Introduction
Herpes simplex virus (HSV), including types HSV-1
and HSV-2, is the primary agent of genital herpes (GH) and is a
globally prevalent virus. HSV-1 and HSV-2 are large double-stranded
DNA viruses that belong to the Herpesviridae family, which
can be found in several populations, in both urban and remote areas
(1,2). HSV-1 commonly causes painful
orofacial herpes, as well as life-threatening meningitis,
keratitis, encephalitis and neonatal infections (3). GH, caused by HSV-1 or HSV-2, is a
prevalent and highly communicable sexually transmitted infection,
which is commonly characterized by recurrent, self-limited genital
ulcers (4,5). Notably, GH is associated with notable
psychological and physiological burdens on affected individuals. In
addition to the acute symptoms, GH commonly results in emotional,
social, relational and sexual distress. However, evidence regarding
the most effective interventions for improving health-related
quality of life is still lacking (6). Both HSV-2 and HSV-1 can cause genital
ulcers, with HSV-2 being the predominant sexually transmitted type
and a major risk factor for increased human immunodeficiency virus
1 (HIV-1) acquisition during co-infection (7–9).
Although there is currently no cure, antiviral therapies such as
acyclovir can improve symptoms, decrease the frequency of
outbreaks, and reduce the risk of transmission to partners
(10). Therefore, a comprehensive
understanding of the biological mechanisms underlying GH recurrence
is essential for improving clinical management and treatment
strategies.
Metabolism, encompassing glycolysis, oxidative
phosphorylation (OXPHOS) and fatty acid oxidation metabolic
pathways, can affect the phenotypes and functions of immune cells.
Metabolic regulation of the immune system serves a crucial role in
the development and progression of numerous diseases, including
cancer, autoimmune disorders and metabolic conditions (11–13).
Alterations in metabolite profiles reflect physiological states,
and also provide critical insights into cellular functions, tissue
metabolism and disease progression (14). Advancements in metabolomics, the
comprehensive study of metabolites within biological systems, have
enabled the identification of disease-specific pathophysiological
characteristics (14–17). Therefore, the present study aimed
to investigate serum metabolite changes in patients with recurrent
HSV-2 GH, thus offering novel insights into its pathogenesis.
Based on the aforementioned evidence linking
metabolism, immunity and disease pathogenesis (10–13),
it was hypothesized that metabolic alterations could contribute to
GH recurrence via modulating immune cell function. To evaluate this
hypothesis, a comprehensive analysis integrating metabolomics,
transcriptomics and single-cell RNA-sequencing (scRNA-seq) was
carried out to identify metabolic pathways associated with HSV-2 GH
recurrence. Through these comprehensive analyses, the current study
aimed to uncover the potential molecular mechanisms underlying GH
pathogenesis, and to identify novel biomarkers and therapeutic
targets.
Materials and methods
Sample preparation
Blood samples were collected from 14 patients with
recurrent HSV-2 GH and 14 healthy volunteers at Sir Run Run Shaw
Hospital (Hangzhou, China) between March and June 2024. All
individuals provided written informed consent prior to study
enrollment. For untargeted metabolomics analysis, serum samples
were collected after centrifugation at 12,000 × g and 4°C for 15
min, and were stored at −80°C. Peripheral blood mononuclear cells
(PBMCs) were isolated through density gradient centrifugation at
400 × g and 25°C for 30 min, and were utilized for RNA-seq and
scRNA-seq. To control for potential hormonal confounding factors,
this study exclusively enrolled female participants. The inclusion
criteria for patients with GH were as follows: Female participants,
aged 18-35 years (mean ± SD, 25.9±4.8 years; median, 25 years);
clinically confirmed recurrent GH with HSV-2 PCR positivity and ≥3
annual recurrences; absence of comorbidities; and no prior HSV
vaccination. The exclusion criteria were as follows: Pregnant
women; treatment with antiviral medications or immunosuppressants
within the past 2 weeks; and those with any history of HSV
vaccination, diabetes, metabolic syndrome or autoimmune diseases.
The inclusion criteria for the participants in the healthy control
(HC) group were: Female participants, aged 18-35 years (mean ± SD,
26.6±4.9 years; median, 27 years); no history of GH; negative for
HSV infection; and no prior HSV vaccination. The demographic and
clinical characteristics of the study participants are listed in
Table SI.
Untargeted metabolomics profiling
Serum samples were stored at −80°C and untargeted
metabolomics profiling was performed by LC-Bio Technologies
(Hangzhou) Co., Ltd. Samples were randomized based on GH and HC.
Following centrifugation at 13,800 × g for 15 min at 4°C, the
supernatant was carefully collected into fresh glass vials for
subsequent analysis. For quality control (QC) purposes, an aliquot
from each biological replicate was pooled to create a
representative QC sample.
Chromatographic separation was performed using a
Thermo Vanquish Flex UPLC system (Thermo Fisher Scientific, Inc.)
equipped with an ACQUITY UPLC T3 column (100×2.1 mm, 1.8 µm; Waters
Corporation) maintained at 40°C with a constant flow rate of 0.35
ml/min, using mobile phase A (5 mmol/l ammonium acetate and 5
mmol/l acetic acid in water) and B (LC-MS grade acetonitrile) with
the following gradient program: 0.0-0.5 min (99-1% B), 0.5-6.5 min
(1-99% B), 6.5-8.0 min (99% B), 8.0-8.5 min (99-1% B) and 8.5-10.0
min (1% B). High-resolution mass spectrometry analysis was
conducted on an Orbitrap Exploris 120 instrument (Thermo Fisher
Scientific, Inc.) operating in alternating positive (+3,800 V) and
negative (−3,400 V) ionization modes with optimized source
parameters [sweep gas 1, auxiliary gas 12, sheath gas 50 (arbitrary
units), source temperature 350°C]. Data-dependent acquisition was
performed with full MS scans (70-1,050 m/z) at 60,000 resolution
(200 m/z) using standard automatic gain control (AGC) and automatic
maximum injection time, selecting the top four most intense
precursor ions (intensity threshold >5,000) for MS/MS
fragmentation at 15,000 resolution with custom AGC and injection
time settings (6 sec dynamic exclusion). System stability was
monitored by analyzing quality control samples every 10 injections,
with inter-QC mass deviation used for batch correction.
The analysis was carried out on the Triple TOF 5,600
Plus high-resolution tandem mass spectrometer system (MS/MS; SCIEX)
in both positive and negative ion modes. Chromatographic separation
was achieved using an ultra-performance liquid chromatography
system (SCIEX ExionLC 2.0+ system), and the resulting LC-MS data
were preprocessed using XCMS software version 3.8.5 (https://github.com/sneumann/xcms). All data from
the untargeted platform (semi-quantitative) were normalized based
on the minimum level detected for each metabolite/run and were
reported as log2 signal-to-noise ratios. Kyoto Encyclopedia of
Genes and Genomes (KEGG) metabolic pathway enrichment analysis were
performed using the MetaboAnalyst (https://www.metaboanalyst.ca/) database. The
metabolomics data are listed in Table
SII.
Total RNA extraction, library
construction and bulk-RNA sequencing
Blood samples were collected from patients with
recurrent HSV-2 GH and healthy volunteers. After density gradient
centrifugation, the PBMC were immediately immersed in liquid
nitrogen for freezing. The frozen samples were then thoroughly
ground using a pre-cooled mortar. Subsequently, cells were lysed
with 1 ml TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). Following the addition of 200 µl chloroform, the
mixture was vigorously shaken for 15 sec, followed by incubation at
room temperature for 2 min. The samples were then centrifuged at
12,000 × g for 15 min at 4°C, and the supernatant was transferred
to a new tube. To precipitate RNA, each tube was supplemented with
0.5 ml isopropyl alcohol, followed by gently mixing prior to
incubation at room temperature for 10 min. After a second
centrifugation at 12,000 × g for 10 min at 4°C, the supernatant was
discarded. The RNA pellet was then washed with 1 ml 75% ethanol and
the supernatant was discarded following centrifugation at 7,500 × g
for 5 min at 4°C. After air drying, total RNA was dissolved in
DEPC-treated water.
For second-generation sequencing, libraries were
prepared as previously described (18). In brief, RNA quantity and integrity
were assessed using the Qubit™ 3.0 Fluorometer (cat. no. Q33216;
Thermo Fisher Scientific, Inc.) and Agilent 5300 Fragment Analyzer
System (cat. no. M5310AA; Agilent Technologies) respectively. RNA
samples demonstrating high purity (A260/A280 ratio 1.8-2.2) and RNA
integrity numbers >7.0 were selected for subsequent library
preparation using Illumina-compatible protocols. mRNA was purified
from 2 µg total RNA using mRNA Capture Beads 2.0 (cat. no. 12629ES;
Shanghai Yeasen Biotechnology Co., Ltd.) through two rounds of
poly(A) selection.
Purified mRNA was fragmented in magnesium-containing
fragmentation buffer (cat. no. 12340ES97; Shanghai Yeasen
Biotechnology Co., Ltd.) at 94°C for 5 min. First-strand cDNA was
synthesized using random hexamer-primed reverse transcription
(SuperScript™ IV Reverse Transcriptase; Thermo Fisher Scientific,
Inc.). Second-strand cDNA was generated using a dUTP incorporation
strategy with E. coli DNA Polymerase I, RNase H and dUTP
solution (cat. no. 12340ES97; Shanghai Yeasen Biotechnology Co.,
Ltd.). Blunt-ended cDNA fragments were adenylated at 3′ ends using
Klenow Fragment (3′-5′ exo-) and ligated to Illumina-compatible
forked adapters containing T-overhangs. PCR products were
size-selected (400±50 bp inserts) using Hieff NGS DNA Selection
Beads (cat. no. 12601ES75; Shanghai Yeasen Biotechnology Co.,
Ltd.). Strand specificity was maintained through dUTP-based strand
marking and uracil excision. Libraries were sequenced in 2×150 bp
paired-end mode on an Illumina NovaSeq™ X Plus platform (LC-bio
Technologies Hangzhou Co. Ltd.) following manufacturer-recommended
protocols.
Differentially expressed gene (DEG)
analysis
To identify DEGs between patients with GH and
healthy volunteers, the ‘DESeq2’ R package (version 4.5; http://bioconductor.org/packages/release/bioc/html/DESeq2.html)
in R (version 4.4.3; http://www.R-project.org/) was utilized. Genes showing
log2 fold change of ≥1.0 and an adjusted P-value of <0.05 were
considered significantly differentially expressed. The RNA-seq data
are presented in Table SIII.
KEGG pathway enrichment analysis
KEGG (http://www.genome.jp/kegg/), a comprehensive database
for understanding biological systems at the genomic and molecular
levels, DEGs of scRNA-seq data was used for pathway enrichment
analysis. This resource integrates genomic, chemical and systematic
information on biological interactions, reactions and disease
pathways.
Gene Set Enrichment Analysis
(GSEA)
GSEA was performed based on the detected metabolites
using KEGG metabolite sets (19).
The analysis aims to identify significant pathways that are
associated with the metabolite profiles. The criteria for selecting
significant pathways were set as follows: Pathways with an absolute
normalized enrichment score (|NES|) >1 and a P-value <0.05
were considered statistically significant. These thresholds help to
filter out the most relevant pathways that show a strong
association with the metabolic differences observed in the
study.
scRNA-seq
scRNA-seq of PBMCs and neutrophils from GH (n=4) and
healthy controls (n=4) was performed using the Chromium Single Cell
3′ Chip and 10× Genomics Chromium Single-Cell 3′ Kit v3 (10×
Genomics, LC-Bio Technologies) as previously described (20). The sequencing data were then
deposited in the Genome Sequence Archive for human database.
Bioinformatics analysis utilized Cell Ranger (v7.1.0; 10× Genomics;
http://www.10×genomics.com/support/software/cell-ranger)
and Seurat (v4.3.0; Satija Lab; http://satijalab.org/seurat) for data processing,
clustering, and differential expression analysis. Nonlinear
dimensionality reduction was performed using t-distributed
Stochastic Neighbor Embedding implemented in Seurat v4.3.0.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was isolated from PBMCs using
TRIzol® Reagent (cat. no. 15596018CN; Invitrogen; Thermo
Fisher Scientific, Inc.). cDNA was synthesized from 1 µg RNA using
oligo dT primer and the PrimeScript RT PCR master mix Kit (cat. no.
RR036A; Takara Biotechnology Co., Ltd.) under the following
conditions: 37°C for 15 min and 85°C for 5 min. qPCR analysis was
carried out using the SYBR Green Master Mix kit (cat. no.
04707516001; Roche Diagnostics) with 95°C for 5 min (initial
denaturation) and 40 cycles of 95°C for 5 sec (denaturation) and
60°C for 1 min (annealing and extension). The relative gene
expression levels are expressed as fold changes relative to control
samples, and were quantified using the 2−ΔΔCq
comparative threshold cycle method (21). ACTB, a housekeeping gene,
served as an endogenous reference to normalize target gene
expression levels across all experimental groups. The primer
sequences are listed in Table
SIV.
Statistical analysis
All data were analyzed using SPSS statistics
software (version 25.0; IBM Corp.). The results are presented as
the mean ± standard error of the mean. The differences were
compared using an unpaired Student's t-test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Metabolic changes in the blood of
patients with recurrent HSV-2 GH
To investigate the alterations in metabolites
associated with recurrent HSV-2 GH, peripheral blood samples were
collected and serum was isolated for comprehensive metabolic
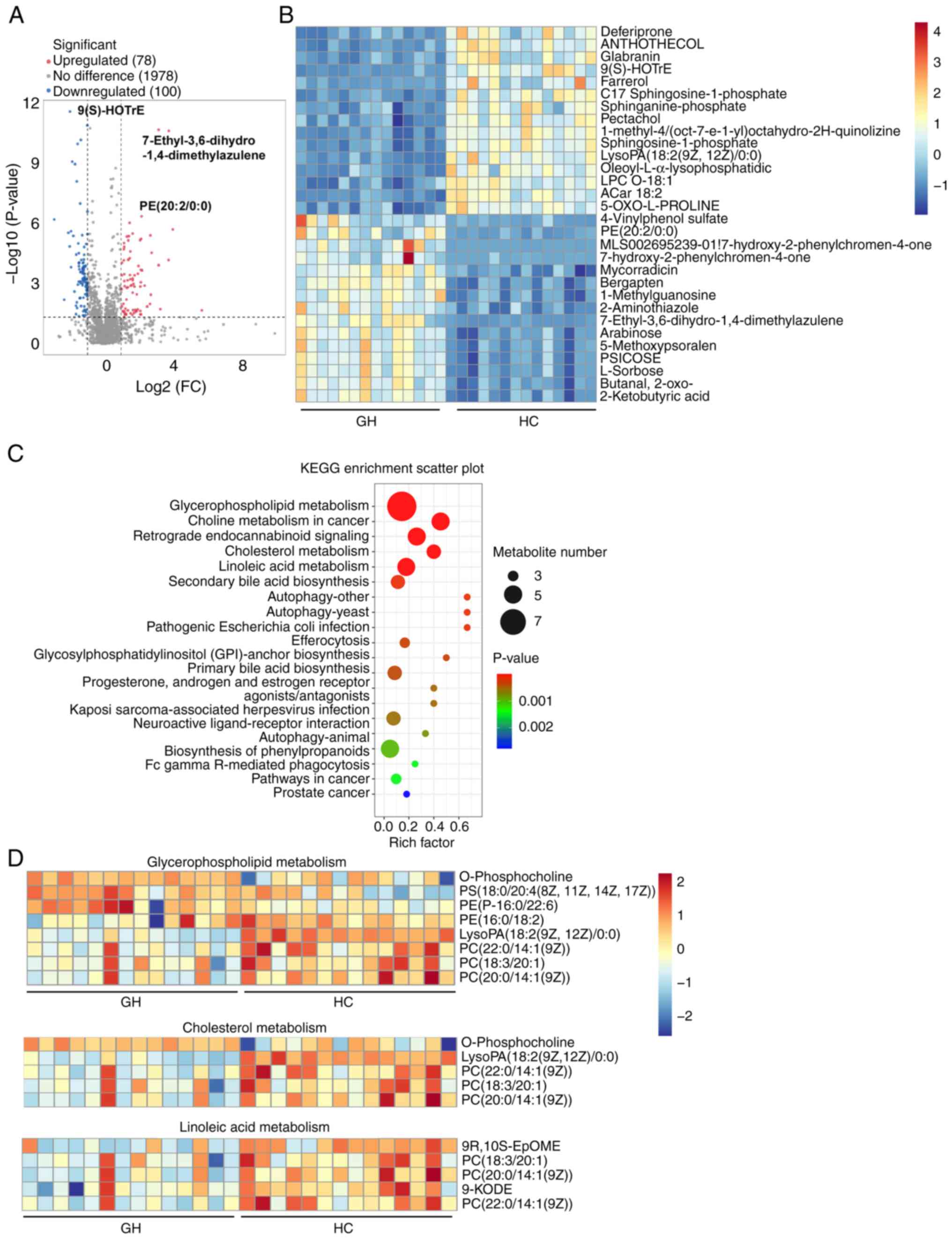
assays. The analysis identified a total of 78 upregulated and 100
downregulated metabolites in patients with GH (Fig. 1A). Notably, the top five
downregulated metabolites were Deferiprone, ANTHOTHECOL Glabranin,
9(S)-HOTrE and Farrerol; conversely, 2-Ketobutyric acid, Butanal,
L-sorbose, PSICOSE and 5-Methoxypsoralen were the five most
significantly upregulated metabolites in the serum of patients with
recurrent GH (Fig. 1B). To further
explore the functional implications of these differentially
expressed metabolites, KEGG enrichment analysis on the metabolomics
data was performed. The top 20 KEGG-enriched pathways in serum
included ‘Glycerophospholipid metabolism’, ‘Choline metabolism in
cancer’, ‘Retrograde endocannabinoid signaling’, ‘Cholesterol
metabolism’ and ‘Linoleic acid metabolism’ (Fig. 1C). Notably, key metabolites in the
Glycerophospholipid metabolism pathway, including O-phosphocholine,
PS (18:0/20:4(8Z,11Z,14Z,17Z)) and PE (16:0/18:2), were markedly
upregulated in the serum of patients with GH compared with HCs
(Fig. 1D). However, the majority
of cholesterol-related metabolites were downregulated in patients
with GH compared with HCs, except O-phosphocholine (Fig. 1D). Additionally, the levels of
metabolites associated with the linoleic acid synthesis pathway,
such as 9R,10S-EpOME, PC (18:3/20:1), PC [20:0/14:1(9Z)], 9-KODE
and PC [22:0/14:1(9Z)], were significantly reduced in the serum of
patients with recurrent GH compared with HCs (Fig. 1D). Overall, these results could
provide novel insights into the metabolic alterations associated
with GH.
GSEA of metabolites in patients with
recurrent HSV-2 GH
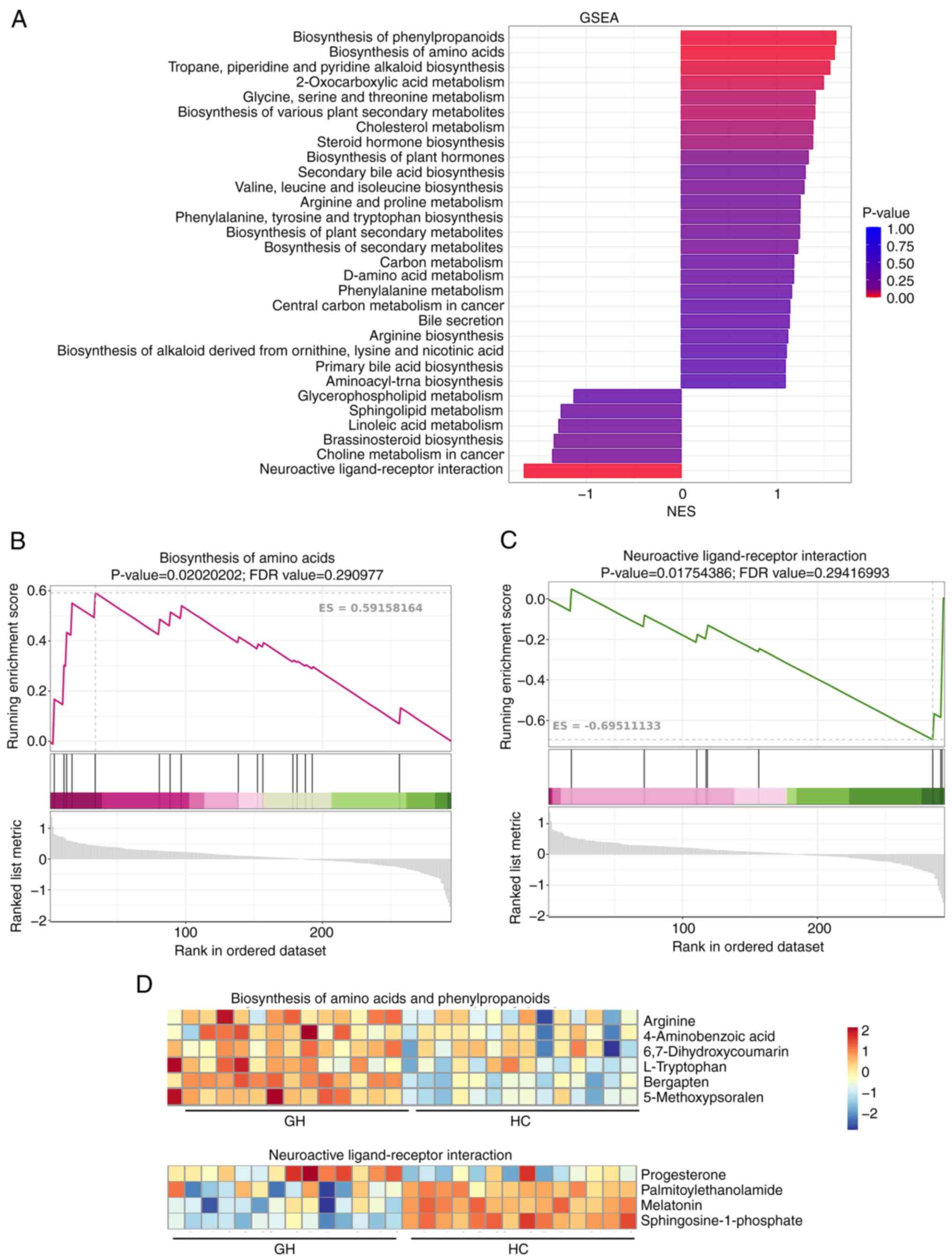
GSEA of the metabolomics data revealed significant
differences in the activation of particular metabolic pathways
between patients with recurrent HSV-2 GH and HCs. As shown in
Fig. 2A, among the top 30
metabolic pathways, 24 were notably enriched in the GH group, while
six were enriched in the normal control group. Notably, pathways,
such as ‘Biosynthesis of amino acids’, ‘Biosynthesis of
phenylpropanoids’, ‘Tropane, piperidine and pyridine alkaloid
biosynthesis’, ‘2-Oxocarboxylic acid metabolism’ and ‘Glycine,
serine and threonine metabolism’ were significantly enriched in the
GH group (Fig. 2A and B).
Conversely, the ‘neuroactive ligand-receptor interaction pathway’
was markedly downregulated in patients with recurrent HSV-2 GH
compared with HCs (Fig. 2A and C).
Metabolomics heatmap data indicated that metabolites associated
with neuroactive ligand-receptor interactions, such as
palmitoylethanolamide, melatonin and sphingosine-1 phosphate, were
all downregulated in the serum samples of patients with recurrent
HSV-2 GH compared with HCs (Fig.
2D). Furthermore, the levels of metabolites involved in amino
acid biosynthesis, including 4-aminobenzoic acid,
6,7-dihydroxycoumarin, L-tryptophan, bergapten and
5-methoxypsoralen, were notably enhanced in the GH group compared
with HCs (Fig. 2D).
RNA-seq analysis of the significant
metabolic pathways in PBMCs of patients with recurrent HSV-2
GH
To elucidate the potential mechanisms underlying
GH-related morbidity and to explore the association between
metabolic changes and transcriptional profiles, RNA-seq analysis
was conducted on blood cells from healthy individuals and patients
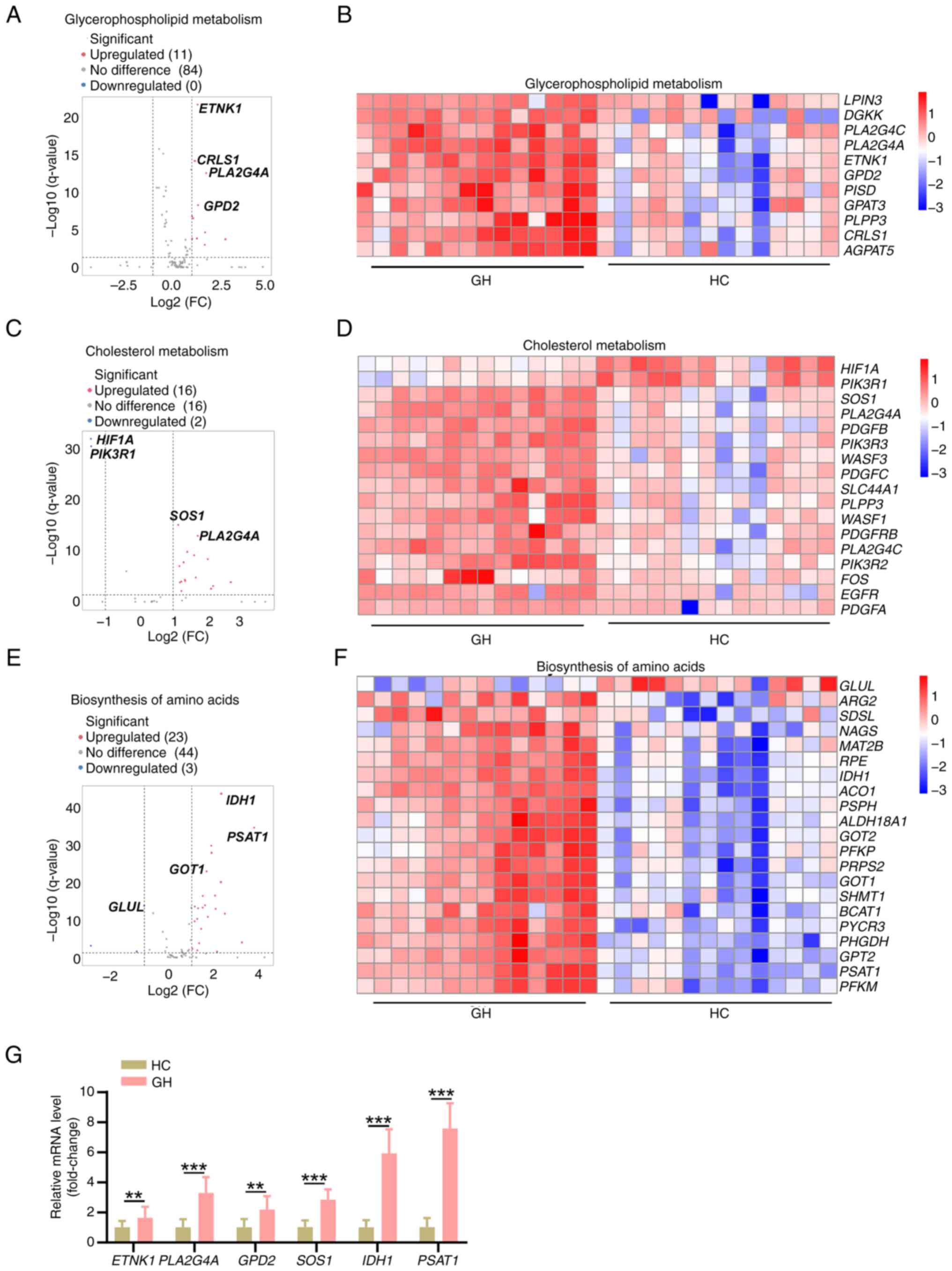
with recurrent HSV-2 GH. The results of RNA-seq analysis revealed
significant gene expression alterations in patients with recurrent
HSV-2 GH, particularly upregulation of genes associated with
glycerophospholipid metabolism, cholesterol metabolism and amino
acid biosynthesis pathways (Fig.
3A-F). These results were consistent with those obtained in the
metabolomics analysis. Several genes associated with the
glycerophospholipid pathway, including ETNK1, CRLS1, PLA2G4A,
GPD2, PISD, AGPAT5, PLPP3, GPAT3, PLA2G4C and DGKK, were
significantly upregulated in the blood cells of patients with
recurrent GH compared with HCs (Fig.
3A and B). Similarly, genes associated with cholesterol
metabolism, such as SOS1, PLA2G4A, PDGFB, PIK3R3, PDGFC, WASF3,
SLC44A1 and PLPP3, were also notably upregulated in the
same samples (Fig. 3C and D). In
terms of the amino acid biosynthesis pathway, the expression of
several genes was also significantly increased in the GH group
compared with in the HC group, including that of IDH1, PSAT1,
ACO1, RPE, GOT1, GPT2, PRPS2, PSPH and SHMT1 (Fig. 3E and F). RT-qPCR analysis also
verified that ETNK1, PLA2G4A, SOS1, GPD2, IDH1 and
PSAT1 were significantly upregulated in blood cells from
patients with recurrent HSV-2 GH compared with in the HC group
(Fig. 3G). These results, combined
with the metabolomic data, revealed significant changes in the
expression of several metabolites and the activation status of
relevant pathways, including those of glycerophospholipid
metabolism, cholesterol metabolism and amino acid biosynthesis, in
patients with recurrent HSV-2 GH.
scRNA-seq analysis of the significant
metabolic pathway-related genes in patients with recurrent HSV-2
GH
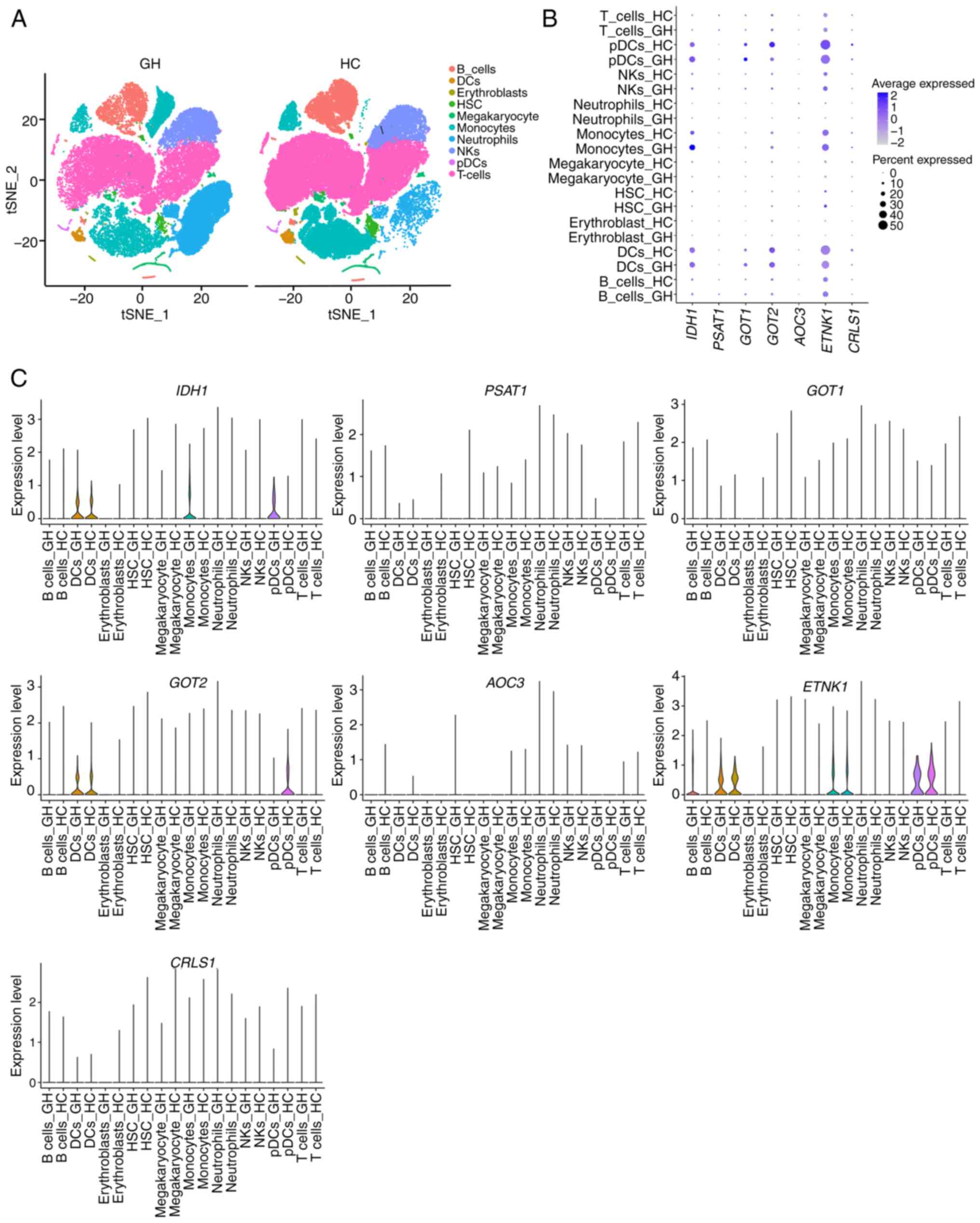
To identify the cell population with enhanced
expression of differentially expressed metabolism-related genes,
the scRNA-seq data in patients with recurrent HSV-2 GH and HC was
analyzed, and the clusters of cells in the PBMCs and neutrophils
were observed (Fig. 4A). The
results demonstrated that IDH1 was highly expressed in
monocytes, dendritic cells (DCs) and plasmacytoid DCs (pDCs), with
notably elevated expression in monocytes from the GH group
(Fig. 4B and C). GOT1 and
GOT2 were also markedly upregulated in pDCs and DCs
(Fig. 4B and C). Additionally,
ETNK1 was highly expressed in B cells, monocytes, DCs and
pDCs (Fig. 4B and C). Overall, the
scRNA-seq analysis revealed distinct metabolic gene expression
patterns across various immune cell subpopulations, including T
cells, B cells, monocytes, neutrophils and DCs.
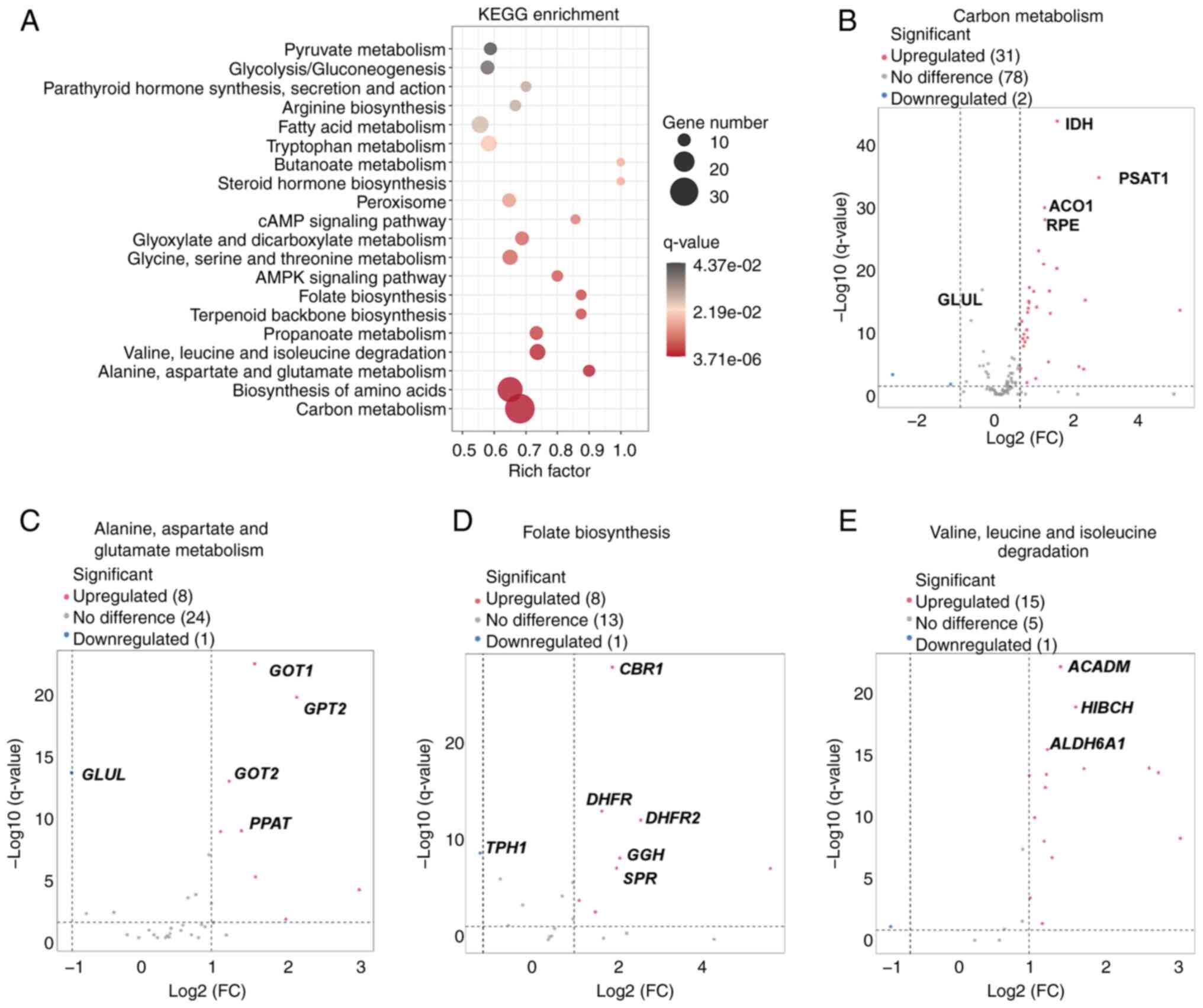
KEGG enrichment analysis of metabolic
genes identified by RNA-seq analysis
Subsequently, KEGG enrichment analysis of the
metabolism-related genes, which were identified by RNA-seq
analysis, was carried out. Several significantly enriched pathways
were detected, including ‘carbon metabolism’, ‘folate
biosynthesis’, ‘valine, leucine and isoleucine degradation’ and
‘alanine, aspartate and glutamate metabolism’ (Fig. 5A). The volcano plot for carbon
metabolism revealed a significant number of upregulated genes
(n=31) and a few downregulated genes (n=2); among the significantly
upregulated genes were IDH1, PSAT1 and RPE, which all
have crucial roles in various metabolic processes (22–24)
(Fig. 5B). The alanine, aspartate
and glutamate metabolism pathway also showed significant changes,
with eight upregulated and one downregulated gene. Key genes, such
as GOT1 and GPT2, were significantly upregulated in
patients with recurrent HSV-2 GH, thus supporting their potential
role in the altered metabolic state associated with GH recurrence
(Fig. 5C). The folate biosynthesis
pathway also exhibited significant changes, with eight upregulated
genes and one downregulated gene. Notably, CBR1 and
DHFR were significantly altered, suggesting their
involvement in the metabolic dysregulation associated with GH
recurrence (Fig. 5D). The valine,
leucine and isoleucine degradation pathway had 15 upregulated
genes, with ACADM and ALDH6A1 being particularly
significant (Fig. 5E). The
pronounced upregulation of these genes could indicate a shift in
amino acid metabolism in patients with recurrent HSV-2 GH. The
aforementioned findings underscored the complex metabolic
reprogramming associated with GH recurrence, with multiple pathways
being significantly affected.
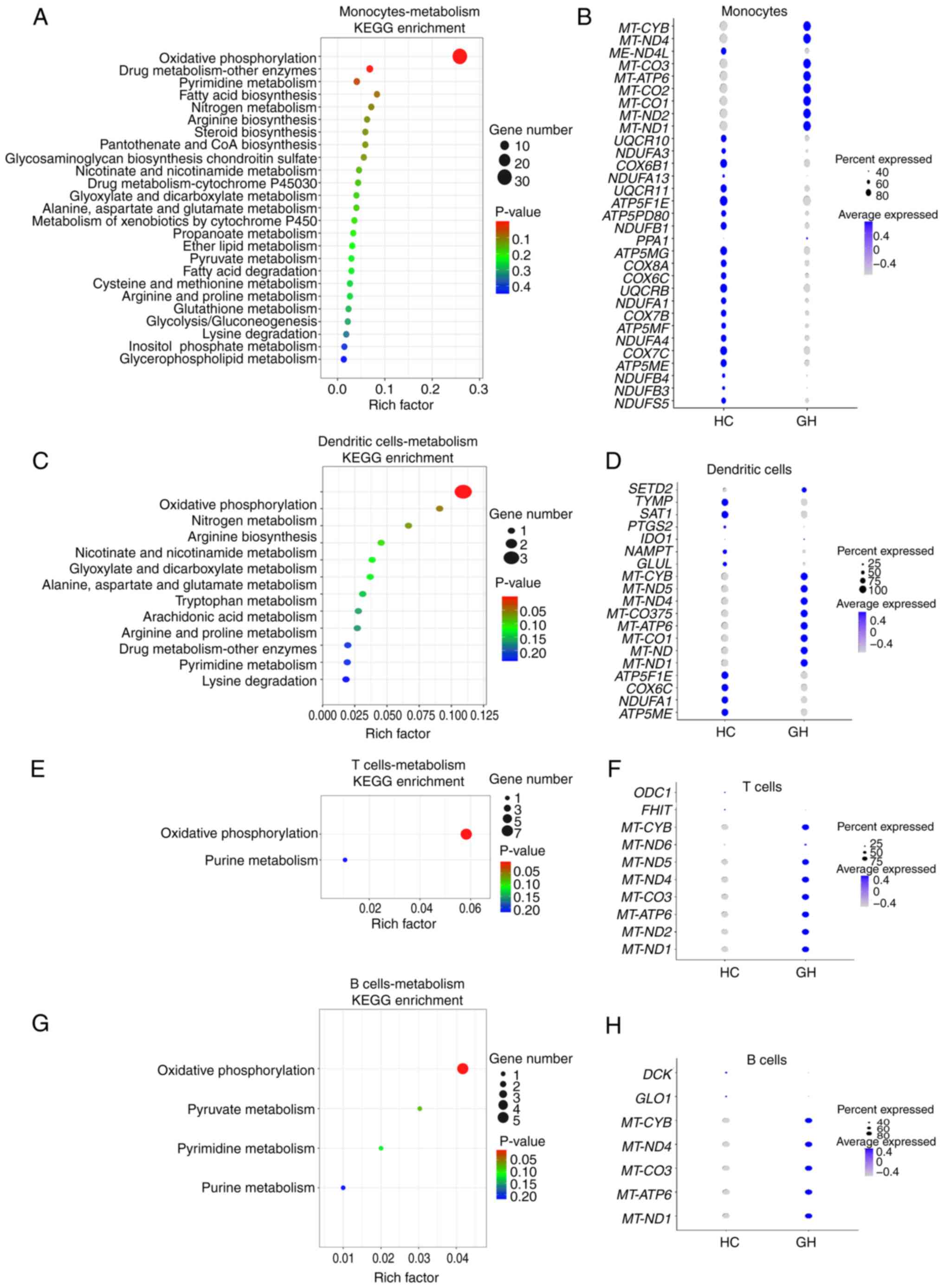
KEGG enrichment analysis of metabolic
genes detected by scRNA-seq analysis
To further analyze the metabolic profiles in
different immune cells, the scRNA-seq data were further assessed
using KEGG enrichment analysis. All immune cells were highly
enriched in the ‘Oxidative phosphorylation’ pathway (Fig. 6A, C, E and G). Monocytes displayed
a significant enrichment in the ‘Oxidative phosphorylation’ and
‘Pyrimidine metabolism’ pathways. Furthermore, almost all of the
mitochondrial genes, including MT-CYB, MT-ND4 and
MT-CO3, were highly expressed in monocytes from patients
with recurrent HSV-2 GH, thus indicating robust metabolic activity,
particularly within the OXPHOS pathway (Fig. 6A and B). In addition, significant
upregulation of mitochondrial genes associated with OXPHOS, such as
MT-CYB and MT-ND4, were recorded in T and B cells,
and DCs (Fig. 6C-H). These
findings suggested that the OXPHOS pathway-associated mitochondrial
genes could display functional roles and responses during this
viral infection.
Discussion
By integrating multiple approaches, including
transcriptomics, metabolomics and scRNA-seq analyses, significant
changes in pathways associated with lipid metabolism, amino acid
biosynthesis and cholesterol metabolism were identified in patients
with GH. Genes associated with the alanine, aspartate and
glutamate, and valine, leucine and isoleucine metabolism pathways
were significantly upregulated in patients with recurrent HSV-2 GH.
In addition, several metabolism-related genes were upregulated in
PBMCs from patients with GH, including IDH1 and
ETNK1, the expression of which was notably increased in DCs
and monocytes. Additionally, mitochondrial genes involved in
OXPHOS, such as MT-CYB and MT-CO3, were significantly
elevated across monocytes, DCs, T cells and B cells. Overall, these
findings indicated metabolic reprogramming in patients with
recurrent HSV-2 GH, thus providing potential biomarkers and
therapeutic targets for the development of future treatment
strategies.
Glycerophospholipid remodeling serves a critical
role in several diseases, including orthoflavivirus infection,
brain injury, Pseudomonas infections and cancer (25–27).
A previous study demonstrated that viral infections can hijack the
host cell glycerophospholipid metabolism to acquire lipid
components for viral envelope synthesis, such as phosphatidylserine
and phosphatidylinositol. Inhibiting related metabolic enzymes,
such as those related to phosphatidylinositol biosynthesis, could
markedly reduce viral titers and cytopathic effects (25,28).
The present study showed that glycerophospholipid metabolism was
upregulated in patients with GH, thus indicating that glycerolipid
biosynthesis could represent a conserved host dependency factor
exploited by this evolving virus. Dysregulated cholesterol
metabolism in arterial macrophages has long been known to be
pathogenic in atherosclerosis, and in an ILC2-driven airway
inflammation model lipid metabolism dysregulation can also be found
(29,30). The present data also found
downregulation of cholesterol-related metabolites, which further
supported the occurrence of a potential lipid metabolism
dysfunction in patients with GH.
Amino acids, the fundamental building blocks of
proteins, and related metabolites, derived from both microorganisms
and host cells, serve a crucial role in regulating immune cell
activation and antibody production, thus markedly affecting several
biological processes, such as inflammation and immune responses
(31,32). Recent studies have identified key
intermediate metabolites in the aromatic amino acid pathways that
undergo alterations during sepsis, influenza and severe acute
respiratory syndrome coronavirus 2 (SARS-CoV-2) infections
(33–35). Herein, the results showed a
significant upregulation of amino acid biosynthesis in patients
with recurrent HSV-2 GH, thus suggesting that aberrant amino acid
biosynthesis could be involved in the pathophysiology of this
disease. A previous Mendelian randomization study revealed that
SARS-CoV-2 infection can affect susceptibility to HSV-1 and HSV-2.
While SARS-CoV-2 infection could suppress HSV-1 reactivation via
inducing a type I interferon response, it could also promote HSV-2
infection via immunosuppression or an inflammatory environment
(36). Based on the results of
this study, the effects between SARS-CoV-2 and GH could also be
mediated by several metabolic pathways.
A previous study reported that serum amino acid
metabolism varies at different time points during influenza virus
infection (35). The levels of
several amino acids, such as valine, proline, citrulline,
isoleucine, asparagine and arginine, and their derivatives may be
altered following viral infection, and these changes could be
associated with disease severity (37). In the present study, the RNA-seq
results also revealed that several amino acid metabolic pathways,
such as alanine, aspartate and glutamate, and valine, leucine and
isoleucine pathways, were enriched in recurrent HSV-2 GH samples.
These amino acid metabolism profiles could indicate GH relapse.
Furthermore, Zhang et al (38) revealed that SARS-CoV-2 exploits
folate and one-carbon metabolism to promote its replication.
Consistently, the present study also revealed that folate
biosynthesis metabolism was upregulated in patients with recurrent
HSV-2 GH. Overall, the aforementioned findings suggested that
targeting host metabolic pathways could be a promising therapeutic
approach against future GH infection.
OXPHOS is a vital mitochondrial metabolic pathway,
which is involved in the generation of adenosine triphosphate
through the electron transport chain and chemiosmosis (39). Emerging evidence has suggested that
viruses can manipulate host OXPHOS to regulate immune responses,
thus enhancing their survival and replication (40). For example, a previous study
demonstrated that HIV can preferentially target glycolytic
CD4+ T cells by relying on elevated OXPHOS activity
regardless of their differentiation and activation status (41). Additionally, liver biopsies from
patients with chronic hepatitis C virus have revealed defects in
OXPHOS, along with increased expression of oxidative stress-related
markers (42). In patients with
SARS-CoV-2, the levels of OXPHOS regulators, such as those of
mitochondrial nuclear retrograde regulator 1, have been shown to be
reduced in heart tissues, potentially contributing to cardiac
complications. Furthermore, the expression levels of core
mitochondrial genes are also decreased during SARS-CoV-2 infection
in both rodent and human hosts (43,44).
Herein, the results showed that the expression levels of
mitochondrial genes involved in OXPHOS, including MT-CYB,
MT-ND4 and MT-CO3 were significantly elevated in
monocytes, T cells and B cells derived from patients with GH.
Another study revealed that HSV could dysregulate lipid synthesis
and induce ferroptosis via enhancing glucose-mediated OXPHOS and
glutamine-dependent reductive carboxylation (45). Additionally, a previous study
demonstrated that following viral infection, pDCs can retain their
ability to secrete type I interferons through lactate dehydrogenase
B-dependent metabolic adaptation (46). Overall, these findings suggested
that OXPHOS modulation could be viral- and cell type-specific.
Although the current study provided valuable
insights, there are several limitations that warrant consideration.
Firstly, the sample size was not sufficient to generalize the
findings across diverse populations, thus highlighting the need for
further research to validate the identified biomarkers.
Additionally, although associations between metabolic changes and
gene expression were recorded, the underlying causal associations
remain to be fully elucidated. Future studies should focus on
investigating the functional roles of these metabolites and genes,
as well as their interactions within the immune system.
Investigating tissue-specific metabolic changes and their effect on
immune cell function could yield a more comprehensive understanding
of the disease. Furthermore, longitudinal studies should be
performed to assess how metabolic alterations evolve over time, and
how these changes could be involved in disease progression and
recurrence. The current study offers compelling evidence of
systemic metabolic reprogramming in recurrent HSV-2 GH and
highlights novel potential therapeutic targets. However, the
research cohort specifically comprised patients diagnosed with
HSV-2-positive GH. Clinically, the recurrence rate of HSV-1 GH is
markedly lower than that of HSV-2, primarily due to a reduced
propensity for neuronal infection (47). Over the past two decades, HSV-1 has
increasingly become the predominant causative agent of
first-episode GH in multiple countries, reflecting a shifting
epidemiological landscape (48,49).
Future studies incorporating rigorous HSV typing are essential to
elucidate potential subtype-specific metabolic vulnerabilities.
In conclusion, the current study revealed
significant metabolic and transcriptional alterations in patients
with recurrent HSV-2 GH, thus providing a foundation for future
studies on metabolic interventions for HSV-2 GH recurrence and
highlighting potential biomarkers for therapeutic
interventions.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the Natural
Science Foundation of China (grant nos. 82373491 and 82471846) and
Peterson's Lab MED Translational Medicine Fund (grant no.
20240131).
Availability of data and materials
The RNA-seq data generated in the present study may
be found in the Sequence Read Archive database under accession
number PRJNA1267039 or at the following URL: (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA1267039).
The metabolomics data generated in the present study may be found
in the National Genomics Data Center database under accession
number PRJCA040378 or at the following URL: (https://ngdc.cncb.ac.cn/search/specific?db=bioproject&q=PRJCA040378).
The scRNA-seq data generated in the present study may be found in
the ScienceDB database (https://www.scidb.cn/en/detail?dataSetId=690475ca10ed4424bdc0f69c3b5cd701).
Authors' contributions
JH, YX, HC and YS conceived and designed the study.
JH, CF, JZ and YC performed the experiments. JH, YX and SC analyzed
the data. JH, HC and JZ wrote and reviewed the manuscript. YS and
HC assume overall responsibility for the manuscript. JH and HC
confirm the authenticity of all the raw data. YS and HC were
responsible for study supervision and funding acquisition. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Sir Run Run Shaw Hospital, affiliated with Zhejiang
University School of Medicine (approval no. 20240106). Written
informed consent was obtained from all participants prior to their
inclusion in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
US Preventive Services Task Force, .
Mangione CM, Barry MJ, Nicholson WK, Cabana M, Chelmow D, Coker TR,
Davis EM, Donahue KE, Jaén CR, et al: Serologic screening for
genital herpes infection: US preventive services task force
reaffirmation recommendation statement. JAMA. 329:502–507. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Havens JL, Calvignac-Spencer S, Merkel K,
Burrel S, Boutolleau D and Wertheim JO: Phylogeographic analysis
reveals an ancient East African origin of human herpes simplex
virus 2 dispersal out-of-Africa. Nat Commun. 13:54772022.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Johnston C, Gottlieb SL and Wald A: Status
of vaccine research and development of vaccines for herpes simplex
virus. Vaccine. 34:2948–2952. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Omarova S, Cannon A, Weiss W, Bruccoleri A
and Puccio J: Genital herpes simplex virus-an updated review. Adv
Pediatr. 69:149–162. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
James C, Harfouche M, Welton NJ, Turner
KM, Abu-Raddad LJ, Gottlieb SL and Looker KJ: Herpes simplex virus:
global infection prevalence and incidence estimates, 2016. Bull
World Health Organ. 98:315–329. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bennett C, Rebafka A, Carrier J, Cook S
and Edwards D: Impact of primary and recurrent genital herpes on
the quality of life of young people and adults: A mixed methods
systematic review. JBI Evid Synth. 20:1406–1473. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Desai DV and Kulkarni SS: Herpes simplex
virus: The interplay between HSV, Host, and HIV-1. Viral Immunol.
28:546–555. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lafferty WE, Downey L, Celum C and Wald A:
Herpes simplex virus type 1 as a cause of genital herpes: Impact on
surveillance and prevention. J Infect Dis. 181:1454–1457. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Masese L, Baeten JM, Richardson BA, Bukusi
E, John-Stewart G, Graham SM, Shafi J, Kiarie J, Overbaugh J and
McClelland RS: Changes in the contribution of genital tract
infections to HIV acquisition among Kenyan high-risk women from
1993 to 2012. AIDS. 29:1077–1085. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schalkwijk HH, Snoeck R and Andrei G:
Acyclovir resistance in herpes simplex viruses: Prevalence and
therapeutic alternatives. Biochem Pharmacol. 206:1153222022.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ning L, Shishi Z, Bo W and Huiqing L:
Targeting immunometabolism against acute lung injury. Clin Immunol.
249:1092892023. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mockler MB, Conroy MJ and Lysaght J:
Targeting T cell immunometabolism for cancer immunotherapy;
Understanding the impact of the tumor microenvironment. Front
Oncol. 4:1072014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fullerton MD, Steinberg GR and Schertzer
JD: Immunometabolism of AMPK in insulin resistance and
atherosclerosis. Mol Cell Endocrinol. 366:224–234. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bone mineral densitometry, . Health and
Public Policy Committee, American College of Physicians. Ann Intern
Med. 109:8461988.PubMed/NCBI
|
|
15
|
Xu H, Zhou S, Tang Q, Xia H and Bi F:
Cholesterol metabolism: New functions and therapeutic approaches in
cancer. Biochim Biophys Acta Rev Cancer. 1874:1883942020.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen D, Zhang X, Li Z and Zhu B: Metabolic
regulatory crosstalk between tumor microenvironment and
tumor-associated macrophages. Theranostics. 11:1016–1030. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Di'Narzo AF, Houten SM, Kosoy R, Huang R,
Vaz FM, Hou R, Wei G, Wang W, Comella PH, Dodatko T, et al:
Integrative analysis of the inflammatory bowel disease serum
metabolome improves our understanding of genetic etiology and
points to novel putative therapeutic targets. Gastroenterology.
162:828–843. e112022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang Z, Gerstein M and Snyder M: RNA-Seq:
A revolutionary tool for transcriptomics. Nat Rev Genet. 10:57–63.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Canzler S and Hackermuller J: multiGSEA: A
GSEA-based pathway enrichment analysis for multi-omics data. BMC
Bioinformatics. 21:5612020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen S, Zhu J, Hua C, Feng C, Wu X, Zhou
C, Chen X, Zhang B, Xu Y, Ma Z, et al: Single-cell RNA sequencing
reveals the diversity of the immunological landscape response to
genital herpes. Virol Sin. 39:860–874. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang C, Yu JJ, Yang C, Yuan ZL, Zeng H,
Wang JJ, Shang S, Lv XX, Liu XT, Liu J, et al: Wild-type IDH1
maintains NSCLC stemness and chemoresistance through activation of
the serine biosynthetic pathway. Sci Transl Med. 15:eade41132023.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang J, Filippakis H, Hougard T, Du H, Ye
C, Liu HJ, Zhang L, Hindi K, Bagwe S, Nijmeh J, et al:
Interleukin-6 mediates PSAT1 expression and serine metabolism in
TSC2-deficient cells. Proc Natl Acad Sci USA. 118:e21012681182021.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jun S, Datta S, Wang L, Pegany R, Cano M
and Handa JT: The impact of lipids, lipid oxidation, and
inflammation on AMD, and the potential role of miRNAs on lipid
metabolism in the RPE. Exp Eye Res. 181:346–355. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hehner J, Schneider L, Woitalla A, Ott B,
Vu KCT, Schöbel A, Hain T, Schwudke D and Herker E:
Glycerophospholipid remodeling is critical for orthoflavivirus
infection. Nat Commun. 15:86832024. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dolce V, Cappello AR, Lappano R and
Maggiolini M: Glycerophospholipid synthesis as a novel drug target
against cancer. Curr Mol Pharmacol. 4:167–175. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kondakova T, D'Heygere F, Feuilloley MJ,
Orange N, Heipieper HJ and Duclairoir Poc C: Glycerophospholipid
synthesis and functions in Pseudomonas. Chem Phys Lipids.
190:27–42. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Farley SE, Kyle JE, Leier HC, Bramer LM,
Weinstein JB, Bates TA, Lee JY, Metz TO, Schultz C and Tafesse FG:
A global lipid map reveals host dependency factors conserved across
SARS-CoV-2 variants. Nat Commun. 13:34872022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yan J and Horng T: Lipid metabolism in
regulation of macrophage functions. Trends Cell Biol. 30:979–989.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Karagiannis F, Masouleh SK, Wunderling K,
Surendar J, Schmitt V, Kazakov A, Michla M, Hölzel M, Thiele C and
Wilhelm C: Lipid-droplet formation drives pathogenic group 2 innate
lymphoid cells in airway inflammation. Immunity. 52:620–634.
e62020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Costantini C, Bellet MM, Renga G,
Stincardini C, Borghi M, Pariano M, Cellini B, Keller N, Romani L
and Zelante T: Tryptophan co-metabolism at the host-pathogen
interface. Front Immunol. 11:672020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dhankhar R, Gupta V, Kumar S, Kapoor RK
and Gulati P: Microbial enzymes for deprivation of amino acid
metabolism in malignant cells: Biological strategy for cancer
treatment. Appl Microbiol Biotechnol. 104:2857–2869. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Maeda R, Seki N, Uwamino Y, Wakui M,
Nakagama Y, Kido Y, Sasai M, Taira S, Toriu N, Yamamoto M, et al:
Amino acid catabolite markers for early prognostication of
pneumonia in patients with COVID-19. Nat Commun. 14:84692023.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Al-Shalan HAM, Zhou L, Dong Z, Wang P,
Nicholls PK, Boughton B, Stumbles PA, Greene WK and Ma B: Systemic
perturbations in amino acids/amino acid derivatives and tryptophan
pathway metabolites associated with murine influenza A virus
infection. Virol J. 20:2702023. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen Q, Liang X, Wu T, Jiang J, Jiang Y,
Zhang S, Ruan Y, Zhang H, Zhang C, Chen P, et al: Integrative
analysis of metabolomics and proteomics reveals amino acid
metabolism disorder in sepsis. J Transl Med. 20:1232022. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yan M, Xiao LY, Gosau M, Friedrich RE,
Smeets R, Fu LL, Feng HC and Burg S: The causal association between
COVID-19 and herpes simplex virus: A Mendelian randomization study.
Front Immunol. 14:12812922023. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bourgin M, Durand S and Kroemer G:
Diagnostic, prognostic and mechanistic biomarkers of COVID-19
identified by mass spectrometric metabolomics. Metabolites.
13:3422023. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang Y, Guo R, Kim SH, Shah H, Zhang S,
Liang JH, Fang Y, Gentili M, Leary CNO, Elledge SJ, et al:
SARS-CoV-2 hijacks folate and one-carbon metabolism for viral
replication. Nat Commun. 12:16762021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ukolova IV and Borovskii GB: OXPHOS
organization and activity in mitochondria of plants with different
life strategies. Int J Mol Sci. 24:152292023. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Purandare N, Ghosalkar E, Grossman LI and
Aras S: Mitochondrial oxidative phosphorylation in viral
infections. Viruses. 15:23802023. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Schank M, Zhao J, Moorman JP and Yao ZQ:
The Impact of HIV- and ART-induced mitochondrial dysfunction in
cellular senescence and aging. Cells. 10:1742021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cortelli P, Mandrioli J, Zeviani M, Lodi
R, Prata C, Pecorari M, Orlando G and Guaraldi G: Mitochondrial
complex III deficiency in a case of HCV related noninflammatory
myopathy. J Neurol. 254:1450–1452. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Guarnieri JW, Dybas JM, Fazelinia H, Kim
MS, Frere J, Zhang Y, Soto Albrecht Y, Murdock DG, Angelin A, Singh
LN, et al: Core mitochondrial genes are down-regulated during
SARS-CoV-2 infection of rodent and human hosts. Sci Transl Med.
15:eabq15332023. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Senthilazhagan K, Sakthimani S, Kallanja D
and Venkataraman S: SARS-CoV-2: Analysis of the effects of
mutations in non-structural proteins. Arch Virol. 168:1862023.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sahu U, Mullarkey MP, Murphy SA, Anderson
JC, Putluri V, Kamal AHM, Park JH, Lee TJ, Ling AL, Kaipparettu BA,
et al: IDH status dictates oHSV mediated metabolic reprogramming
affecting anti-tumor immunity. Nat Commun. 16:38742025. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Greene TT, Jo Y, Chiale C, Macal M, Fang
Z, Khatri FS, Codrington AL, Kazane KR, Akbulut E, Swaminathan S,
et al: Metabolic deficiencies underlie reduced plasmacytoid
dendritic cell IFN-I production following viral infection. Nat
Commun. 16:14602025. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lee AG, Scott JM, Fabbrizi MR, Jiang X,
Sojka DK, Miller MJ, Baldridge MT, Yokoyama WM and Shin H: T cell
response kinetics determines neuroinfection outcomes during murine
HSV infection. JCI Insight. 5:e1342582020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Cairns TM, Ditto NT, Lou H, Brooks BD,
Atanasiu D, Eisenberg RJ and Cohen GH: Global sensing of the
antigenic structure of herpes simplex virus gD using
high-throughput array-based SPR imaging. PLoS Pathog.
13:e10064302017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Rathbun MM and Szpara ML: A holistic
perspective on herpes simplex virus (HSV) ecology and evolution.
Adv Virus Res. 110:27–57. 2021. View Article : Google Scholar : PubMed/NCBI
|