi A: Opposing unfolded-protein-response signals
converge on death receptor 5 to control apoptosis. Science.
345:98–101. 2014. View Article : Google Scholar
|
|
19
|
Li G, Mongillo M, Chin KT, Harding H, Ron
D, Marks AR and Tabas I: Role of ERO1-alpha-mediated stimulation of
inositol 1,4,5-triphosphate receptor activity in endoplasmic
reticulum stress-induced apoptosis. J Cell Biol. 186:783–792. 2009.
View Article : Google Scholar
|
|
20
|
Prieto K, Cao Y, Mohamed E, Trillo-Tinoco
J, Sierra RA, Urueña C, Sandoval TA, Fiorentino S, Rodriguez PC and
Barreto A: Polyphenol-rich extract induces apoptosis with
immunogenic markers in melanoma cells through the ER
stress-associated kinase PERK. Cell Death Discov. 5:1342019.
View Article : Google Scholar
|
|
21
|
Alfaro C, Sanmamed MF, Rodríguez-Ruiz ME,
Teijeira Á, Oñate C, González Á, Ponz M, Schalper KA, Pérez-Gracia
JL and Melero I: Interleukin-8 in cancer pathogenesis, treatment
and follow-up. Cancer Treat Rev. 60:24–31. 2017. View Article : Google Scholar
|
|
22
|
Raghuwanshi SK, Su Y, Singh V, Haynes K,
Richmond A and Richardson RM: The chemokine receptors CXCR1 and
CXCR2 couple to distinct G protein-coupled receptor kinases to
mediate and regulate leukocyte functions. J Immunol. 189:2824–2832.
2012. View Article : Google Scholar
|
|
23
|
Shen T, Yang Z, Cheng X, Xiao Y, Yu K, Cai
X, Xia C and Li Y: CXCL8 induces epithelial-mesenchymal transition
in colon cancer cells via the PI3K/Akt/NF-κB signaling pathway.
Oncol Rep. 37:2095–2100. 2017. View Article : Google Scholar
|
|
24
|
Liubomirski Y, Lerrer S, Meshel T,
Rubinstein-Achiasaf L, Morein D, Wiemann S, Körner C and Ben-Baruch
A: Tumor-stroma-inflammation networks promote pro-metastatic
chemokines and aggressiveness characteristics in triple-negative
breast cancer. Front Immunol. 10:7572019. View Article : Google Scholar
|
|
25
|
Zhu YM, Webster SJ, Flower D and Woll PJ:
Interleukin-8/CXCL8 is a growth factor for human lung cancer cells.
Br J Cancer. 91:1970–1976. 2004. View Article : Google Scholar
|
|
26
|
Yi M, Peng C, Xia B and Gan L: CXCL8
facilitates the survival and paclitaxel-resistance of
triple-negative breast cancers. Clin Breast Cancer. 22:e191–e198.
2022. View Article : Google Scholar
|
|
27
|
Zhai J, Shen J, Xie G, Wu J, He M, Gao L,
Zhang Y, Yao X and Shen L: Cancer-associated fibroblasts-derived
IL-8 mediates resistance to cisplatin in human gastric cancer.
Cancer Lett. 454:37–43. 2019. View Article : Google Scholar
|
|
28
|
Xue J, Song Y, Xu W and Zhu Y: The
CDK1-related lncRNA and CXCL8 mediated immune resistance in lung
adenocarcinoma. Cells. 11:26882022. View Article : Google Scholar
|
|
29
|
Zhang H, Yu QL, Meng L, Huang H, Liu H,
Zhang N, Liu N, Yang J, Zhang YZ and Huang Q: TAZ-regulated
expression of IL-8 is involved in chemoresistance of hepatocellular
carcinoma cells. Arch Biochem Biophys. 693:1085712020. View Article : Google Scholar
|
|
30
|
Liu Q, Li A, Tian Y, Wu JD, Liu Y, Li T,
Chen Y, Han X and Wu K: The CXCL8-CXCR1/2 pathways in cancer.
Cytokine Growth Factor Rev. 31:61–71. 2016. View Article : Google Scholar
|
|
31
|
Ogura M, Takeuchi H, Kawakubo H, Nishi T,
Fukuda K, Nakamura R, Takahashi T, Wada N, Saikawa Y, Omori T, et
al: Clinical significance of CXCL-8/CXCR-2 network in esophageal
squamous cell carcinoma. Surgery. 154:512–520. 2013. View Article : Google Scholar
|
|
32
|
Hosono M, Koma YI, Takase N, Urakawa N,
Higashino N, Suemune K, Kodaira H, Nishio M, Shigeoka M, Kakeji Y
and Yokozaki H: CXCL8 derived from tumor-associated macrophages and
esophageal squamous cell carcinomas contributes to tumor
progression by promoting migration and invasion of cancer cells.
Oncotarget. 8:106071–106088. 2017. View Article : Google Scholar
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
34
|
Hayakawa K, Nakajima S, Hiramatsu N,
Okamura M, Huang T, Saito Y, Tagawa Y, Tamai M, Takahashi S, Yao J
and Kitamura M: ER stress depresses NF-kappaB activation in
mesangial cells through preferential induction of C/EBP beta. J Am
Soc Nephrol. 21:73–81. 2010. View Article : Google Scholar
|
|
35
|
Knall C, Young S, Nick JA, Buhl AM,
Worthen GS and Johnson GL: Interleukin-8 regulation of the
Ras/Raf/mitogen-activated protein kinase pathway in human
neutrophils. J Biol Chem. 271:2832–2838. 1996. View Article : Google Scholar
|
|
36
|
Cheng GZ, Park S, Shu S, He L, Kong W,
Zhang W, Yuan Z, Wang LH and Cheng JQ: Advances of AKT pathway in
human oncogenesis and as a target for anti-cancer drug discovery.
Curr Cancer Drug Targets. 8:2–6. 2008. View Article : Google Scholar
|
|
37
|
Waugh DJJ and Wilson C: The interleukin-8
pathway in cancer. Clin Cancer Res. 14:6735–6741. 2008. View Article : Google Scholar
|
|
38
|
Cheng K and Hao M: Metformin inhibits
TGF-β1-induced epithelial-to-mesenchymal transition via PKM2
relative-mTOR/p70s6k signaling pathway in cervical carcinoma cells.
Int J Mol Sci. 17:20002016. View Article : Google Scholar
|
|
39
|
Hetz C and Papa FR: The unfolded protein
response and cell fate control. Mol Cell. 69:169–181. 2018.
View Article : Google Scholar
|
|
40
|
He J, Zhou Y and Sun L: Emerging
mechanisms of the unfolded protein response in therapeutic
resistance: From chemotherapy to Immunotherapy. Cell Commun Signal.
22:892024. View Article : Google Scholar
|
|
41
|
Dong D, Ni M, Li J, Xiong S, Ye W, Virrey
JJ, Mao C, Ye R, Wang M, Pen L, et al: Critical role of the stress
chaperone GRP78/BiP in tumor proliferation, survival, and tumor
angiogenesis in transgene-induced mammary tumor development. Cancer
Res. 68:498–505. 2008. View Article : Google Scholar
|
|
42
|
Li J and Lee AS: Stress induction of
GRP78/BiP and its role in cancer. Curr Mol Med. 6:45–54. 2006.
View Article : Google Scholar
|
|
43
|
Tan Y, Dourdin N, Wu C, De Veyra T, Elce
JS and Greer PA: Ubiquitous calpains promote caspase-12 and JNK
activation during endoplasmic reticulum stress-induced apoptosis. J
Biol Chem. 281:16016–16024. 2006. View Article : Google Scholar
|
|
44
|
Yuan YJ, Liu S, Yang H, Xu JL, Zhai J,
Jiang HM and Sun B: Acetylshikonin induces apoptosis through the
endoplasmic reticulum stress-activated PERK/eIF2α/CHOP
axis in oesophageal squamous cell carcinoma. J Cell Mol Med.
28:e180302024. View Article : Google Scholar
|
|
45
|
Hetz C, Zhang K and Kaufman RJ:
Mechanisms, regulation and functions of the unfolded protein
response. Nat Rev Mol Cell Biol. 21:421–438. 2020. View Article : Google Scholar
|
|
46
|
Yin X, Zhang P, Xia N, Wu S, Liu B, Weng L
and Shang M: GPx8 regulates apoptosis and autophagy in esophageal
squamous cell carcinoma through the IRE1/JNK pathway. Cell Signal.
93:1103072022. View Article : Google Scholar
|
|
47
|
Wang YM, Xu X, Tang J, Sun ZY, Fu YJ, Zhao
XJ, Ma XM and Ye Q: Apatinib induces endoplasmic reticulum
stress-mediated apoptosis and autophagy and potentiates cell
sensitivity to paclitaxel via the IRE-1α-AKT-mTOR pathway in
esophageal squamous cell carcinoma. Cell Biosci. 11:1242021.
View Article : Google Scholar
|
|
48
|
Mamik MK and Ghorpade A: Chemokine CXCL8
promotes HIV-1 replication in human monocyte-derived macrophages
and primary microglia via nuclear factor-κB pathway. PLoS One.
9:e921452014. View Article : Google Scholar
|
|
49
|
Aarntzen EHJG, Hermsen R, Drenth JPH,
Boerman OC and Oyen WJG: 99mTc-CXCL8 SPECT to monitor disease
activity in inflammatory bowel disease. J Nucl Med. 57:398–403.
2016. View Article : Google Scholar
|
|
50
|
Amrouche L, Desbuissons G, Rabant M,
Sauvaget V, Nguyen C, Benon A, Barre P, Rabaté C, Lebreton X,
Gallazzini M, et al: MicroRNA-146a in human and experimental
ischemic AKI: CXCL8-dependent mechanism of action. J Am Soc
Nephrol. 28:479–493. 2017. View Article : Google Scholar
|
|
51
|
Barnes PJ: New treatments for chronic
obstructive pulmonary disease. Ann Ist Super Sanita. 39:573–582.
2003.
|
|
52
|
Traves SL, Smith SJ, Barnes PJ and
Donnelly LE: Specific CXC but not CC chemokines cause elevated
monocyte migration in COPD: A role for CXCR2. J Leukoc Biol.
76:441–450. 2004. View Article : Google Scholar
|
|
53
|
Long X, Ye Y, Zhang L, Liu P, Yu W, Wei F,
Ren X and Yu J: IL-8, a novel messenger to cross-link inflammation
and tumor EMT via autocrine and paracrine pathways (Review). Int J
Oncol. 48:5–12. 2016. View Article : Google Scholar
|
|
54
|
Sheshadri N, Poria DK, Sharan S, Hu Y, Yan
C, Koparde VN, Balamurugan K and Sterneck E: PERK signaling through
C/EBPδ contributes to ER stress-induced expression of
immunomodulatory and tumor promoting chemokines by cancer cells.
Cell Death Dis. 12:10382021. View Article : Google Scholar
|
|
55
|
Zhang L, Xu S, Cheng X, Wu J, Wang Y, Gao
W, Bao J and Yu H: Inflammatory tumor microenvironment of thyroid
cancer promotes cellular dedifferentiation and silencing of
iodide-handling genes expression. Pathol Res Pract. 246:1544952023.
View Article : Google Scholar
|
|
56
|
Püschel F, Favaro F, Redondo-Pedraza J,
Lucendo E, Iurlaro R, Marchetti S, Majem B, Eldering E, Nadal E,
Ricci JE, et al: Starvation and antimetabolic therapy promote
cytokine release and recruitment of immune cells. Proc Natl Acad
Sci USA. 117:9932–9941. 2020. View Article : Google Scholar
|
|
57
|
MacManus CF, Pettigrew J, Seaton A, Wilson
C, Maxwell PJ, Berlingeri S, Purcell C, McGurk M, Johnston PG and
Waugh DJJ: Interleukin-8 signaling promotes translational
regulation of cyclin D in androgen-independent prostate cancer
cells. Mol Cancer Res. 5:737–748. 2007. View Article : Google Scholar
|
|
58
|
Matsuo Y, Raimondo M, Woodward TA, Wallace
MB, Gill KR, Tong Z, Burdick MD, Yang Z, Strieter RM, Hoffman RM
and Guha S: CXC-chemokine/CXCR2 biological axis promotes
angiogenesis in vitro and in vivo in pancreatic cancer. Int J
Cancer. 125:1027–1037. 2009. View Article : Google Scholar
|
|
59
|
Wang J, Hu W, Wu X, Wang K, Yu J, Luo B,
Luo G, Wang W, Wang H, Li J and Wen J: CXCR1 promotes malignant
behavior of gastric cancer cells in vitro and in vivo in AKT and
ERK1/2 phosphorylation. Int J Oncol. 48:2184–2196. 2016. View Article : Google Scholar
|
|
60
|
Urbantat RM, Blank A, Kremenetskaia I,
Vajkoczy P, Acker G and Brandenburg S: The CXCL2/IL8/CXCR2 pathway
is relevant for brain tumor malignancy and endothelial cell
function. Int J Mol Sci. 22:26342021. View Article : Google Scholar
|
|
61
|
Knall C, Worthen GS and Johnson GL:
Interleukin 8-stimulated phosphatidylinositol-3-kinase activity
regulates the migration of human neutrophils independent of
extracellular signal-regulated kinase and p38 mitogen-activated
protein kinases. Proc Natl Acad Sci USA. 94:3052–3057. 1997.
View Article : Google Scholar
|
|
62
|
Luppi F, Longo AM, de Boer WI, Rabe KF and
Hiemstra PS: Interleukin-8 stimulates cell proliferation in
non-small cell lung cancer through epidermal growth factor receptor
transactivation. Lung Cancer. 56:25–33. 2007. View Article : Google Scholar
|
|
63
|
Brandl M, Seidler B, Haller F, Adamski J,
Schmid RM, Saur D and Schneider G: IKK(α) controls canonical
TGF(ß)-SMAD signaling to regulate genes expressing SNAIL and SLUG
during EMT in panc1 cells. J Cell Sci. 123:4231–4239. 2010.
View Article : Google Scholar
|
|
64
|
Xiong M, Jiang L, Zhou Y, Qiu W, Fang L,
Tan R, Wen P and Yang J: The miR-200 family regulates
TGF-β1-induced renal tubular epithelial to mesenchymal transition
through Smad pathway by targeting ZEB1 and ZEB2 expression. Am J
Physiol Renal Physiol. 302:F369–F379. 2012. View Article : Google Scholar
|