Introduction
Myocardial infarction (MI) is associated with a high
incidence of morbidity and mortality. Despite the beneficial
effects of prompt reperfusion therapy on survival rates in the
initial stages of MI, post-MI cardiac remodeling remains a
significant contributing factor to heart failure and later
mortality (1). Cardiac remodeling
is a complex process involving functional and structural
disturbances in the myocardium, primarily driven by myocardial
injury and overload (2). This
process is typically characterized by three distinct stages:
Inflammation, proliferation and maturation (3). Single-cell RNA sequencing (scRNA-seq)
represents a novel technological advancement that facilitates
transcriptome analysis at the single-cell level, thereby offering
comprehensive insights into the heterogeneity of diverse cell
populations within tissues. This technology enables the
identification of novel marker genes, cellular clusters and
cellular heterogeneity, as well as the inference of cellular
lineage trajectories. Furthermore, it can elucidate intercellular
interactions and facilitates comparisons between healthy and
diseased cells (4,5). In the context of MI, scRNA-seq can
elucidate transcriptional variability among similar cell types in
post-infarction tissues in comparison to normal tissues.
Furthermore, it can detect gene expression differences and dynamic
changes over time.
The present study reviewed the application of
scRNA-seq for the analysis of temporal changes in cardiomyocytes
and non-cardiomyocytes following MI. It aimed to provide a
theoretical basis for identifying new therapeutic targets for
preventing and treating post-infarction cardiac remodeling by
classifying different cell clusters and elucidating their roles in
post-infarction tissue remodeling.
Single-cell sequencing
Emergence of single-cell
sequencing
Traditional gene expression analysis techniques,
such as quantitative PCR, microarrays and high-throughput RNA
sequencing, permit the analysis of cell populations that are
heterogeneous at the gene expression level. These methods provide
only an averaged representation of gene expression across the
group, often overlooking low-abundance transcripts and masking the
unique characteristics of individual cells. As a result,
intercellular heterogeneity remains largely undetected (6–8). To
address these limitations, single-cell sequencing technology was
developed, enabling a shift from population-level analyses to the
study of individual cells. This technological advancement allows
for the in-depth exploration of tissues, organs and even entire
systems at the cellular level, markedly enhancing the current
understanding of cellular diversity and function (9–11).
Single-cell sequencing technology
Single-cell sequencing technology can be broadly
summarized as the amplification and sequencing of DNA or RNA at the
level of a single cell. The process primarily involves single-cell
isolation, nucleic acid processing, DNA amplification, library
construction, sequencing and data analysis (12–15).
Currently, single-cell sequencing techniques are categorized into
several types, including single-cell genome sequencing, single-cell
transcriptome sequencing (scRNA-seq), single-cell epigenome
sequencing, single-cell multiomics sequencing and spatial
transcriptome sequencing. Common single-cell sequencing methods
include Smart-seq (16),
Smart-seq2 (17), Drop-seq
(18), STRT-seq (19,20),
MARS-seq (21), MARS-seq2
(22) and Fluidigm C1 (23), which can be selected based on
specific experimental goals. Single-cell sequencing has been
applied across various fields, such as stem cell and developmental
biology, oncology and immunology and has become increasingly
important in both clinical and basic medical research. The present
review focused on the application of single-cell sequencing for
analyzing individual cells within post-infarction myocardial
tissues. Specifically, it highlighted the exploration of the
heterogeneity and enrichment of cardiomyocytes and
non-cardiomyocytes in these tissues, demonstrating the clustering
of these cells at different time points (for example, days 3, 7, 8
and 21 post-infarction) compared with control groups.
Advantages and disadvantages of
different single-cell sequencing methods to study
cardiomyocytes
Cardiomyocytes in the heart can reach ≤100 µm in
length and 25 µm in width. This presents challenges for current
commercial single-cell transcriptome sequencing (scRNA-seq)
platforms, which have limitations regarding cell size. For
instance, the 10X Genomics platform has a microtubule diameter of
50 µm in its chip and it is recommended that the captured cells not
exceed 40 µm. As a result, cells must be filtered using a 30–40 µm
sieve to remove larger cells prior to capture. However, this
filtration step excludes large cardiomyocytes and prevents the
generation of scRNA-seq data from adult primary heart tissues
(24), To address this issue, the
majority of studies on cardiomyocytes have adopted single-nucleus
RNA sequencing (snRNA-seq), as it targets the nuclei of cells
rather than the entire cell. While snRNA-seq offers the advantage
of detecting nuclear-localized transcripts and maintaining membrane
integrity, it has limitations. Specifically, the method reduces
assay sensitivity and omits cytoplasmic mRNA information.
Additionally, isolated nuclei tend to adhere more than isolated
whole cells, increasing the risk of aggregation and duplex rate
expansion (25,26). However, snRNA-seq allows for the
transcriptomic analysis of tens of thousands of nuclei isolated
from fresh or frozen tissues, including mature mammalian hearts. It
facilitates the isolation of intact single cells from complex
tissues, reduces bias from easily recoverable cell types and
mitigates aberrant gene expression during cytokinesis (27,28).
Single-cell sequencing to analyze changes in
cardiomyocytes following MI
Cluster analysis and changes in
cardiomyocytes following MI
There are two key studies that focus on the cluster
analysis of cardiomyocytes following MI, as outlined in Table I. In 2020, Litviňuková et al
(29) identified five ventricular
cardiomyocyte populations (vCM1-vCM5) and five atrial cardiomyocyte
populations (aCM1-aCM5) by isolating single cells, nuclei and
CD45+ cells from the right and left ventricular free
walls, right and left atria, left ventricular apices and
interventricular septum using both scRNA-seq and snRNA-seq.
Detailed information about the marker genes for each cluster is
provided in Fig. 1A. vCM1 and vCM2
showed minimal differences, with vCM2 highly expressing PRELID2 and
the myosin genes MYH6 and CDH13. vCM3 expressed stress-responsive
genes, including ANKRD1 (30),
FHL1 (31), DUSP27 (32), XIRP1 and XIRP2. vCM4 was enriched
in nuclear-encoded mitochondrial genes such as NDUFB11, NDUFA4,
COX7C and COX5B. vCM5 highly expressed DLC1 and EBF2. Regarding the
atrial myocytes, aCM1 expressed basal atrial myocyte genes and
lower levels of neurological function molecules such as ADGRL2,
NFXL1 and ROBO2, while aCM2 was enriched in the right atrium and
predominantly expressed HAMP, SLIT3, ALDH1A2 (33), BRINP3 and GRXCR2. aCM3-5 and vCM3-5
shared similar transcriptional profiles. The present study
systematically mapped the molecular typing of atrial and
ventricular myocytes for the first time in adults, laying the
foundation for understanding cellular diversity in the
physiological state of the heart.
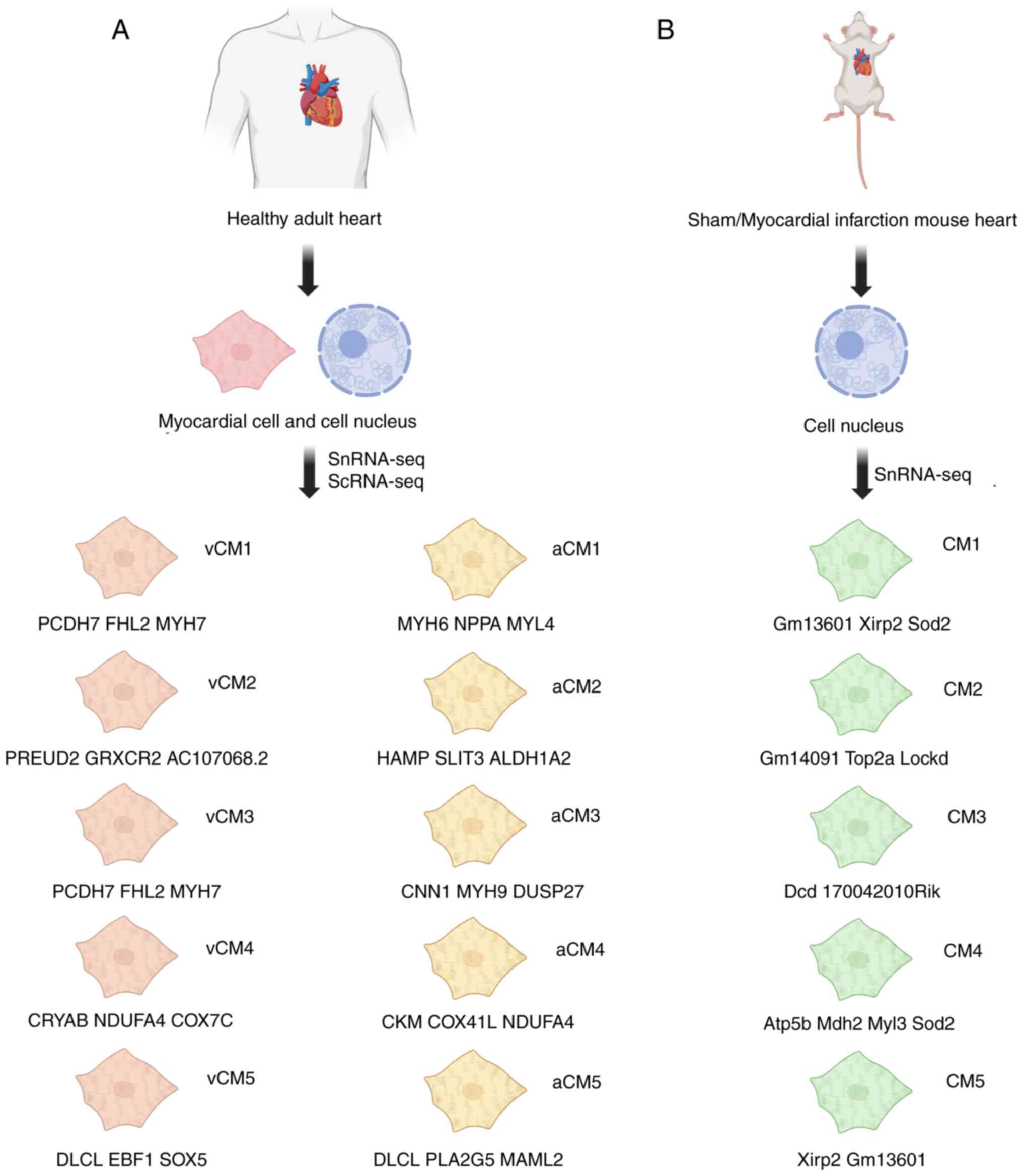 | Figure 1.Cardiomyocyte cluster typing and
marker genes for each cluster. vCM1-vCM5 represent myocardial cell
populations under different ventricular conditions, which are
determined by characteristics such as stress response, metabolic
changes and proliferation. Marker genes such as MYH6 are notably
expressed in vCM2, indicating that they are closely related to
metabolic activities in myocardial repair. (A) Adult cardiomyocyte
cluster typing and marker genes for each cluster. (B) Mouse
cardiomyocyte cluster typing and marker genes for each cluster.
SnRNA-seq, single-nucleus RNA sequencing; ScRNA-seq, single-cell
RNA sequencing; vCM, ventricular cardiomyocyte clusters; aCM,
atrial cardiomyocyte clusters; vCM1, functionally maintained
cardiomyocytes; vCM2, myocardial cells enriched in the right
ventricle (with high expression of MYH6); vCM3, stress-responsive
cardiomyocytes (expressing ANKRD1, etc.); vCM4, high-energy
metabolic cardiomyocytes (enriched mitochondrial genes, CRYAB);
vCM5, electrophysiologically associated cardiomyocytes (high
expression of DLC1 and EBF2); aCM1, basic functional
cardiomyocytes; aCM2, metabolic regulation of cardiomyocytes in the
right atrium (predominantly expressed ALDH1A2, etc.); aCM3, smooth
muscle-like cardiomyocytes; aCM4, cardiomyocytes with high
metabolic activity; aCM5, electrophysiological regulation of
cardiomyocytes; CM1, steady-state contractile mature
cardiomyocytes; CM2, injury stress-response cardiomyocytes (high
expression of Top2A and Casc5); CM3, metabolism-adaptive
cardiomyocytes; CM4, inflammatory/fibrotic regulatory
cardiomyocytes (expressing Atp5b, Sod2 and Mb); CM5, terminally
differentiated/senescent cardiomyocytes (expressing Xirp2, Ankrd1
and CD44). |
 | Table I.A typical study on the analysis of
myocardial cell subsets by ScRNA-seq. |
Table I.
A typical study on the analysis of
myocardial cell subsets by ScRNA-seq.
| Source of
organization | Right and left
ventricular free walls, right and left atria, left ventricular apex
and interventricular septum in adults | Ventricular tissue
in mice |
|---|
| Methods | 1. Fresh tissue for
ScRNA-seq | SnRNA-seq |
|
| 2. Frozen tissue
for SnRNA-seq |
|
| Number of cells
captured | 45,870 cells;
78,023 CD45+ enriched cells and 363,213 nucleus | 21,737 myocardial
cell nucleus |
| Clusters | vCM1-vCM5;
aCM1-aCM5 | CM1-CM5 |
Cui et al (34) performed snRNA-seq on both
regenerating and non-regenerating cardiomyocytes in neonatal mice
on postnatal days 1 and 8, as well as on days 1 and 3 following MI
[1 and 3 days post-infarction (dpi)], comparing these with
sham-operated mice. The authors identified five main cardiomyocyte
populations, named CM1-CM5, based on the expression of Myh6. The
marker genes for each cluster are detailed in Fig. 1B. CM2 was characterized by high
expression of cell cycle genes such as Top2A and Casc5. CM4 highly
expressed metabolic genes involved in oxygen reduction (such as
Atp5b, Sod2 and Mb) and mechanistically, overexpression of NFYA and
NFE2L1 promoted cardiomyocyte proliferation and survival through
the activation of CM4-associated injury response pathways. CM5
exhibited high expression of actin filament-regulated genes (such
as Xirp2, Ankrd1 and CD44) with a gene expression profile
suggestive of decreased cardiac function (downregulation of
contractile/calcium channel genes), activation of stress/apoptotic
pathways and association with pathological remodeling (upregulation
of dilated cardiomyopathy and muscle hypertrophy-related genes).
Additionally, CM4 was found to be associated with cardiac
regeneration, as it upregulated cell cycle genes, particularly
during the G2/M phase. The proportion of CM4
cardiomyocytes increased following cardiac injury, with a marked
increase in regenerating hearts at 3 dpi, suggesting that CM4 may
be a key source of cardiomyocyte proliferation after injury. By
contrast, the CM5 population was markedly increased in
non-regenerating hearts at 1 and 3 dpi, which was potentially
linked to hypertrophic remodeling and cardiomyocyte apoptosis after
infarction (34). Together, the
aforementioned two studies revealed significant molecular
heterogeneity in cardiomyocytes and identified key functional
subpopulations. In particular, the study of Cui et al
(34) highlighted the value of CM4
as a potential target for regenerative therapies, as well as the
importance of CM5 as a marker of pathological remodeling.
Single-cell sequencing to investigate
the regenerative properties of cardiomyocytes
Kretzschmar et al (35) found that circulating cardiomyocytes
were observed only during the early postnatal growth phase by using
single-cell mRNA sequencing and genetic lineage tracing in two Ki67
knock-in mouse models. While a large number of proliferating
cardiomyocytes were detected in neonatal hearts, they were absent
in damaged adult hearts. Similarly, Cui et al (34) analyzed the number of proliferating
cardiomyocytes in regenerating hearts at 1, 3 and 7 dpi, revealing
that cardiomyocyte proliferation commenced as early as 3 dpi.
In other studies, fluorescence-activated cell
sorting was used to isolate cardiomyocyte nuclei with an antibody
targeting the myocardial nuclear membrane protein PCM1, which then
facilitated snRNA-seq analysis of cardiomyocyte nuclei (24,36,37).
Analysis of nuclear RNAs revealed that 23% of unique molecular
identifiers were mapped to introns and 52% to exons, confirming
cardiomyocyte regeneration following MI in neonatal mice (34). Additionally, Tani et al
(38) identified the mechanisms
underlying cardiac reprogramming for in vivo repair through
microarray and scRNA-seq. That study demonstrated that cardiac
reprogramming could repair chronic MI by promoting myocardial
regeneration and reducing fibrosis.
Heterogeneity of non-cardiomyocytes and
changes following MI
Endothelial cell (EC) heterogeneity
and changes following MI
The heterogeneity of ECs following MI has been
extensively studied using single-cell sequencing, with three key
articles providing valuable insights, as summarized in Table II. In 2020, Wang et al
(39) performed scRNA-seq on
ventricular tissues from timed-pregnant ICR/CD-1 regenerating and
non-regenerating mice, as well as age-matched sham controls, at 1
and 3 days following MI. Their analysis revealed that neonatal 1-
and 8-days mouse hearts exhibited heterogeneous EC clusters. A
total of six EC clusters were identified through cluster analysis,
namely Art.EC, VEC1-3, Endo.EC and Pro.EC. The marker genes for
each subcluster are shown in Fig.
2A. However, changes in the number of post-infarction
subclusters were not discussed in detail. All six clusters
exhibited high expression of the EC marker genes Cdh5 and Pecam1.
The Cxcl12 gene and others were enriched in the Art.EC population
(40), while H19 and Cpe were
enriched in the Endo.EC population (41,42).
Enrichment of cell cycle genes (Hmgb2, Birc5) in Pro.EC may provide
a cellular source for vascular regeneration.
![Endothelial cell cluster typing and
marker genes for each cluster. The high expression of capillary
marker Gpihbp1 in VEC1 and VEC2, as well as the enrichment of
macrovascular genes such as Plvap and Vwf in VEC3, indicate that
they are related to the maintenance of vascular activity and
angiogenesis. Endothelial cells have multiple functions in blood
vessels, including vascular stability, blood flow regulation,
material exchange and immune response. Following myocardial
infarction, endothelial cells have the functions of repair,
vascular regeneration and remodeling, as well as coagulation. (A)
Endothelial cell clusters of ventricular tissue origin and marker
genes. (B) Endothelial cell clusters of TIP origin and marker
genes. TIP, cardiac interstitial cell population; ScRNA-seq,
single-cell RNA sequencing; VEC1-3, venous endothelial cells (VEC1
highly expresses Gpihbp1, VEC2 cluster highly expresses Cxcl1 and
Icam1, VEC3 highly expresses Plvap and Vwf); Art.EC, arterial
endothelial cells (enriched with artery-related genes such as
Cxcl12); Endo, metabolism-related endothelial cells (specifically
expressing metabolic genes such as H19 and Cpe); Pro.EC,
proliferative active endothelial cells [highly expressing
proliferation genes Hmgb2 and Birc5]; VEC1, microvascular
endothelial cells [expressing Ly6a (encoding SCA1) and vascular
transcription factor Sox17]; VEC2, arterial endothelial cells
(involved in NOTCH signaling pathways, such as Sox17, Hey1); VEC3,
venous endothelial cells.](/article_images/mmr/32/6/mmr-32-06-13680-g01.jpg) | Figure 2.Endothelial cell cluster typing and
marker genes for each cluster. The high expression of capillary
marker Gpihbp1 in VEC1 and VEC2, as well as the enrichment of
macrovascular genes such as Plvap and Vwf in VEC3, indicate that
they are related to the maintenance of vascular activity and
angiogenesis. Endothelial cells have multiple functions in blood
vessels, including vascular stability, blood flow regulation,
material exchange and immune response. Following myocardial
infarction, endothelial cells have the functions of repair,
vascular regeneration and remodeling, as well as coagulation. (A)
Endothelial cell clusters of ventricular tissue origin and marker
genes. (B) Endothelial cell clusters of TIP origin and marker
genes. TIP, cardiac interstitial cell population; ScRNA-seq,
single-cell RNA sequencing; VEC1-3, venous endothelial cells (VEC1
highly expresses Gpihbp1, VEC2 cluster highly expresses Cxcl1 and
Icam1, VEC3 highly expresses Plvap and Vwf); Art.EC, arterial
endothelial cells (enriched with artery-related genes such as
Cxcl12); Endo, metabolism-related endothelial cells (specifically
expressing metabolic genes such as H19 and Cpe); Pro.EC,
proliferative active endothelial cells [highly expressing
proliferation genes Hmgb2 and Birc5]; VEC1, microvascular
endothelial cells [expressing Ly6a (encoding SCA1) and vascular
transcription factor Sox17]; VEC2, arterial endothelial cells
(involved in NOTCH signaling pathways, such as Sox17, Hey1); VEC3,
venous endothelial cells. |
 | Table II.A typical study on ScRNA-seq of
endotheliocyte subsets. |
Table II.
A typical study on ScRNA-seq of
endotheliocyte subsets.
| Source of
organization | Ventricular tissue
of mice | Cardiac
interstitial cell populations in mice | Non-cardiac muscle
tissue from mice |
|---|
| Methods | 10X Genomics | 10X Genomics | ScRNA-seq |
|
| scRNA-seq | ScRNA-seq |
|
| Number of cells
captured | 17,320 cells | 13,331 cells | 35,312 cells |
| Clusters | Art.EC, VEC1-3,
Endo.EC, Pro.EC | EC1, EC2, EC3 | 5 endothelial cell
clusters |
In the VEC clusters, Gpihbp1 was highly expressed,
with VEC3 showing a higher expression level among the three
subclusters (VEC1-3) (40). The
VEC2 cluster overexpressed Cxcl1 and Icam1 and activated the
ROS/TNF signaling pathway to drive an inflammatory response.
In 2019, Farbehi et al (43) conducted sham and MI surgeries on
mice at 3 and 7 days post-infarction. The authors extracted the
cardiac interstitial cell population from 8-week-old male
PdgfraGFP/+ mice and performed single-cell expression
profiling. A total of three major EC clusters (EC1, EC2 and EC3)
were identified, The EC1 population expressed Ly6a (encoding SCA1)
and the vascular transcription factor Sox17, as indicated by marker
genes, which may represent microvascular ECs. EC2 expressed Sox17
and Hey1, which function downstream of the Notch signaling pathway
and are important for arterial ECs. In addition to the marker
genes, a number of EC3 cells expressed Prox1 and Lyve1. The EC1
cluster showed a rapid decrease at 3 dpi and a subsequent increase
at 7 dpi, although it remained below control levels.
In 2021, Tombor et al (44) conducted single-cell sequencing and
trajectory analysis on non-myocardial tissues from C57BL/6J
Cdh5-CreERT2 mice at various time points (days 0, 1, 3, 5, 7, 14
and 28 post-infarction). The authors identified five EC clusters,
with Cdh5 and Pecam1 serving as key marker genes. Subcluster 3
activated neutrophil chemotaxis and cytokine signaling to
exacerbate inflammatory damage. Subcluster 4 showed an increase in
number between days 1 and 7, returning to baseline levels after day
14. This subcluster expressed mesenchymal genes and proliferation
markers, while fatty acid metabolism genes were downregulated,
suggesting dysregulated energy metabolism.
Trajectory analysis confirmed that ECs underwent an
‘interstitial’ transient transition with partial recovery after 14
days. A separate study (45) found
that circFndc3b was markedly downregulated in heart following MI.
However, overexpression of circFndc3b in cardiac ECs increased
vascular endothelial growth factor expression, enhanced angiogenic
activity and reduced apoptosis in cardiomyocytes and ECs.
Additionally, the LPA-LPA2 signaling pathway was shown to promote
vascular neogenesis and maintain vascular homeostasis, which is
essential for restoring blood flow and repairing ischemic tissues
(46). EC clusters show a
sequential response of inflammation, proliferation and
mesenchymalization following MI and targeted intervention of
specific subpopulations (including inhibition of VEC2 inflammation,
amplification of Pro.EC and blockade of mesenchymal transformation)
is expected to break through the current bottleneck in vascular
regeneration therapy.
Fibroblast heterogeneity and
associated changes following MI
Cardiac fibroblasts are transformed into
myofibroblasts (MYOs) following injury, which plays a critical role
in mediating healing after acute MI (AMI) and contributes to
long-term fibrosis in chronic disease (47). Fibroblasts are essential for
cardiac remodeling following MI. Several studies have analyzed the
characterization and changes in various fibroblast clusters
following MI using single-cell sequencing, as detailed in Table III.
 | Table III.A typical study on ScRNA-seq of
fibroblast subsets. |
Table III.
A typical study on ScRNA-seq of
fibroblast subsets.
| Source of
organization | Cardiac
interstitial cell populations in mice | Non-cardiomyocytes
from mice | Epicardial
interstitial cells in mice | Ventricular tissue
in mice | Ventricular tissue
in mice | Ventricular tissue
in mice | Mouse heart
tissue |
|---|
| Methods | 10XGenomics | ScRNA-seq | 10X Genomics | 10X Genomics | 10X Genomics | ATAC- | 10X Genomics |
|
| ScRNA-seq |
| ScRNA-seq | ScRNA-seq | ScRNA-seq | Seq | ScRNA-seq |
| Number of cells
captured | 13,331 cells | 27,349 cells | 38,600 cells | 17,320 cells | 10,487 cells | 29,176 cells | - |
| Clusters | MYO, F-Act, F-Cyc,
F-CI, F-SH, F-SL, F-WntX, F-Trans | Fibro-1, Fibro-2,
Fibro-3, Fibro-4, Fibro-5, Fibro-Myo | I, II, III; (three
main Myofb clusters: Myofb, ProlifMyofb, MFCs) | FB1-FB4,
Pro.FB | CF1-7 | A-J | F-SL, F-SH,
F-Trans, F-WntX, F-Act, F-CI, F-Cyc, F-IFNS |
In 2019, Farbehi et al (43) classified the fibroblast population
into eight clusters: MYOs, activated fibroblasts (F-Act), a unique
GFP+ proliferating population (F-Cyc),
fibroblasts-circulating intermediates (F-CI), fibroblast Sca1 high
cluster (F-SH), fibroblast Sca1 low cluster (F-SL),
fibroblast-Wnt-expressing cluster (F-WntX) and
fibroblast-transitional cluster (F-Trans). The marker genes for
each cluster were not detailed.
MYOs expressed fibrillogenic proteins, such as
periostin (Postn) and contractile proteins such as smooth muscle
actin (48), along with collagen
genes, including Col1a1, Col3a1 and Col5a2 (49), which can drive collagen deposition
with scarring, with a significant increase in numbers on day 7
post-MI. F-Cyc specifically expressed the cell cycle genes Cdk1 and
Mki67 (Ki67), a source of reparative cells that could inhibit
overproliferation through targeted regulation, which peaked on day
3 and decreased on day 7 following MI, which is consistent with
previous studies showing that the peak of fibroblast proliferation
occurs on days 2–4 following MI (48,50).
F-CI showed upregulated expression of genes involved in fibroblast
activation, such as Postn, Cthrc1 and Acta2, although it did not
express cell cycle markers, which is suggestive of a potentially
activated fibroblast population that could enter the cell cycle.
This cluster also exhibited upregulated protein translation genes,
a feature not observed in F-Act. High expression of Ly6a (Sca1) and
Pdgfra was observed in F-SH, with multidirectional differentiation
capacity or involved in tissue repair, which was reduced on day 3
and partially restored on day 7 following MI. The newly identified
F-WntX and F-Trans were present in both pseudo and myocardial
infarcted hearts and F-WntX specifically showed overexpression of
the Wnt signaling pathway inhibitor Wif1, which affected the repair
process and it was markedly reduced at day 7 post-MI.
A single-cell sequencing study by Zhuang et
al (51) involving
non-cardiomyocytes from healthy and post-infarction male mice (aged
8–10 weeks) at days 0, 3 and 7 identified six fibroblast clusters
named Fibro-1, Fibro-2, Fibro-3, Fibro-4, Fibro-5 and Fibro-Myo.
Marker genes for Fibro-4 and Fibro-5 were not markedly expressed.
Marker genes for the remaining subgroups are shown in Fig. 3A. The proportion of fibroblasts
decreased at 3 dpi compared with the sham group, while Fibro-Myo
markedly increased at 7 dpi relative to other fibroblast clusters,
indicating its crucial role in ischemic response and healing
process. Cthrc1 and Ddah1 were both markedly upregulated in the
Fibro-Myo cluster. Cthrc1 is a known marker for myofibroblasts
(52), while Ddah1 can also serve
as a myofibroblast marker. Thus, Fibro_Myo plays an important role
in promoting cardiac healing and may act as a relevant
subpopulation of intervening cells after improved cardiac
remodeling.
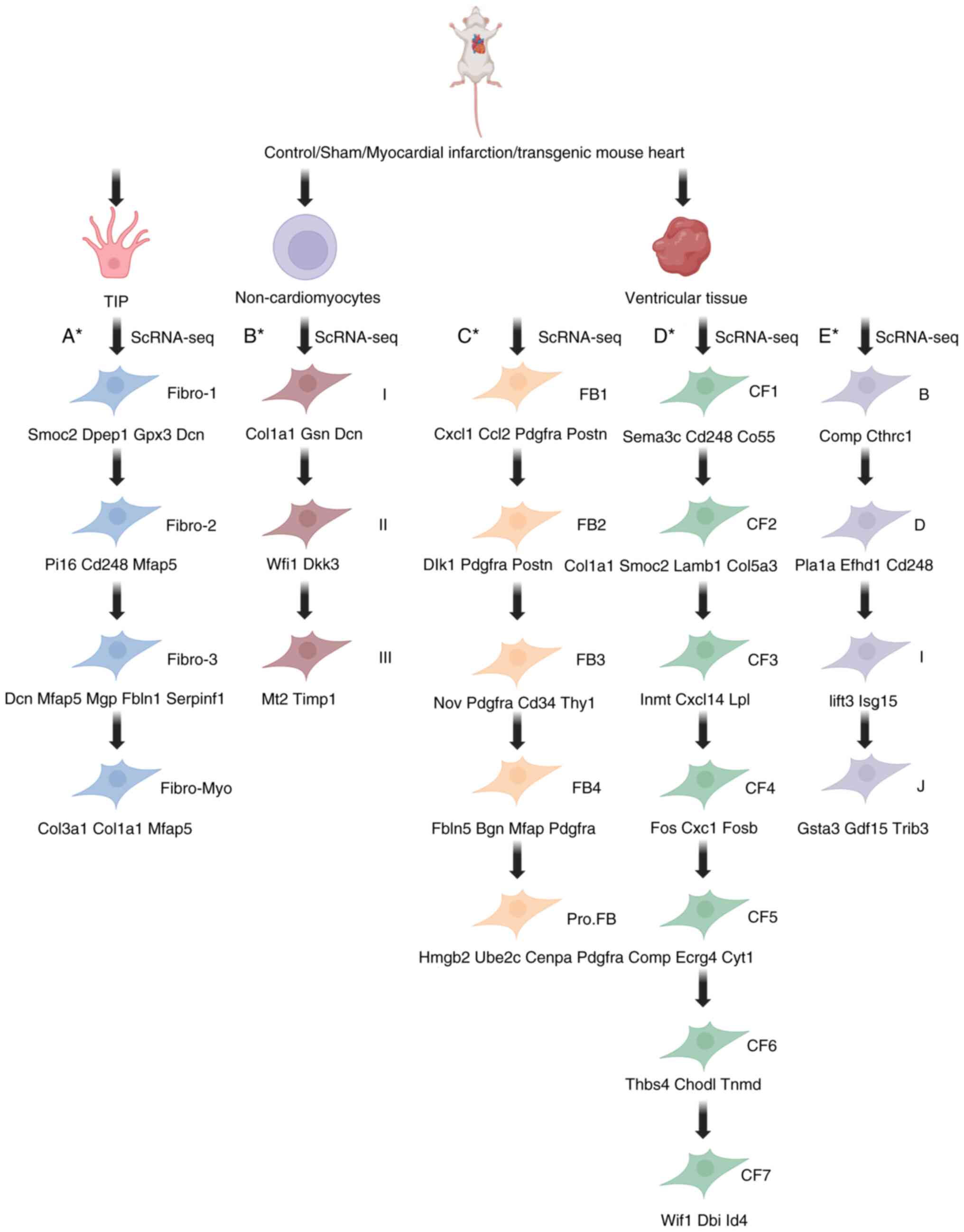 | Figure 3.Fibroblast cluster typing and marker
genes for each cluster. The high expression of myofibroblast marker
genes Acta2, Tagln and Cthrc1 in FB1/B group/Fibro-Myo; enrichment
of the differentiation inhibitory gene Dlk1 in FB2; the specific
expression of the proliferation gene Mki67 in Pro.FB; the
upregulation of matrix homeostasis genes Fbln5 and Bgn in FB4; the
significant expression of sclerosis-related genes Comp, Cilp and
Angptl7 in CF5/CF6 and the enrichment of osteogenic genes Adamtsl2
and paracrine factor SFRP2 in MFCs. This indicates that they are
respectively involved in fibrotic driving, differentiation
inhibition, proliferation repair, matrix remodeling and chronic
scar hardening. (A*) Fibroblast clusters of cardiac interstitial
cell origin and marker genes. (B*) Fibroblast clusters of
non-cardiomyocyte origin and marker genes. (C*-E*) Fibroblast
clusters of cardiac ventricular tissue origin and marker genes. The
CF1-CF4 subsets highly express genes related to the degradation of
homeostasis fibroblasts (Hsd11b1, Lpl and Dpt) and ECM; CF5 and CF6
express genes related to the activation of fibroblasts, cartilage
development and ossification, including Angptl7, Cilp, Comp, Ecrg4,
Fmod, Postn, Meox1 and Thbs4. ScRNA-seq, single-cell RNA
sequencing; Fibro1, the matrix generates fibroblasts; ibro2,
damage-responsive fibroblasts; Fibro3, precursor fibroblasts;
Fibro-myo, myofibroblasts (highly expressing Cthrc1 and Ddah1);
Type I, quiescent interstitial progenitor cells; Type II, cells
responding to transitional state damage; Type III, terminally
differentiated profibrotic cells; FB1, stromal homeostasis
fibroblasts; FB2, inflammatory injury response fibroblasts
(specifically expressing Dlk1); FB3, precursor of myofibroblasts
(specifically expressing Nov, Thy1, Pi16, Axl and Cd34); FB4,
perivascular repair fibroblasts (highly expressing Fbln5, Bgn and
Mfap); Pro.FB, regenerative potential progenitor cells; CF1,
resting-state progenitor cells; CF2, immunomodulatory fibroblasts;
CF3, stromal homeostasis fibroblasts; CF4, transition state
precursor fibroblasts; CF5, contractile myofibroblasts; CF6,
perivascular repair fibroblasts; CF7, lipid metabolism-related
fibroblasts (responsive growth factor); B, the stromal
microenvironment maintains fibroblasts; D, antigen-presenting
fibroblasts; I, myofibroblast precursor cells; J, terminal
contractile myofibroblasts. |
In 2020, Forte et al (53) performed scRNA-seq on epicardial
mesenchymal cells from seven male Wt1Cre transgenic mice,
identifying three fibroblast populations (Type I, II and III). The
marker genes for each population are shown in Fig. 3B. The marker gene ZsGreen was
expressed in all Type I–III fibroblasts. The ratio of Type I
fibroblasts was markedly higher on days 1 and 3 post-infarction,
with the highest contribution ratio on day 3. Type III fibroblasts
were markedly higher on day 3 post-infarction.
Within the Type I fibroblast population, three
closely related subpopulations of ZsGreen+ epicardial-derived
fibroblasts (EpiDs) were identified: steady-state
epicardial-derived fibroblasts (HEpiDs), progenitor-like state
fibroblasts (PLS) and late-phase fibroblasts (LR). The HEpiD
population highly expressed genes involved in the response to
organic substances and metabolic effectors, such as Dpep1, Lpl,
Hsd11b1 and Cxcl14. The PLS population remained relatively stable
across all time points, with comparatively high expression of genes
related to cell migration and morphogenesis such as Cd248 and Pi16.
The LR population, which was predominantly present during the
post-infarction maturation phase (days 14–18), showed high
expression of genes involved in cellular differentiation,
osteogenesis and matrix remodeling, such as Adamtls2.
Type II fibroblasts specifically expressed genes
closely related to valve leaflet development and were involved in
endochondral ossification, Wnt signaling and structural
morphogenesis. However, Type III fibroblasts have not been fully
characterized to date.
Additionally, three main myofibroblast (Myofb)
populations were observed after subclustering: Myofb (Acta2,
Cthrc1), proliferative myofibroblasts (ProlifMyofb; Acta2, Cthrc1
and cell cycle genes) and a group of stromal fibroblasts (MFCs)
similar to those found in mature scars (48). Myofb expressed Acta2 and Cthrc1 and
their proportion increased markedly on days 3, 5 and 7
post-infarction (peaking at day 5). ProlifMyofb expressed Acta2,
Cthrc1, the pro-repair paracrine factors SFRP2 and CLU and cell
cycle genes. The proportions increased markedly on days 3 and 5
post-infarction, with a decreasing trend on day 7. Stromal
fibroblast-like cells MFCs increased markedly in proportion on days
14–28 post-infarction (mature scar phase). The aforementioned key
populations (such as early activated FI/EpiDs, proliferating
ProlifMyofb, MFCs contributing to bone/chondrocyte-like ECM) and
their specific marker genes (Pi16, Adamtsl2, Comp and Cilp) and
pathways (Wnt signaling) provide new potential targets for
anti-fibrotic and amelioration of scar remodeling therapies.
In 2020, Wang et al (39) identified five cardiac fibroblast
(FB) subclusters, labeled FB1-FB4 and proliferating fibroblasts
(Pro.FB), all of which highly expressed FB marker genes such as
Postn, Col1a1 and Pdgfra. Specific genes defining each cluster were
identified using differential expression analyses (the marker gene
expression for each subcluster is shown in Fig. 3C). Partial FB1 cells expressed high
levels of myofibroblast marker genes, including Acta2 and Tagln,
but low levels of the static FB marker Pdgfra, reflecting the early
steps in the activation and differentiation of cardiac FBs into
myofibroblasts (48,53). FB2 expressed Dlk1, an inhibitor of
myofibroblast differentiation (54). The specific genes expressed by FB3
may be related to normal fibroblast physiological functions
(55–58). High expression of extracellular
matrix organization-related genes such as Fbln5, Bgn and Mfap in
FB4 may be associated with the regulation of matrix structure and
homeostasis and could be considered as a regulatory target for
improving scar elasticity. By comparing the number dynamics of FB
subclusters between 1- and 8-day-old neonatal mice following MI, it
was found that the percentage of Pro.FB was markedly higher in
8-day-old neonatal mice at 3 days post-infarction, indicating that
FB proliferation precedes cardiac fibrosis. This observation aligns
with previous findings FB proliferation occurred following MI in
8-day-old neonatal mice (55). FB1
showed the highest increase in P8-injured hearts, demonstrating it
as a core fibrotic effector group. By temporally regulating
specific subgroups (inhibiting early amplification of Pro.FB and
blocking the fibrotic transformation of FB1), the fibrotic process
after infarction can be precisely intervened.
In 2023, Tani et al (38) classified fibroblasts into seven
clusters by performing scRNA-seq on left ventricular tissues from
control mice (Tcf21iCre/Tomato) and TTg (Tcf21iCre/Tomato/MGTH2A)
mice 3 months following MI. These clusters were labeled CF1 to CF7,
with their respective marker genes shown in Fig. 3D. CF5 and CF6 numbers increased in
Ctrl-MI and decreased in TTg-MI mice compared with controls.
Clusters CF1-CF4 expressed genes associated with steady-state
fibroblasts (Hsd11b1, Lpl and Dpt) and ECM degradation, while CF5
and CF6 expressed genes related to activated fibroblasts,
chondrogenesis and ossification, including Angptl7, Cilp, Comp,
Ecrg4, Fmod, Postn, Meox1 and Thbs4. CF7 expressed genes responsive
to growth factors. Future studies may reveal a direct association
between CF5/CF6 and its genetic signature
(Comp+/Cilp+/Postn+) and elevated
cardiac stiffness in advanced stages. Angptl7, Thbs4 or downstream
signaling pathways could be used as targets for the development of
novel antifibrotic drugs. CF5/CF6 signature profiles or expression
levels could be used as non-invasive indicators (serum COMP assay)
to assess the severity of chronic fibrosis and response to therapy.
Non-invasive indicators include serum COMP and CILP test. Patients
with high CF5/CF6 ratios may require targeted antisclerotic
therapy.
Ruiz-Villalba et al (52) performed single-cell sequencing on
cardiac tissues from healthy and post-infarction Col1α1-GFP mice at
7 and 14 days post-infarction, identifying 10 fibroblast clusters
(A-J). Clusters B, D, I and J showed distinct expression profiles,
as shown in Fig. 3E. Clusters F
and H-K remained unchanged across different time points. Cluster I
decreased to a minimum and then increased at 14 dpi, while the
percentage of cluster B markedly increased following MI, peaking at
day 14 and then declining. Cluster B was markedly enriched in the
infarct zone at 7 days post-MI, decreasing by day 30 (52). Cluster B had pathways and gene
ontologies related to ECM organization, cell proliferation and
cell-matrix adhesion (56,57). The top marker of subcluster B,
Cthrc1, has been associated with vascular remodeling and fibrosis
(58–61). The highly specific expression of
ECM-related genes in subcluster B suggests a strong association
with fibrotic scar formation, which exhibits intense necrotic
features after infarction and localizes to damaged tissue.
Precision targeted intervention of Cthrc1 or its downstream pathway
inhibits cluster B activation and blocks early fibrotic outbreak.
The development of relevant assays targeting serum profiles of
genes characteristic of group B (Cthrc1) to assess fibrosis
activity and therapeutic efficacy following MI should be
considered.
In 2022, Janbandhu et al (62) performed scRNA-seq on tdTomato+
CD31−CD45− fibroblasts from male magnetic
mice 8–12 weeks after a sham operation or MI. The fibroblasts were
classified into eight clusters: F-SL, F-SH, F-Trans, F-WntX, F-Act,
F-CI, F-Cyc and F-IFNS (IF Stimulated). The F-Act cluster was
markedly expanded following MI; the F-CI cluster was markedly
increased in cKO (Hif-1a Δflox/-;PdgfraMCM/+) hearts
following MI; and the number of F-SLs was markedly increased in cKO
and HET (Hif1a flox/+; PdgfraMCM/+) hearts
following MI. The F-Act cluster specifically exhibited
downregulated expression of genes encoding negative regulators of
cell signaling, particularly genes involved in key pathways
regulating CF proliferation such as MAPK-ERK1/2 and fibroblast
growth factor (FGF). This suggests that hypoxia-inducible factor 1
(HIF-1)α deletion may deregulate the inhibition of the
proliferative pathway and promote the conversion of F-Act to the
proliferative state (F-CI). The proliferation-associated subgroups
F-CI (proliferative intermediate state) and F-Cyc (proliferative
state) also showed downregulation of HIF-1 target genes, directly
confirming the central role of HIF-1α signaling in the regulation
of CF proliferative program. The HIF-1α pathway and its downstream
regulation of key subgroups (F-Act, F-CI) and signaling nodes
(MAPK-ERK, FGF pathway negative regulatory factors) are potential
targets for intervention. Modulating HIF-1α activity or interfering
with its downstream effector molecules may precisely control
pathological CF proliferation and activation and attenuate
deleterious fibrosis. The identified specific subgroups (markedly
expanded F-Act, accumulated F-CI and F-SL in cKO) and their
characteristic gene expression profiles are expected to serve as
novel molecular markers to assess the development of fibrosis.
Monocyte/macrophage heterogeneity and
associated changes following MI
Macrophages are key players in inflammatory response
and post-infarction cardiac remodeling. The literature detailing
their detection via single-cell sequencing is summarized in
Table IV. In 2019, Farbehi et
al (43) classified monocytes
and macrophages into several clusters through single-cell RNA-seq,
as follows: M1MO, M1MΦ, M2MΦ, MAC-IFNIC, MAC-TR, MAC8, MAC7 and
MAC6. The M1MΦ cluster was identified as a derivative of classical
monocytes and was characterized by Ccr2high+Adgre1
(F4/80)+ Ly6c2+ H2-Aa (MHC-II)+,
driving the early inflammatory response. Non-classical M2MΦ cells,
which were characterized by Ccr2high Adgre1 (F4/80)+
H2-Aa (MHC-II) high-Ly6c2, participate in repair through the
non-arginase pathway (Arg1low), providing a novel immunomodulatory
mechanism After MI, the M1MΦ and M1MO populations increased at 3
dpi, while the M2MΦ population expanded at 7 dpi. On the third day
following MI, targeted inhibition of M1MΦ (such as anti-CCR2
antibody) was carried out to alleviate inflammatory damage; on the
7th day following MI, the M2MΦ/Cx3cr1+ population (such
as Cx3cr1 agonists) was enhanced to promote tissue repair.
 | Table IV.A typical study on ScRNA-seq of
monocyte subsets. |
Table IV.
A typical study on ScRNA-seq of
monocyte subsets.
| Source of
organization | Cardiac
interstitial cell populations in mice | Ventricular tissue
in mice | Myocardial
CD45+ leukocytes in mice | Non-cardiomyocytes
from mice | CD45+
non-cardiomyocytes from mice |
|---|
| Methods | 10X | 10X | 10X | ScRNA-seq | ScRNA-seq |
|
| Genomics | Genomics | Genomics |
|
|
|
| ScRNA-seq | ScRNA-seq | ScRNA-seq |
|
|
| Number of cells
captured | 13,331 cells | 17,320 cells | 30,135 cells | 27,349 cells | 26,275 cells |
| Clusters | M1MO, M1MΦ, M2MΦ,
MAC-IFNIC, MAC-TR, MAC8, MAC7, MAC6 | M1, M2 | MAC-TRc1-3,
Mo-Ly6c, Mo-Ear2, MAC-Olr1, MAC-Gpnmb, MAC-Lars2, IFNIC | IFNIC, MAC-Mo/M1,
MAC-M2, MAC-TR, MAC-3, MAC-APC, MAC-4, MAC-Fib, MAC-Endo | MΦ1, MΦ2, MΦ3 |
In 2020, Wang et al (39) used Adgre1 as a specific marker for
mononuclear macrophages and classified macrophages into M1 and M2
clusters through higher-resolution sub-clustering analyses (the
marker genes for these clusters are shown in Fig. 4A). An increase in macrophage and
monocyte percentages was observed at all analyzed time points
post-MI. However, the timing and magnitude of macrophage elevation
differed depending on the neonatal stage, suggesting that
macrophages may play a role in post-infarction cardiac remodeling
and myocardial regeneration. Hearts from P1 mice exhibited higher
percentages of macrophages and monocytes 1 dpi, while hearts from
P8 showed elevated macrophage and monocyte percentages at 3 dpi. At
1 dpi, P1 mice demonstrated a rapid increase in the M1 and M2
macrophage populations compared with controls, with a unique level
of M2 infiltration not observed at other time points. Additionally,
both P1 and P8 neonatal mice showed a pronounced increase in M1 and
M2 macrophage composition at 3 dpi compared with controls (39). M2 macrophages secrete
anti-inflammatory cytokines, promoting wound healing and tissue
repair, which may support cardiac regeneration in neonatal mice and
it provides a theoretical basis for promoting cardiac repair via
enhancing the polarization or function of the M2 phenotype.
Furthermore, the macrophage-secreted factor CLCF1 was found to
enhance cardiomyocyte proliferation (39), which may provide options for
regenerative therapy.
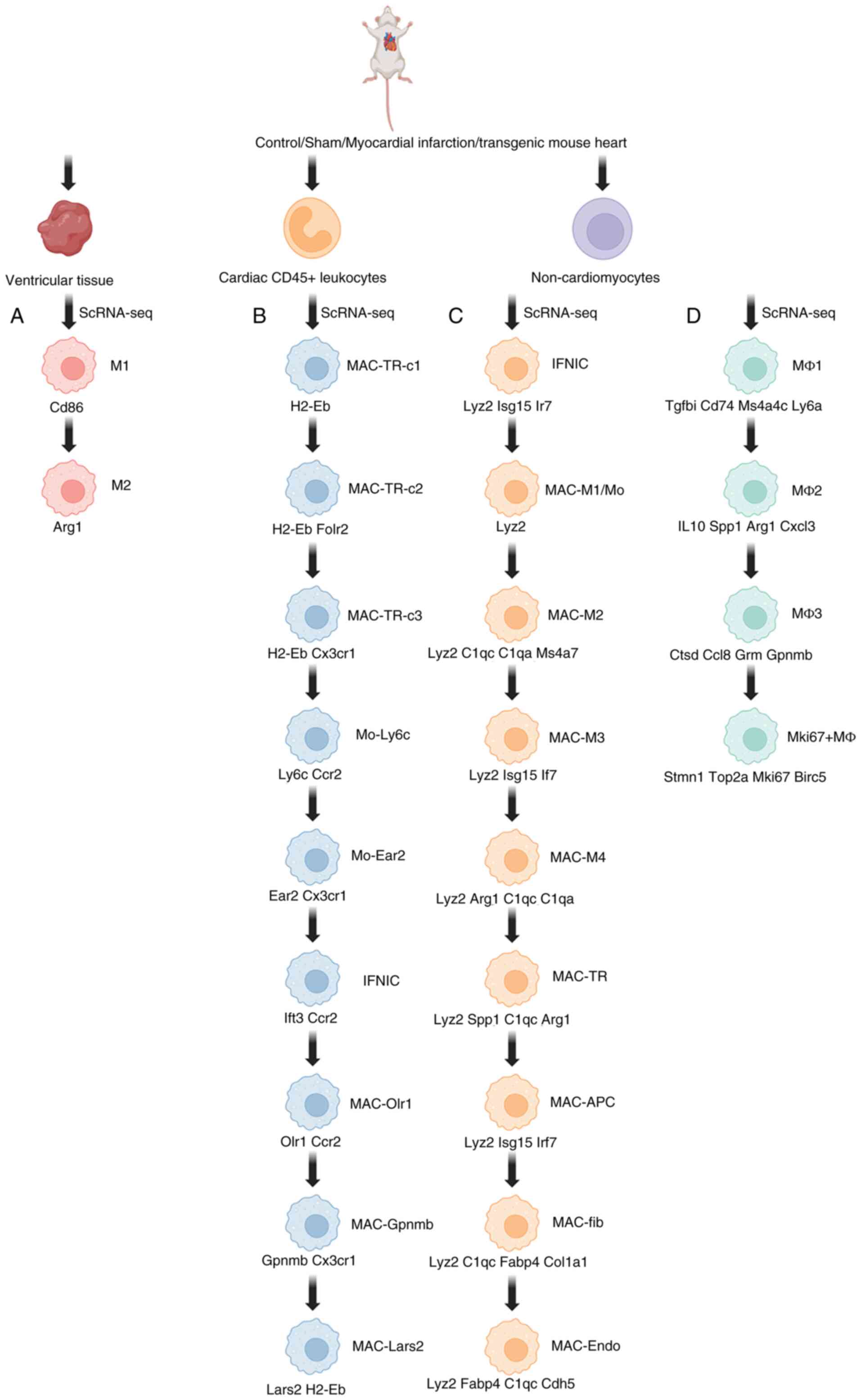 | Figure 4.Monocyte macrophage clusters typing
and marker genes for each cluster. High expression of
pro-inflammatory genes Ly6c2, S100a8 and the C1q family
(*C1qa/b/c*) in M1/MAC-Mo; enrichment of repair genes Spp1, Cd163
and Mrc1 in M2/MAC-TR; the significant upregulation of
anti-fibrotic genes Fabp5 and Gpnmb in Bhlhe41+MΦ; the activation
of phagocyte-related gene Folr2 in MAC-Gpnmb; and the co-expression
of the cross-lineage genes Col1a1 (fibroblast) and Cdh5
(endothelial) in MAC-Fib/MAC-Endo indicates that they are
respectively involved in inflammation initiation, tissue repair,
fibrosis inhibition, fragment clearance and lineage plasticity. (A)
Monocyte cell clusters of Cardiac ventricular tissue origin and
marker genes. (B) Monocyte cell clusters of Cardiac
CD45+ leukocytes origin and marker genes. (C) Monocyte
cell clusters of non-cardiomyocyte origin and marker genes. (D)
Monocyte cell clusters of non-cardiomyocyte (temporary cardiac
resident macrophage subset) origin and marker genes. The Ccr2_hi
cluster contains subgroups such as MAC_Mo/M1 and IFNIC and enriches
pro-inflammatory transcription factors such as Stat1 and Irf7. The
Ccr2_lo cluster contains subgroups such as MAC-TR, MAC-Fib and
MAC-Endo and is characterized by highly expressing Lyve1 and
Cx3cr1. ScRNA-seq, single-cell RNA sequencing; M1,
pro-inflammatory/damage-responsive macrophages (highly expressing
C1qa, C1qb, C1qc, Pf4); M2, repair/regeneration-promoting
macrophages (highly expressing C1qa, C1qb, C1qc, Spp1); MAC-TR-c1,
resting-state tissue-resident macrophages; MAC-TR-c2,
damage-responsive macrophages; MAC-TR-c3, ECM remodeling
macrophages; Mo-Ly6c, inflammatory mononuclear derived monocytes;
Mo-Ear2, anti-inflammatory readiness monocytes; IFNIC,
interferon-responsive macrophages; MAC-Olr1, lipid phagocytic
macrophages; MAC-Gpnmb, profibrotic macrophages; MAC-Lars2,
metabolic repair macrophages; IFNIC, interferon-responsive
macrophages; MAC-Mo/M1, classic pro-inflammatory macrophages;
MAC-M2, repair macrophages; MAC-TR, tissue-resident macrophages;
MAC-3, lipid-clearing macrophages; MAC-APC, antigen-presenting
macrophages; MAC-4, profibrotic macrophages; MAC-Fib, profibrotic
macrophages; MAC-Endo, promotes angiogenesis macrophages; MΦ1,
homeostatic tissue-resident macrophages (expressing
Timd4+ and Lyve1); MΦ2, repair hub macrophages
(expressing Bhlhe41+, Vegfa+); MΦ3,
profibrotic macrophages (expressing Spp1+ and
Mmp9+); Mki67+MΦ, proliferative and migratory
macrophages (expressing Mki67+ and
Cxcr4+). |
In 2022, Zhuang et al (63) identified nine subclusters of
mononuclear macrophages through single-cell RNA-seq: 3 intratissue
macrophage clusters (MAC-TRc1-3), 2 monocyte clusters (Mo-Ly6c and
Mo-Ear2) and 4 ischemia-associated macrophage subclusters with
differentially expressed marker genes (MAC-Olr1, MAC-Gpnmb,
MAC-Lars2 and IFNIC), with specific marker genes detailed in
Fig. 4B. MAC-Olr1 numbers
increased at 1 dpi and were characterized by the upregulation of
SASP and glycolysis pathways. The MAC-Gpnmb subcluster markedly
increased at 7 dpi and showed a powerful devouring ability (it was
positively correlated with phagocytosis and cellular FAO levels).
The Mo-Ly6c cluster was related to pro-inflammatory responses and
angiogenesis. The Mo-Ear2 cluster may be involved in adaptive
immune regulation following MI, providing a theoretical basis for
precisely regulating the differentiation of monocytes into
beneficial phenotypes (such as inhibiting pro-inflammatory Mo-Ly6c
and enhancing regulatory Mo-Ear2), which is helpful for balancing
inflammation and repair. Among the three MAC-TRs clusters,
MAC-TR-c2 expresses Folr2 and hematopoietic cell lineages,
lysosomes and endocytic signals are enriched in Folr2+
macrophages, making it an ideal target for promoting cardiac
repair. The proportion of Folr2+ macrophages decreased
on the first day following MI and recovered 7 days following
MI.
Zhuang et al (51) identified seven macrophage clusters:
IFNIC, MAC-Mo/M1, MAC-M2, MAC-TR, MAC-3, MAC-APC and MAC-4. The
marker genes for the newly identified clusters are detailed in
Fig. 4C. The proportions of
MAC-Mo/M1 and MAC-M2 increased at 3 dpi, with a particularly rapid
rise in MAC-M2, while MAC-TR showed greater proliferation by 7 dpi
(51). Pseudotemporal analysis of
macrophage populations revealed the presence of fibroblast-like
macrophages (MAC-Fib) and endothelial-like macrophages (MAC-Endo),
which expressed fibroblast markers (Col1a1) and endothelial markers
(Cdh5), respectively. Tissue-resident macrophages (MAC-TR) showed
increased expression of Cx3cr1 and H2-Aa, while downregulating the
pro-inflammatory gene Ly6c2. Some of these cells also expressed
Cd163 and Mrc1 while lacking H2-Aa, suggesting an anti-inflammatory
role in ischemic injury.
Further exploration of the core transcriptional
regulation driving macrophage differentiation was conducted using
single-cell regulatory network inference and clustering analysis
(64). A total of two clusters
were identified: The Ccr2_hi cluster contains subgroups, including
MAC_Mo/M1 and IFNIC; is enriched in pro-inflammatory transcription
factors such as Stat1 and Irf7; and it was positively associated
with myeloid cell activation, immune response and extracellular
secretion pathways (51). Previous
research has found that excessive activation of Irf3 is associated
with excessive interferon production and adverse inflammatory
responses (65). The Ccr2_lo
cluster contains subgroups such as MAC-TR, MAC-Fib and MAC-Endo and
is characterized by highly expressing Lyve1 and Cx3cr1 (66). This cluster is markedly enriched in
the gene set involved in the regulation of cell proliferation and
the response to growth factor HGF/TGF-β and is regulated by
transcription factors such as Egr1 and Jund (51). Functional comparative analysis
clearly supported the Ccr2_lo cluster (especially MAC-TR) to exert
protective effects of self-renewal and anti-inflammation following
MI (51). Enhancing the
anti-inflammatory function of MAC-TR and the angiogenesis ability
of MAC-Endo by using small molecule agonists is a direction for
treating cardiac remodeling following MI. In addition, in
vitro expansion of Ccr2_lo characteristic cells and
transplantation into the infarcted area is also a means to achieve
anti-inflammatory and vascular regeneration simultaneously.
In 2023, Xu et al (67) identified two major tissue
macrophage populations at steady state (Sham) using scRNA-seq in
8–10 week-old male C57BL/6J mice:
LYVE1+MHCII−CCR2− and
LYVE1−MHCII+CCR2−. The two
populations were markedly reduced at 3 days post-MI. Four
additional macrophage clusters showed a significant increase in
response to MI: MΦ1, MΦ2, MΦ3 (referred to as
Bhlhe41+MΦ) and Mki67+MΦ. MΦ3, which
exhibited higher Bhlhe41 activation. The specific marker genes for
each cluster are detailed in Fig.
4D. The number of MΦ3 cells markedly increased on day 3
post-MI, peaked on 7 dpi and returned to baseline by 14 dpi,
clearly indicating the key intervention time window targeting this
repair macrophage. In addition, Fabp5, Gpnmb and Ccl8 were markedly
upregulated in MΦ3 at 3 dpi, whereas Il1b, Il10, Tgfb1 and Nfkb1
were downregulated. Cell death-related markers, including those for
autophagy, apoptosis and necroptosis, were also downregulated,
along with O-GlcNAc modification, ubiquitylation levels and hypoxic
injury markers in Bhlhe41+ MΦ cells. These macrophages
played roles in collagen catabolic processes, positive regulation
of tissue remodeling and lipid metabolism pathways. Notably,
Bhlhe41+ MΦs were the only macrophage cluster enriched
in cardiac fibroblast lineage cells (Pdgfr-GFP+). It is
suggested that this subgroup plays a core protective role in
restricting the dilation of the infarcted area and preventing
pathological fibrosis by inhibiting the activation of
myofibroblasts and excessive collagen deposition. The
Bhlhe41+ m4 subgroup demonstrates a strong repair
ability. In the future, therapeutic strategies can be focused on
promoting its expansion and recruitment, maintaining its phenotype
and function and simulating its secretory factors/functions,
etc.
T-cell and B-cell heterogeneity and
associated changes following MI
T-cells and B-cells are critical participants in the
post-infarction inflammatory response. The literature on their
role, as identified by single-cell sequencing, is summarized in
Table V. In 2019, Farbehi et
al (43) identified two T-cell
clusters through single-cell RNA-seq: TC1-Cd8 (Cd8a+),
representing cytotoxic T-cells and TC2-Cd4 (Cd+4
Lef1+), which likely represents helper T-cells.
 | Table V.A typical study on ScRNA-seq of T/B
cell subsets. |
Table V.
A typical study on ScRNA-seq of T/B
cell subsets.
| Source of
organization | Cardiac
interstitial cell populations in mice | Non-cardiomyocytes
from mice | Cardiac and
mediastinal lymph node B cells in mice | Myocardial
CD45+ leukocytes in mice |
|---|
| Methods | 10X Genomics | ScRNA-seq | 10X Genomics | 10X Genomics |
|
| ScRNA-seq |
| ScRNA-seq, S2
Cartridge | ScRNA-seq |
|
|
|
| ScRNA-seq |
|
| Number of cells
captured | 13,331 cells | 27,349 cells | 6,588 cells | 30,135 cells |
| Clusters | TC1-Cd8
(Cd8a+), TC2Cd4 (Cd4+ Lef1+) | Naïve T-cell,
effector T-cell, regulatory T-cell, NK-cell | B1, B2, MZ, GC, hB,
CD74+ cluster, IFNR, Cyc | CD4-C1-CCR7 and
CD8-C1-CCR7 represent naïve T cells and CD4-C2-CXCR3 and
CD8-C2-Gzmk represent effector T cells; B cell-Cd20, B
cell-Cd23 |
In 2020, Zhuang et al (51) performed single-cell sequencing on
non-cardiomyocytes from 8–10 week-old male mice, comparing
sham-operated mice and those at 3, 7 and 10 days post-infarction.
The authors classified T-cells into four clusters: Naïve, effector,
regulatory and NK cells. The marker genes for each cluster are
detailed in Fig. 5A. A significant
infiltration of T-cells was observed 7 days following MI. The
effector T-cell population showed upregulation of Ccl6 and Il1b and
was enriched for pathways associated with cell activation and
immune system regulation. The regulatory T-cell population
positively associated with inflammatory pathways of humoral immune
responses and myeloid leukocyte differentiation. Effector T-cells
were enriched for pro-inflammatory regulators such as Irf5 and
Fosb, while regulatory T-cells were enriched for activated Sp1
regulators. Exploring drugs targeting transcriptional regulatory
factors such as Irf5 or Fosb can regulate the activation and
function of effector T cells. Regulating the immunomodulatory
properties and Sp1-related stability exhibited by T cell subsets is
a potential therapeutic strategy for promoting immune tolerance and
improving the cardiac repair environment.
![T and B cell clusters typing and
marker genes for each cluster [MZ marker genes related literature].
High expression of pro-inflammatory factor IL-1β, chemokine Ccl6
and transcription factor Irf5/Fosb in effector T cells regulate the
enrichment of immune regulatory pathways (such as Sp1) in T cells;
co-activation of the dual-chemokine receptor Cxcr5/Ccr7 and the
tissue repair factor Tgfb1 in hB cells; and upregulation of the
cell cycle gene Trp53/Cdc27 in Cyc B cells indicates that they are
respectively involved in inflammatory drive, immunosuppression,
repair chemotaxis and proliferation responses. (A) T cell clusters
typing and marker genes for each cluster. (B) Cell clusters typing
and marker genes for each cluster. ScRNA-seq, single-cell RNA
sequencing; Naive T cells, immune response preparatory T cells;
effector T cells, pro-inflammatory injury type T cells (expressing
Ccl6 and IL-1β); regulatory T cells, immunosuppressive T cells
(rich in Sp1 regulators); NK cells, direct killer T cells; B1
cells, natural immune barrier type B cells; B2 cells, lymphoid
tissue localization type B cells; marginal zone B cells (MZ), rapid
antibody-responsive B cells; germinal center B cells (GC),
antibody-affinity mature B cells; hB, heart-associated B cells,
repair of core-type B cells (expressing Tgfb1, Cd69, Cxcr5 and
Ccr7); CD74+, antigen-presenting type B fine (expressing
Cd74+); IFNR cells, interferon-responsive B cells; Cyc
cells, proliferation type B cells (expressing Trp53, Cdc27, Mrto4,
Nhp2, Ranbp1 and Ncl Gnl3).](/article_images/mmr/32/6/mmr-32-06-13680-g04.jpg) | Figure 5.T and B cell clusters typing and
marker genes for each cluster [MZ marker genes related literature].
High expression of pro-inflammatory factor IL-1β, chemokine Ccl6
and transcription factor Irf5/Fosb in effector T cells regulate the
enrichment of immune regulatory pathways (such as Sp1) in T cells;
co-activation of the dual-chemokine receptor Cxcr5/Ccr7 and the
tissue repair factor Tgfb1 in hB cells; and upregulation of the
cell cycle gene Trp53/Cdc27 in Cyc B cells indicates that they are
respectively involved in inflammatory drive, immunosuppression,
repair chemotaxis and proliferation responses. (A) T cell clusters
typing and marker genes for each cluster. (B) Cell clusters typing
and marker genes for each cluster. ScRNA-seq, single-cell RNA
sequencing; Naive T cells, immune response preparatory T cells;
effector T cells, pro-inflammatory injury type T cells (expressing
Ccl6 and IL-1β); regulatory T cells, immunosuppressive T cells
(rich in Sp1 regulators); NK cells, direct killer T cells; B1
cells, natural immune barrier type B cells; B2 cells, lymphoid
tissue localization type B cells; marginal zone B cells (MZ), rapid
antibody-responsive B cells; germinal center B cells (GC),
antibody-affinity mature B cells; hB, heart-associated B cells,
repair of core-type B cells (expressing Tgfb1, Cd69, Cxcr5 and
Ccr7); CD74+, antigen-presenting type B fine (expressing
Cd74+); IFNR cells, interferon-responsive B cells; Cyc
cells, proliferation type B cells (expressing Trp53, Cdc27, Mrto4,
Nhp2, Ranbp1 and Ncl Gnl3). |
In 2022, Zhuang et al (63) conducted scRNA-seq of live
myocardial CD45+ leukocytes. Samples were collected at 7
days post-sham surgery and at 1 or 7 dpi. The authors identified
CD4-C1-CCR7 and CD8-C1-CCR7 as markers for naïve T-cells, while
CD4-C2-CXCR3 and CD8-C2-Gzmk were markers for effector T-cells.
These markers were positively associated with the differentiation
of Th17, Th1 and Th2 cells, as well as the PD-L1 signaling pathway.
CD4-C2-CXCR3 and CD8-C2-Gzmk T-cell populations both markedly
increased at 7 dpi. Cxcr3+ CD4 T-cells and Gzmk+ CD8
T-cells activated key adaptive immune responses via Fos and Fosb
activity on 7 dpi. Targeting specific effector T cell subsets or
their activation pathways (such as Fos/Fosb) may serve as a new
strategy for regulating cardiac inflammation and repair following
MI.
In 2022, Qian et al (68) observed that CD8+
effector T-cells secreted effector molecules such as GZMB, GNLY and
PRF1, which promoted apoptosis and EC shedding, leading to plaque
erosion. This was demonstrated via scRNA-seq of peripheral blood
mononuclear cells from 10 patients with AMI (5 with plaque rupture
and 5 without) (69). Upregulation
of CXCR4, B4GALT1 and TNFAIP3 was associated with angiogenesis and
wound healing, thereby promoting plaque healing (70). It may help evaluate the plaque
stability or healing potential of patients with AMI and provide a
basis for individualized intervention.
In 2021, Heinrichs et al (71) performed single-cell RNA and B-cell
receptor sequencing on 20 B-cells purified from the heart and
mediastinal lymph nodes on day 5 post-MI, revealing extensive
phenotypic diversity among B-cells infiltrating the infarcted
heart. scRNA-seq of 6,588 B-cells using UMAP revealed nine B-cell
subpopulations in the heart, including B1, B2 (including follicular
B-cells), marginal zone B-cells (MZ), germinal center B-cells (GC)
and heart-associated B-cells (hB). Additionally, three smaller
clusters were identified: a CD74+ cluster, an
IFN-responsive transcript-rich subpopulation and a cluster enriched
in cell cycle-related and ribosome biogenesis transcripts (Trp53,
Cdc27, Mrto4, Nhp2, Ranbp1, Ncl and Gnl3). This last cluster likely
represents cycling B-cell subpopulations (Fig. 5B).
With the exception of the hB population, all other
clusters underwent significant numerical expansion post-MI. B cells
are the only subpopulation expressing Cxcr5. The hB cell population
upregulates the expression of transforming growth factor β1 (Tgfb1)
and the activation marker Cd69, suggesting that it is in an
activated state and may be involved in the process of immune
regulation or tissue repair. It is also the only subpopulation that
simultaneously highly expressed both Cxcr5 and Ccr7 (double
positive). Flow cytometry confirmed that
CCR7+CXCR5+ B cells were specifically
enriched in the infarcted scar tissue. Their numbers peaked at 7
dpi (71). The pro-repair
properties of hB cells (such as secreting Tgfb1) and their specific
localization in scar tissue make them potential therapeutic targets
for promoting myocardial repair or inhibiting adverse remodeling.
Regulating its chemotaxis (such as the CXCR5/CCR7 pathway) or
functions (such as the Tgfb1 signaling) may have therapeutic value.
The expansion of different B cell subsets (such as pro-inflammatory
Cyc B that may regulate/repair hB) reflects the complex immune
response following MI, providing ideas for developing the
regulation of immune balance and optimizing the repair process.
In 2022, Zhuang et al (63) identified two B-cell clusters with
differential expression of Cd20 (Ms4a1) and Cd23 (Fcer1g):
B-cell-Cd20 and B-cell-Cd23. B-cell-Cd20 showed a slight expansion
at 1 dpi, while B-cell-Cd23 expanded at 7 dpi. B cell-CD20 highly
expressed CD20 (regulating B cell maturation, differentiation and
BCR signal transduction) and CD69 (an early activation marker). B
cell-Cd23 specifically and highly expresses CD23 (a low-affinity
IgE receptor involved in the activation regulation of B cells). The
CD20+ or CD23+ B cell subsets can be used as
markers for evaluating the immune response following MI.
Interactions between cells in the heart
following MI
Increased intercellular communication is a hallmark
of cardiac repair and plays a critical role in cardiac remodeling
(72). The analysis of cellular
interactions through single-cell sequencing has deepened the
understanding of post-infarction cellular interactions, as
demonstrated in the following studies.
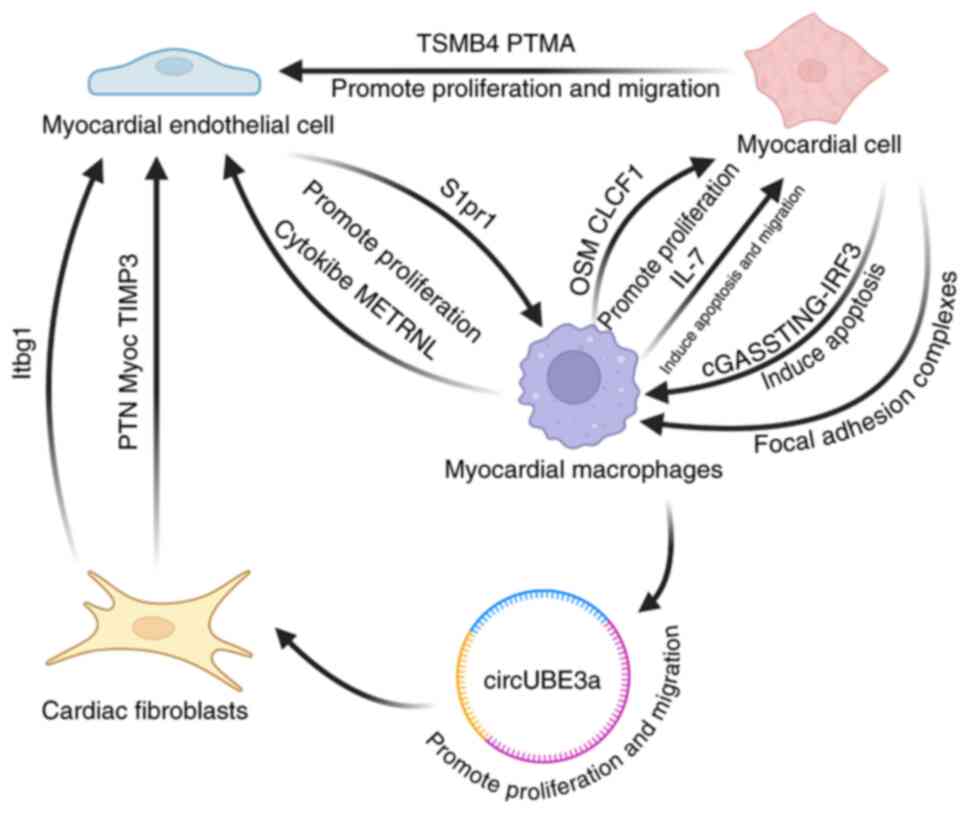
Cardiomyocytes and macrophages
In 2020, Li et al (73) found that macrophage secretion of
oncostatin M (OSM) promoted cardiomyocyte (CM) proliferation and
cardiac regeneration after infarction. Wang et al (39) observed that mononuclear macrophages
secreted CLCF1, which promoted neonatal CM proliferation. Yan et
al (74) in 2021 showed that
post-infarction macrophages could induce CM apoptosis and enhance
CM migration in vitro via IL-7. Wong et al (75) concluded that post-infarction
CCR2+ macrophages interact with CMs via adhesion plaque
complex markers such as β-integrins. Hu et al (76) demonstrated that, after infarction,
CMs that had undergone death could interact with macrophages
through the cGAS-STING-IRF3 pathway, potentially inducing apoptosis
in healthy CMs.
Cardiomyocytes and ECs
In 2021, Gladka et al (77) found that ZEB2 induced
cardiomyocytes to produce TMSB4 and PTMA, which drove EC migration
and proliferation, thereby promoting angiogenesis and improving
cardiac function.
Macrophages and ECs
In 2021, Kuang et al (78) demonstrated that ECs interacted with
macrophages in a contact-dependent manner via the
S1P/S1PR1/ERK/CSF1 signaling pathway, promoting the proliferation
of anti-inflammatory macrophages in damaged cardiac tissues and
reducing adverse cardiac remodeling post-MI. Alonso-Herranz et
al (79) found that post-MI
cardiac macrophages increased MMP14 (MT1-MMP) expression, activated
TGF-β1 and triggered paracrine SMAD2-mediated signaling in ECs.
Reboll et al (80) reported
that macrophages interact with ECs via the cytokine metrnl.
Fibroblasts and ECs
In 2019, Farbehi et al (43) revealed that myofibroblasts
interacted closely with ECs through an interaction network.
Fibroblast populations (F-SH, F-SL, F-Act and F-WntX) frequently
interacted with ECs and PDGFRA-GFP+ fibroblasts and
CD31+ ECs showed close spatial associations or direct
contact, with F-WntX upregulating ligands such as PTN, MyoC and
TIMP3. In 2020, Zhuang et al (51) utilized a selected collection of
human ligand-receptor pairs (81)
and the STRING database (82) to
demonstrate that Fibro-Myo and Endo_1 interacted via Itgb1.
Fibroblasts and macrophages
In 2021, Wang et al (83) identified that post-infarction
macrophages release endosomal membrane-derived vesicles enriched
with circUbe3a, which promotes fibroblast proliferation, migration
and phenotypic transformation, potentially exacerbating myocardial
fibrosis post-MI. Wang et al (39) identified RSPO1 as a mediator of
cellular crosstalk between epicardial cells and ECs, promoting the
angiogenic capacity of ECs during the regeneration of
post-infarction neonatal hearts.
In conclusion, interactions between cardiomyocytes
and non-cardiomyocytes, as well as between non-cardiomyocyte
populations, play an essential role in post-infarction cardiac
remodeling. These interactions are summarized in Fig. 6.
Summary and prospects
Through single-cell sequencing technology, mainly
scRNA-seq and snRNA-seq, different subsets of cardiomyocytes and
changes in the proportion of subsets and the expression of
different genes following MI have been explored. It has been
reported that cardiomyocytes following MI have renewability, which
is maintained within 1 week after birth. The aforementioned changes
of interstitial cardiomyocytes indicate their different functions.
Different interactions occur between cardiomyocytes and stromal
cells through regulatory factors, also demonstrating the response
of cardiomyocytes and cardiomyocyte stromal cells to MI.
Single-cell sequencing can help researchers precisely distinguish
the specific roles and changes of different cell subsets in these
processes, analyze the signaling pathways and interaction networks
of different cell subsets and identify the key molecules and
pathways that affect cardiac remodeling, thereby providing more
precise targets for treatment following MI.
However, the differences in the time points
selected across studies may affect the interpretation and
conclusion: Sparse time point designs (such as analyzing only 3 and
28 days) may miss key transitions. The peak of inflammation in
mouse models is within 72 h, while in humans it lasts several
weeks, resulting in limited functional studies on repair phase cell
subsets (such as VEC2 ECs). Technical method differences (such as
the sensitivity of 10X Genomics and Smart-seq2) can affect the
capture of rare subgroups (such as VEC3). To reduce bias, studies
need to adopt a dense time series design, combine spatial
transcriptome verification and correct the cross-species time axis
to comprehensively analyze the spatio-temporal specific functions
of cell subsets. In addition, single-cell sequencing can reveal
significant differences and provide a complementary value in cell
responses between animal models and clinical samples. Regarding
temporal dynamics differences. animal models (such as mouse models)
can help analyze h-level cell transformation after injury (such as
Bhlhe41+ macrophages expanding at 72 h to mediate
repair), while clinical samples (human autopsies/transplanted
hearts) only provide single-point snapshots and are difficult to
employ for capturing dynamic processes. In animal models, the
mononuclear-macrophage transformation is predominant, while in
clinical samples, B/T cells are more involved (for example,
CXCR5+hB cells secrete IL-10 to promote repair). In
animal models, there is regeneration in the heart, but in adult
human hearts, there is almost no regeneration. The fibroblast
subsets in the scar area are more complex (such as the enrichment
of the FAP+/DDR2+ pro-fibrotic subsets).
Future research should more deeply identify and
functionally verify the key rare cell subpopulations involved in
repair (such as angiogenesis, anti-inflammation and the transition
from pro-fibrotic to anti-fibrotic) and regeneration. The
combination of scRNA-seq with lineage tracing, CRISPR screening and
space technology may help precisely reveal the specific roles of
these cells in dynamic changes, fate determination and
intercellular communication after injury. Design individualized
targeted therapeutic strategies (such as specific antibodies, small
interfering RNA and small molecule inhibitors) should be employed
for analyzing key dysregulated pathways or harmful cell populations
in specific patients. The integration of scRNA-seq and spatial
transcriptomics is the core direction in the future, as it can not
only identify cell types, but also accurately reveal their spatial
distribution, neighborhood associations and local ecological niche
characteristics in the infarct, boundary and distal areas.
Combining space technology and stromal omics, studying how
different cells sense, reshape and respond to regional-specific
extracellular matrix changes, is at the core of fibrosis
regulation. Inferring cell interactions (such as CellChat and
NicheNet) using scRNA-seq data will depict the signal networks that
change dynamically following MI in greater depth. In the future, it
will be necessary to use secretomics to verify and quantify the
activities of key signaling pathways. Deeply integrating multimodal
data, utilizing artificial intelligence to mine hidden patterns in
complex data, establishing a comprehensive human heart cell atlas
(health and disease status), developing minimally invasive
biomarkers and intelligent targeted delivery systems and verifying
single-cell guided strategies in clinical trials should be the
focus of future studies. scRNA-seq is expected to markedly change
the current understanding and management of MI within the next
decade, moving from the traditional ‘one-size-fits-all’ treatment
to a new era of precise diagnosis and targeted intervention based
on individual cellular and molecular characteristics.
Acknowledgements
Not applicable.
Funding
The present study was supported by National Natural Science
Foundation Youth Fund (grant no. 81700230), China Postdoctoral
Science Foundation (grant no. 2022M711321) and Jining Medical
University Research Fund for Academician Lin He New Medicine (grant
no. JYHL2022FZD03).
Availability of data and materials
Not applicable.
Authors' contributions
XB, HG, CG, NL, LG and XC contributed to the study
conception and design. The first draft of the manuscript was
written by XB. Writing was supervised and guided by XC and LG. Data
authentication is not applicable. All authors commented on previous
versions of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Heusch G: Myocardial ischaemia-reperfusion
injury and cardioprotection in perspective. Nat Rev Cardiol.
17:773–789. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
van der Bijl P, Abou R, Goedemans L, Gersh
BJ, Holmes DR Jr, Ajmone Marsan N, Delgado V and Bax JJ: Left
ventricular post-infarct remodeling: Implications for systolic
function improvement and outcomes in the modern era. JACC Heart
Fail. 8:131–140. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu Y, Xu J, Wu M, Kang L and Xu B: The
effector cells and cellular mediators of immune system involved in
cardiac inflammation and fibrosis after myocardial infarction. J
Cell Physiol. 235:8996–9004. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zheng GXY, Terry JM, Belgrader P, Ryvkin
P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J,
et al: Massively parallel digital transcriptional profiling of
single cells. Nat Commun. 8:140492017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hao Y, Hao S, Andersen-Nissen E, Mauck WM
III, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al:
Integrated analysis of multimodal single-cell data. Cell.
184:3573–3587.e29. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rondini EA and Granneman JG: Single cell
approaches to address adipose tissue stromal cell heterogeneity.
Biochem J. 477:583–600. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mackay IM, Arden KE and Nitsche A:
Real-time PCR in virology. Nucleic Acids Res. 30:1292–1305. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kalisky T, Blainey P and Quake SR: Genomic
analysis at the single-cell level. Annu Rev Genet. 45:431–445.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
van der Leun AM, Thommen DS and Schumacher
TN: CD8+ T cell states in human cancer: Insights from
single-cell analysis. Nat Rev Cancer. 20:218–232. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Stewart BJ, Ferdinand JR and Clatworthy
MR: Using single-cell technologies to map the human immune
system-implications for nephrology. Nat Rev Nephrol. 16:112–128.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zaragosi LE, Deprez M and Barbry P: Using
single-cell RNA sequencing to unravel cell lineage relationships in
the respiratory tract. Biochem Soc Trans. 48:327–336. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ziegenhain C, Vieth B, Parekh S, Reinius
B, Guillaumet-Adkins A, Smets M, Leonhardt H, Heyn H, Hellmann I
and Enard W: Comparative analysis of single-cell RNA sequencing
methods. Mol Cell. 65:631–643.e4. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gao S: Data analysis in single-cell
transcriptome sequencing. Methods Mol Biol. 1754:311–326. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Saliba AE, Westermann AJ, Gorski SA and
Vogel J: Single-cell RNA-seq: Advances and future challenges.
Nucleic Acids Res. 42:8845–8860. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tang F, Barbacioru C, Wang Y, Nordman E,
Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, et al: mRNA-Seq
whole-transcriptome analysis of a single cell. Nat Methods.
6:377–382. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hou Y, Song L, Zhu P, Zhang B, Tao Y, Xu
X, Li F, Wu K, Liang J, Shao D, et al: Single-cell exome sequencing
and monoclonal evolution of a JAK2-negative myeloproliferative
neoplasm. Cell. 148:873–885. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Picelli S, Faridani OR, Björklund AK,
Winberg G, Sagasser S and Sandberg R: Full-length RNA-seq from
single cells using Smart-seq2. Nat Protoc. 9:171–181. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Macosko EZ, Basu A, Satija R, Nemesh J,
Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck
EM, et al: Highly parallel genome-wide expression profiling of
individual cells using nanoliter droplets. Cell. 161:1202–1214.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Klein CA, Seidl S, Petat-Dutter K, Offner
S, Geigl JB, Schmidt-Kittler O, Wendler N, Passlick B, Huber RM,
Schlimok G, et al: Combined transcriptome and genome analysis of
single micrometastatic cells. Nat Biotechnol. 20:387–392. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sasagawa Y, Nikaido I, Hayashi T, Danno H,
Uno KD, Imai T and Ueda HR: Quartz-Seq: A highly reproducible and
sensitive single-cell RNA sequencing method, reveals non-genetic
gene-expression heterogeneity. Genome Biol. 14:R312013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jaitin DA, Kenigsberg E, Keren-Shaul H,
Elefant N, Paul F, Zaretsky I, Mildner A, Cohen N, Jung S, Tanay A
and Amit I: Massively parallel single-cell RNA-seq for marker-free
decomposition of tissues into cell types. Science. 343:776–779.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Keren-Shaul H, Kenigsberg E, Jaitin DA,
David E, Paul F, Tanay A and Amit I: MARS-seq2.0: An experimental
and analytical pipeline for indexed sorting combined with
single-cell RNA sequencing. Nat Protoc. 14:1841–1862. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang X, Li T, Liu F, Chen Y, Yao J, Li Z,
Huang Y and Wang J: Comparative analysis of droplet-based
ultra-high-throughput single-cell RNA-Seq systems. Mol Cell.
73:130–142.e5. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yekelchyk M, Guenther S, Preussner J and
Braun T: Mono- and multi-nucleated ventricular cardiomyocytes
constitute a transcriptionally homogenous cell population. Basic
Res Cardiol. 114:362019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Habib N, Avraham-Davidi I, Basu A, Burks
T, Shekhar K, Hofree M, Choudhury SR, Aguet F, Gelfand E, Ardlie K,
et al: Massively parallel single-nucleus RNA-seq with DroNc-seq.
Nat Methods. 14:955–958. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fischer J and Ayers T: Single nucleus
RNA-sequencing: How it's done, applications and limitations. Emerg
Top Life Sci. 5:687–690. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Selewa A, Dohn R, Eckart H, Lozano S, Xie
B, Gauchat E, Elorbany R, Rhodes K, Burnett J, Gilad Y, et al:
Systematic comparison of high-throughput single-cell and
single-nucleus transcriptomes during cardiomyocyte differentiation.
Sci Rep. 10:15352020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hu P, Liu J, Zhao J, Wilkins BJ, Lupino K,
Wu H and Pei L: Single-nucleus transcriptomic survey of cell
diversity and functional maturation in postnatal mammalian hearts.
Genes Dev. 32:1344–1357. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Litviňuková M, Talavera-López C, Maatz H,
Reichart D, Worth CL, Lindberg EL, Kanda M, Polanski K, Heinig M,
Lee M, et al: Cells of the adult human heart. Nature. 588:466–472.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mikhailov AT and Torrado M: The enigmatic
role of the ankyrin repeat domain 1 gene in heart development and
disease. Int J Dev Biol. 52:811–821. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kwapiszewska G, Wygrecka M, Marsh LM,
Schmitt S, Trösser R, Wilhelm J, Helmus K, Eul B, Zakrzewicz A,
Ghofrani HA, et al: Fhl-1, a new key protein in pulmonary
hypertension. Circulation. 118:1183–1194. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Geng T, Liu Y, Xu Y, Jiang Y, Zhang N,
Wang Z, Carmichael GG, Taylor HS, Li D and Huang Y: H19 lncRNA
promotes skeletal muscle insulin sensitivity in part by targeting
AMPK. Diabetes. 67:2183–2198. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lee JH, Protze SI, Laksman Z, Backx PH and
Keller GM: Human pluripotent stem cell-derived atrial and
ventricular cardiomyocytes develop from distinct mesoderm
populations. Cell Stem Cell. 21:179–194.e4. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cui M, Wang Z, Chen K, Shah AM, Tan W,
Duan L, Sanchez-Ortiz E, Li H, Xu L, Liu N, et al: Dynamic
transcriptional responses to injury of regenerative and
non-regenerative cardiomyocytes revealed by single-nucleus RNA
sequencing. Dev Cell. 53:102–116.e8. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kretzschmar K, Post Y, Bannier-Hélaouët M,
Mattiotti A, Drost J, Basak O, Li VSW, van den Born M, Gunst QD,
Versteeg D, et al: Profiling proliferative cells and their progeny
in damaged murine hearts. Proc Natl Acad Sci USA.
115:E12245–E12254. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li Y, Lui KO and Zhou B: Reassessing
endothelial-to-mesenchymal transition in cardiovascular diseases.
Nat Rev Cardiol. 15:445–456. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Libby P, Pasterkamp G, Crea F and Jang IK:
Reassessing the mechanisms of acute coronary syndromes. Circ Res.
124:150–160. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tani H, Sadahiro T, Yamada Y, Isomi M,
Yamakawa H, Fujita R, Abe Y, Akiyama T, Nakano K, Kuze Y, et al:
Direct reprogramming improves cardiac function and reverses
fibrosis in chronic myocardial infarction. Circulation.
147:223–238. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang Z, Cui M, Shah AM, Tan W, Liu N,
Bassel-Duby R and Olson EN: Cell-type-specific gene regulatory
networks underlying murine neonatal heart regeneration at
single-cell resolution. Cell Rep. 33:1084722020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kalucka J, de Rooij LPMH, Goveia J,
Rohlenova K, Dumas SJ, Meta E, Conchinha NV, Taverna F, Teuwen LA,
Veys K, et al: Single-cell transcriptome atlas of murine
endothelial cells. Cell. 180:764–779.e20. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang H, Pu W, Li G, Huang X, He L, Tian
X, Liu Q, Zhang L, Wu SM, Sucov HM and Zhou B: Endocardium
minimally contributes to coronary endothelium in the embryonic
ventricular free walls. Circ Res. 118:1880–1893. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Artap S, Manderfield LJ, Smith CL,
Poleshko A, Aghajanian H, See K, Li L, Jain R and Epstein JA:
Endocardial Hippo signaling regulates myocardial growth and
cardiogenesis. Dev Biol. 440:22–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Farbehi N, Patrick R, Dorison A, Xaymardan
M, Janbandhu V, Wystub-Lis K, Ho JW, Nordon RE and Harvey RP:
Single-cell expression profiling reveals dynamic flux of cardiac
stromal, vascular and immune cells in health and injury. Elife.
8:e438822019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tombor LS, John D, Glaser SF, Luxán G,
Forte E, Furtado M, Rosenthal N, Baumgarten N, Schulz MH, Wittig J,
et al: Single cell sequencing reveals endothelial plasticity with
transient mesenchymal activation after myocardial infarction. Nat
Commun. 12:6812021. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Garikipati VNS, Verma SK, Cheng Z, Liang
D, Truongcao MM, Cimini M, Yue Y, Huang G, Wang C, Benedict C, et
al: Circular RNA CircFndc3b modulates cardiac repair after
myocardial infarction via FUS/VEGF-A axis. Nat Commun. 10:43172019.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Pei J, Cai L, Wang F, Xu C, Pei S, Guo H,
Sun X, Chun J, Cong X, Zhu W, et al: LPA2 contributes to
vascular endothelium homeostasis and cardiac remodeling after
myocardial infarction. Circ Res. 131:388–403. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kanisicak O, Khalil H, Ivey MJ, Karch J,
Maliken BD, Correll RN, Brody MJ, J Lin SC, Aronow BJ, Tallquist MD
and Molkentin JD: Genetic lineage tracing defines myofibroblast
origin and function in the injured heart. Nat Commun. 7:122602016.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Fu X, Khalil H, Kanisicak O, Boyer JG,
Vagnozzi RJ, Maliken BD, Sargent MA, Prasad V, Valiente-Alandi I,
Blaxall BC and Molkentin JD: Specialized fibroblast differentiated
states underlie scar formation in the infarcted mouse heart. J Clin
Invest. 128:2127–2143. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Tallquist MD and Molkentin JD: Redefining
the identity of cardiac fibroblasts. Nat Rev Cardiol. 14:484–491.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ivey MJ, Kuwabara JT, Pai JT, Moore RE,
Sun Z and Tallquist MD: Resident fibroblast expansion during
cardiac growth and remodeling. J Mol Cell Cardiol. 114:161–174.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhuang L, Lu L, Zhang R, Chen K and Yan X:
Comprehensive integration of single-cell transcriptional profiling
reveals the heterogeneities of non-cardiomyocytes in healthy and
ischemic hearts. Front Cardiovasc Med. 7:6151612020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Ruiz-Villalba A, Romero JP, Hernández SC,
Vilas-Zornoza A, Fortelny N, Castro-Labrador L, San Martin-Uriz P,
Lorenzo-Vivas E, García-Olloqui P, Palacio M, et al: Single-Cell
RNA sequencing analysis reveals a crucial role for CTHRC1 (collagen
triple helix repeat containing 1) cardiac fibroblasts after
myocardial infarction. Circulation. 142:1831–1847. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Forte E, Skelly DA, Chen M, Daigle S,
Morelli KA, Hon O, Philip VM, Costa MW, Rosenthal NA and Furtado
MB: Dynamic interstitial cell response during myocardial infarction
predicts resilience to rupture in genetically diverse mice. Cell
Rep. 30:3149–3163.e6. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Rodriguez P, Sassi Y, Troncone L, Benard
L, Ishikawa K, Gordon RE, Lamas S, Laborda J, Hajjar RJ and Lebeche
D: Deletion of delta-like 1 homologue accelerates
fibroblast-myofibroblast differentiation and induces myocardial
fibrosis. Eur Heart J. 40:967–978. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang Z, Cui M, Shah AM, Ye W, Tan W, Min
YL, Botten GA, Shelton JM, Liu N, Bassel-Duby R and Olson EN:
Mechanistic basis of neonatal heart regeneration revealed by
transcriptome and histone modification profiling. Proc Natl Acad
Sci USA. 116:18455–18465. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Horn MA and Trafford AW: Aging and the
cardiac collagen matrix: Novel mediators of fibrotic remodelling. J
Mol Cell Cardiol. 93:175–185. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
González A, Schelbert EB, Díez J and
Butler J: Myocardial interstitial fibrosis in heart failure:
Biological and translational perspectives. J Am Coll Cardiol.
71:1696–1706. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wang Y, Lee M, Yu G, Lee H, Han X and Kim
D: CTHRC1 activates pro-tumorigenic signaling pathways in
hepatocellular carcinoma. Oncotarget. 8:105238–105250. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Binks AP, Beyer M, Miller R and LeClair
RJ: Cthrc1 lowers pulmonary collagen associated with
bleomycin-induced fibrosis and protects lung function. Physiol Rep.
5:e131152017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Stohn JP, Perreault NG, Wang Q, Liaw L and
Lindner V: Cthrc1, a novel circulating hormone regulating
metabolism. PLoS One. 7:e471422012. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
LeClair RJ, Durmus T, Wang Q, Pyagay P,
Terzic A and Lindner V: Cthrc1 is a novel inhibitor of transforming
growth factor-beta signaling and neointimal lesion formation. Circ
Res. 100:826–833. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Janbandhu V, Tallapragada V, Patrick R, Li
Y, Abeygunawardena D, Humphreys DT, Martin EMMA, Ward AO, Contreras
O, Farbehi N, et al: Hif-1a suppresses ROS-induced proliferation of
cardiac fibroblasts following myocardial infarction. Cell Stem
Cell. 29:281–297.e12. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Zhuang L, Wang Y, Chen Z, Li Z, Wang Z,
Jia K, Zhao J, Zhang H, Xie H, Lu L, et al: Global characteristics
and dynamics of single immune cells after myocardial infarction. J
Am Heart Assoc. 11:e0272282022. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Aibar S, González-Blas CB, Moerman T,
Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine JC,
Geurts P, Aerts J, et al: SCENIC: Single-cell regulatory network
inference and clustering. Nat Methods. 14:1083–1086. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
King KR, Aguirre AD, Ye YX, Sun Y, Roh JD,
Ng RP Jr, Kohler RH, Arlauckas SP, Iwamoto Y, Savol A, et al: IRF3
and type I interferons fuel a fatal response to myocardial
infarction. Nat Med. 23:1481–1487. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Dick SA, Macklin JA, Nejat S, Momen A,
Clemente-Casares X, Althagafi MG, Chen J, Kantores C, Hosseinzadeh
S, Aronoff L, et al: Self-renewing resident cardiac macrophages
limit adverse remodeling following myocardial infarction. Nat
Immunol. 20:29–39. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Xu Y, Jiang K, Su F, Deng R, Cheng Z, Wang
D, Yu Y and Xiang Y: A transient wave of Bhlhe41+
resident macrophages enables remodeling of the developing infarcted
myocardium. Cell Rep. 42:1131742023. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Qian J, Gao Y, Lai Y, Ye Z, Yao Y, Ding K,
Tong J, Lin H, Zhu G, Yu Y, et al: Single-cell RNA sequencing of
peripheral blood mononuclear cells from acute myocardial
infarction. Front Immunol. 13:9088152022. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Leistner DM, Kränkel N, Meteva D,
Abdelwahed YS, Seppelt C, Stähli BE, Rai H, Skurk C, Lauten A,
Mochmann HC, et al: Differential immunological signature at the
culprit site distinguishes acute coronary syndrome with intact from
acute coronary syndrome with ruptured fibrous cap: Results from the
prospective translational OPTICO-ACS study. Eur Heart J.
41:3549–3560. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Vergallo R and Crea F: Atherosclerotic
plaque healing. N Engl J Med. 383:846–857. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Heinrichs M, Ashour D, Siegel J, Büchner
L, Wedekind G, Heinze KG, Arampatzi P, Saliba AE, Cochain C,
Hofmann U, et al: The healing myocardium mobilizes a distinct
B-cell subset through a CXCL13-CXCR5-dependent mechanism.
Cardiovasc Res. 117:2664–2676. 2021.PubMed/NCBI
|
|
72
|
Zhang Y, Kanter EM and Yamada KA:
Remodeling of cardiac fibroblasts following myocardial infarction
results in increased gap junction intercellular communication.
Cardiovasc Pathol. 19:e233–e240. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Li Y, Feng J, Song S, Li H, Yang H, Zhou
B, Li Y, Yue Z, Lian H, Liu L, et al: gp130 controls cardiomyocyte
proliferation and heart regeneration. Circulation. 142:967–982.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Yan M, Yang Y, Zhou Y, Yu C, Li R, Gong W
and Zheng J: Interleukin-7 aggravates myocardial
ischaemia/reperfusion injury by regulating macrophage infiltration
and polarization. J Cell Mol Med. 25:9939–9952. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Wong NR, Mohan J, Kopecky BJ, Guo S, Du L,
Leid J, Feng G, Lokshina I, Dmytrenko O, Luehmann H, et al:
Resident cardiac macrophages mediate adaptive myocardial
remodeling. Immunity. 54:2072–2088.e7. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Hu S, Gao Y, Gao R, Wang Y, Qu Y, Yang J,
Wei X, Zhang F and Ge J: The selective STING inhibitor H-151
preserves myocardial function and ameliorates cardiac fibrosis in
murine myocardial infarction. Int Immunopharmacol. 107:1086582022.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Gladka MM, Kohela A, Molenaar B, Versteeg
D, Kooijman L, Monshouwer-Kloots J, Kremer V, Vos HR, Huibers MMH,
Haigh JJ, et al: Cardiomyocytes stimulate angiogenesis after
ischemic injury in a ZEB2-dependent manner. Nat Commun. 12:842021.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Kuang Y, Li X, Liu X, Wei L, Chen X, Liu
J, Zhuang T, Pi J, Wang Y, Zhu C, et al: Vascular endothelial S1pr1
ameliorates adverse cardiac remodelling via stimulating reparative
macrophage proliferation after myocardial infarction. Cardiovasc
Res. 117:585–599. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Alonso-Herranz L, Sahún-Español Á, Paredes
A, Gonzalo P, Gkontra P, Núñez V, Clemente C, Cedenilla M,
Villalba-Orero M, Inserte J, et al: Macrophages promote
endothelial-to-mesenchymal transition via MT1-MMP/TGFβ1 after
myocardial infarction. Elife. 9:e579202020. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Reboll MR, Klede S, Taft MH, Cai CL, Field
LJ, Lavine KJ, Koenig AL, Fleischauer J, Meyer J, Schambach A, et
al: Meteorin-like promotes heart repair through endothelial KIT
receptor tyrosine kinase. Science. 376:1343–1347. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Ramilowski JA, Goldberg T, Harshbarger J,
Kloppmann E, Lizio M, Satagopam VP, Itoh M, Kawaji H, Carninci P,
Rost B and Forrest AR: A draft network of ligand-receptor-mediated
multicellular signalling in human. Nat Commun. 6:78662015.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45(D1): D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Wang Y, Li C, Zhao R, Qiu Z, Shen C, Wang
Z, Liu W, Zhang W, Ge J and Shi B: CircUbe3a from M2
macrophage-derived small extracellular vesicles mediates myocardial
fibrosis after acute myocardial infarction. Theranostics.
11:6315–6333. 2021. View Article : Google Scholar : PubMed/NCBI
|
















![Endothelial cell cluster typing and
marker genes for each cluster. The high expression of capillary
marker Gpihbp1 in VEC1 and VEC2, as well as the enrichment of
macrovascular genes such as Plvap and Vwf in VEC3, indicate that
they are related to the maintenance of vascular activity and
angiogenesis. Endothelial cells have multiple functions in blood
vessels, including vascular stability, blood flow regulation,
material exchange and immune response. Following myocardial
infarction, endothelial cells have the functions of repair,
vascular regeneration and remodeling, as well as coagulation. (A)
Endothelial cell clusters of ventricular tissue origin and marker
genes. (B) Endothelial cell clusters of TIP origin and marker
genes. TIP, cardiac interstitial cell population; ScRNA-seq,
single-cell RNA sequencing; VEC1-3, venous endothelial cells (VEC1
highly expresses Gpihbp1, VEC2 cluster highly expresses Cxcl1 and
Icam1, VEC3 highly expresses Plvap and Vwf); Art.EC, arterial
endothelial cells (enriched with artery-related genes such as
Cxcl12); Endo, metabolism-related endothelial cells (specifically
expressing metabolic genes such as H19 and Cpe); Pro.EC,
proliferative active endothelial cells [highly expressing
proliferation genes Hmgb2 and Birc5]; VEC1, microvascular
endothelial cells [expressing Ly6a (encoding SCA1) and vascular
transcription factor Sox17]; VEC2, arterial endothelial cells
(involved in NOTCH signaling pathways, such as Sox17, Hey1); VEC3,
venous endothelial cells.](/article_images/mmr/32/6/mmr-32-06-13680-g01.jpg)


![T and B cell clusters typing and
marker genes for each cluster [MZ marker genes related literature].
High expression of pro-inflammatory factor IL-1β, chemokine Ccl6
and transcription factor Irf5/Fosb in effector T cells regulate the
enrichment of immune regulatory pathways (such as Sp1) in T cells;
co-activation of the dual-chemokine receptor Cxcr5/Ccr7 and the
tissue repair factor Tgfb1 in hB cells; and upregulation of the
cell cycle gene Trp53/Cdc27 in Cyc B cells indicates that they are
respectively involved in inflammatory drive, immunosuppression,
repair chemotaxis and proliferation responses. (A) T cell clusters
typing and marker genes for each cluster. (B) Cell clusters typing
and marker genes for each cluster. ScRNA-seq, single-cell RNA
sequencing; Naive T cells, immune response preparatory T cells;
effector T cells, pro-inflammatory injury type T cells (expressing
Ccl6 and IL-1β); regulatory T cells, immunosuppressive T cells
(rich in Sp1 regulators); NK cells, direct killer T cells; B1
cells, natural immune barrier type B cells; B2 cells, lymphoid
tissue localization type B cells; marginal zone B cells (MZ), rapid
antibody-responsive B cells; germinal center B cells (GC),
antibody-affinity mature B cells; hB, heart-associated B cells,
repair of core-type B cells (expressing Tgfb1, Cd69, Cxcr5 and
Ccr7); CD74+, antigen-presenting type B fine (expressing
Cd74+); IFNR cells, interferon-responsive B cells; Cyc
cells, proliferation type B cells (expressing Trp53, Cdc27, Mrto4,
Nhp2, Ranbp1 and Ncl Gnl3).](/article_images/mmr/32/6/mmr-32-06-13680-g04.jpg)