Introduction
Various musculoskeletal disorders develop with age.
In bones, the progressive increase in receptor activator of NF-κB
ligand and sclerostin levels (1,2), as
well as a decline in normal type I collagen (3), lead to a reduction in bone mass and
density that ultimately results in osteoporosis (4). In cartilage, an increase in matrix
metalloproteinases and disintegrin and metalloproteinase with
thrombospondin motifs, coupled with a reduction in normal type II
collagen, contributes to cartilage degeneration, leading to
osteoarthritis (5,6). In peripheral nerves, axonal
regenerative capacity is reported to decline with age; however, the
pathology in peripheral nerves remains to be elucidated (7).
Repressor element 1 silencing transcription factor
(REST) is a transcriptional factor that regulates neuron-specific
gene expression in the nervous system (8). Our previous studies demonstrated that
REST expression increases with age in peripheral nerves, leading to
a decline in axonal regenerative capacity (9,10).
However, the role of REST in peripheral nerve axonal regeneration
with age remains to be elucidated. Nuclear transport mechanisms are
essential for transcription factors to perform their functions
within cells (11). Under healthy
aging, REST is transported to the nucleus in brain cells, where it
provides neuroprotection against oxidative stress and apoptosis
(12,13). On the other hand, oxidative stress
has been shown to induce abnormal modifications in nuclear proteins
such as histones, leading to immune responses associated with
disease (14). Moreover, REST is
not transported to the nucleus in neurodegenerative diseases such
as Alzheimer's disease and Parkinson's disease (12,13).
Thus, understanding the role of REST in peripheral nerves requires
elucidation of its nuclear transport mechanism.
Several studies have reported that hydrogen
(H2) has neuroprotective effects. In mice, oral
administration of H2-rich water (HRW) suppresses
oxidative stress markers and provides protective effects against
cognitive impairment (15). A
previous study using a cerebral ischemia mouse model reported that
intraperitoneal administration of HRW promoted neuronal recovery
through autophagy (16).
Furthermore, using a diabetic rat model with peripheral neuropathy,
intraperitoneal administration of HRW improved axonal degeneration
(17). However, the mechanism of
neuroprotective effects of H2 administration on
peripheral nerves remains to be elucidated. This study investigated
the effects of H2 administration on the decline in
axonal regenerative capacity with aging. Additionally, to elucidate
the pathology of age-related decline in axonal regeneration, the
molecular mechanisms involved in REST nuclear transport were
analyzed.
Materials and methods
Animals
This study was approved by the Animal Care Committee
of Juntendo University, Tokyo, Japan (2-1-1 Hongo, Bunkyo-ku,
Tokyo, 113-8421, Japan; Registration No. 1555; Approval No.
2024183, Date of Approval: March 12th, 2024).
Eighteen male C57BL/6J mice (Young group: 8-week-old
mice, n=6; Aged group: 70-week-old mice, n=6; Aged + H2
group: 70-week-old mice, n=6) for quantitative polymerase chain
reaction (qPCR) and western blotting, and 15 male C57BL6J mice
(Young group: 8-week-old mice, n=5; Aged group: 70-week-old mice,
n=5; Aged + H2 group: 70-week-old mice, n=5) for
immunofluorescence staining were purchased from JAPN SLC, Inc.
(Shizuoka, Japan). The body weight of the mice at the start of the
experiment was 40.0±6.4 g. Mice were housed in sterile cages with
five animals per cage, under controlled conditions of 22±2°C,
40–60% humidity, and a 12-h light/dark cycle. The mice were given
water that was CRF-1 gamma-ray-irradiated (15 kGy) (Oriental Yeast
Co. Ltd., Tokyo, Japan) ad libitum. As estrogen levels are
known to influence peripheral neuropathy and naturally decrease
with age, male mice with less estrogen fluctuations and
susceptibility to estrogen were used in this study. The health
status and behavior of the animals were monitored daily throughout
the experiment period. Humane endpoints were defined as a loss of
20% of body weight, difficulty breathing, coughing, wheezing,
severe diarrhea, vomiting, flaccid or spastic paralysis,
convulsions, coupled with body temperature significantly below
normal, and continuously assessed to determine whether animals
should be euthanized before the scheduled end of the experiment. No
animals reached these humane endpoints during the study.
Hydrogen treatment
H2-containing physiological saline was
prepared using the H2 gas generator Suilive®
SS-150 (SUISO JAPAN Co. Ltd., Osaka, Japan). The Aged +
H2 group received daily intraperitoneal injections of
H2-containing physiological saline (10 ml/kg,
H2 concentration: 800 ppb) for 2 weeks. The Aged group
received an equivalent volume of normal saline (10 ml/kg).
Analysis of REST and GAP43 expression
in young and aged mice
The Young group (n=6), the Aged group (n=6), and the
Aged + H2 group (n=6) were sacrificed by cervical
dislocation and their sciatic nerves (SN) were collected. The
expression levels of REST and growth-associated protein 43 (GAP43),
a neuronal protein known for its important role in axon
regeneration, in SN were measured by qPCR, western blotting, and
immunofluorescence staining and compared among the three
groups.
Cell culture
Mouse embryonic fibroblast cell line NIH3T3 (Cell
line service, Eppelheim, Germany) was cultured in a humidified
incubator with 5% CO2 at 37°C. The culture medium consisted of
Dulbecco's modified Eagle's medium/Ham's F-12 (Sigma-Aldrich),
supplemented with 10% fetal bovine serum and 100 U/ml penicillin.
H2-containing culture medium was prepared using
Suilive® SS-150. H2 concentration was
confirmed to be 800 ppb at the start of administration and 500 ppb
after 2 h according to our preliminary experience (data not shown).
Based on the results of these preliminary experiments, the cell
culture time in the H2-containing culture medium was set
to 2 h.
Construction of REST-overexpressed
cells
Using cultured cell lines, REST-overexpressed
(REST-OE) cells were constructed (REST-OE group). REST was
overexpressed using a lentiviral vector (VectorBuilder Inc.,
Chicago, IL, USA; vector ID: VB900006-3284rup), while a mock
plasmid served as the negative control (Control group). The
plasmids were amplified in Escherichia coli DH5α, and plasmid DNA
was purified from the amplified lentiviral vectors using the
QUIAGEN® Plasmid Maxi kit. To make REST-OE cells, cells
were transfected with the REST plasmid using Lipofectamine 3000
(Thermo Fisher Scientific), according to the manufacturer's
instructions. REST-OE cells were cultured in
H2-containing medium (H2 concentration: 800
ppb) for 2 h in the cells of the REST-OE + H2 group.
Quantitative PCR
Total RNA was extracted from SN of animal models and
from cultured cell lines using in vitro models with the
RNeasy Micro kit (Qiagen, Tokyo, Japan), following the
manufacturer's instructions. Complementary DNA was synthesized
using the PrimeScript™ RT Reagent Kit (Takara, Shiga,
Japan). Next, qPCR was performed with SYBR Green real-time PCR
assay (Thermo Fisher Scientific) according to the ΔΔCT method. The
expression levels of targets (REST and GAP43) were normalized to
glyceraldehyde-3-phosphate dehydrogenase (GAPDH). qPCR reactions
were performed in triplicate, and the ΔΔCt method was used for
relative quantification (18). The
primer sequences used are listed in Table I.
 | Table I.Primer sequences used for
quantitative PCR in the present study. |
Table I.
Primer sequences used for
quantitative PCR in the present study.
| Gene | Forward primer
sequences (5′-3′) | Reverse primer
sequences (5′-3′) |
|---|
| Rest |
ACCTGCAGCAAGTGCAACTA |
CCGCATGTGTCGCGTTAGA |
| Gap43 |
AAGGCAGGGGAAGATACCAC |
TTGTTCAATCTTTTGGTCCTCAT |
| Gapdh |
TGTGTCCGTCGTGGATCTG |
TTGCTGTTGAAGTCGCAGG |
Western blotting
Proteins were extracted from SN of animal models and
from cultured cell lines using 1 × radio immunoprecipitation assay
(RIPA) buffer. Equal amounts of protein were loaded for each sample
as determined by BCA assay and separated by sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and
transferred onto a polyvinylidene difluoride (PVDF) membranes by
Trans-Blot Turbo system (BIORAD, CA, USA). Non-specific binding
sites were blocked with PVDF Blocking Reagent (Toyobo Co. Ltd.,
Osaka, Japan) for 1 h at room temperature. The membranes were then
washed three times with Tris-buffered saline containing 0.1% Tween
20 (TBST), 10 min per wash. Subsequently, membranes were incubated
overnight at 4°C with primary antibodies diluted in Solution 1
(Toyobo Co. Ltd.), according to the manufacturer's instructions.
Although each target protein and its corresponding loading control
were detected on separate membranes, equal amounts of protein (20
µg) were loaded per lane as determined by BCA assay, and all
membranes were quantified based on relative band intensities using
standardized loading across samples.
The following primary antibodies were used: rabbit
polyclonal anti-REST (1:1,000, 22242-1-AP; ProteinTech), rabbit
polyclonal anti-GAP43 (1:1,000, 16971-1-AP; ProteinTech), rabbit
polyclonal anti-PRICKLE1 (RILP) (1:1,000, 22589-1-AP; ProteinTech),
rabbit monoclonal anti-Huntingtin (1:5,000, ab109115; Abcam),
rabbit monoclonal anti-DCTN1/p150Glued (1:1,000, ab246505; Abcam),
rabbit polyclonal anti-LC3 (1:1,000, 14600-1-AP; ProteinTech),
mouse monoclonal anti-GAPDH (1:2,000, sc-32233; Santa Cruz, CA,
USA), and rabbit polyclonal anti-Lamin B1 (1:5,000, 12987-1-AP;
ProteinTech).
After incubation, membranes were washed with TBST
and incubated with appropriate secondary antibodies for 2 h at room
temperature. Following additional washes, signals were detected
using the Amersham Imager 680 system (GE Healthcare Life Sciences,
IL, USA). Image Lab 3.0 software was used to measure the relative
optical density of the protein bands. GAPDH was used as a loading
control for the total cell and cytoplasmic fractions, and Lamin B1
as a loading control for the nuclear fractions.
Immunofluorescence staining
Immunofluorescence staining was performed to assess
the expression of REST and GAP43 in both animal models and cultured
cell lines. SN was fixed in 4% paraformaldehyde at room temperature
for 72 h and subsequently embedded in paraffin. Tissue sections
were prepared by slicing cutting the harvested SN into 3 µm-thick
sections. Samples were deparaffinized and subjected to antigen
retrieval by autoclaving at 121°C for 10 min. Cell lines were
cultured using the Nunc™ Lab-Tek™ II Chamber
Slide™ System to create three group. Then, the cultured
medium was discarded, and were fixed in 4% paraformaldehyde at room
temperature for 15 min. To suppress autofluorescence sections were
treated with True View™ (SP-8400, Vector, CA, USA),
followed by blocking in PBS containing 0.05% Tween 20 and 2% bovine
serum albumin (BSA) (A2153, Sigma-Aldrich, MO, USA) for 30 min.
Sections were then incubated with primary antibodies against the
target proteins at 4°C for 15 h. After washing with TBST, a goat
anti-mouse IgG antibody conjugated to Alexa Fluor 488 (A11001,
Thermo Fisher Scientific) was used as the secondary antibody. A
rabbit IgG monoclonal antibody was used as a negative control.
Fluorescence intensity was visualized using a fluorescence imaging
microscope (Leica, TCSSP5, Wetzlar, Germany) in photon-counting
mode. The primary antibodies were rabbit polyclonal anti-REST
(1:100, 22242-1-AP, ProteinTech, IL, USA), and rabbit polyclonal
anti-GAP43 (1:100, 16971-1-AP, ProteinTech) in this study.
Fluorescence intensity was measured at 10 randomly
selected sites within the perikaryon in the region of interest
(ROI) showing fluorescence emission. The mean fluorescence
intensity was then calculated. Fluorescence levels detected by each
antibody were compared among the groups: for the in vivo
experiment (Young group, Aged group, Aged + H2 group)
and for the in vitro experiment (Control group, REST-OE
group, and REST-OE + H2 group).
Isolation of cytoplasmic and nuclear
fractions from REST-regulated cells
To demonstrate that REST, functioning as a
transcriptional regulator, is transported to the nucleus from the
cytoplasm, we used NE-PER® Nuclear and Cytoplasmic
Extraction Kit (Thermo Fisher Scientific), following the
manufacturer's instructions. Using these fractions, REST and the
proteins reported to be involved in REST nuclear transport,
including RILP, Huntingtin, and DCTN1/p150Glued, and LC3
were quantified by western blotting.
Statistical analysis
Data are expressed as the mean ± standard deviation
(SD). Statistical analysis was performed using one-way analysis of
variance (ANOVA), followed by Sidak's multiple comparisons test for
pairwise group comparisons, with Prism 7 software (GraphPad
Software, San Diego, CA, USA). A P-value of less than 0.05 was
considered statistically significant.
Results
REST expression and GAP43 in SN
To examine differences in the expression of REST and
GAP43 among the Young group, the Aged group, and the Aged +
H2 group, their expression in SN were quantified by
qPCR, western blotting, and immunofluorescence staining.
qPCR analysis revealed a significant upregulation of
REST in the Aged group compared with the Young group (Young group:
1.00±0.00-fold; Aged group: 2.37±0.35-fold, P=0.0014), whereas it
was significantly decreased in the Aged + H2 group
compared with the Aged group (Aged group: 2.37±0.35-fold; Aged +
H2 group: 0.98±0.02-fold, P=0.0013) (Fig. 1A). GAP43 expression in SN was
significantly decreased in the Aged group compared with the Young
group (Young group: 1.00±0.00-fold; Aged group: 0.70±0.04-fold,
P=0.0274), whereas it was significantly increased in the Aged +
H2 group compared with the Aged group (Aged group:
0.70±0.04-fold, P=0.0274; Aged + H2 group:
1.00±0.13-fold, P=0.0249) (Fig.
1B).
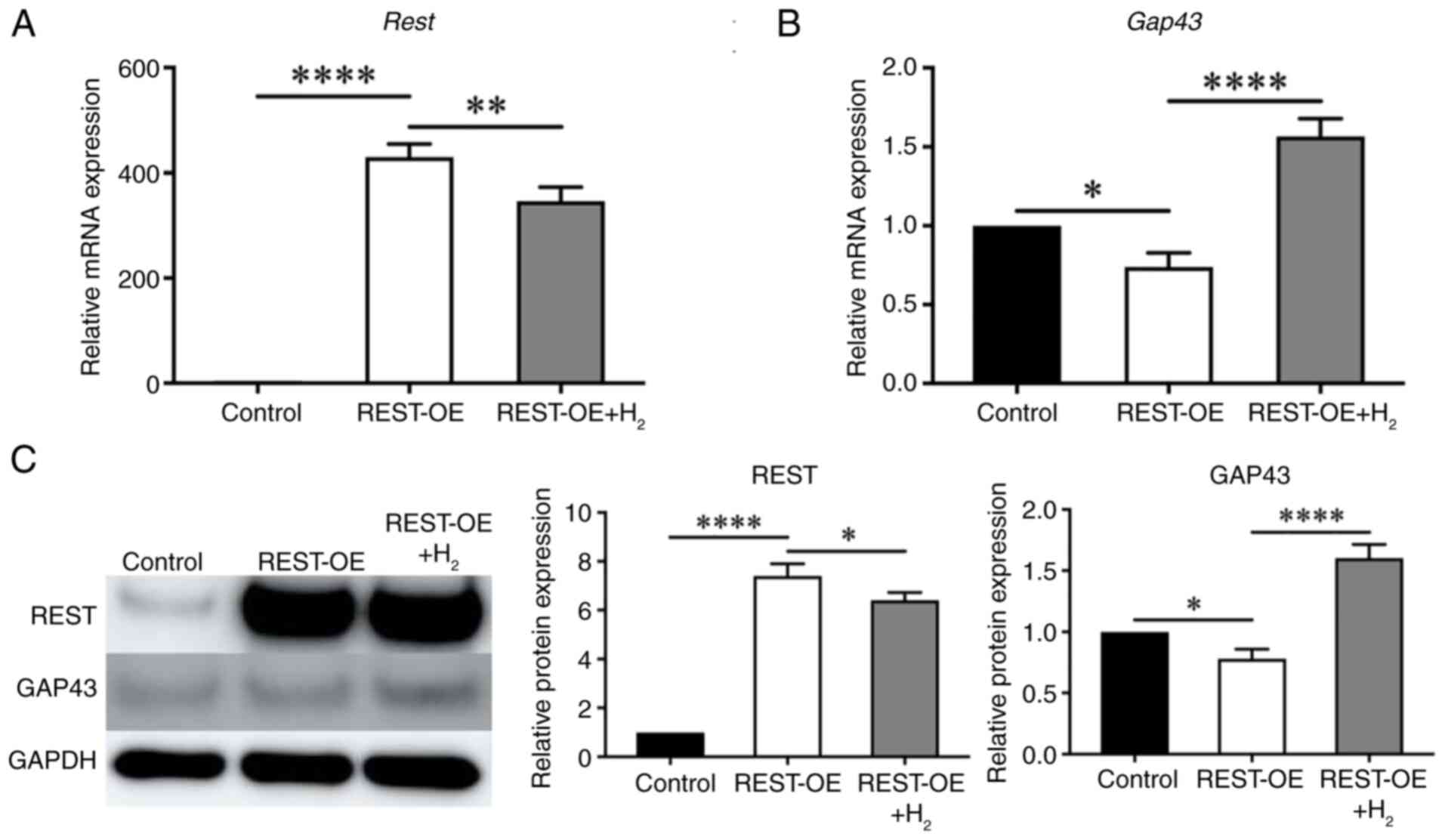
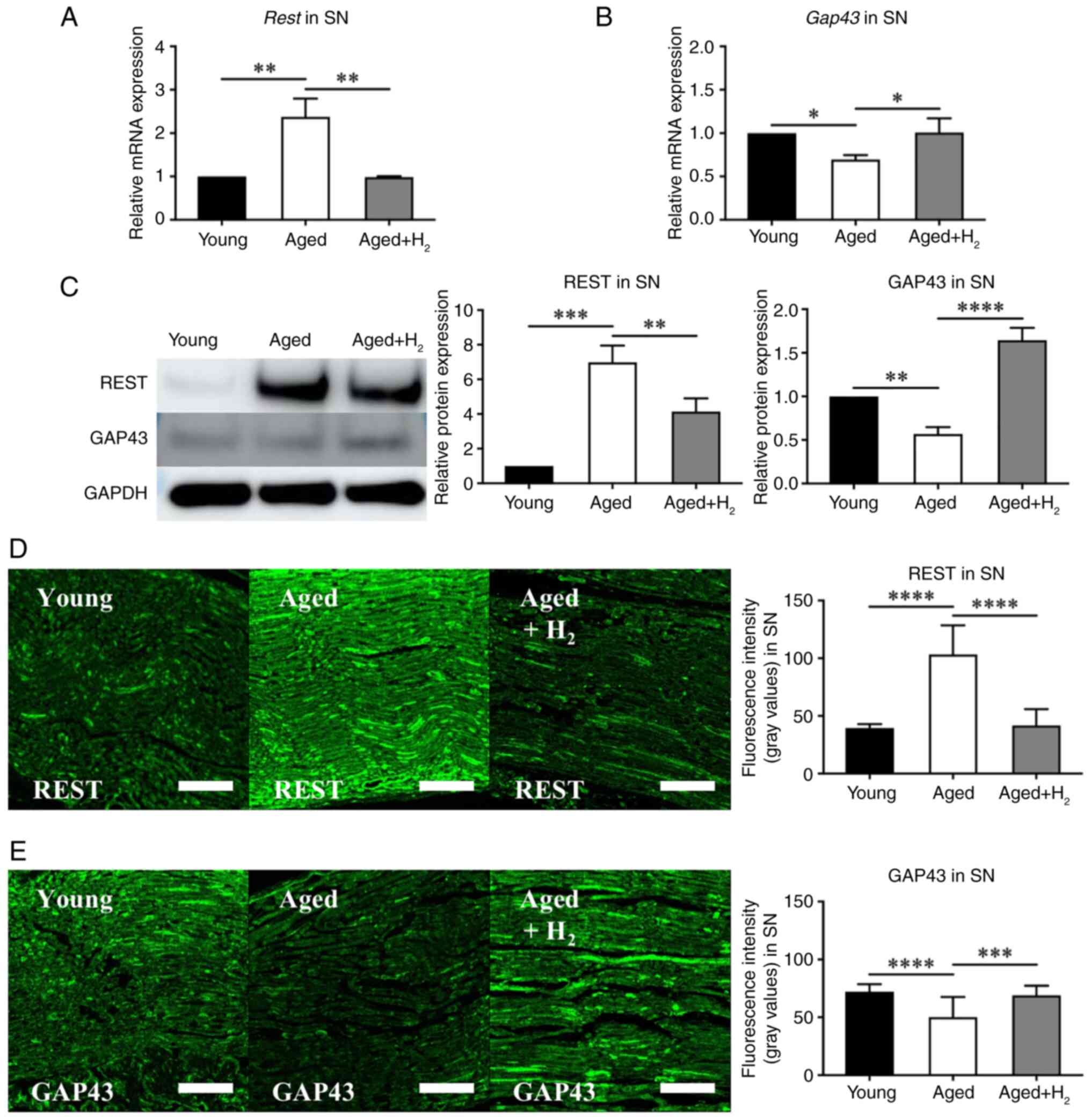 | Figure 1.REST and GAP43 expression in SN of
the young, aged and aged + H2 groups. The aged group was
compared with the young group, and the aged + H2 group
was compared with the aged group. The graphs show the
quantification of relative protein and mRNA abundance. Mean ± SD,
n=6 mice per group. *P<0.05, **P<0.01, ***P<0.001 and
****P<0.0001 (one-way ANOVA). (A) qPCR analysis of Rest
expression. (B) qPCR analysis of Gap43 expression. (C)
Western blot analysis of REST and GAP43 expression. (D)
Histochemical assessment of REST expression by immunofluorescence
staining. Scale bar, 100 µm. (E) Histochemical assessment of GAP43
expression by immunofluorescence staining. Scale bar, 100 µm.
GAP43, growth-associated protein 43; H2, hydrogen; qPCR,
quantitative PCR; REST, repressor element 1 silencing transcription
factor; SN, sciatic nerves. |
Western blotting further supported these results.
REST expression was markedly increased in the Aged group compared
with the Young group (Young group: 1.00±0.00-fold; Aged group:
6.98±0.79-fold, P=0.0001), whereas it was significantly decreased
in the Aged + H2 group compared with the Aged group
(Aged group: 6.98±0.79-fold; Aged + H2 group:
4.14±0.62-fold, P=0.0082) (Fig.
1C). GAP43 expression in SN was significantly decreased in the
Aged group compared with the Young group (Young group:
1.00±0.00-fold; Aged group: 0.57±0.06-fold, P=0.0040), whereas it
was significantly increased in the Aged + H2 group
compared with the Aged group (Aged group: 0.57±0.06-fold; Aged +
H2 group: 1.64±0.12-fold, P<0.0001) (Fig. 1C).
Immunofluorescence staining revealed that the
fluorescence intensity of REST in SN was significantly increased in
the Aged group compared with the Young group (Young group:
39.57±3.19; Aged group: 103.30±6.48, P<0.0001), whereas it was
significantly decreased in the Aged + H2 group compared
with the Aged group (Aged group: 103.30±6.48; Aged + H2
group: 41.75±3.64, P<0.0001) (Fig.
1D). On the other hand, the fluorescence intensity of GAP43 in
SN was significantly decreased in the Aged group compared with the
Young group (Young group: 72.15±6.26; Aged group: 50.17±4.51,
P<0.0001), whereas it was significantly increased in the Aged +
H2 group compared with the Aged group (Aged group:
50.17±4.51; Aged + H2 group: 68.96±2.17, P=0.0002)
(Fig. 1E).
These results suggest that REST is upregulated with
aging and inhibits axonal regeneration capacity; however,
H2 may have the potential to recover this reduced
capacity for axonal regeneration in mice.
REST expression and GAP43 expression
in REST-OE cells
To verify whether the results obtained in SN can be
reproduced in cultured cells, REST-OE cells were constructed and
compared among the Control, REST-OE, and REST-OE + H2
groups.
qPCR showed that REST expression was markedly
increased in the REST-OE group compared with the Control group
(Control group: 1.00±0.00-fold; REST-OE group: 430.0±20.51-fold,
P<0.0001), whereas it was significantly decreased in the REST-OE
+ H2 group compared with the REST-OE group (REST-OE
group: 430.0±20.51-fold; REST-OE + H2 group:
346.6±21.95-fold, P=0.0059) (Fig.
2A). On the other hand, GAP43 expression was significantly
decreased in the REST-OE group compared with the Control group
(Control group: 1.00±0.00-fold; REST-OE group: 0.74±0.07-fold,
P=0.0248), whereas it was significantly increased in the REST-OE +
H2 group compared with the REST-OE group (REST-OE group:
0.74±0.07-fold; REST-OE + H2 group: 1.60±0.09-fold,
1.57±0.09-fold, P<0.0001) (Fig.
2B).
At the protein level, REST expression followed a
similar pattern. REST expression was significantly increased in the
REST-OE group compared with the Control group (Control group:
1.00±0.00-fold; REST-OE group: 7.40±0.41-fold, P<0.0001),
whereas it was significantly decreased in the REST-OE +
H2 group compared with the REST-OE group (REST-OE group:
7.40±0.41-fold; REST-OE + H2 group: 6.40±0.26-fold,
P=0.0240) (Fig. 2C). On the other
hand, GAP43 expression was significantly decreased in the REST-OE
group compared with the Control group (Control group:
1.00±0.00-fold; REST-OE group: 0.78±0.06-fold, P=0.0409), whereas
it was significantly increased in the REST-OE + H2 group
compared with the REST-OE group (REST-OE group: 0.78±0.06-fold;
REST-OE + H2 group: 1.60±0.09-fold, P<0.0001)
(Fig. 2C).
These results are consistent with those observed in
mice, suggesting that these cells are a suitable model for
evaluating changes in SN of animal models.
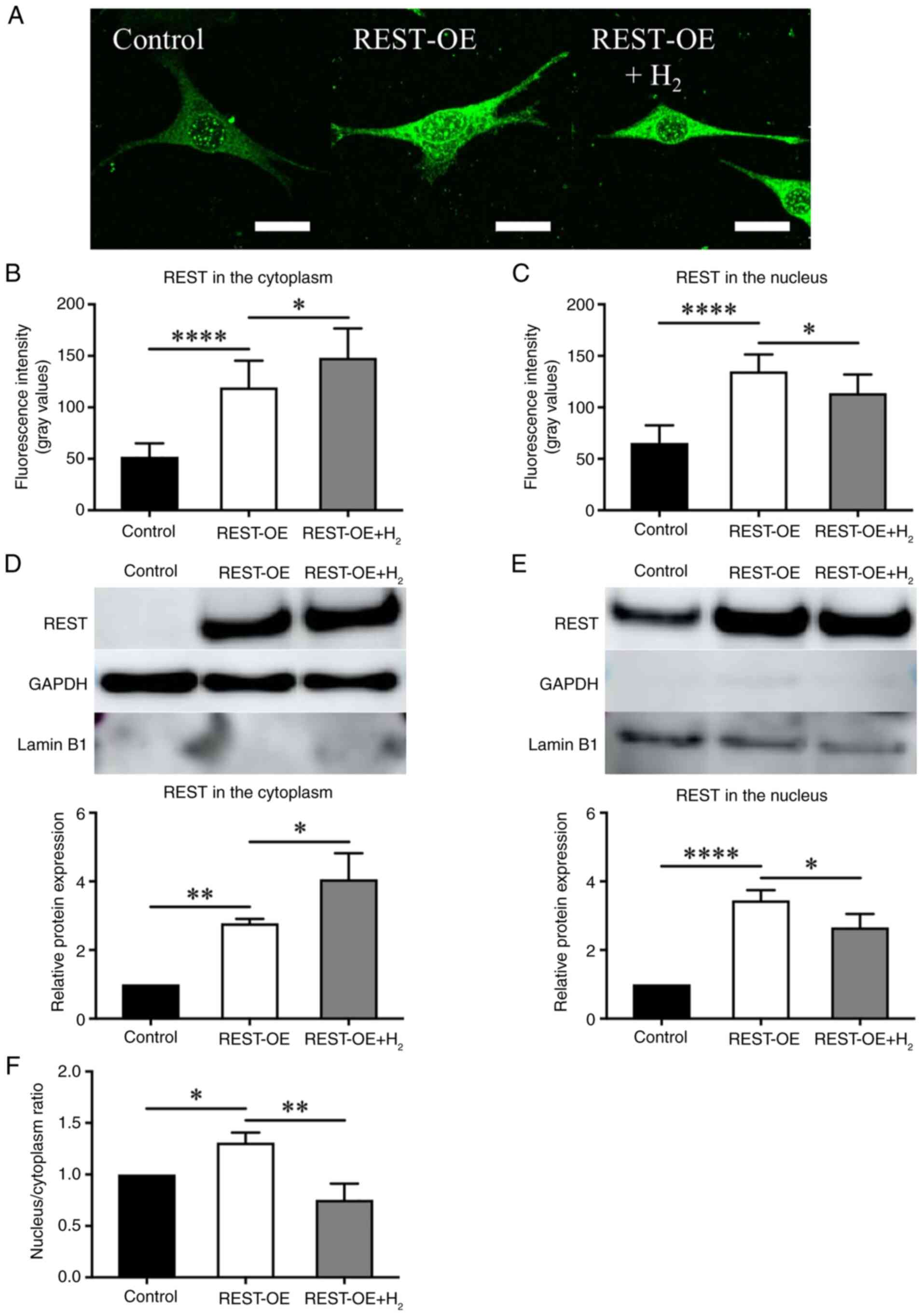
Differences in the localization of
REST expression between the cytoplasm and the nucleus
H2 reduces the intracellular expression
of REST (Fig. 2A and C);
therefore, to examine differences in the localization of REST
expression, in vitro cell lines were separated into
cytoplasmic and nuclear fractions, and REST expression was
investigated by immunofluorescence staining and western
blotting.
Immunofluorescence staining revealed that the
fluorescence intensity of REST in the cytoplasm was significantly
elevated in the REST-OE group compared to controls (52.01±12.40 in
Control vs. 119.33±24.81 in REST-OE, P<0.0001), and further
increased following H2 treatment (119.33±24.81 in
REST-OE vs. 148.08±27.25 in REST-OE + H2,
P=0.0226) (Fig. 3A and B).
On the other hand, the fluorescence intensity of REST in the
nucleus was significantly increased in the REST-OE group compared
with the Control group (Control group: 65.39±16.39; REST-OE group:
135.03±15.61, P<0.0001), whereas it was significantly decreased
in the REST-OE + H2 group compared with the REST-OE
group (REST-OE group: 135.03±15.61; REST-OE + H2 group:
113.86±17.26, P=0.0144) (Fig. 3A and
C).
Western blotting supported these findings. REST
protein levels in the cytoplasm were significantly increased in the
REST-OE group compared to controls (1.00±0.00-fold in Control vs.
2.78±0.11-fold in REST-OE, P=0.0053), and further increased
with H2 treatment (2.78±0.11-fold in REST-OE vs.
4.07±0.62-fold in REST-OE + H2, P=0.0238)
(Fig. 3D). On the other hand, REST
expression in the nucleus was significantly increased in the
REST-OE group compared with the Control group (Control group:
1.00±0.00-fold; REST-OE group: 3.45±0.24-fold, P<0.0001),
whereas it was decreased in the REST-OE + H2 group
compared with the REST-OE group (REST-OE group: 3.45±0.24-fold;
REST-OE + H2 group: 2.66±0.32-fold, P=0.0281)
(Fig. 3E). Furthermore, the ratio
of REST expression in the nucleus to that in the cytoplasm was
calculated. As a result, the REST-OE group showed a significant
increase compared to controls (Control group: 1.00±0.00-fold;
REST-OE group: 1.31±0.08-fold, P=0.0355) (Fig. 3F). In contrast, the REST-OE +
H2 group showed a significant decrease compared to the
REST-OE group (REST-OE group: 1.31±0.08-fold; REST-OE +
H2 group: 0.75±0.13-fold, P=0.0021) (Fig. 3F).
Although H2 reduces REST expression in
the whole cell (Fig. 2A and C), it
increases REST expression in the cytoplasm and decreases it in the
nucleus in REST-OE cells. This suggests that H2 affects
the degradation within the cytoplasm and nuclear transport of
REST.
REST nuclear transport proteins and
autophagy-related protein LC3 in the cytoplasm and nucleus of
REST-OE cells
REST in the cytoplasm is either degraded by
autophagy or transported into the nucleus. To investigate these
processes, REST nuclear transport proteins, such as
REST-interacting LIM domain protein (RILP), Huntingtin, and
DCTN1/p150Glued, and the autophagy-related protein,
light chain 3 protein (LC3), were quantified by western
blotting.
RILP levels were significantly higher in the REST-OE
group compared with the Control group (Control group:
1.00±0.00-fold; REST-OE group: 1.42±0.04-fold, P=0.0003);
however, it was significantly decreased following H2
treatment (1.42±0.04-fold in REST-OE vs. 1.16±0.08-fold in REST-OE
+ H2, P=0.0040) (Fig. 4A). In contrast, there was no
significant difference in the expression of Huntingtin between the
REST-OE group and the Control group (Control group: 1.00±0.00-fold;
REST-OE group: 1.17±0.18-fold, p=0.7587), as well as between the
REST-OE + H2 group and the REST-OE group (REST-OE group:
1.17±0.18-fold; REST-OE + H2 group: 1.15±0.37-fold, P=0.9970)
(Fig. 4B). There was no
significant difference in the expression of
DCTN1/p150Glued between the REST-OE group and the
Control group (Control group: 1.00±0.00-fold; REST-OE group:
0.97±0.07-fold, P=0.9126), as well as between the REST-OE +
H2 group and the REST-OE group (REST-OE group:
0.97±0.07-fold; REST-OE + H2 group: 0.95±0.09-fold,
P=0.9421) (Fig. 4C). The
expression of LC3 was significantly decreased in the REST-OE group
compared with the Control group (Control group: 1.00±0.00-fold;
REST-OE group: 0.70±0.08-fold, P<0.0001), whereas it was
significantly increased in the REST-OE + H2 group
compared with the REST-OE group (REST-OE group: 0.70±0.08-fold;
REST-OE + H2 group: 0.91±0.08-fold, P=0.0022)
(Fig. 4D).
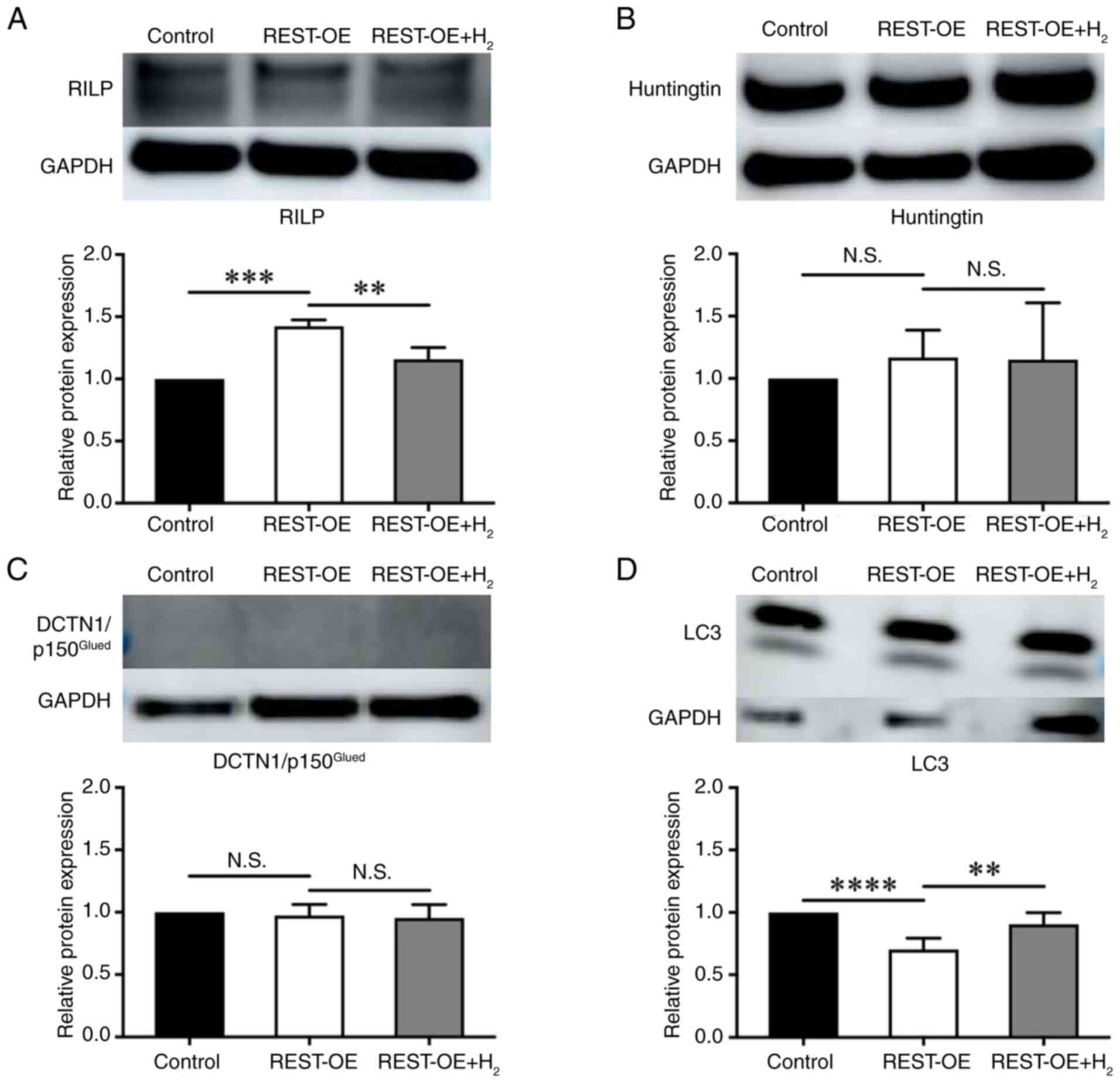 | Figure 4.RILP, Huntingtin,
DCTN1/p150Glued and LC3 expression in the control,
REST-OE and REST-OE + H2 groups evaluated by western
blotting. The REST-OE group was compared with the control group,
and the REST-OE + H2 group was compared with the REST-OE
group. The graphs show the semi-quantification of relative protein
expression. Mean ± SD, n=3 per group. **P<0.01, ***P<0.001
and ****P<0.0001 (one-way ANOVA). (A) RILP expression. (B)
Huntingtin expression. (C) DCTN1/p150Glued expression.
(D) LC3 expression. DCTN1, dynactin subunit 1; H2,
hydrogen; N.S., no significant difference; REST, repressor element
1 silencing transcription factor; REST-OE, REST-overexpressed;
RILP, REST-interacting LIM domain protein. |
To further evaluate the effects of H2 on
REST in nuclear transport and cytoplasm degradation, the expression
of RILP, Huntingtin, DCTN1/p150Glued, and LC3 in
cytoplasmic and nuclear extracts was quantified by western
blotting. In the cytoplasm, the expression of RILP in the cytoplasm
was significantly increased in the REST-OE group compared with the
Control group (Control group: 1.00±0.00-fold; REST-OE group:
1.43±0.04-fold, P<0.0001), whereas it was significantly
decreased in the REST-OE + H2 group compared with the
REST-OE group (REST-OE group: 1.43±0.04-fold; REST-OE +
H2 group: 0.97±0.06-fold, P<0.0001) (Fig. 5A). The expression of Huntingtin in
the cytoplasm was significantly increased in the REST-OE group
compared with the Control group (Control group: 1.00±0.00-fold;
REST-OE group: 1.54±0.16-fold, P=0.0105), whereas it was
significantly decreased in REST-OE + H2 group compared
with the REST-OE group (REST-OE group: 1.54±0.16-fold; REST-OE +
H2 group: 0.77±0.16-fold, P=0.0018) (Fig. 5B). The expression of
DCTN1/p150Glued in the cytoplasm was not significantly
different between the REST-OE group and the Control group (Control
group: 1.00±0.00-fold; REST-OE group: 0.92±0.07-fold, P=0.4667),
and between the REST-OE + H2 group and the REST-OE group
(REST-OE group: 0.92±0.07-fold; REST-OE + H2 group:
1.02±0.08-fold, P=0.3248) (Fig.
5C). The expression of LC3 in the cytoplasm was significantly
decreased in the REST-OE group compared with the Control group
(Control group: 1.00±0.00-fold; REST-OE group: 0.68±0.04-fold,
P=0.0017), whereas it was significantly increased in the REST-OE +
H2 group compared with the REST-OE group (REST-OE group:
0.68±0.04-fold; REST-OE + H2 group: 1.03±0.08-fold,
P=0.0011) (Fig. 5D). In the
nucleus, there was no significant difference between the expression
of RILP, Huntingtin, DCTN1/p150Glued, and LC3 between
the REST-OE group and the Control group (RILP: 1.19±0.09-fold,
P=0.0827; Huntingtin: 1.20±0.26-fold, P=0.5167;
DCTN1/p150Glued: 1.02±0.27-fold, p=0.9953; LC3:
0.98±0.10-fold, P=0.9079), as well as between the REST-OE +
H2 group and the REST-OE group (RILP: 1.10±0.08-fold,
P=0.4874; Huntingtin: 1.18±0.16-fold, P=0.9923;
DCTN/p150Glued: 0.94±0.10-fold, P=0.8998; LC3:
0.99±0.06-fold, P=0.9458) (Fig.
6A-D).
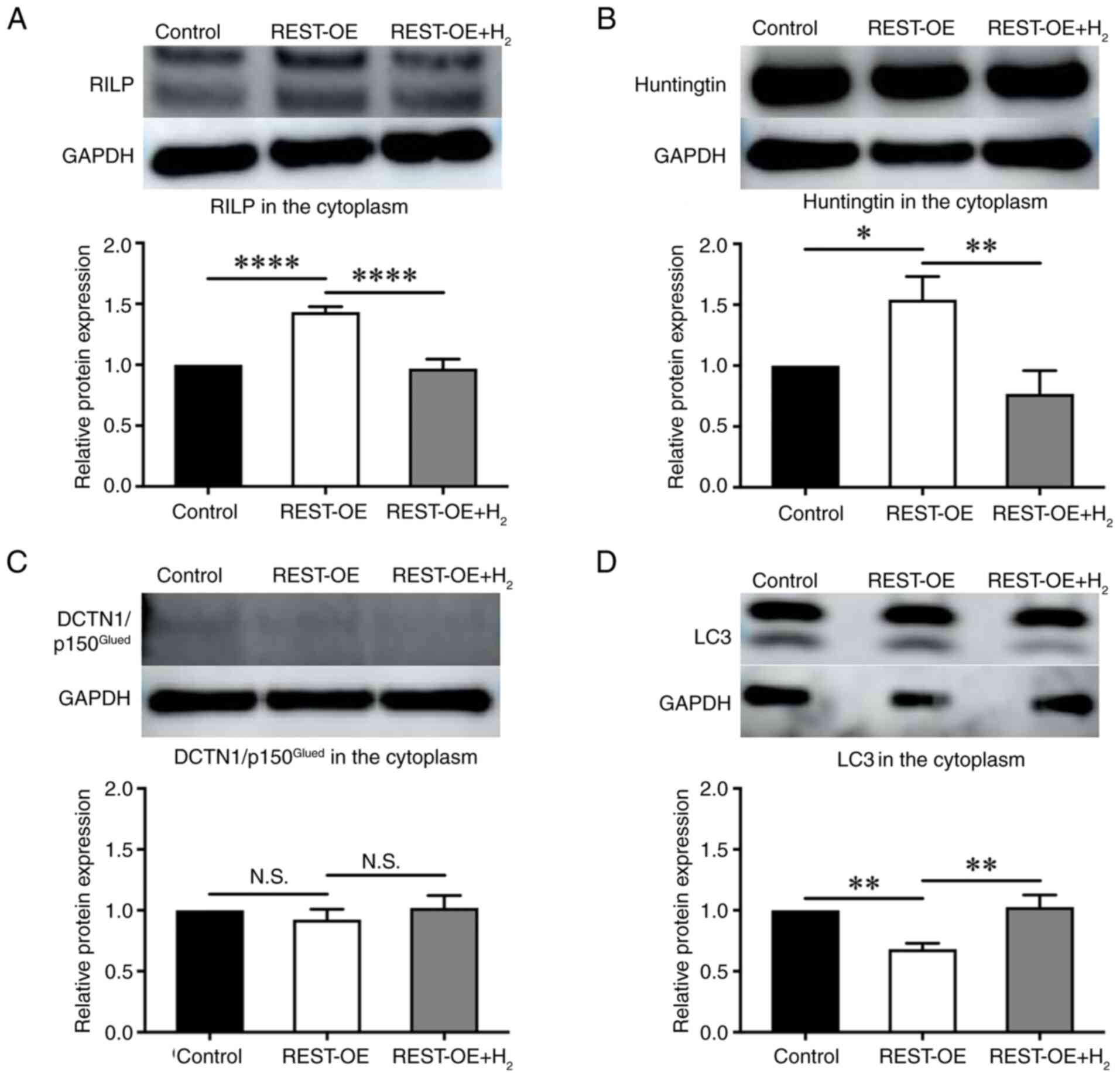 | Figure 5.RILP, Huntingtin,
DCTN1/p150Glued and LC3 expression in the cytoplasm of
the control, REST-OE and REST-OE + H2 groups evaluated
by western blotting. The REST-OE group was compared with the
control group, and the REST-OE + H2 group was compared
with the REST-OE group. The graphs show the semi-quantification of
relative protein expression. Mean ± SD, n=3 per group. *P<0.05,
**P<0.01 and ****P<0.0001 (one-way ANOVA). (A) RILP
expression. (B) Huntingtin expression. (C)
DCTN1/p150Glued expression. (D) LC3 expression. DCTN1,
dynactin subunit 1; H2, hydrogen; N.S., no significant
difference; REST, repressor element 1 silencing transcription
factor; REST-OE, REST-overexpressed; RILP, REST-interacting LIM
domain protein. |
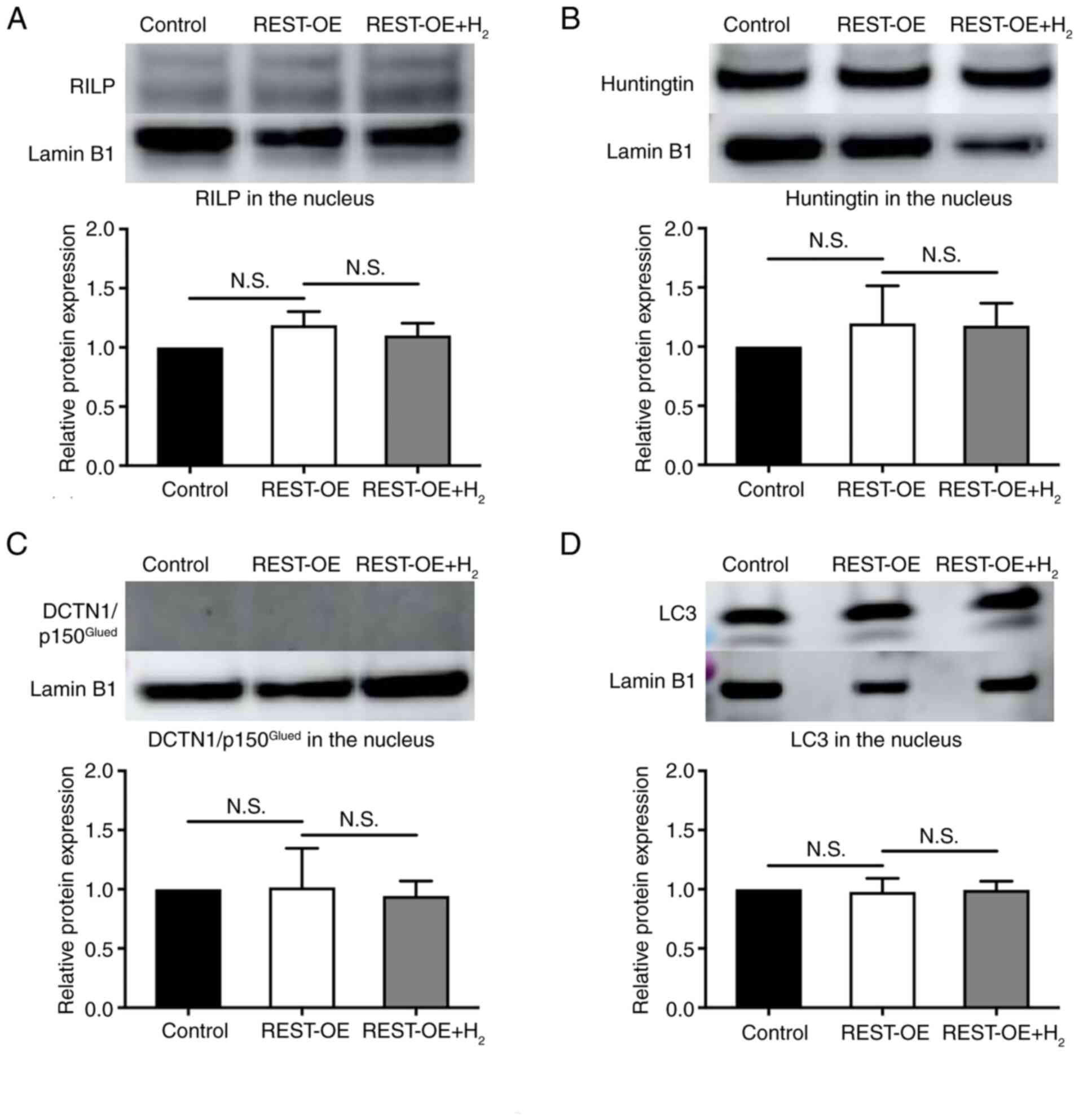 | Figure 6.RILP, Huntingtin,
DCTN1/p150Glued and LC3 expression in the nucleus of the
control, REST-OE and REST-OE + H2 groups evaluated by
western blotting. The REST-OE group was compared with the control
group, and the REST-OE + H2 group was compared with the
REST-OE group. The graphs show the semi-quantification of relative
protein expression. Mean ± SD, n=3 per group. The data were
analyzed using one-way ANOVA. (A) RILP expression. (B) Huntingtin
expression. (C) DCTN1/p150Glued expression. (D) LC3
expression. DCTN1, dynactin subunit 1; H2, hydrogen;
N.S., no significant difference; REST, repressor element 1
silencing transcription factor; REST-OE, REST-overexpressed; RILP,
REST-interacting LIM domain protein. |
Together these results suggest that under REST-OE
conditions, REST nuclear transport proteins RILP and Huntingtin
were upregulated but autophagy was suppressed in the cytoplasm,
which promotes the nuclear transport of REST from the cytoplasm. On
the other hand, H2 inhibited the nuclear transport of
REST due to suppression of REST nuclear transport-related proteins
such as RILP and Huntingtin.
Discussion
In this study, REST expression was decreased, and
GAP43 expression was increased following H2
administration to aged animal models and REST-OE cells (Figs. 1 and 2). Next, we examined the intracellular
localization of REST expression. We found that REST expression was
increased in the cytoplasm and decreased in the nucleus following
H2 addition in REST-OE cells (Fig. 3). Furthermore, the REST expression
in the cytoplasm was investigated, focusing on nuclear transport
mechanisms and autophagy. We found that H2 addition led
to a decrease in several REST nuclear transport proteins, and
nuclear translocation of REST was suppressed (Fig. 5A-C). H2 administration
restored the impaired autophagic capacity (Fig. 5D).
Various types of stress promote nuclear transport of
proteins from the cytoplasm to the nucleus. Hikeshi, which is not a
member of the importin family, is increased during heat shock
stress and promotes the nuclear transport of heat shock protein 70s
(19). On the other hand, in
vitro studies using human bronchial epithelial-like cells found
that the antioxidant vitamin E inhibits the nuclear transport of
phospho-signal transducer and activator of transcription 3 and
suppresses the induction of inflammatory cells (20). These findings suggest that the
nuclear transport of transcriptional factors depends on biological
stress. Previous studies reported that H2 has a
neuroprotective effect through antioxidant ability (17). Furthermore, it may affect nuclear
transport of transcriptional factors via its antioxidant ability.
Since both cytoplasmic and nuclear expression of REST were changed
by H2 administration in this study, our findings suggest
that H2 influences the nuclear transport of REST.
Moreover, H2 administration led to an increase in the
expression of axon regeneration markers through suppression of the
nuclear transport of REST. Therefore, nuclear transport of REST was
investigated through a molecular approach.
REST is transported into the nucleus by forming a
complex with RILP, Huntingtin, and DCTN1/p150Glued
(21). In this study, under high
REST expression conditions, the expression of RILP and Huntingtin
in the cytoplasm were increased, promoting the nuclear transport of
REST (Fig. 5A and B). On the other
hand, the expression of RILP and Huntingtin was decreased, and the
nuclear transport of REST was suppressed by H2 addition
(Fig. 5A and B). It is reported
that RILP is activated by reactive oxygen species (ROS) and
inhibited by the antioxidant N-acetylcysteine (22,23).
Melatonin is an antioxidant that provides protective effects
against Huntington's disease (24). Additionally, melatonin decreases
with aging, leading to a decrease in antioxidant activity and an
increase in the expression of Huntingtin (24). It is likely that the antioxidant
ability of H2 led to a decrease in the expression of
RILP and Huntingtin. This study reveals new findings about the
regulatory roles of RILP and Huntingtin in the nuclear transport of
REST. On the other hand, among the REST nuclear transport proteins,
only DCTN1/p150Glued in the cytoplasm remained
unaffected by changes in REST expression.
DCTN1/p150Glued has other functions than the nuclear
transport of REST, such as retrograde axonal transport, and
microtubule stabilization (25,26).
Therefore, the change in the expression of
DCTN1/p150Glued in the cytoplasm was not detectable,
although the nuclear transport of REST was suppressed.
Autophagy is an important process for maintaining
cellular homeostasis (27);
however, its activity decreases with aging (28,29).
Furthermore, REST expression in neurons increases with age, leading
to decreased autophagy activity (30). On the other hand, myocardial
protective effects of H2 on sepsis-induced
cardiomyopathy via autophagy activation (31). Thus, H2 may have the
capacity to promote autophagy activity according to these reports.
In this study, the expression of LC3 in both whole-cell and
cytoplasmic fractions are increased in REST-OE cells by
H2 addition (Figs. 4D
and 5D). Therefore,
H2-induced changes in autophagy activity in the
cytoplasm may be upregulated.
This study has some limitations. First, a
comprehensive analysis of the nuclear transport proteins related to
REST, which have various functions in neurons, was not performed.
RILP, Huntingtin, and DCTN1/p150Glued were selected as
they form a complex with REST and that protein complex plays
essential roles in the nuclear transport of REST (21). Second, the optimal duration of
H2 administration and addition was not evaluated. The
primary methods of H2 administration include inhalation
of H2 gas, drinking HRW, and injection of
H2-containing physiological saline (32). In this study, the duration of
intraperitoneal injection of H2-containing physiological
saline was determined based on a study evaluating autophagy in a
rat neurodegeneration model (33).
Additionally, in the in vitro study using H9C2 cells in a
hypoxia-reoxygenation model, the H2 administration
period was set to 2 h based on a previous study that assessed cell
survival (34). Third, REST-OE
cells used in this study were generated by fibroblasts, which are
non-neuronal cells. Although Schwann cells were initially
considered due to their physiological relevance to the peripheral
nervous system, stable overexpression of REST in these cells was
not achieved. This limitation was likely attributable to a
combination of REST-induced growth inhibition and cytotoxic effects
associated with Lipofectamine 3000-based transfection.
Consequently, fibroblasts were selected as an alternative model due
to their genetic stability and widespread use in studies of
nuclear-cytoplasmic transport mechanisms. Although primary cultured
neurons can be maintained for a limited period, their application
in long-term experiments is constrained. In fact, Xue et al
(30) considered a 10-day culture
duration as ‘long-term’ in studies using neuronal cell.
Furthermore, gene delivery efficiency in primary neurons is
generally low, and specialized transfection methods are often
required (35). On the other hand,
a previous study reported the use of NIH3T3 cells, the cells used
in the present study, in peripheral nerve research (36). Furthermore, the nuclear transport
mechanism of megakaryoplastic leukemia 1 protein, a co-activator of
the transcription factor serum response factor, has been analyzed
using the NIH3T3 cell line (37).
Thus, the use of NIH3T3 cells is considered scientifically valid
for studying the nuclear transport mechanisms of cytoplasmic
proteins. Nevertheless, this approach has a limitation in
physiological relevance, as fibroblasts differ from neuronal cells
and cannot replicate neuron-specific functions or the neural
microenvironment (38).
Consequently, the current findings may not fully reflect the in
vivo role of REST. To address this limitation, future studies
are planned using peripheral nerve-specific conditional REST
knockout mice. In vivo axonal regeneration will be evaluated
using a sciatic nerve crush model, and axon regeneration will be
assessed in primary cultured dorsal root ganglion neurons. These
approaches are expected to complement the current in vitro
results and provide a more comprehensive understanding of the
physiological role of REST in peripheral nerve regeneration.
Previous studies reported that axon regenerative
capacity declines with aging, however, the underlying pathology
remains unclear. In this study, to elucidate the pathology of
age-related decline in axonal regeneration, the molecular
mechanisms involved in REST nuclear transport were analyzed. The
results of this study suggest that the regulation of nuclear
transport of REST may improves the decline in age-related axon
regeneration.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Japan Society of the
Promotion of Science KAKENHI (grant no. 22K09342).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
KN, DK and YU conceptualized the study. TS, TNK and
YY designed the methodology. TS performed most of the experiments.
SK, NI and KK assisted with the experiments. TS, KN, NH and MI
analyzed and interpreted the data. TS and KN prepared the original
draft, while TS, KN and MI reviewed and edited the manuscript. KN
secured funding. TS, KN, DK and NH confirm the authenticity of all
the raw data. All authors have read and approved the final version
of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Animal Care
Committee of Juntendo University, Tokyo, Japan (registration no.
1555; approval no. 2024183; date of approval, March 12, 2024).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Murayama M, Hirata H, Shiraki M, Iovanna
JL, Yamaza T, Kukita T, Komori T, Moriishi T, Ueno M, Morimoto T,
et al: Nupr1 deficiency downregulates HtrA1, enhances SMAD1
signaling, and suppresses age-related bone loss in male mice. J
Cell Physiol. 238:566–581. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ke HZ, Richards WG, Li X and Ominsky MS:
Sclerostin and Dickkopf-1 as therapeutic targets in bone diseases.
Endocr Rev. 33:747–783. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Creecy A, Brown KL, Rose KL, Voziyan P and
Nyman JS: Post-translational modifications in collagen type I of
bone in a mouse model of aging. Bone. 143:1157632021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang TL, Shen H, Liu A, Dong SS, Zhang L,
Deng FY, Zhao Q and Deng HW: A road map for understanding molecular
and genetic determinants of osteoporosis. Nat Rev Endocrinol.
16:91–103. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rim YA, Nam Y and Ju JH: The role of
chondrocyte hypertrophy and senescence in osteoarthritis initiation
and progression. Int J Mol Sci. 21:23582020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fujii Y, Liu L, Yagasaki L, Inotsume M,
Chiba T and Asahara H: Cartilage homeostasis and osteoarthritis.
Int J Mol Sci. 23:63162022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Verdú E, Ceballos D, Vilches JJ and
Navarro X: Influence of aging on peripheral nerve function and
regeneration. J Peripher Nerv Syst. 5:191–208. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mampay M and Sheridan GK: REST: An
epigenetic regulator of neuronal stress responses in the young and
ageing brain. Front Neuroendocrinol. 53:1007442019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Goto K, Naito K, Nakamura S, Nagura N,
Sugiyama Y, Obata H, Kaneko A and Kaneko K: Protective mechanism
against age-associated changes in the peripheral nerves. Life Sci.
253:1177442020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kaneko A, Naito K, Nakamura S, Miyahara K,
Goto K, Obata H, Nagura N, Sugiyama Y and Kaneko K: Influence of
aging on the peripheral nerve repair process using an artificial
nerve conduit. Exp Ther Med. 21:1682021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rodriguez-Bravo V, Pippa R, Song WM,
Carceles-Cordon M, Dominguez-Andres A, Fujiwara N, Woo J, Koh AP,
Ertel A, Lokareddy RK, et al: Nuclear pores promote lethal prostate
cancer by increasing POM121-driven E2F1, MYC, and AR nuclear
import. Cell. 174:1200–1215.e20. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kawamura M, Sato S, Matsumoto G, Fukuda T,
Shiba-Fukushima K, Noda S, Takanashi M, Mori N and Hattori N: Loss
of nuclear REST/NRSF in aged-dopaminergic neurons in Parkinson's
disease patients. Neurosci Lett. 699:59–63. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lu T, Aron L, Zullo J, Pan Y, Kim H, Chen
Y, Yang TH, Kim HM, Drake D, Liu XS, et al: REST and stress
resistance in ageing and Alzheimer's disease. Nature. 507:448–454.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mir AR and Moinuddin Islam S: Circulating
autoantibodies in cancer patients have high specificity for
glycoxidation modified histone H2A. Clin Chim Acta. 453:48–55.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nagata K, Nakashima-Kamimura N, Mikami T,
Ohsawa I and Ohta S: Consumption of molecular hydrogen prevents the
stress-induced impairments in hippocampus-dependent learning tasks
during chronic physical restraint in mice. Neuropsychopharmacology.
34:501–508. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang J, Zhang XJ, Du YY, Shi G, Zhang CC
and Chen R: Hydrogen-rich saline promotes neuronal recovery in mice
with cerebral ischemia through the AMPK/mTOR signal-mediated
autophagy pathway. Acta Neurobiol Exp (Wars). 83:317–330. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Han XC, Ye ZH, Hu HJ, Sun Q and Fan DF:
Hydrogen exerts neuroprotective effects by inhibiting oxidative
stress in experimental diabetic peripheral neuropathy rats. Med Gas
Res. 13:72–77. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2 (−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kose S, Furuta M and Imamoto N: Hikeshi, a
nuclear import carrier for Hsp70s, protects cells from heat
shock-induced nuclear damage. Cell. 149:578–589. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhao H, Gong J, Li L, Zhi S, Yang G, Li P,
Li R and Li J: Vitamin E relieves chronic obstructive pulmonary
disease by inhibiting COX2-mediated p-STAT3 nuclear translocation
through the EGFR/MAPK signaling pathway. Lab Invest. 102:272–280.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shimojo M: Huntingtin regulates
RE1-silencing transcription factor/neuron-restrictive silencer
factor (REST/NRSF) nuclear trafficking indirectly through a complex
with REST/NRSF-interacting LIM domain protein (RILP) and dynactin
p150 Glued. J Biol Chem. 283:34880–34886. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Daulat AM, Wagner MS, Audebert S,
Kowalczewska M, Ariey-Bonnet J, Finetti P, Bertucci F, Camoin L and
Borg JP: The serine/threonine kinase MINK1 directly regulates the
function of promigratory proteins. J Cell Sci. 135:jcs2593472022.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fu G, Xu Q, Qiu Y, Jin X, Xu T, Dong S,
Wang J, Ke Y, Hu H, Cao X, et al: Suppression of Th17 cell
differentiation by misshapen/NIK-related kinase MINK1. J Exp Med.
214:1453–1469. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kim J, Li W, Wang J, Baranov SV, Heath BE,
Jia J, Suofu Y, Baranova OV, Wang X, Larkin TM, et al: Biosynthesis
of neuroprotective melatonin is dysregulated in Huntington's
disease. J Pineal Res. 75:e129092023. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Moughamian AJ and Holzbaur EL: Dynactin is
required for transport initiation from the distal axon. Neuron.
74:331–343. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lazarus JE, Moughamian AJ, Tokito MK and
Holzbaur EL: Dynactin subunit p150(Glued) is a neuron-specific
anti-catastrophe factor. PLoS Biol. 11:e10016112013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mizushima N and Komatsu M: Autophagy:
Renovation of cells and tissues. Cell. 147:728–741. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Aman Y, Schmauck-Medina T, Hansen M,
Morimoto RI, Simon AK, Bjedov I, Palikaras K, Simonsen A, Johansen
T, Tavernarakis N, et al: Autophagy in healthy aging and disease.
Nat Aging. 1:634–650. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kaushik S, Tasset I, Arias E, Pampliega O,
Wong E, Martinez-Vicente M and Cuervo AM: Autophagy and the
hallmarks of aging. Ageing Res Rev. 72:1014682021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xue WJ, He CF, Zhou RY, Xu XD, Xiang LX,
Wang JT, Wang XR, Zhou HG and Guo JC: High glucose and palmitic
acid induces neuronal senescence by NRSF/REST elevation and the
subsequent mTOR-related autophagy suppression. Mol Brain.
15:612022. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cui Y, Li Y, Meng S, Song Y and Xie K:
Molecular hydrogen attenuates sepsis-induced cardiomyopathy in mice
by promoting autophagy. BMC Anesthesiol. 24:722024. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Iketani M and Ohsawa I: Molecular hydrogen
as a neuroprotective agent. Curr Neuropharmacol. 15:324–331. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang H, Huo X, Chen H, Li B, Liu J, Ma W,
Wang X, Xie K, Yu Y and Shi K: Hydrogen-rich saline activated
autophagy via HIF-1α pathways in neuropathic pain model. Biomed Res
Int. 2018:46708342018.PubMed/NCBI
|
|
34
|
Song D, Liu X, Diao Y, Sun Y, Gao G, Zhang
T, Chen K and Pei L: Hydrogen-rich solution against myocardial
injury and aquaporin expression via the PI3K/Akt signaling pathway
during cardiopulmonary bypass in rats. Mol Med Rep. 18:1925–1938.
2018.PubMed/NCBI
|
|
35
|
Palomino SM, Gabriel KA, Mwirigi JM,
Cervantes A, Horton P, Funk G, Moutal A, Martin LF, Khanna R, Price
TJ and Patwardhan A: Genetic editing of primary human dorsal root
ganglion neurons using CRISPR-Cas9. Sci Rep. 15:111162025.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tanaka H, Yamashita T, Asada M, Mizutani
S, Yoshikawa H and Tohyama M: Cytoplasmic p21(Cip1/WAF1) regulates
neurite remodeling by inhibiting Rho-kinase activity. J Cell Biol.
158:321–329. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Rajakylä EK, Viita T, Kyheröinen S, Huet
G, Treisman R and Vartiainen MK: RNA export factor Ddx19 is
required for nuclear import of the SRF coactivator MKL1. Nat
Commun. 6:59782015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Vierbuchen T, Ostermeier A, Pang ZP,
Kokubu Y, Südhof TC and Wernig M: Direct conversion of fibroblasts
to functional neurons by defined factors. Nature. 463:1035–1041.
2010. View Article : Google Scholar : PubMed/NCBI
|