Introduction
At present, cancer ranks as either the first or
second leading cause of premature mortality among most populations
worldwide, and the global number of individuals affected by cancer
is expected to continue to rise in the years ahead (1). Cancer is a complex disorder with
multiple contributing factors and developmental stages, involving
processes such as the activation of proto-oncogenes and the
inactivation of tumor suppressor genes. Despite the current
advancements in comprehensive cancer treatment, the complex
pathogenesis of the disease still merits in-depth exploration to
provide a novel direction for personalized cancer therapy (2). Proteins, such as organic
macromolecules, are not only the fundamental organic matter
constituting cells and the material basis of life, but are also
associated with life phenomena (such as maintaining cell survival,
proliferation, differentiation and homeostasis, and activating and
regulating intracellular signaling pathways and material metabolic
balance). Notably, the ubiquitin-proteasome system (UPS) is
responsible for catalyzing the repair and degradation of a large
number of key protein substrates. Dysregulation of the UPS may thus
be implicated in the onset and progression of numerous diseases,
including cancer. During the process of ubiquitination, three
enzymes are essential for attaching ubiquitin proteins to their
target proteins: i) Ubiquitin-activating enzyme (E1); ii)
ubiquitin-conjugating enzyme (E2); and iii) ubiquitin ligase (E3)
(3–5). Specifically, E3 binds to the
substrate protein, and this interaction ultimately results in the
degradation of the substrate through the 26S proteasome (3). Research has shown that the modulation
of E3 activity constitutes one of the key factors influencing the
development and progression of cancer (4). Among the various types of E3 ligases,
Cullin-RING ligases (CRL) are the ones with the largest number of
members and the most diverse types, and they play a role in cancer
development (5,6). One of these CRL1 ligases, also
referred to as the S-phase kinase-associated protein 1
(SKP1)/Cullin 1 (CUL1)/F-box protein (FBP) complex (SCF complex),
has been extensively investigated (7–9). A
total of 80–90% of proteins within cells undergo degradation via
the UPS (2). The SCF complex
consists of three invariant core components and variable FBPs.
RING-box 1 is responsible for the recruitment of E2; CUL1, a
scaffolding protein, also serves as the catalytic core; and SKP1
links the SCF complex to variable FBPs and their corresponding
target proteins to recruit substrates for ubiquitination (9). To date, 69 FBPs have been identified
in humans. These proteins can be further categorized into the
following three groups: i) Those containing leucine-rich repeats
(FBXL); ii) those with WD-40 repeats (FBXW); and iii) those
featuring only uncharacterized structural domains (FBXO) (10,11).
A growing body of evidence indicates that abnormal expression of
FBPs is linked to the occurrence, proliferation, angiogenesis and
metastasis of various malignant tumors. For instance, FABP5 can
alter the fatty acid metabolism pathways and products of tumor
cells, providing them with more energy and nutritional support,
thereby promoting the growth and spread of tumors (12). FABP4 is induced in endothelial
cells by the NOTCH1 signaling pathway and plays a significant role
in the formation of the tumor vascular system. In in situ mouse
ovarian tumor models, FABP4 is essential for angiogenesis and is an
important target in tumor angiogenesis, especially in tumors of
low-grade, interstitial and free fatty acid-rich tissues (13). Within the FBP family, F-box protein
2 (FBXO2) is abnormally expressed in multiple types of cancer. It
shows abnormal expression in various cancers, such as gastric
cancer (GC) (14), colorectal
cancer (CRC) (15), ovarian cancer
(OV) (16), endometrial cancer
(EC) (17), osteosarcoma (OS)
(18), glioblastoma (GB) (19), papillary thyroid carcinoma (PTC)
(20) and oral squamous cell
carcinoma (OSCC) (21).
Specifically, it regulates EMT, mediates substrate degradation
through the UPS, activates related signaling pathways (such as
STAT3, PI3K-Akt), affects cell cycle and autophagy, thereby
promoting the malignant processes of corresponding cancers such as
cell proliferation, migration and invasion, and chemotherapy
resistance. Furthermore, its expression level is often associated
with clinical features such as cancer prognosis and metastasis
(14–21).
Composition and role of the UPS
It is widely acknowledged that protein homeostasis
is essential for maintaining normal cellular physiological
activities, among which the timely degradation or repair of
misfolded proteins is particularly crucial (22,23).
The degradation pathways mainly encompass autophagy (such as
lysosomal degradation pathway) and the UPS (24). The relationship between ubiquitin
and protein regulation was initially investigated by Hershko et
al (25) in the 1980s.
Subsequently, to gain a deeper understanding of ubiquitin and its
associated protein hydrolysis, Hershko et al (22) elaborated on ubiquitinating enzymes
further. This allowed the clarification of the biological
characteristics of the UPS, highlighting its essential role in
protein degradation, as well as its specific physiological
functions (including roles in the cell cycle, DNA repair, protein
synthesis, transcription and stress responses). The UPS selective
basis (such as short-term signals involved in protein-specific
degradation) and key mechanistic traits such as polyubiquitin
chains and the subunit selectivity of protein degradation were also
identified. These findings triggered a substantial expansion of
research in the ubiquitin field during the 1990s (26,27).
The UPS acts as a major system for the intracellular non-lysosomal
degradation of proteins, and regulates and eliminates aberrant
proteins in a highly specific manner. The UPS has been demonstrated
to be aberrantly activated in a variety of biologically notable
processes, including cell viability, proliferation and invasion,
colonization and metastasis, recurrence, vascular invasion,
immunomodulation, and chemotherapy resistance, in a wide range of
cancer types (28,29). In OV, the UPS is abnormally
activated (16). On one hand, it
promotes cell proliferation and inhibits apoptosis through the
SOX6-FBXO2-Sad1 and UNC84 domain-containing protein 2 (SUN2) axis.
On the other hand, it is associated with chemotherapy resistance
through FBXO2. In PTC, UPS promotes the ubiquitination and
degradation of p53 through abnormal expression of FBXO2, promoting
cell proliferation and being related to tumor size and metastasis
(20). In OSCC, UPS regulates the
transformation of tumor cells to highly malignant clones through
FBXO2, promoting malignant processes such as proliferation and
invasion (21). The basic
components of the UPS comprise ubiquitin, E1, E2, E3, the 26 S
proteasome and deubiquitinating enzyme (DUB) (3). The process by which ubiquitin binds
to a substrate and causes it to be degraded is referred to as
ubiquitination (3). Ubiquitination
is a protein degradation process involving a multi-step cascade
reaction, primarily mediated by three types of ubiquitinating
enzymes: E1, E2 and E3. Initially, E1 utilizes the energy released
from ATP hydrolysis to form a thioester bond between the C-terminal
of ubiquitin and the cysteine residue at the active site of E1,
thereby activating ubiquitin, which constitutes the first step of
ubiquitination. Subsequently, the activated ubiquitin is
transferred to the cysteine residue located at the active site of
E2. After which, the activated ubiquitin is transferred to a
cysteine residue located at the active site of the E2, leading to
the formation of a thioester bond between the E2 enzyme and
ubiquitin. Then, in a two-step reaction, the E2 enzyme facilitates
the attachment of ubiquitin to the substrate with the assistance of
E3 that recognizes a specific target. In this process, the
specificity for substrates is guaranteed by 500–1,000 specific E3
enzymes encoded in the human genome (3,4,7,29).
Eventually, the substrate proteins marked with ubiquitin are broken
down by the 26S proteasome, which in turn regulates DNA repair,
influences stress responses and modifies cell proliferation
(30) (Fig. 1). When mutated or overexpressed in
various malignancies, ubiquitinating enzymes affect and regulate
various cellular pathways, such as protein transport, chromatin
remodeling, the cell cycle and apoptosis. Overexpression of NEDD4-1
in liver cancer can affect protein transport-related signals by
regulating AKT ubiquitination, promoting the cell cycle and
inhibiting apoptosis (31). In
multiple myeloma, it can also mediate AKT degradation to accelerate
apoptosis (32). MDM2
overexpression in cancer can degrade p53 and interfere with
apoptosis and the cell cycle (33). Mutation will cause loss of
substrate degradation ability, leading to uncontrolled cell cycle
and abnormal proliferation (34).
Therefore, the UPS serves a vital role in oncogenic signaling and
the development of malignant tumors (7). Furthermore, it has been demonstrated
that the UPS also exerts a notable role in neurodegenerative
diseases and various inflammatory reactions (23).
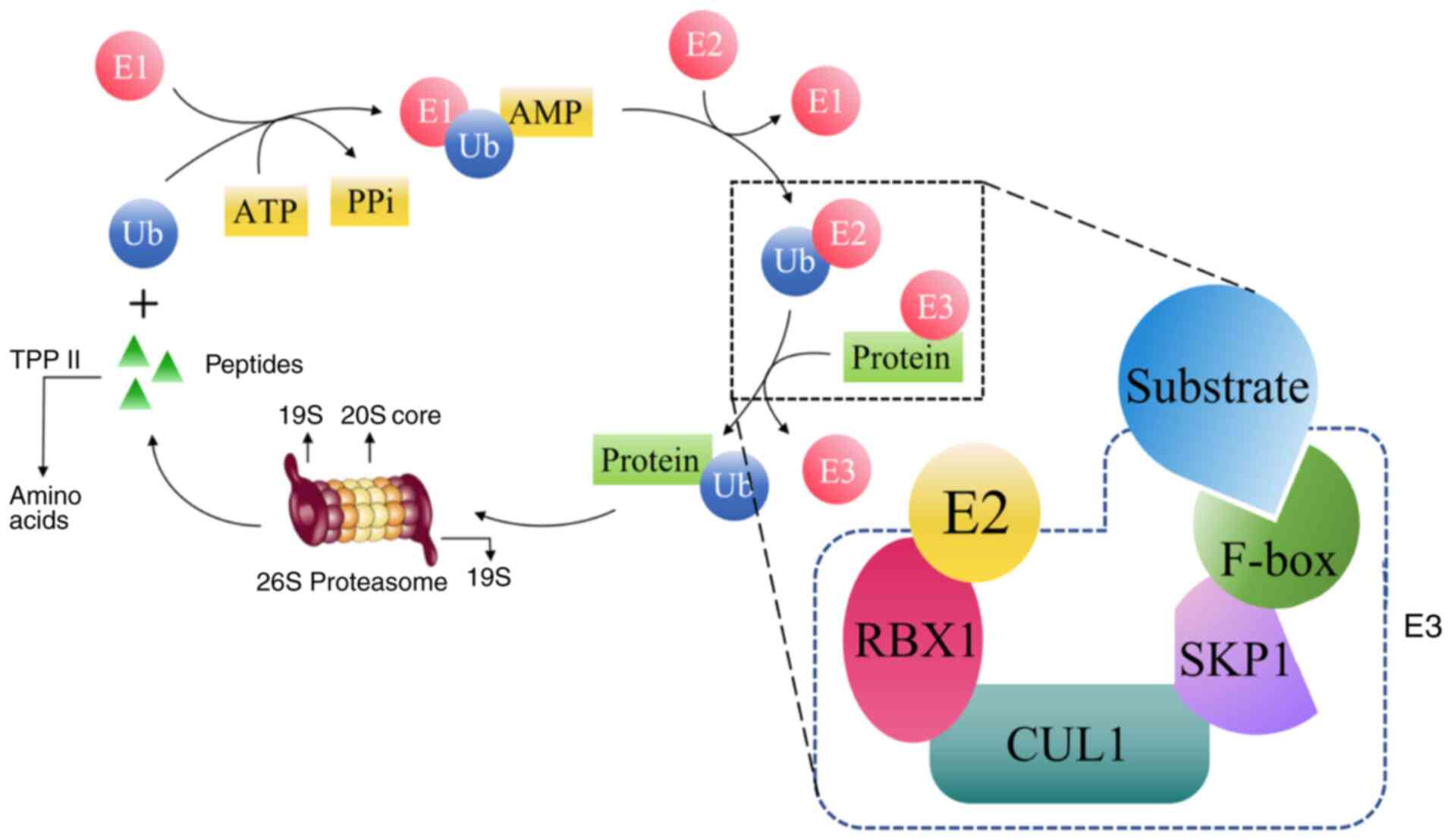 | Figure 1.Schematic diagram of the
ubiquitination process. E1, ubiquitin-activating enzyme; E2,
ubiquitin-conjugating enzyme; E3, ubiquitin ligase; TPP II,
tripeptidyl peptidase II; RBX1, ubiquitin ligase with zinc finger
structure; CUL1, Cullin-1 proteins; SKP1, S-phase kinase-related
protein 1; F-box, F-box protein; Ub, ubiquitin; PPi,
pyrophosphate. |
Among ubiquitinating enzymes, the selectivity of the
UPS is determined by the E3 ligase (7,28). A
number of E3 ligases, when dysfunctional, can impair DNA damage
repair, cause cell cycle disturbances, lead to abnormal gene
expression regulation and disrupt or continuously interrupt signal
transduction, ultimately exerting carcinogenic or tumor-promoting
effects. For instance, anaphase-promoting complex (APC/C) is a
representative E3 ligase that is indispensable for mitotic
progression (35). Research has
revealed that the abnormal regulation of cell division cycle 20
(CDC20) and cadherin 1 (CDH1) is linked to cancer (36). Inhibition of CDC20 can disrupt
mitosis and subsequently induce cell apoptosis, which suggests that
CDC20 possesses oncogenic characteristics (36,37).
The depletion of CDC20 can suppress the Wnt signaling pathway, in
turn reducing the proliferation of cancer cells in colon cancer
(38,39). CDH1 facilitates the
ubiquitination-mediated hydrolysis process initiated by BRAF either
in an APC-dependent or APC-independent manner, thereby contributing
to tumor formation (35).
Neurologically expressed developmentally downregulated 4-1
(NEDD4-1) has been demonstrated to exert a vital function in
modulating cancer progression. Specifically, NEDD4-1 stabilizes
MDM2 via Lys63-linked polyubiquitination, a process that
facilitates p53 degradation and thereby enhances cell proliferation
(31). Furthermore, NEDD4-1 binds
to N-Myc and strengthens the polyubiquitination of this protein,
thereby promoting the proliferation of neuroblastoma cells
(40). Silencing of NEDD4-1
results in reduced AKT phosphorylation and increased PTEN
expression, which in turn inhibits the proliferation and migration
of hepatocellular carcinoma cells (41). In addition, NEDD4-1 mediates the
ubiquitination of AKT at the Ser473 site, which triggers
phosphorylation at this position. This process promotes the
proteasomal degradation of AKT, thereby inhibiting AKT signaling
and accelerating apoptosis in multiple myeloma cells (32). MDM2, a key regulator of the tumor
suppressor p53, mediates the ubiquitination of p53 and triggers its
degradation via the proteasome. Mutations or deletions of p53 are
present in approximately half of all types of cancer (33). MDM2 expression is upregulated in
numerous cancer types, such as lung cancer and liver cancer, and
the chemotherapeutic drug cisplatin induces apoptosis in cancer
cells by phosphorylating and activating the p53 protein (42). However, increased MDM2 expression,
combined with p53 mutations or downregulation, leads to the
development of cisplatin resistance in epidermoid carcinoma
(42).
The E3 ligases that regulate the
ubiquitination-mediated degradation of proteins can be mainly
classified into two types (30).
HECT ligases are characterized by a C-terminal domain. They first
form a thioester bond to receive ubiquitin molecules from E2
ligases, after which the ubiquitin undergoes refolding and is
transferred to substrate proteins (43). RING ligase, which consists of RING
and RING-like ligases and their accompanying proteins, has a zinc
finger that transfers ubiquitin directly to the substrate via the
ubiquitin conjugating enzyme E2 (44). CRL is the most extensive E3 ligase
and serves a role in cancer. CRL1 ligase, otherwise referred to as
the SCF complex, comprises FBPs and FBXW7 as key components. These
elements serve a role in regulating the stability of diverse
cellular factors, including cell cycle regulators, oncogenic
transcription factors, cell surface receptors and signaling
molecules (34,45). Consequently, impairment of the
SCF-FBXW7 complex leads to unregulated cell proliferation and
survival, genomic instability and disordered signaling pathways
that promote cancer invasion and metastasis (46–48).
In summary, the substrate specificity of the SCF
complex is governed by FBPs, with each type of FBP capable of
recognizing and binding to a unique range of substrates (49). In addition to being components of
the SCF complex, FBPs are involved in DNA replication,
transcription, cell differentiation and cell death (50).
Composition and role of the FBP family
According to their specific structural domains, FBPs
can be further divided into three subfamilies: i) FBXL; ii) FBXW;
and iii) FBXO (10,11). FBPs directly interact with
post-translationally modified substrates and mediate the
ubiquitination and degradation of target proteins in various
cellular biological processes, such as the cell cycle,
epithelial-mesenchymal transition (EMT), apoptosis and multiple
tumor-related signaling pathways (such as the PI3K-AKT-mTOR, p53
and nuclear factor erythroid 2-related factor 2 pathways), thus
affecting tumor progression (51).
Numerous members of the FBP family serve notable
roles in tumor development (Table
SI). For instance, SKP2 engages with multiple signaling
cascades, including but not limited to the PI3K/Akt (52), ERK (53), peroxisome proliferator-activated
receptor γ (54), insulin-like
growth factor 1 (55) and mTOR
(10) signaling pathways. Through
the aforementioned signaling pathways and their potential
interactions, SKP2 exhibits high expression in breast cancer,
melanoma, pancreatic cancer, GC, lymphoma, prostate cancer and
nasopharyngeal cancer; notably, its high expression is associated
with poor prognosis in these malignancies (56). The dysregulation of the
SKP2/mH2A1/CDK8 pathway and the hydrolysis of mH2A1 by
SKP2-targeted proteins are crucial for breast cancer progression
and prognosis (57,58). In gliomas, FBXL18 exerts oncogenic
effects by curbing apoptosis, which is achieved through promotion
of K63-linked ubiquitination of AKT (59). In OV, FBXO6 (60) and FBXO16 (61) function as proto-oncogenes through
ribonuclease T2 and heterogeneous nuclear ribonucleoprotein L
ubiquitination and degradation, respectively. Additionally, FBXO32
promotes lung adenocarcinoma progression by targeting PTEN and
promoting its degradation to regulate the cell cycle, promote the
PI3K/AKT/mTOR pathway and ultimately accelerate EMT (62). FBXO32 acts as a target gene for
melanocyte inducing transcription factor (a major transcription
factor in melanocytes), facilitating melanoma progression in
vivo, and the knockdown of FBXO32 induces global changes in
melanoma gene expression profiles (63).
On the other hand, FBXW7, as an oncogene, mediates
tumor suppression by negatively regulating a number of oncogenic
proteins (50). The inactivation
of this protein in lung cancer, hepatocellular carcinoma, CRC,
breast cancer and hematopoietic system tumors holds importance for
the initiation and progression of these malignancies (64). Among the five types of tumors
mentioned above [lung cancer, hepatocellular carcinoma, CRC, breast
cancer and hematopoietic system tumors], microRNA-223 influences
the proliferation and apoptosis of CRC cells by activating the
NOTCH and AKT/mTOR pathways (65).
Thus, it can be inferred that the loss of FBXW7 may serve as an
independent prognostic marker for tumors (64). It has also been shown that FBXO4
acts as another tumor suppressor within the FBP family, and it
facilitates ubiquitin-mediated degradation of cyclin D1 through
phosphorylation at the Thr286 site (8). Consequently, dysfunction of FBXO4
causes cyclin D1 to accumulate in vivo and drives the
progression of malignant tumors, including melanoma (66) and esophageal cancer (67,68).
The absence of FBXO4 expression in mice caused cyclin D1
accumulation, and thus, malignant transformation of BrafV600E
melanoma, suggesting that FBXO4 deficiency serves a role in
melanoma development (66). These
studies have substantiated the tumor-suppressive effects of FBXO4
(8,66–68).
FBXO11 and FBXO31 are also recognized tumor suppressors, and both
target a variety of oncogenic substrates. For instance, FBXO11
mediates the ubiquitination-dependent degradation of the
proto-oncogene product B-cell lymphoma 6 protein, and it is often
absent or mutated in diffuse large B-cell lymphomas (69). FBXO31 orchestrates its regulatory
role in tumor-related processes by triggering the ubiquitination
and degradation of distinct substrates, with the underlying
mechanism exhibiting substrate-specific divergence. For cyclinD1,
FBXO31 exerts its function by recognizing phosphorylation
modifications (e.g., at the Thr286 site) of cyclinD1; it then
associates with the SCF (SKP1-Cullin-F-box) E3 ubiquitin ligase
complex, thereby facilitating the ubiquitination and subsequent
proteasomal degradation of cyclinD1 to preclude aberrant cell cycle
progression (70). In the case of
SNAIL, a key transcription factor governing epithelial-mesenchymal
transition (EMT), FBXO31 binds to specific structural domains of
SNAIL, which disrupts the interaction between SNAIL and its
stabilizing factors (e.g., SIP1) and induces conformational changes
in SNAIL. These events collectively promote the ubiquitination and
degradation of SNAIL, ultimately suppressing tumor cell invasion
and metastasis (71). Regarding
MDM2, the E3 ubiquitin ligase of the tumor suppressor p53, FBXO31
interacts with the RING domain of MDM2 to trigger MDM2
self-ubiquitination. This self-ubiquitination leads to MDM2
degradation, which alleviates the inhibitory effect of MDM2 on p53
and restores the tumor-suppressive functions of p53 (e.g., cell
cycle arrest and apoptosis induction) (72). For mitogen-activated protein kinase
kinase 6 (MAPKK6), a pivotal upstream kinase in the MAPK signaling
pathway, FBXO31 is capable of recognizing the unique conformational
state of hyperactivated MAPKK6. It then mediates the ubiquitination
and degradation of hyperactivated MAPKK6 via the SCF complex,
preventing excessive activation of the MAPK pathway and the
consequent abnormal cell proliferation (73). Collectively, FBXO31 achieves the
goal of inhibiting the occurrence and development of various
malignant tumors such as breast cancer (74), OV (75), liver cancer (76) and prostate cancer (77) by triggering the ubiquitination
degradation of specific substrates. Additionally, FBXL14 is capable
of inducing the ubiquitination-mediated degradation of the key
oncogene c-Myc. However, in glioma stem cells, this ubiquitination
process can be reversed by the DUB ubiquitin specific peptidase 13
(USP13), which suggests that an antagonistic relationship exists
between FBXL14 and USP13 (78).
Furthermore, FBXL14 can target and degrade CUB domain-containing
protein 1 (CDCP1), thereby reducing the stability of CDCP1 and
inhibiting breast cancer metastasis (79).
Taken together, all of the aforementioned FBP family
members influence various cellular signaling pathways via UPS, and
thereby affect the progression of malignant tumors.
Introduction of FBXO2
FBXO2, alternatively referred to as Fbx2, Fbg1,
Fbs1, neural F-box 42 kDa (NFB42) and organ of Corti protein 1, is
an FBP. It is highly abundant in the cytoplasm of eukaryotic cells
and functions as a subunit of the E3 ligase (80,81).
Research on FBXO2 began with the proteomic analysis of rat neural
tissues. As early as 1998, researchers identified an FBP referred
to as NFB42, with a molecular weight of ~42 kDa, in rat brain
tissues. This protein was highly expressed in neurons and served a
role in sustaining the non-dividing state of cells (82). Subsequently, through homology
sequence cloning technology, it was revealed that human FBXO2
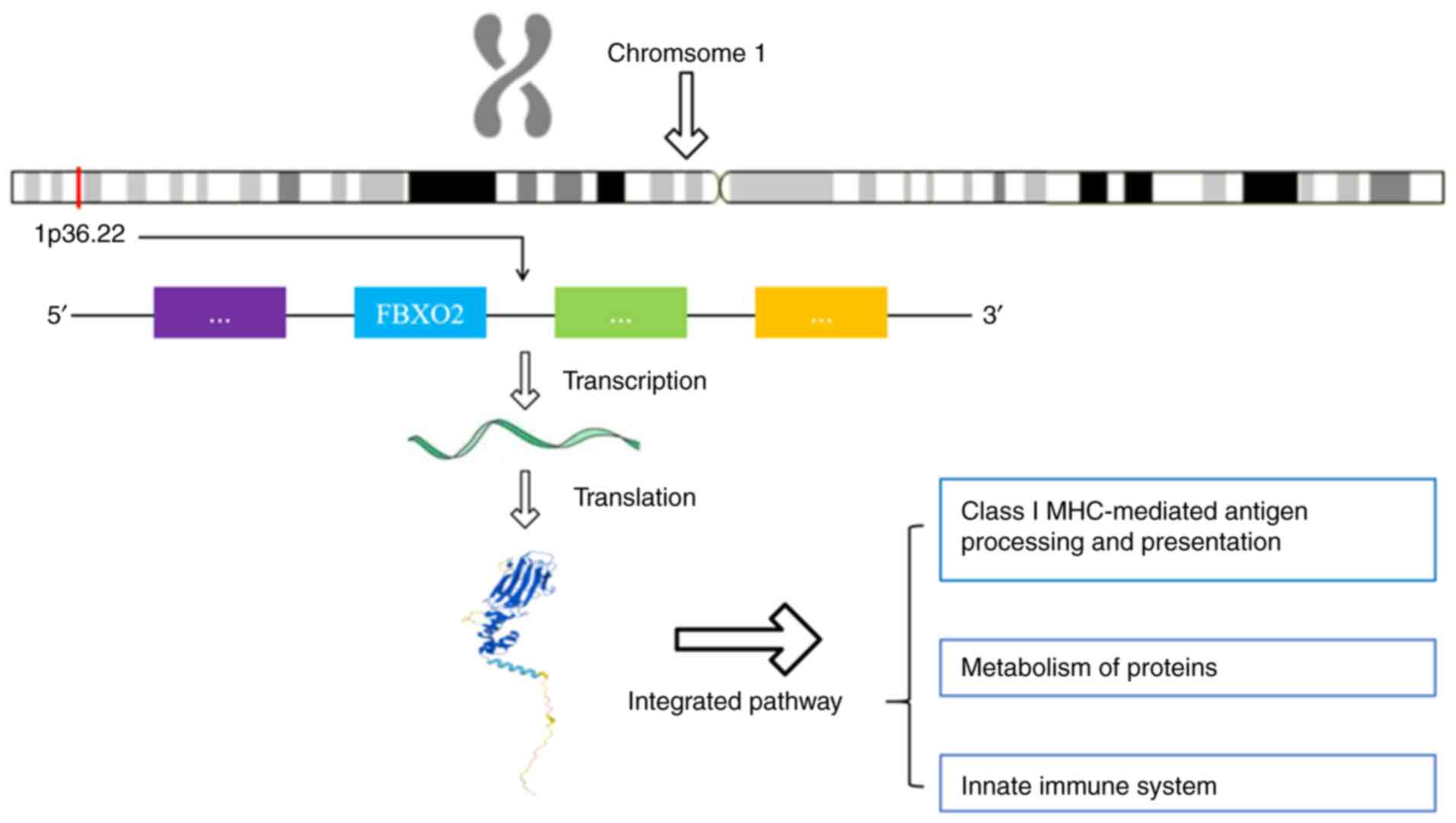
(Online Mendelian Inheritance in Man 607112) is situated on
chromosome 1p36.22 and comprises six exons encoding a cytoplasmic
protein containing 296 amino acids (83) (Fig.
2). The human FBXO2 consists of an F-box domain (FBA) and a
glycoprotein-binding domain (SBD). The FBA domain is situated at
the N-terminal and exhibits a triple-helical bundle conformation
(84). It binds to the SKP1
protein within the SCF complex and serves as the core module for E3
ligase activity (83). The SBD is
located at the C-terminal and contains multiple protein interaction
modules, such as hydrophobic pockets and sugar recognition motifs
(84,85). For instance, FBXO2 recognizes the
Epstein-Barr virus glycoprotein B (gB) through glycosylation sites
and mediates its endoplasmic reticulum-associated degradation
(86). Research into the
functionality of FBXO2 has indicated that it functions as the
substrate recognition subunit of the SCF ubiquitin ligase complex,
facilitating the ubiquitination and degradation of a variety of
proteins (87,88). Further research has shown that
FBXO2 is abnormally highly expressed in various cancer types (such
as OV and thyroid cancer), and promotes tumor progression by
regulating key signaling molecules such as p53 (16,20).
Comparative genomics studies have shown that FBXO2
has similar domain composition and substrate recognition abilities
in species such as mammals, birds and fish, but its regulatory
network shows certain adaptive differentiation among different
species (89–91). FBXO2 is specifically expressed in
tissues such as the brain and adrenal glands of mice, and its
absence leads to the accumulation of amyloid precursor protein in
neurons, suggesting its potential role in Alzheimer's disease (AD)
(92). Additionally, FBXO2
participates in the mechanism of liver fibrosis induced by
excessive exercise in mice by regulating the formation of muscle
lactate vesicles (89). In fish,
the homologous genes may be involved in ubiquitination regulation
during liver development. For instance, the abnormal expression of
FBXO32 in fish skeletal muscle tissues is associated with muscle
atrophy (90), suggesting the
conserved function of F-box family genes in organ development.
FBXO2 exhibits conservation during the evolution of insect sex
chromosomes, and its biological function may be associated with the
dosage compensation effect of insect sex chromosomes (i.e.,
balancing the expression dosage of genes on sex chromosomes between
male and female individuals through specific mechanisms to maintain
normal physiological functions) (91). Current studies have not yet
identified the homologous gene of FBXO2 in fruit flies, but FBPs
serve a key role in the asymmetric division of neural stem cells,
indicating the cross-species conservation of ubiquitination
regulation in stem cell biology (93).
FBXO2, as an important member of the FBP family,
exhibits both evolutionary conservation and functional diversity,
making it a hot topic in interdisciplinary research. From neural
development to disease regulation, FBXO2 precisely regulates
protein homeostasis through the ubiquitin network. Future studies
need to further analyze the regulatory differences of FBXO2 in
various diseases and explore its potential as a therapeutic target
for diseases.
Role and mechanisms of FBXO2 in
non-tumorigenic diseases
FBXO2 holds considerable importance in metabolic
disorders. FBXO2 is a functional E3 ligase for insulin receptors
(IRs) in the liver, and FBXO2 protein and mRNA levels are increased
in the livers of obese individuals. FBXO2 mediates the
ubiquitin-dependent degradation of IRs, thereby inhibiting the
PI3K-AKT pathway and impairing the integrity of insulin signaling
(87). This leads to insulin
resistance and abnormal glucose metabolism, offering a novel
therapeutic target for type 2 diabetes and associated metabolic
disorders (87,94). The expression levels of FBXO2 are
elevated in the liver tissues of patients suffering from
non-alcoholic fatty liver disease (NAFLD). The upregulated FBXO2
aggravates NAFLD by targeting the α subunit of hydroxy-COA
dehydrogenase and promoting its proteasome degradation in HepG2 and
293T cells. This suggests that FBXO2 may serve as a potential
therapeutic target for NAFLD (88). Additionally, FBXO2 exerts
considerable effects in neurological diseases. A study has shown
that there is an association between FBXO2 and the occurrence and
development of Parkinson's disease (PD), and variations in FBXO2
may reduce the risk of PD among Han individuals in mainland China
and serve as a biomarker for risk assessment of PD (95). Cognitive dysfunction is one of the
notable features of AD (96). The
pathogenesis of AD is associated with multiple mechanisms such as
transcriptional dysregulation, erroneous protein degradation and
synaptic dysfunction (97). A
previous study has shown that the upregulation of FBXO2 induced by
histone methyltransferase Smyd3 in AD mouse models (P301S Tau mice)
affected the function of the N-methyl-D-aspartate receptor (NMDAR)
and the cognitive behavior of mice, which provides a potential
therapeutic strategy for AD (97).
Niemann-Pick C (NPC) disease is an autosomal recessive genetic
disorder that ultimately leads to neurodegeneration and premature
mortality. In the central nervous system, FBXO2 localizes to
damaged lysosomes and facilitates their degradation. Deficiency of
FBXO2 delays the clearance of impaired lysosomes, thereby
exacerbating motor function impairments in NPC, intensifying
neurodegeneration, and ultimately reducing survival (98). In osteoarthritis (OA), Tang et
al (99) demonstrated that RNA
binding motif protein 47 stabilized FBXO2 mRNA by activating the
STAT3 signaling pathway, thereby promoting the inflammatory,
apoptotic and extracellular matrix (ECM) degradation of
chondrocytes treated with IL-1β, ultimately progressing OA. Another
study (100) employed single-cell
transcriptomic analysis to pinpoint novel specific biomarkers for
nucleus pulposus and intrafibrous annulus cells, while also
defining the cell populations in non-degenerative and degenerative
human intervertebral discs (IVD) from the same individual.
According to gene expression profiles at the single-cell resolution
and Gene Ontology and Kyoto Encyclopedia of Genes and Genomes
analyses, FBXO2 was identified as one of the genes that could
predict IVD degeneration, which offers novel perspectives on the
biomarkers of IVD degeneration, and thus, enhances diagnostic and
therapeutic strategies. Furthermore, previous research (101) has demonstrated that inner ear
organoids are derived from embryonic stem cells, by targeting the
lineage-specific ear gene FBXO2 with the multi-allelic reporter
cassette (Venus/Hygro/CreER), researchers revealed the gradient
characteristics exhibited by cochlear tissue during development
(these characteristics are associated with cell differentiation or
structural formation) through the expression of Venus and the
activity of CreER. This approach also marked the sensory lineage,
confirmed the enrichment of the type I vestibular hair cell
subpopulation and revealed strong expression in adult cerebellar
granule cells. Subsequently, McGovern et al (102) described a novel tamoxifen-induced
CreER mouse strain, referred to as FBXO2CreERT2 mice. In adult
mice, the induction of FBXO2CreERT2 occurred more frequently in
type I hair cells than in type II hair cells. Therefore,
FBXO2CreERT2 mice were considered a novel tool for specifically
manipulating inner ear epithelial cells and targeting the type I
hair cells of the vestibule. The FBXO2/SCF ubiquitin ligase complex
enhances ubiquitin-mediated phagocytosis of group A streptococcus
(GAS) by recognizing the N-acetylglucosamine on GAS side chains,
promoting ubiquitin-mediated ligase activity (103). Additionally, gB in the
Epstein-Barr virus binds to the sugar-binding structural domain of
FBXO2 through its glycosylation site. Through the
ubiquitin-proteasome pathway, gB is degraded to limit viral
infectivity (86).
Role and mechanisms of FBXO2 in tumorigenic
diseases
FBXO2 exerts a cancer-promoting effect in multiple
types of malignancies, such as GC, OV, OS and thyroid cancer, with
its mechanism of action differing according to the specific cancer
type (Table SII). In GC, it
enhances cell proliferation, migration and invasion through the
regulation of EMT (14). In CRC,
it may inhibit tumor suppressor proteins through EMT and UPS
degradation, affecting the Wnt signaling pathway (104). Specifically, FBXO2 regulates key
EMT transcription factors such as SNAIL and TWIST, initiating the
EMT process in CRC cells. Subsequently, these key EMT transcription
factors can bind to the promoter of tumor suppressor genes to
inhibit their transcription, or affect the subcellular localization
and function of tumor suppressor proteins by altering the cell
state. At the same time, EMT may also collaborate with the UPS,
changing the kinase activity to phosphorylate tumor suppressor
proteins, which are then recognized by the UPS and degraded.
Eventually, the inhibition of tumor suppressor proteins will
release the negative regulation on Wnt and other pathways,
promoting the progression of CRC (43). In OV, it promotes cancer
development through the SOX6-FBXO2-SUN2 axis (among them, SUN2 is
fully known as Sad1 and UNC84 domain-containing protein 2, which is
a transmembrane protein located on the nuclear membrane. Its core
function is related to the maintenance of the nuclear membrane
structure, nuclear localization, and the transport of nuclear and
cytoplasmic substances. Additionally, it plays a role in the
connection between the cytoskeleton and the nuclear skeleton) and
related pathways, and is associated with chemotherapy resistance
(16). In addition, in EC it
degrades fibrillin 1 (FBN1) to regulate the cell cycle and
autophagy. In OS, it stabilizes IL-6R to activate the STAT3
signaling pathway (17). In GB, it
participates in tumor-microenvironment interaction (19), and in PTC, it degrades p53 to
promote proliferation (20).
Finally, in oral OSCC, it promotes the transformation of cells to
malignant clones (21). Therefore,
FBXO2 has the potential to act as a biomarker and therapeutic
target in a variety of cancer types.
Effect of FBXO2 on GC and its
mechanisms
GC is a common malignant tumor globally and stands
as the fourth primary cause of cancer-associated deaths. The
incidence of GC rises steadily with age, and the median survival
time for patients with advanced GC is <12 months (105,106). Therefore, it is of great
importance to explore approaches for the management of this disease
(106). Sun et al
(14) used four GC cell lines
(MGC-80-3, AGS, SGC-7901 and MKN-28) to conduct comparisons in
terms of their transcription and translation levels, as well as
migratory and invasive abilities, using reverse
transcription-quantitative PCR, western blotting, Transwell assays
and wound healing assays. To further substantiate the experimental
conclusions, MGC 80-3 cells were transfected with FBXO2 small
interfering RNA (siRNA), and the aforementioned experiments were
repeated. The findings showed that in siRNA-FBXO2 MGC 80-3 cells,
the mRNA expression of FBXO2 was downregulated, and the ability of
cells to proliferate, migrate and invade was notably suppressed.
Additionally, the expression of epithelial markers (E-cadherin
protein) was elevated, and the expression of mesenchymal markers
(N-cadherin and vimentin protein) was notably reduced. All these
results suggested that low FBXO2 expression could inhibit the
proliferation, migration and invasion of GC cells by reducing EMT.
In summary, FBXO2 has the ability to promote the proliferation,
migration and invasion of GC cells, and FBXO2-mediated EMT serves a
notable role in the migration and invasion of GC, and may emerge as
a novel target for the diagnosis and treatment of this
malignancy.
Effect of FBXO2 on CRC and its
mechanisms
CRC is the second and third most prevalent cancer
type in women and men, respectively, accounting for 9.2% of global
deaths, with incidence and mortality rates being 25% higher in men
than in women (107). Wei et
al (15) observed that FBXO2
was mainly expressed in the cytoplasm of CRC cells, and that the
overexpression of FBXO2 increased the expression of N-cadherin.
This indicates that FBXO2 is implicated in CRC metastasis through
EMT. In this article (15), the
team further investigated and found that the mechanism by which
FBXO2 promotes the development of CRC may be through the
degradation of N-cadherin by UPS, which regulates the tumor
proliferative activity of CRC. In addition, the excessive
expression of N-cadherin in CRC can further impact the expression
and localization of β-catenin. Furthermore, Zhao et al
(104) demonstrated that
β-catenin serves a notable role in the Wnt signaling pathway and
colon cancer development. These studies further suggested that the
upregulation of FBXO2 expression is associated with proliferation,
infiltration and distant metastasis of CRC, and that the inhibition
of FBXO2 expression may constitute a therapeutic strategy to
improve the prognosis of patients with CRC. Therefore, expression
levels of FBXO2 can serve as a potential biomarker for CRC
metastasis.
Effect of FBXO2 on OV and its
mechanisms
OV is the fifth most lethal cancer among women
globally, with an incidence and mortality rate of 3.4 and 4.7%,
respectively. This indicates that it poses a serious threat to the
health and survival of women (108). Ji et al (16) found, through analysis of multiple
genetic databases, that FBXO2 was highly expressed in OV tissues
and cells. The underlying mechanisms may involve the transcription
factor SOX6 binding to FBXO2, which leads to the abnormal
upregulation of FBXO2 expression. Subsequently, FBXO2 binds to the
glycosylated SUN2 protein and functions as an E3 ligase to mediate
the ubiquitination-dependent degradation of SUN2. This process
inhibits apoptosis, promotes cell proliferation and ultimately
accelerates the progression of OV. Further silencing of FBXO2
expression using short hairpin RNA revealed that cells with FBXO2
silenced showed reduced proliferative, migratory and invasive
abilities compared with previously. Furthermore, in in vivo
experiments, the knockdown of FBXO2 led to a notable reduction in
the volume, size and weight of subcutaneous tumors. Thus, the
aforementioned findings revealed a novel SOX6-FBXO2-SUN2 axis that
serves a role in OV development, and targeting this axis could
serve as an effective therapeutic approach for OV. Recent research
(109) has indicated that FBXO2
is capable of impacting chemotherapy resistance. It achieves this
by affecting the PI3K-Akt signaling pathway, as well as the
interactions between focal adhesions and ECM receptors, while also
regulating tumorigenesis. In vitro experiments showed that,
in A2780 and SKOV3 OV cell lines where FBXO2 was silenced, the 50%
maximum inhibitory concentration of cisplatin was reduced. This
suggests that FBXO2 is a potential biomarker linked to chemotherapy
resistance in high-grade serous OV (HGSOC), and it can act as a
valuable prognostic indicator as well as a potential target in
HGSOC (109).
Effect of FBXO2 on EC and its
mechanisms
EC accounts for >90% of all uterine malignancies,
and its incidence is increasing. The majority of patients diagnosed
with EC are postmenopausal, with the median age at diagnosis being
60 years (110). Therefore,
improving the early diagnosis and prognostic assessment of patients
with EC is of critical importance (110). Che et al (17) revealed that FBXO2 binds to and
causes the degradation of FBN1 via polyubiquitination, which is
further enhanced by the regulation of cell cycle proteins (CDK4,
CyclinD1, CyclinD2 and CyclinA1) and the inhibition of autophagy
signaling pathways [autophagy related 4A cysteine peptidase (ATG4A)
and ATG4D], to promote EC proliferation. Subsequently, a knockdown
mouse model, verification via immunohistochemistry and other
techniques confirmed that FBXO2 knockdown led to elevated FBN1
expression and reduced Ki67 expression. Additionally, either FBN1
knockdown alone or the combined knockdown of both FBXO2 and FBN1
resulted in decreased Ki67 expression. These results implied that
the deletion of FBN1 blocks the proliferative effect of FBXO2. In
conclusion, the aforementioned experimental study demonstrated that
targeting FBXO2 may treat EC through the regulation of cell cycle
and autophagy signaling pathways, and that patients with high FBXO2
expression may be potential candidates for treatment with CDK4/6
inhibitors in EC (17).
Effect of FBXO2 on OS and its
mechanisms
OS is the most common primary bone malignancy,
characterized by strong local invasiveness and metastatic
potential, with the 5-year survival rate of patients with
metastases being <20% (111).
Research into improving the survival of patients with OS has proved
challenging. Zhao et al (18) confirmed that the persistent
activation of the IL-6 signaling pathway is harmful to bone tissue
and hypothesized that this ultimately contributes to the
development of OS. Through further research, it was identified that
upregulated FBXO2 in OS cells binds to the C-terminal SBD of IL-6R.
This binding gives rise to a stable complex. Once the stable IL-6R
binds to IL-6, it recruits Janus kinase, which in turn causes the
continuous phosphorylation of the Y705 site on STAT3. This, in
turn, activates the STAT3 signaling pathway. A luciferase reporter
gene assay showed that the overexpression of FBXO2 tripled the
transcriptional activity of STAT3, and the mRNA levels of its
downstream target genes [such as the anti-apoptotic proteins Mcl-1
and X-linked inhibitor of apoptosis (XIAP)] were notably
upregulated. Chromatin immunoprecipitation further confirmed that
FBXO2 strengthened the binding of STAT3 to the promoters of Mcl-1
and XIAP, promoting their transcription and ultimately promoting
the multiplication of OS cells. In a nude mouse xenograft model,
the expression levels of IL-6R and phosphorylated STAT3 in the
tumors formed by U2OS cells with FBXO2 knockout were notably
reduced, and the protein levels of Mcl-1 and XIAP decreased by
~50%. This directly demonstrated the core role of the
FBXO2-IL-6R-STAT3 axis in tumor growth. Treatment of cells
overexpressing FBXO2 (such as MG63 cells) with STAT3 inhibitors
(such as Stattic) could reverse their promoting effect, and the
cell count returned to the level of the control group. Furthermore,
the addition of IL-6R neutralizing antibodies (such as CNTO328)
also notably inhibited STAT3 activation mediated by FBXO2 and cell
proliferation. According to the aforementioned research, in OS, the
function of FBXO2 undergoes a functional reversal, whereby its
glycoprotein recognition domain specifically binds to IL-6R,
inhibits its degradation to stabilize the receptor and thereby
activates the STAT3 signaling pathway, instead of following the
traditional ubiquitination degradation pathway. Therefore,
inhibiting the expression or activity of FBXO2 may represent an
effective therapeutic strategy for OS (112). At present, biotherapies aimed at
the IL-6/STAT3 signaling pathway primarily center on monoclonal
antibodies. Based on the aforementioned study, the application of
IL-6 monoclonal antibodies may prove to be an effective approach
for the treatment of OS.
Effect of FBXO2 on GB and its
mechanisms
GB is the most common and highly malignant primary
brain tumor among adults. The median survival is 14–24 months and
surgical treatment is the main therapeutic modality (113). However, the recurrence rate
remains high and the prognosis remains poor (113). Buehler et al (19) revealed that FBXO2 expression was
increased in recurrent GB, and that knockdown of FBXO2 enhanced
in vivo survival and mitigated the invasive growth of glioma
cells in brain sections. FBXO2 was also found to be concentrated in
the infiltration zone, with FBXO2-positive cancer cells being
linked to synaptic signaling processes. In addition, Atkin et
al (114) found that FBXO2 in
the brain governs the abundance and localization of specific NMDAR
subunits, GluN1 and GluN2A, and affects synapse formation and
maintenance to a certain degree. The aforementioned research
illustrates the potential function of FBXO2-dependent interactions
within the glioma microenvironment in facilitating tumor growth.
Thus, inhibition of FBXO2 may be used to treat recurrent GB.
Effect of FBXO2 on PTC and its
mechanisms
PTC accounts for ~84% of all thyroid cancers. It is
categorized as a well-differentiated thyroid cancer, with the
5-year relative survival rate reaching 98.5%. The majority of cases
of well-differentiated thyroid cancer are asymptomatic and are
identified during physical examinations or incidentally during
diagnostic imaging studies. Surgery is effective for the majority
of cases of well-differentiated thyroid cancer, and post-surgical
radioactive iodine treatment is capable of enhancing the overall
survival rate in patients at a high risk of recurrence (115). Anti-angiogenic tyrosine kinase
inhibitors and targeted therapies against the genetic mutations
that cause thyroid cancer are increasingly being used in the
treatment of metastatic diseases (115). Guo et al (20) performed in vitro experiments
to examine FBXO2 expression in PTC tissues. The findings indicated
that FBXO2 expression was upregulated in both PTC tissues and cell
lines. The expression levels of FBXO2 were positively associated
with the tumor size of PTC, as well as lymph node metastasis and
extracapsular invasion. Furthermore, silencing of FBXO2 suppressed
the proliferation of PTC cells and induced cell apoptosis, while
the overexpression of FBXO2 notably boosted the proliferation of
PTC cells. A mechanistic study demonstrated that FBXO2 is capable
of directly binding to p53 and facilitating its ubiquitination and
degradation (20). Knocking down
the p53 gene to a certain extent weakened the inhibitory effect of
FBXO2 knockdown on the progression of PTC cells, allowing the
proliferation, invasion, and metastasis processes of the originally
blocked tumor cells to recover. Knockdown of FBXO2 suppressed the
proliferation of PTC cells and promoted their apoptosis by
targeting p53 for ubiquitination and degradation (20). The aforementioned research findings
lay the groundwork for the diagnosis and treatment of PTC.
Effect of FBXO2 on OSCC and its
mechanisms
OSCC accounts for >90% of all malignant tumors in
the oral cavity (116). Due to
the extensive genetic susceptibility of the oral mucosa of patients
with OSCC to carcinogenic factors, the risk of developing
concurrent tumors in the oral cavity notably increases (116). Cheng et al (21) established the evolutionary
trajectory of tumor cells by relying on single-cell RNA sequencing
data, identified the dynamic clonal characteristics of OSCC and
further clarified that FBXO2 is a key gene that determines the
specific transcriptional state leading to the transformation of
tumor cells into clones with notably high malignant
characteristics. The expression levels of FBXO2 in OSCC are notably
higher than those in normal samples, particularly in patients with
more advanced clinical stages. In OSCC cells, knockdown of FBXO2
leads to a corresponding inhibition of cell proliferation,
G1-S phase transition, migration, invasion, EMT and
anti-apoptotic capacity, whereas its overexpression results in the
promotion of these processes (117). These findings offer novel
perspectives on clonal heterogeneity and lay the foundation for
more effective therapeutic approaches against OSCC, as well as for
overcoming the resistance of OSCC to treatment.
Role of FBXO2 in clinical tumor
treatment
FBXO2 is involved in the specific recognition and
degradation of substrate proteins within the UPS. Impairment of its
function is closely linked to the onset and progression of various
tumors such as GC (14), OV
(16), OS (18) and thyroid cancer (20), thereby endowing it with the
potential to act as a diagnostic or prognostic biomarker. In OV, Ji
et al (16) demonstrated
that the transcription factor SOX6 promotes the expression of FBXO2
by recognizing the potential response elements located in the
promoter region of FBXO2., Abnormally highly expressed FBXO2
targeted and degraded the tumor suppressor gene SUN2, inhibited
cell apoptosis and activated the proliferation-promoting signaling
pathway, thereby promoting the progression of OV. Knockdown of
FBXO2 notably inhibited the proliferation, metastasis and apoptosis
of OV cells, suggesting that its high expression may serve as an
independent indicator of poor prognosis in OV. In related research
on GB (19), it was found that
FBXO2 was continuously upregulated in recurrent GB, and its
expression levels exhibited a negative association with patient
survival rates. In addition, it was demonstrated that knockout of
FBXO2 could prolong the survival period of mice in an orthotopic
xenograft model and reduce the invasive growth of tumors in
organoid brain slices. Further analysis showed that high FBXO2
expression was related to the enrichment of tumor infiltration
areas and the interaction between glioma and the microenvironment,
and may promote malignant progression by regulating the interaction
between tumor cells and surrounding tissues (19). Through immunohistochemical studies
on CRC, it was revealed that high expression of FBXO2 is closely
related to distant metastasis and the American Joint Committee on
Cancer clinical stage of CRC, and is an independent influencing
factor for poor prognosis of patients (15,104). In addition, its carcinogenic
mechanisms may be related to regulating tumor proliferation
activity (positively associated with Ki-67 expression) and
angiogenesis formation (positively associated with VEGF expression)
(104). When exploring the
potential of FBXO2 as a diagnostic biomarker, a new patented study
(Chinese patent no. CN111381047A) proposed that detecting the
autoantibodies of FBXO2 in serum could be used for lung cancer
screening. Clinical data indicated that the levels of FBXO2
autoantibodies in the serum of patients with lung cancer were
notably lower compared with those of healthy controls, with a
specificity of 96.6% but a sensitivity of 20.0%. It is hypothesized
that combination with other markers may further improve the
diagnostic efficacy. Currently, this method still needs to be
verified by a larger-scale cohort study.
At present, the development of specific
small-molecule compounds targeting FBXO2 remains in the early
stages. Some proteasome inhibitors (such as bortezomib and MG-132)
(32,118) are not specifically targeted at
FBXO2, but can inhibit the degradation function of its substrates
by blocking the UPS. For example, treatment with MG-132 can
increase the accumulation of FBXO2 substrate proteins, thereby
indirectly inhibiting its oncogenic activity (118). Salidroside (a metabolic
regulator) can reduce the secretion of lactate vesicles carrying
FBXO2 by muscles in an over-exercise -induced liver fibrosis model,
thereby alleviating liver cell apoptosis and fibrosis (89). This suggests that metabolic
intervention may affect the intercellular transfer of FBXO2,
providing novel ideas for cancer treatment.
Summary and future prospects
In conclusion, the relationship between the FBP
family and tumors has attracted significant attention from
researchers. As research progresses, a more comprehensive
understanding of the interactions among the FBP family members is
required. Future drug development should focus on the complex
interactions and regulatory mechanisms of the FBP family to target
drugs at the FBP family. Among them, FBXO2 is a key component of
the ubiquitin ligase SCF complex. Besides serving a notable role in
hepatic glucose metabolism and cerebral degenerative diseases,
abnormally expressed FBXO2 is closely linked to the progression of
diverse tumors, such as GC, CRC, OV, EC, GB, thyroid cancer and
OSCC, and is also regarded to be one of the proto-oncogenes. In the
present review, the association between FBXO2 and tumorigenesis and
development was analyzed by reviewing the composition of the UPS
and FBP families, the roles they serve in malignant tumors, and the
progress of research on the relationship between FBXO2 and tumors.
Nevertheless, the complex mechanisms through which upregulated
FBXO2 promotes the growth, migration and invasion of various tumors
needs to be further studied in depth, which will help to more
clearly elucidate the pathogenic mechanisms of tumors and offer
novel avenues for the treatment of human malignant neoplasms.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by The Human Resources and
Social Security Department System of Shanxi Province, the Fund
Program for the Scientific Activities of Selected Returned Overseas
Professionals in Shanxi Province (grant no. 20210001), the Research
Project Supported by Shanxi Scholarship Council of China (grant no.
2021-116), Shanxi ‘136’ Leading Clinical Key Specialty (grant no.
2019XY002) and Shanxi Provincial Key Laboratory of Hepatobiliary
and Pancreatic Diseases (under construction).
Availability of data and materials
Not applicable.
Authors' contributions
JZ was responsible for drafting and revising the
manuscript; JY, XZ, YY, SS and SZ collected relevant literature and
assisted in manuscript revision. JZ and JY designed the tables and
figures. JH revised the manuscript. Data authentication is not
applicable. All authors have read and approved the final version of
the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
FBXL
|
F-box protein with leucine-rich amino
acid repeats
|
|
FBXW
|
F-box protein with WD-40 amino acid
repeats
|
|
FBXO
|
F-box protein with only
uncharacterized structural domains
|
References
|
1
|
Soerjomataram I and Bray F: Planning for
tomorrow: Global cancer incidence and the role of prevention
2020–2070. Nat Rev Clin Oncol. 18:663–672. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gong J, Cao J, Liu G and Huo JR: Function
and mechanism of F-box proteins in gastric cancer (Review). Int J
Oncol. 47:43–50. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Çetin G, Klafack S, Studencka-Turski M,
Krüger E and Ebstein F: The ubiquitin-proteasome system in immune
cells. Biomolecules. 11:602021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zheng N, Zhou Q, Wang Z and Wei W: Recent
advances in SCF ubiquitin ligase complex: Clinical implications.
Biochim Biophys Acta. 1866:12–22. 2016.PubMed/NCBI
|
|
5
|
Kim YJ, Lee Y, Shin H, Hwang S, Park J and
Song EJ: Ubiquitin-proteasome system as a target for anticancer
treatment-an update. Arch Pharm Res. 46:573–597. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Luo Z, Pan Y, Jeong LS, Liu J and Jia L:
Inactivation of the cullin (CUL)-RING E3 ligase by the
NEDD8-activating enzyme inhibitor MLN4924 triggers protective
autophagy in cancer cells. Autophagy. 8:1677–1679. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Park J, Cho J and Song EJ:
Ubiquitin-proteasome system (UPS) as a target for anticancer
treatment. Arch Pharm Res. 43:1144–1161. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lin DI, Barbash O, Kumar KGS, Weber JD,
Harper JW, Klein-Szanto AJP, Rustgi A, Fuchs SY and Diehl JA:
Phosphorylation-dependent ubiquitination of cyclin D1 by the SCF
(FBX4-alphaB crystallin) complex. Mol Cell. 24:355–366. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu P, Cong X, Liao S, Jia X, Wang X, Dai
W, Zhai L, Zhao L, Ji J, Ni D, et al: Global identification of
phospho-dependent SCF substrates reveals a FBXO22 phosphodegron and
an ERK-FBXO22-BAG3 axis in tumorigenesis. Cell Death Differ.
29:1–13. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shapira M, Kakiashvili E, Rosenberg T and
Hershko DD: The mTOR inhibitor rapamycin down-regulates the
expression of the ubiquitin ligase subunit Skp2 in breast cancer
cells. Breast Cancer Res. 8:R462006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tekcham DS, Chen D, Liu Y, Ling T, Zhang
Y, Chen H, Wang W, Otkur W, Qi H, Xia T, et al: F-box proteins and
cancer: An update from functional and regulatory mechanism to
therapeutic clinical prospects. Theranostics. 10:4150–4167. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Warren WG, Osborn M, Yates A, Wright K and
O'Sullivan SE: The emerging role of fatty acid binding protein 5
(FABP5) in cancers. Drug Discov Today. 28:1036282023. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mukherjee A, Chiang CY, Daifotis HA,
Nieman KM, Fahrmann JF, Lastra RR, Romero IL, Fiehn O and Lengyel
E: Adipocyte-induced FABP4 expression in ovarian cancer cells
promotes metastasis and mediates carboplatin resistance. Cancer
Res. 80:1748–1761. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sun X, Wang T, Guan ZR, Zhang C, Chen Y,
Jin J and Hua D: FBXO2, a novel marker for metastasis in human
gastric cancer. Biochem Biophys Res Commun. 495:2158–2164. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wei X, Bu J, Mo X, Lv B, Wang X and Hou B:
The prognostic significance of FBXO2 expression in colorectal
cancer. Int J Clin Exp Pathol. 11:5054–506. 2018.PubMed/NCBI
|
|
16
|
Ji J, Shen J, Xu Y, Xie M, Qian Q, Qiu T,
Shi W, Ren D, Ma J, Liu W and Liu B: FBXO2 targets glycosylated
SUN2 for ubiquitination and degradation to promote ovarian cancer
development. Cell Death Dis. 13:4422022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Che X, Jian F, Wang Y, Zhang J, Shen J,
Cheng Q, Wang X, Jia N and Feng W: FBXO2 promotes proliferation of
endometrial cancer by ubiquitin-mediated degradation of FBN1 in the
regulation of the cell cycle and the autophagy pathway. Front Cell
Dev Biol. 8:8432020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhao X, Guo W, Zou L and Hu B: FBXO2
modulates STAT3 signaling to regulate proliferation and
tumorigenicity of osteosarcoma cells. Cancer Cell Int. 20:2452020.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Buehler M, Yi X, Ge W, Blattmann P,
Rushing E, Reifenberger G, Felsberg J, Yeh C, Corn JE, Regli L, et
al: Quantitative proteomic landscapes of primary and recurrent
glioblastoma reveal a protumorigeneic role for FBXO2-dependent
glioma-microenvironment interactions. Neuro Oncol. 25:290–302.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Guo W, Ren Y and Qiu X: FBXO2 promotes the
progression of papillary thyroid carcinoma through the p53 pathway.
Sci Rep. 14:225742024. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cheng J, Liu O, Bin X and Tang Z: FBXO2 as
a switch guides a special fate of tumor clones evolving into a
highly malignant transcriptional subtype in oral squamous cell
carcinoma. Apoptosis. 30:167–184. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hershko A, Heller H, Elias S and
Ciechanover A: Components of ubiquitin-protein ligase system.
Resolution, affinity purification, and role in protein breakdown. J
Biol Chem. 258:8206–8214. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lyu L, Chen Z and McCarty N: TRIM44 links
the UPS to SQSTM1/p62-dependent aggrephagy and removing misfolded
proteins. Autophagy. 18:783–798. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Paudel RR, Lu D, Chowdhury SR, Monroy EY
and Wang J: Targeted protein degradation via lysosomes.
Biochemistry. 62:564–579. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hershko A, Ciechanover A, Heller H, Haas
AL and Rose IA: Proposed role of ATP in protein breakdown:
Conjugation of protein with multiple chains of the polypeptide of
ATP-dependent proteolysis. Proc Natl Acad Sci USA. 77:1783–1786.
1980. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hochstrasser M: Ubiquitin and
intracellular protein degradation. Curr Opin Cell Biol.
4:1024–1031. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hochstrasser M: Ubiquitin-dependent
protein degradation. Annu Rev Genet. 30:405–439. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jacopo M: Unconventional protein secretion
(UPS): Role in important diseases. Mol Biomed. 4:22023. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Aliabadi F, Sohrabi B, Mostafavi E,
Pazoki-Toroudi H and Webster TJ: Ubiquitin-proteasome system and
the role of its inhibitors in cancer therapy. Open Biol.
11:2003902021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li W, Bengtson MH, Ulbrich A, Matsuda A,
Reddy VA, Orth A, Chanda SK, Batalov S and Joazeiro CA: Genome-wide
and functional annotation of human E3 ubiquitin ligases identifies
MULAN, a mitochondrial E3 that regulates the organelle's dynamics
and signaling. PLoS One. 3:e14872008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xu C, Fan CD and Wang X: Regulation of
Mdm2 protein stability and the p53 response by NEDD4-1 E3 ligase.
Oncogene. 34:281–289. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Huang X, Gu H, Zhang E, Chen Q, Cao W, Yan
H, Chen J, Yang L, Lv N, He J, et al: The NEDD4-1 E3 ubiquitin
ligase: A potential molecular target for bortezomib sensitivity in
multiple myeloma. Int J Cancer. 146:1963–1978. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shangary S and Wang S: Targeting the
MDM2-p53 interaction for cancer therapy. Clin Cancer Res.
14:5318–5324. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fujii Y, Yada M, Nishiyama M, Kamura T,
Takahashi H, Tsunematsu R, Susaki E, Nakagawa T, Matsumoto A and
Nakayama KI: Fbxw7 contributes to tumor suppression by targeting
multiple proteins for ubiquitin-dependent degradation. Cancer Sci.
97:729–736. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wan L, Chen M, Cao J, Dai X, Yin Q, Zhang
J, Song SJ, Lu Y, Liu J, Inuzuka H, et al: The APC/C E3 ligase
complex activator FZR1 restricts BRAF oncogenic function. Cancer
Discov. 7:424–441. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Huang HC, Shi J, Orth JD and Mitchison TJ:
Evidence that mitotic exit is a better cancer therapeutic target
than spindle assembly. Cancer Cell. 16:347–358. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Manchado E, Guillamot M, de Cárcer G,
Eguren M, Trickey M, García-Higuera I, Moreno S, Yamano H, Cañamero
M and Malumbres M: Targeting mitotic exit leads to tumor regression
in vivo: Modulation by Cdk1, Mastl, and the PP2A/B55α,δ
phosphatase. Cancer Cell. 18:641–654. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kidokoro T, Tanikawa C, Furukawa Y,
Katagiri T, Nakamura Y and Matsuda K: CDC20, a potential cancer
therapeutic target, is negatively regulated by p53. Oncogene.
27:1562–1571. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hadjihannas MV, Bernkopf DB, Brückner M
and Behrens J: Cell cycle control of Wnt/β-catenin signalling by
conductin/axin2 through CDC20. EMBO Rep. 13:347–354. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu PY, Xu N, Malyukova A, Scarlett CJ,
Sun YT, Zhang XD, Ling D, Su SP, Nelson C, Chang DK, et al: The
histone deacetylase SIRT2 stabilizes Myc oncoproteins. Cell Death
Differ. 20:503–514. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Huang ZJ, Zhu JJ, Yang XY and Biskup E:
NEDD4 promotes cell growth and migration via PTEN/PI3K/AKT
signaling in hepatocellular carcinoma. Oncol Lett. 14:2649–2656.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hayashi S, Ozaki T, Yoshida K, Hosoda M,
Todo S, Akiyama S and Nakagawara A: p73 and MDM2 confer the
resistance of epidermoid carcinoma to cisplatin by blocking p53.
Biochem Biophys Res Commun. 347:60–66. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liu L, Wong CC, Gong B and Yu J:
Functional significance and therapeutic implication of ring-type E3
ligases in colorectal cancer. Oncogene. 37:148–159. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lipkowitz S and Weissman AM: RINGs of good
and evil: RING finger ubiquitin ligases at the crossroads of tumour
suppression and oncogenesis. Nat Rev Cancer. 11:629–643. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yang H, Lu X, Liu Z, Chen L, Xu Y, Wang Y,
Wei G and Chen Y: FBXW7 suppresses epithelial-mesenchymal
transition, stemness and metastatic potential of cholangiocarcinoma
cells. Oncotarget. 6:6310–6325. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Mao JH, Perez-Losada J, Wu D, Delrosario
R, Tsunematsu R, Nakayama KI, Brown K, Bryson S and Balmain A:
Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor
gene. Nature. 432:775–779. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Rajagopalan H, Jallepalli PV, Rago C,
Velculescu VE, Kinzler KW, Vogelstein B and Lengauer C:
Inactivation of hCDC4 can cause chromosomal instability. Nature.
428:77–81. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Grim JE, Knoblaugh SE, Guthrie KA, Hagar
A, Swanger J, Hespelt J, Delrow JJ, Small T, Grady WM, Nakayama KI
and Clurman BE: Fbw7 and p53 cooperatively suppress advanced and
chromosomally unstable intestinal cancer. Mol Cell Biol.
32:2160–2167. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Thompson LL, Rutherford KA, Lepage CC and
McManus KJ: The SCF complex is essential to maintain genome and
chromosome stability. Int J Mol Sci. 22:85442021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Uddin S, Bhat AA, Krishnankutty R, Mir F,
Kulinski M and Mohammad RM: Involvement of F-BOX proteins in
progression and development of human malignancies. Semin Cancer
Biol. 36:18–32. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Liu Y, Pan B, Qu W, Cao Y, Li J and Zhao
H: Systematic analysis of the expression and prognosis relevance of
FBXO family reveals the significance of FBXO1 in human breast
cancer. Cancer Cell Int. 21:1302021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Gao D, Inuzuka H, Tseng A, Chin RY, Toker
A and Wei W: Phosphorylation by Akt1 promotes cytoplasmic
localization of Skp2 and impairs APCCdh1-mediated Skp2 destruction.
Nat Cell Biol. 11:397–408. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Lin YW and Yang JL: Cooperation of ERK and
SCFSkp2 for MKP-1 destruction provides a positive feedback
regulation of proliferating signaling. J Biol Chem. 281:915–926.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zaytseva YY, Wang X, Southard RC, Wallis
NK and Kilgore MW: Down-regulation of PPARgamma1 suppresses cell
growth and induces apoptosis in MCF-7 breast cancer cells. Mol
Cancer. 7:902008. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Lu Y, Zi X and Pollak M: Molecular
mechanisms underlying IGF-I-induced attenuation of the
growth-inhibitory activity of trastuzumab (Herceptin) on SKBR3
breast cancer cells. Int J Cancer. 108:334–341. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Asmamaw MD, Liu Y, Zheng YC, Shi XJ and
Liu HM: Skp2 in the ubiquitin-proteasome system: A comprehensive
review. Med Res Rev. 40:1920–1949. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Sistrunk C, Kim SH, Wang X, Lee SH, Kim Y,
Macias E and Rodriguez-Puebla ML: Skp2 deficiency inhibits chemical
skin tumorigenesis independent of p27(Kip1) accumulation. Am J
Pathol. 182:1854–1864. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Fuchs SY, Spiegelman VS and Kumar KGS: The
many faces of beta-TrCP E3 ubiquitin ligases: Reflections in the
magic mirror of cancer. Oncogene. 23:2028–2036. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhang J, Yang Z, Ou J, Xia X, Zhi F and
Cui J: The F-box protein FBXL18 promotes glioma progression by
promoting K63-linked ubiquitination of Akt. FEBS Lett. 591:145–154.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Ji M, Zhao Z, Li Y, Xu P, Shi J, Li Z,
Wang K, Huang X and Liu B: FBXO6-mediated RNASET2 ubiquitination
and degradation governs the development of ovarian cancer. Cell
Death Dis. 12:3172021. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Ji M, Zhao Z, Li Y, Xu P, Shi J, Li Z,
Wang K, Huang X, Ji J, Liu W and Liu B: FBXO16-mediated hnRNPL
ubiquitination and degradation plays a tumor suppressor role in
ovarian cancer. Cell Death Dis. 12:7582021. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Wu J, Wen T, Marzio A, Song D, Chen S,
Yang C, Zhao F, Zhang B, Zhao G, Ferri A, et al: FBXO32-mediated
degradation of PTEN promotes lung adenocarcinoma progression. Cell
Death Dis. 15:2822024. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Habel N, El-Hachem N, Soysouvanh F,
Hadhiri-Bzioueche H, Giuliano S, Nguyen S, Horák P, Gay AS, Debayle
D, Nottet N, et al: FBXO32 links ubiquitination to epigenetic
reprograming of D cells. Cell Death Differ. 28:1837–1848. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Fan J, Bellon M, Ju M, Zhao L, Wei M, Fu L
and Nicot C: Clinical significance of FBXW7 loss of function in
human cancers. Mol Cancer. 21:872022. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Liu Z, Ma T, Duan J, Liu X and Liu L:
MicroRNA-223-induced inhibition of the FBXW7 gene affects the
proliferation and apoptosis of colorectal cancer cells via the
notch and akt/mTOR pathways. Mol Med Rep. 23:1542021. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Lee EK, Lian Z, D'Andrea K, Letrero R,
Sheng W, Liu S, Diehl JN, Pytel D, Barbash O, Schuchter L, et al:
The FBXO4 tumor suppressor functions as a barrier to
BRAFV600E-dependent metastatic melanoma. Mol Cell Biol.
33:4422–4433. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Lian Z, Lee EK, Bass AJ, Wong KK,
Klein-Szanto AJ, Rustgi AK and Diehl JA: FBXO4 loss facilitates
carcinogen induced papilloma development in mice. Cancer Biol Ther.
16:750–755. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Barbash O, Zamfirova P, Lin DI, Chen X,
Yang K, Nakagawa H, Lu F, Rustgi AK and Diehl JA: Mutations in Fbx4
inhibit dimerization of the SCF(Fbx4) ligase and contribute to
cyclin D1 overexpression in human cancer. Cancer Cell. 14:68–78.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Duan S, Cermak L, Pagan JK, Rossi M,
Martinengo C, di Celle PF, Chapuy B, Shipp M, Chiarle R and Pagano
M: FBXO11 targets BCL6 for degradation and is inactivated in
diffuse large B-cell lymphomas. Nature. 481:90–93. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Santra MK, Wajapeyee N and Green MR: F-box
protein FBXO31 mediates cyclin D1 degradation to induce G1 arrest
after DNA damage. Nature. 459:722–725. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Zou S, Ma C, Yang F, Xu X, Jia J and Liu
Z: FBXO31 suppresses gastric cancer EMT by targeting snail1 for
proteasomal degradation. Mol Cancer Res. 16:286–295. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Malonia SK, Dutta P, Santra MK and Green
MR: F-box protein FBXO31 directs degradation of MDM2 to facilitate
p53-mediated growth arrest following genotoxic stress. Proc Natl
Acad Sci USA. 112:8632–8637. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Liu J, Han L, Li B, Yang J, Huen MSY, Pan
X, Tsao SW and Cheung AL: F-box only protein 31 (FBXO31) negatively
regulates p38 mitogen-activated protein kinase (MAPK) signaling by
mediating lysine 48-linked ubiquitination and degradation of
mitogen-activated protein kinase kinase 6 (MKK6). J Biol Chem.
289:21508–21518. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Liu D, Xia H, Wang F, Chen C and Long J:
MicroRNA-210 interacts with FBXO31 to regulate cancer proliferation
cell cycle and migration in human breast cancer. OncoTargets Ther.
9:5245–5255. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Islam S, Dutta P, Sahay O, Gopalakrishnan
K, Muhury SR, Parameshwar P, Shetty P and Santra MK:
Feedback-regulated transcriptional repression of FBXO31 by c-myc
triggers ovarian cancer tumorigenesis. Int J Cancer. 150:1512–1524.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Ma Y, Sun WL, Ma SS, Zhao G, Liu Z, Lu Z
and Zhang D: LincRNA ZNF529-AS1 inhibits hepatocellular carcinoma
via FBXO31 and predicts the prognosis of hepatocellular carcinoma
patients. BMC Bioinformatics. 24:542023. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Duan S, Moro L, Qu R, Simoneschi D, Cho H,
Jiang S, Zhao H, Chang Q, de Stanchina E, Arbini AA and Pagano M:
Loss of FBXO31-mediated degradation of DUSP6 dysregulates ERK and
PI3K-AKT signaling and promotes prostate tumorigenesis. Cell Rep.
37:1098702021. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Fang X, Zhou W, Wu Q, Huang Z, Shi Y, Yang
K, Chen C, Xie Q, Mack SC, Wang X, et al: Deubiquitinase USP13
maintains glioblastoma stem cells by antagonizing FBXL14-mediated
Myc ubiquitination. J Exp Med. 214:245–267. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Cui YH, Kim H, Lee M, Yi JM, Kim RK, Uddin
N, Yoo KC, Kang JH, Choi MY, Cha HJ, et al: FBXL14 abolishes breast
cancer progression by targeting CDCP1 for proteasomal degradation.
Oncogene. 37:5794–5809. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Yoshida Y, Chiba T, Tokunaga F, Kawasaki
H, Iwai K, Suzuki T, Ito Y, Matsuoka K, Yoshida M, Tanaka K and Tai
T: E3 ubiquitin ligase that recognizes sugar chains. Nature.
418:438–442. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Nelson RF, Glenn KA, Zhang Y, Wen H,
Knutson T, Gouvion CM, Robinson BK, Zhou Z, Yang B, Smith RJ and
Paulson HL: Selective cochlear degeneration in mice lacking the
F-box protein, Fbx2, a glycoprotein-specific ubiquitin ligase
subunit. J Neurosci. 27:5163–5171. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Erhardt JA, Hynicka W, DiBenedetto A, Shen
N, Stone N, Paulson H and Pittman RN: A novel F box protein, NFB42,
is highly enriched in neurons and induces growth arrest. J Biol
Chem. 273:35222–35227. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Yoshida Y, Tokunaga F, Chiba T, Iwai K,
Tanaka K and Tai T: Fbs2 is a new member of the E3 ubiquitin ligase
family that recognizes sugar chains. J Biol Chem. 278:43877–43884.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Mizushima T, Yoshida Y, Kumanomidou T,
Hasegawa Y, Suzuki A, Yamane T and Tanaka K: Structural basis for
the selection of glycosylated substrates by SCF(Fbs1) ubiquitin
ligase. Proc Natl Acad Sci USA. 104:5777–5781. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Nelson RF, Glenn KA, Miller VM, Wen H and
Paulson HL: A novel route for F-box protein-mediated ubiquitination
links CHIP to glycoprotein quality control. J Biol Chem.
281:20242–20251. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Zhang HJ, Tian J, Qi XK, Xiang T, He GP,
Yu X, Zhang X, Zhao B, Feng QS, Chen MY, et al: Epstein-Barr virus
activates F-box protein FBXO2 to limit viral infectivity by
targeting glycoprotein B for degradation. PLoS Pathog.
14:e10072082018. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Liu B, Lu H, Li D, Xiong X, Gao L, Wu Z
and Lu Y: Aberrant expression of FBXO2 disrupts glucose homeostasis
through ubiquitin-mediated degradation of insulin receptor in obese
mice. Diabetes. 66:689–698. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Liu Z, Chen NY, Zhang Z, Zhou S and Hu SY:
F-box only protein 2 exacerbates non-alcoholic fatty liver disease
by targeting the hydroxyl CoA dehydrogenase alpha subunit. World J
Gastroenterol. 29:4433–4450. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Liu Y, Zhou R, Guo Y, Hu B, Xie L, An Y,
Wen J, Liu Z, Zhou M, Kuang W, et al: Muscle-derived small
extracellular vesicles induce liver fibrosis during overtraining.
Cell Metab. 37:824–841. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Menard R, Morin E, Morse D, Halluin C,
Pende M, Baanannou A, Grendler J, Fuqua H, Li J, Lancelot L, et al:
Zebrafish genetic model of neuromuscular degeneration associated
with atrogin-1 expression. BioRxiv Prepr Serv Biol.
9:2025.03.07.642048. 2025.
|
|
91
|
Li X, Mank JE and Ban L: The grasshopper
genome reveals long-term gene content conservation of the X
chromosome and temporal variation in X chromosome evolution. Genome
Res. 34:997–1007. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Atkin G, Hunt J, Minakawa E, Sharkey L,
Tipper N, Tennant W and Paulson HL: F-box only protein 2 (Fbxo2)
regulates amyloid precursor protein levels and processing. J Biol
Chem. 289:7038–7048. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Stephenson SEM, Costain G, Blok LER, Silk
MA, Nguyen TB, Dong X, Alhuzaimi DE, Dowling JJ, Walker S, Amburgey
K, et al: Germline variants in tumor suppressor FBXW7 lead to
impaired ubiquitination and a neurodevelopmental syndrome. Am J Hum
Genet. 109:601–617. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Chen N, Cao R, Zhang Z, Zhou S and Hu S:
Sleeve gastrectomy improves hepatic glucose metabolism by
downregulating FBXO2 and activating the PI3K-AKT pathway. Int J Mol
Sci. 24:55442023. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Yuan L, Song Z, Deng X, Yang Z, Yang Y,
Guo Y, Lu H and Deng H: Genetic analysis of FBXO2, FBXO6, FBXO12,
and FBXO41 variants in Han Chinese patients with Sporadic
Parkinson's disease. Neurosci Bull. 33:510–514. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Twarowski B and Herbet M: Inflammatory
processes in alzheimer's disease-pathomechanism, diagnosis and
treatment: A review. Int J Mol Sci. 24:65182023. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Williams JB, Cao Q, Wang W, Lee YH, Qin L,
Zhong P, Ren Y, Ma K and Yan Z: Inhibition of histone
methyltransferase Smyd3 rescuesNMDARand cognitivedeficits in a
tauopathy mouse model. Nat Commun. 14:912023. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Liu EA, Schultz ML, Mochida C, Chung C,
Paulson HL and Lieberman AP: Fbxo2 mediates clearance of damaged
lysosomes and modifies neurodegeneration in the Niemann-Pick C
brain. JCI Insight. 5:e1366762020. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Tang Z, Li J and Li C:
Post-transcriptional regulator RBM47 stabilizes FBXO2 mRNA to
advance osteoarthritis development: WGCNA analysis and experimental
validation. Biochem Genet. 62:3092–3110. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Cherif H, Mannarino M, Pacis AS, Ragoussis
J, Rabau O, Ouellet JA and Haglund L: Single-cell RNA-seq analysis
of cells from degenerating and non-degenerating intervertebral
discs from the same individual reveals new biomarkers for
intervertebral disc degeneration. Int J Mol Sci. 23:39932022.
View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Hartman BH, Bӧscke R, Ellwanger DC,
Keymeulen S, Scheibinger M and Heller S: Fbxo2VHC mouse and
embryonic stem cell reporter lines delineate in vitro-generated
inner ear sensory epithelia cells and enable otic lineage selection
and Cre-recombination. Dev Biol. 443:64–77. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
McGovern MM, Hartman B, Thawani A,
Maunsell H, Zhang H, Yousaf R, Heller S, Stone J and Groves AK:
Fbxo2CreERT2: A new model for targeting cells in the
neonatal and mature inner ear. Hear Res. 428:1086862023. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Yamada A, Hikichi M, Nozawa T and Nakagawa
I: FBXO2/SCF ubiquitin ligase complex directs xenophagy through
recognizing bacterial surface glycan. EMBO Rep. 22:e525842021.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Zhao H, Ming T, Tang S, Ren S, Yang H, Liu
M, Tao Q and Xu H: Wnt signaling in colorectal cancer: Pathogenic
role and therapeutic target. Mol Cancer. 21:1442022. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Norwood DA, Montalvan-Sanchez E, Dominguez
RL and Morgan DR: Gastric cancer: Emerging trends in prevention,
diagnosis, and treatment. Gastroenterol Clin North Am. 51:501–518.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Machlowska J, Baj J, Sitarz M, Maciejewski
R and Sitarz R: Gastric cancer: Epidemiology, risk factors,
classification, genomic characteristics and treatment strategies.
Int J Mol Sci. 21:40122020. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Li J, Ma X, Chakravarti D, Shalapour S and
DePinho RA: Genetic and biological hallmarks of colorectal cancer.
Genes Dev. 35:787–820. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Zhang R, Siu MKY, Ngan HYS and Chan KKL:
Molecular biomarkers for the early detection of ovarian cancer. Int
J Mol Sci. 23:120412022. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Lai W, Xie R, Chen C, Lou W, Yang H, Deng
L, Lu Q and Tang X: Integrated analysis of scRNA-seq and bulk
RNA-seq identifies FBXO2 as a candidate biomarker associated with
chemoresistance in HGSOC. Heliyon. 10:e284902024. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Yen TT, Wang TL, Fader AN, Shih IM and
Gaillard S: Molecular classification and emerging targeted therapy
in endometrial cancer. Int J Gynecol Pathol. 39:26–35. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Shoaib Z, Fan TM and Irudayaraj JMK:
Osteosarcoma mechanobiology and therapeutic targets. Br J
Pharmacol. 179:201–217. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Zhao X, Guo W, Zou L and Hu B: FBXO2
modulates STAT3 signaling to regulate proliferation and
tumorigenicity of osteosarcoma cells. Cancer Cell Int. 20:2452020.
View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Ma R, Taphoorn MJB and Plaha P: Advances
in the management of glioblastoma. J Neurol Neurosurg Psychiatry.
92:1103–1111. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Atkin G, Moore S, Lu Y, Nelson RF, Tipper
N, Rajpal G, Hunt J, Tennant W, Hell JW, Murphy GG and Paulson H:
Loss of F-box Only protein 2 (Fbxo2) disrupts levels and
localization of Select nmda receptor subunits, and promotes
aberrant synaptic connectivity. J Neurosci. 35:6165–6178. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Boucai L, Zafereo M and Cabanillas ME:
Thyroid cancer: A review. JAMA. 331:425–435. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Badwelan M, Muaddi H, Ahmed A, Lee KT and
Tran SD: Oral squamous cell carcinoma and concomitant primary
tumors, what do we know? A review of the literature. Curr Oncol.
30:3721–3734. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Cheng J, Liu O, Bin X and Tang Z: FBXO2 as
a switch guides a special fate of tumor clones evolving into a
highly malignant transcriptional subtype in oral squamous cell
carcinoma. Apoptosis. 30:167–184. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Zhang DM, He T, Katusic ZS, Lee HC and Lu
T: Muscle-specific f-box only proteins facilitate bk channel β(1)
subunit downregulation in vascular smooth muscle cells of diabetes
mellitus. Circ Res. 107:1454–1459. 2010. View Article : Google Scholar : PubMed/NCBI
|