Cardiovascular diseases (CVDs), including heart
failure (HF), hypertension, coronary heart disease (CHD), vascular
calcification and others, are among the primary causes of health
deterioration in humans worldwide. The Global Health Assessment
2019 report by the World Health Organization (1) indicated that heart disease has
remained the leading cause of mortality globally for the past two
decades. Improving the prognosis and course of treatment for CVDs
requires early detection and diagnosis. Every year, there is an
increase in the prevalence of CVDs despite advancements in therapy
for controlling traditional risk factors. Furthermore, the number
of CVD-associated mortalities is predicted to increase to ~23.6
million by 2030 as a result of aging and changing lifestyles
(2,3). Thus, in-depth studies to broaden the
current understanding of CVDs mechanisms, diagnosis and treatment
are imperative.
Research on exosomes is part of a newly emerged
field, which has attracted great attention due to their role in
regulating intercellular communication. Exosomes are a subtype of
extracellular vesicles with a typical size of ~100 nm, which can
carry a range of lipids, carbohydrates, nucleic acids and proteins
(4). Numerous CVDs are directly
impacted by different circular RNAs (circRNAs), long noncoding RNAs
(lncRNAs), messenger RNAs, short nucleolar RNAs and microRNAs
(miRNA) transported by exosomes. Due to these properties, exosomes
have also been proposed as potential diagnostic and/or prognostic
biomarkers for CVD, as well as novel potential therapeutic
modalities (5). Epigenetics
regulates the function and expression levels of CVD-associated
genes primarily through ncRNA regulation and DNA methylation,
thereby influencing the progression of CVD (6). Exosome-mediated epigenetic regulation
participates in the interaction between blood vessels and
circulating cells, as well as in intercellular communication
processes, and exosomes serve as biomarkers of cell activation
(7).
The aim of the present study was to examine the
molecular mechanism behind exosome-mediated epigenetic regulation
in the development of CVDs, and to explore the potential roles of
exosome-mediated epigenetic regulation in the diagnosis and
treatment of CVDs.
Epigenetics is considered chromosomal changes that
lead to stable genetic phenotypes in the absence of changes in the
DNA sequence (8). Epigenetic
markers are typically present in the first stages of CVD, and are
valuable molecular markers for the diagnosis, evaluation and
prediction of CVD response to treatment (9). Controlling these changed genes and
proteins could also provide a novel focus for the management of
heart disease, which is of considerable relevance for certain
diseases that are becoming more common as the population ages and
are yet challenging to treat clinically. This is because epigenetic
modifications are reversible. In CVDs, through DNA methylation and
ncRNA regulation, epigenetics primarily controls the function and
expression level of associated genes, thus influencing the
development of CVD (6,10).
Exosomes participate in intercellular communication
among cells via direct communication with neighboring cells or via
secretion of soluble factors under physiological and pathological
conditions, which can extend the interaction over long distances
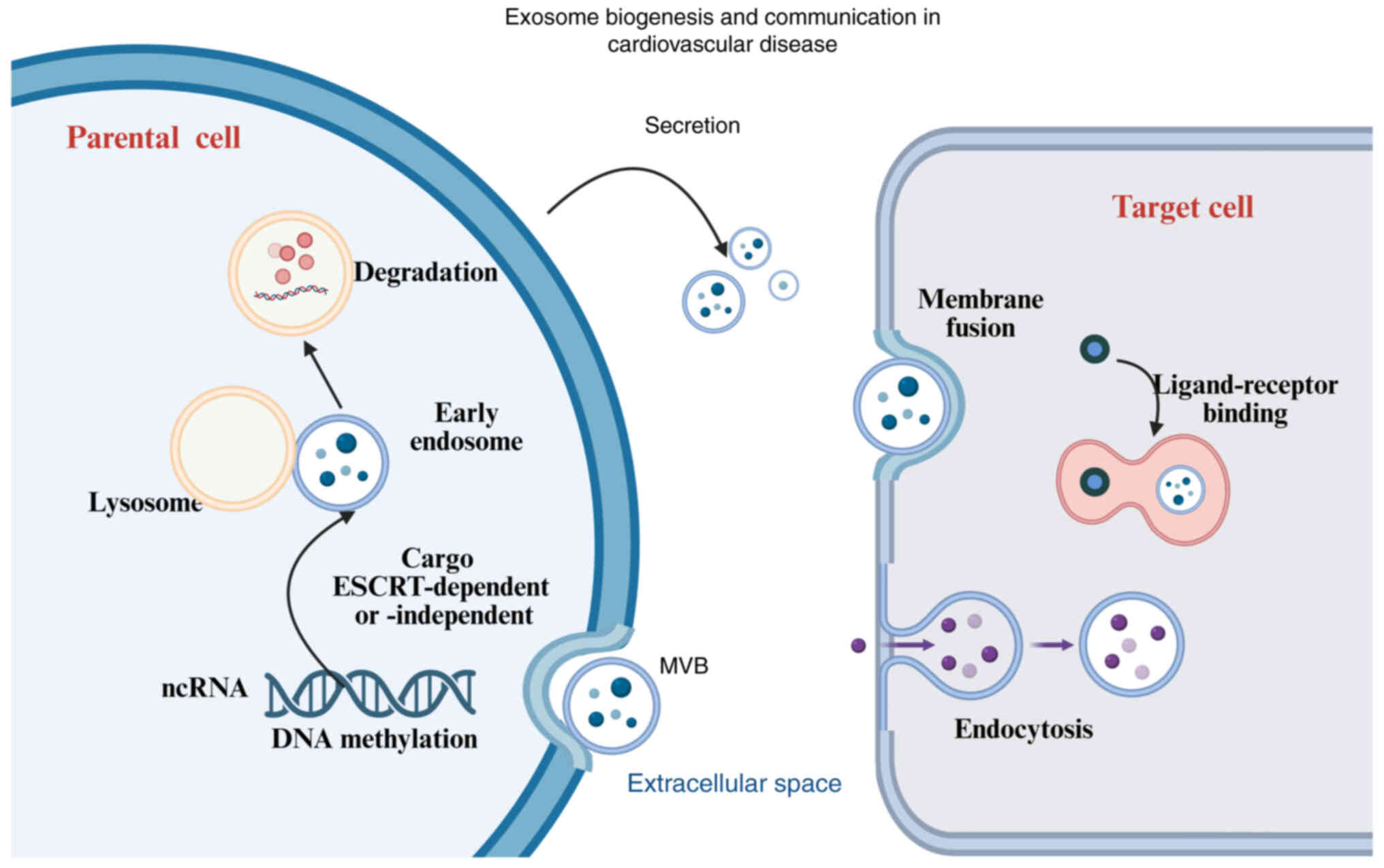
(11,12). Exosome formation involves a number
of processes, including synthesis of intraluminal vesicles,
secretion and fusion with target cells, and cargo loading. The
process of exosomal biogenesis commences with endosomal membrane
invagination, resulting in the formation of multivesicular bodies
(MVBs) in vivo (13).
During this step, MVBs integrate proteins and cytoplasmic nucleic
acids (14). On the one hand, when
MVBs fuse with lysosomes, the encapsulated contents are degraded.
When endosomes fuse with parental cell membranes, they are secreted
in exosomes (15), as illustrated
in Fig. 1. Since endosomes are the
result of plasma membrane budding, the membrane protein orientation
of the exosomes generated by this second invagination process is
identical to that of the parental cells (16). Cargo sorting to exosomes is
generally handled by endosomal sorting complexes required for
transport (ESCRT) or sphingolipids (ESCRT-dependent or
ESCRT-independent pathways). Cargo, such as proteins and RNA, can
remain in the extracellular space unbroken if it is not part of the
exosome lipid bilayer. Depending on the source, target and
environment, target cells absorb exosomes through a variety of
methods, such as membrane fusion, ligand receptor binding and
endocytosis. Exosomes can enter adjacent target cells and bodily
fluids, and even reach distant cells once they are secreted
(8,9). This diagram illustrates the process
of exosome biogenesis, including endosomal invagination,
multivesicular body formation, cargo sorting, and exosome release.
Exosomes facilitate intercellular communication by transferring
genetic material (such as ncRNAs) to target cells through fusion,
receptor binding or endocytosis. This process plays a key role in
regulating gene expression and epigenetic changes in CVDs.
LncRNAs, circRNAs and miRNAs are examples of ncRNAs,
which are generally not translated into proteins but function
through the regulation of gene expression. Instead, they carry out
their specific biological roles at the RNA level (6). MiRNAs are ~22 nucleotides in length,
and are mainly transcribed by large primary miRNAs that are
involved in the post-transcriptional regulation of gene expression,
and are produced by RNA polymerase II. Pri-miRNAs comprise one or
more hairpin (stem-loop) structures, each of which is consists of
~70 nucleotides (17). In addition
to being the targets of epigenetic changes such as DNA methylation,
miRNAs also function as histone deacetylase and DNA
methyltransferase (DNMT) regulators (18). LncRNAs are noncoding RNAs that
control gene expression patterns by modifying the accessibility of
DNA and the structure of chromatin via molecular mechanisms such as
scaffolding, decoy, guiding and signal transduction (19). CircRNAs are long, noncoding
endogenous RNA molecules that have no 5′cap or 3′cluster (A) end,
and have single-stranded covalently closed RNA rings that can be
used as templates for protein synthesis. CircRNAs interact with
miRNAs to regulate target gene expression and mRNA translation, and
bind to functional proteins or RNA-binding proteins in order to
control their transport and function (20,21),
as illustrated in Table I.
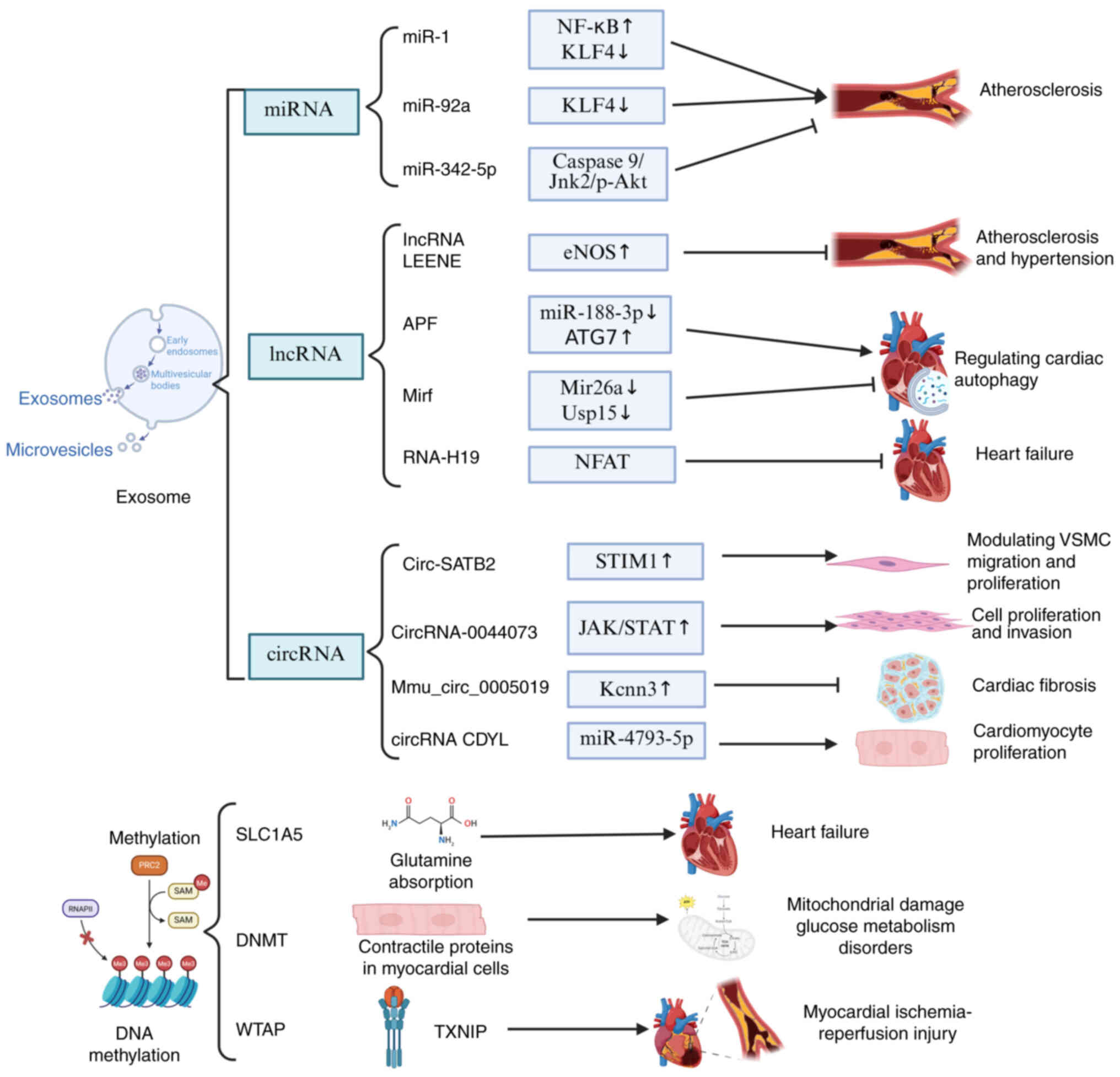
The role of ncRNAs in CVDs has attracted great
attention, particularly due to their potential in early disease
detection and prevention. MiRNAs, lncRNAs and circRNAs offer new
perspectives for understanding the progression of CVDs. Notably,
lncRNAs can exert both protective and detrimental effects in CVD,
and this dual regulatory role makes them a key focus on
cardiovascular research, as illustrated in Fig. 2. First, miRNAs act as essential
regulatory factors in diseases such as coronary heart disease,
myocardial infarction and vascular calcification (22,23).
Furthermore, miRNAs also play a critical role in atherosclerosis.
In endothelial cells (ECs), miR-342-5p exerts an
anti-atherosclerotic effect by suppressing inflammatory responses
(24), whereas miR-92a promotes
the progression of atherosclerosis (25). Similarly, in pulmonary arterial
hypertension, miR-181a-5p and miR-324-5p confer protective effects
by inhibiting pulmonary vascular remodeling, thereby slowing
disease progression (26). Similar
to miRNAs, lncRNAs in CVDs can exert both protective effects and
promote disease progression. For instance, lncRNA LEENE has been
shown to inhibit atherosclerosis in vitro through its
interaction with miR-1 (23,27).
In addition, specific lncRNAs, including APF, CAIF and Mirf, have
been demonstrated to regulate cardiac autophagy, which may play a
critical role during myocardial infarction-induced injury (28,29).
Additionally, lncRNAs RNA-H19, LIPCAR and COL1A1 have been proposed
as biomarkers for risk assessment and prediction of HF (30–32).
Furthermore, circRNAs, as novel regulatory molecules, also play an
important role in CVDs. For example, circRNAs can modulate the
migration and proliferation of vascular smooth muscle cells (VSMCs)
by interacting with miRNAs such as miRNA-939 and miRNA-221, thereby
contributing to the development of atherosclerosis (33,34).
In cardiomyocytes, circRNAs also play a crucial role, and are
considered novel regulatory elements involved in hypertrophy,
fibrosis, autophagy and apoptosis (35). Among them, mmu_circ_0005019 and
circRNA CDYL have been identified as key regulators in the
progression of HF (36,37). Notably, the overexpression of
certain miRNAs may also lead to adverse effects. For example,
excessive levels of miRNA-765, miRNA-483-3p and miRNA-143/145 can
disrupt the renin-angiotensin system and increase the blood
pressure (38). Similarly,
circACTA2 interacts with miRNA-548F-5p, which targets the
3′-untranslated region of α-SMA mRNA to promote VSMC contraction
and regulate vascular tone, thereby contributing to the development
of hypertension (39). In summary,
miRNAs, lncRNAs and circRNAs play multifaceted roles in CVDs.
In-depth research on these RNA molecules may offer novel insights
and potential biomarkers for the early diagnosis, treatment and
prevention of CVDs.
miRNAs are the most abundant ncRNAs packaged within
exosomes. Exosomal miRNAs derived from the heart and vascular
system exhibit distinct expression patterns under various
pathophysiological conditions. Their abundance and composition
depend not only on the state of CVD but also on specific stimuli
(40). The incorporation of miRNAs
into exosomes relies on distinct selective packaging mechanisms, a
process that is critical for their functional roles within the
cardiovascular system (41).
Previous studies in mouse models have further advanced the
understanding of the role of miRNAs in cardiac development.
Notably, cardiac-specific downregulation of the Dicer gene markedly
disrupts miRNA synthesis in neonatal mice, resulting in atrial
hypertrophy, cardiac dysfunction, ventricular remodeling and
dilation, and ultimately early mortality (42,43).
These findings underscore the critical importance of miRNAs in
cardiac development and functional regulation. Among the numerous
cardiac-specific miRNAs (44),
miR-1 is particularly abundant, representing ~24% of the total
miRNA population in the human heart and exhibiting pronounced
enrichment in cardiac tissue (45). The miR-1 sequence is composed of
two almost identical transcripts known as miR-1-1 and miR-1-2.
While the loss of both transcripts results in mortality, the
deletion of any of these transcripts causes defective cardiomyocyte
differentiation and impaired cardiac electrical conduction
(42,46). These findings indicate that miR-1
plays an indispensable role in normal cardiac development and
functional maintenance. However, dysregulated expression of miRNAs,
particularly abnormal levels within exosomes, may contribute to the
onset of CVDs. For instance, excessive expression of certain
miRNAs, such as miR-195 and miR-208, in vitro can lead to
concentric ventricular hypertrophy and the development of heart
failure (47,48). Therefore, measuring circulating
miRNA levels and identifying specific exosomal miRNA signatures may
provide critical information for the early diagnosis, monitoring
and treatment of CVDs. Dysregulation of specific miRNAs, such as
upregulation of miR-21 and miR-155 (49,50),
or downregulation of miR-126 (51), is closely associated with the
progression of cardiac atherosclerosis, and these miRNAs hold
promise as potential biomarkers for cardiac defects, as summarized
in Table I.
DNA methylation is a common modification in
eukaryotic cells, which, by controlling DNA methyltransferases
(DNMT), can transfer genetic information to the DNA of the
offspring (52). Accumulating
evidence has suggested that aberrant methylation status of
candidate genes, such as AHRR (53), F2RL3 (54) and CDKN2B-AS1 (55), can be used as indicators to assess
the progression of cardiovascular disorders, as illustrated in
Fig. 2. The expression of
potential genes linked to CHD, HF hypertension and other CVDs has
been reported to be strongly correlated with DNA methylation
(56), as illustrated in Table II.
Specifically, alterations at multiple gene
methylation sites have been demonstrated to be strongly associated
with CVDs. Previous studies have identified that three DNA
methylation areas, including the SLC9A1 (57), SLC1A5 (58) and TNRC6C genes (59), are associated with CVDs. Among
them, the methylation of one CpG (CG22304262) in SLC1A5 is strongly
associated with incident CHD, and shows significant associations
with diabetes, blood pressure and all-cause mortality (60–62).
The primary role of SLC1A5 in the heart is to regulate glutamine
uptake. However, in patients with HF, reduced SLC1A5 expression
leads to impaired glutamine absorption, resulting in disrupted
cardiac glutamine storage and an imbalance in glutamine
homeostasis, which ultimately compromises myocardial function and
health (63). In addition to
SLC1A5 methylation, previous epigenomic studies revealed broader
associations between differential DNA methylation patterns and
atherosclerosis as well as CHD (64). For instance, specific loci such as
AHRR (cg05575921) and F2RL3 (cg03636183) in blood-derived DNA have
been linked to both smoking exposure and increased risk of incident
CHD (65,66). Similarly, epigenome-wide studies
have identified methylation signatures related to chronic
inflammation and lipid metabolism that correlate with
atherosclerosis progression (67,68).
Importantly, these epigenetic markers (e.g., methylation of AHRR
and LINE-1 elements) have been shown to provide greater predictive
value for CHD than traditional risk factors alone (69). Additionally, alterations in the
methylation levels of genes associated with acute myocardial
infarction (AMI), such as Csf1r, Ptpn6, Map3k14 and Col6a1, further
support the mechanistic role of DNA methylation in cardiovascular
pathophysiology (70). Previous
studies have shown that upregulation of thioredoxin-interacting
protein is associated with myocardial ischemia/reperfusion (MI/R)
injury (71), potentially through
aberrant activation of the m6A methyltransferase complex component
WTAP (72). In vivo
experiments further demonstrated that exosome-mediated delivery of
WTAP small interfering RNA effectively alleviated MI/R injury
(73).
Furthermore, DNA methylation mediated by DNMT3a
plays a critical role in maintaining the structural and metabolic
homeostasis of human cardiomyocytes (74), thus underscoring its relevance in
CVDs. Previous research has shown that DNMT3a deficiency alters the
expression of contractile protein genes in cardiomyocytes, leading
to mitochondrial damage and glucose metabolism disorders (75), which may represent a critical
therapeutic target for cardiac diseases. Furthermore, extracellular
vesicles in the circulation of patients with acute coronary
syndrome are enriched in DNMTs, particularly showing markedly
increased expression of the de novo methyltransferases
DNMT3A and DNMT3B. These enzymes can modulate the methylome and
signaling pathways of recipient cells, thereby contributing to CVDs
(76). Further in vitro
studies revealed that downregulation of DNMTs not only participates
in the physiological regulation of cardiomyocytes but also plays a
crucial role in pathological conditions such as cardiac hypertrophy
(77). Notably, the DNMT inhibitor
RG108 has been shown to attenuate pressure overload-induced cardiac
hypertrophy in animal models, possibly by modulating epigenetic
mechanisms in non-myocyte cells (78). A previous study showed that
selenium supplementation exerts beneficial effects in alleviating
HF symptoms (79). By reducing
cardiomyocyte death and reactive oxygen species production,
selenium not only helps mitigate HF, but also indirectly improves
the epigenetic state of cardiomyocytes by inhibiting the
DNMT2-mediated methylation of the glutathione peroxidase 1 gene
promoter (80).
Beyond genes that directly affect cardiac health,
DNA methylation also plays a critical role in other physiological
systems. In mouse models, the loss of the Wnt receptor LRP6 in
aortic VSMCs leads to increased methylation of GTPase-activating
protein-binding proteins, which may be linked to the
transcriptional responses that trigger arterial calcification
(81). Furthermore, DNA
methylation patterns in the placenta have also been found to be
associated with maternal blood pressure and CVDs. A previous study
revealed that mothers with a family history of hypertension
exhibited lower global placental DNA methylation levels in
placental samples, which as accompanied by higher mean arterial
pressure, thus highlighting the potential role of placental
epigenetics in the development of CVDs (82). Additionally, in the field of cancer
research, DNMTs have been shown to play a crucial role in mediating
tumor drug resistance and progression through exosome-based
delivery mechanisms (83–85).
In summary, DNA methylation, as a key epigenetic
regulatory mechanism, is likely to play an important role in the
onset and progression of CVDs. However, the mechanisms underlying
exosome-mediated DNA methylation in the cardiovascular system
remain to be fully elucidated. Although these pathways have been
extensively studied in cancer, such as hepatocellular carcinoma
(86), colorectal cancer (87) and acute myeloid leukemia (88), as well as in other diseases
including Alzheimer's disease (89), type 2 diabetes (90) and systemic lupus erythematosus
(91), direct evidence in CVDs is
relatively limited, highlighting the urgent need for systematic
investigations to clarify their specific roles and clinical
implications. Future research focusing on the role of exosomes in
epigenetic regulation may provide novel insights into the
pathogenesis of CVDs.
The homeostasis of the cardiovascular system is
maintained through the interplay of various cell types, including
cardiac fibroblasts, ECs, VSMCs, cardiomyocytes, immune cells and
resident stem cells (92). These
cells release exosomes that mediate intercellular signaling,
thereby contributing to tissue repair, remodeling and the
regulation of pathological responses in the cardiovascular system.
Notably, exosomes are not merely structural vesicles, but
functional carriers that transport epigenetic information and
influence gene expression in recipient cells. Consequently,
exosome-mediated epigenetic regulatory mechanisms are increasingly
recognized as critical contributors to the onset and progression of
CVDs. Previous studies have demonstrated that exosomes of different
cellular origins, such as ECs, erythrocytes, platelets and
leukocytes, exhibit donor cell-specific profiles of RNAs, proteins
and modifying enzymes, thereby reflecting the physiological or
pathological state of their source cells (93,94).
For example, exosomes derived from ECs are essential for the
phenotypic transition of VSMCs (95), while exosomal miRNA profiles in the
plasma of atherosclerotic patients have been investigated as
potential early biomarkers (59,60).
These findings suggest that pathological alterations in the
cellular microenvironment not only influence exosome abundance and
secretion dynamics, but also profoundly reshape their epigenetic
regulatory cargo.
CVDs, including HF, hypertension, atherosclerosis,
atrial fibrillation and AMI, continue to be the primary causes
worldwide rates of morbidity and mortality, despite continuous
advancements in the clinical diagnosis and treatment of heart
disease. Exosomes are involved in the movement and interchange of
molecules that signal, which makes them a crucial component in
controlling the course of CVDs, as shown in Table III. Furthermore, ncRNAs found in
exosomes function as novel regulators in dyslipidemia, which raises
the risk of CVD caused by atherosclerosis. For example, miR-26a is
markedly downregulated in exosomes from obese mice and overweight
individuals, and its restoration reverses multiple markers
associated with dysregulated lipid metabolism, suggesting a pivotal
role for exosomal miRNAs in the cardiometabolic axis. Importantly,
exosome-mediated epigenetic mechanisms exhibit substantial
heterogeneity across different CVD subtypes. For instance, in HF,
exosomal ncRNAs may contribute to the regulation of myocardial
autophagy and fibrosis, whereas, in atherosclerosis, they are more
likely to influence inflammatory cell infiltration and endothelial
repair processes. Exosomes associated with hypertension are often
enriched in miRNAs involved in vascular tone regulation, whereas
those related to AMI tend to carry factors that modulate
cardiomyocyte apoptosis and stress responses. These differences
underscore the importance of analyzing exosomal epigenetic
functions in a disease-specific context, which represents a
promising direction for precision cardiovascular therapy.
Accordingly, the following section aimed to systematically explore
the roles of exosomal ncRNAs and DNA methylation-related factors in
the pathophysiological mechanisms of four major CVD subtypes,
namely HF, hypertension, atherosclerosis and myocardial infarction,
with the aim of providing a theoretical basis for understanding
their regulatory networks and clinical potential.
In HF, exosomal miRNAs play a pivotal role in
regulating myocardial remodeling, and exhibit potential as both
biomarkers and therapeutic targets, as shown in Table III. In a rat model of chronic HF,
exosomal miR-124 derived from human umbilical cord mesenchymal stem
cells was found to target PRSS23 and suppress associated fibrotic
signaling pathways, thereby reducing the expression of the
endothelial marker CD31, alleviating myocardial ischemic injury,
and promoting angiogenesis and cellular repair (96). A clinical study also highlighted
the critical role of exosomal miRNAs in HF (97). Previous studies have shown that
patients with HF and reduced ejection fraction exhibit elevated
levels of exosomal miR-92b-5p (97,98).
This miRNA correlates positively with left atrial diameter,
systolic blood pressure and left ventricular end-diastolic
diameter, while it correlates negatively with fractional shortening
and left ventricular ejection fraction (97), suggesting its potential involvement
in the structural and functional remodeling of the left-side of the
heart. In addition, miR-425 and miR-744 have also exhibited
beneficial effects in mitigating myocardial fibrosis. In
angiotensin-induced models, overexpression of these two miRNAs
effectively suppresses collagen production and fibrotic processes
in fibroblasts, whereas their downregulation is accompanied by
significant upregulation of α-SMA and COL1, further confirming
their critical regulatory role in cardiac fibrosis (99). Therefore, miR-425 and miR-744, as
exosome-delivered epigenetic regulators, represent promising
therapeutic targets and potential novel tools for reversing cardiac
remodeling in HF. Notably, the regulatory effects of exosomal
miRNAs may be bidirectional. Previous research has reported that
serum exosomal miR-27a levels are significantly reduced in patients
with HF, and this decrease is closely associated with poor
prognosis (100). Additionally,
aberrant expression of miR-126 in peripheral blood mononuclear
cells of patients with chronic HF has been shown to exacerbate
cardiac injury (101).
Collectively, exosomal miRNAs modulate myocardial fibrosis through
multiple targets and mechanisms, thus playing a critical role in
the structural and functional remodeling of the heart.
Hypertension is a major risk factor for
cardiovascular events such as coronary artery disease, HF, stroke
and myocardial infarction (102,103). In addition, its onset and
progression are also governed by a variety of molecular and
cellular processes. In recent years, exosome-mediated ncRNA
signaling has been recognized as a key epigenetic regulatory
mechanism linking these pathological processes (104,105). According to previous studies,
numerous cellular and molecular processes, such as the
renin-angiotensin-aldosterone system, endothelial dysfunction,
vascular remodeling, oxidative stress, angiogenesis, VSMC
proliferation and inflammation, are regulated by a number of
exosomal ncRNAs, which ultimately lead to the development of
hypertension (77,106).
Exosomal miRNAs can influence the development and
occurrence of hypertension in a positive or negative manner. In
spontaneously hypertensive rats (SHRs), compared with Wistar-Kyoto
rats, 23 exosomal miRNAs were upregulated and 4 were downregulated,
indicating a systemic alteration in circulating exosomal miRNA
profiles under hypertensive conditions (107). For example, miR-155-5p was
significantly downregulated in exosomes derived from adventitial
fibroblasts of SHR aortas (1).
This miRNA could suppress ACE/Ang II signaling, reduce oxidative
stress and inflammation, and thereby inhibit VSMCs migration in
SHRs (108). On the other hand,
certain exosomal miRNAs have been shown to exert pro-hypertensive
effects. Exosomal miR-27a derived from THP-1 monocytes could
inhibit the phosphorylation of endothelial nitric oxide synthase in
mesenteric arteries, and reduce Mas receptor expression in ECs,
thereby impairing vasodilation and elevating blood pressure
(109). Following Ang II
treatment, the levels of intercellular adhesion molecule-1 in THP-1
cells increased, whereas exosomal miR-17 could downregulate its
expression, thus modulating endothelial inflammatory responses and
indirectly affecting the vascular tone (110). In addition, exosomal miR-106b-5p,
released from macrophages, could be transferred to glomerular
mesangial cells, where it suppressed the transcription factors
Pde3b and E2f1, thus mediating tissue responses associated with
inflammatory hypertension (111).
A previous epidemiological study reported that extracellular
vesicle-derived miRNAs, particularly miR-199a/b and miR-223-3p,
exhibited significant interactions with elevated systolic blood
pressure, thus potentially contributing to hypertension through
mechanisms involving vascular function, inflammatory responses and
oxidative stress (112). In
summary, miRNAs play a critical regulatory role in the onset and
progression of hypertension. They modulate vascular tone and
remodeling through multiple pathways, such as exosomal miR-155
(113), which impairs endothelial
nitric oxide signaling and promotes vascular inflammation, and
exosomal miR-505 (114), which
enhances vascular smooth muscle cell proliferation and migration,
reflecting their unique and complex functional characteristics in
hypertensive pathology. However, current research on miRNAs in
hypertension remains largely preclinical (115), with a lack of systematic
validation in large populations and limited progress in
translational applications.
Atherosclerosis is a long-term immunoinflammatory
disease characterized by the accumulation of lipid-rich particles
in arterial plaques, which is one of the main causes of CVDs, with
serious clinical consequences such as myocardial infarction and
stroke. In recent years (116,117), exosome-mediated regulation of
ncRNAs has been recognized as one of the key epigenetic mechanisms
driving the development and progression of atherosclerosis (AS), as
shown in Table III. Exosomal
ncRNAs primarily regulate lipid metabolism, vascular inflammation
and cell survival, which have crucial regulatory functions in the
onset and advancement of atherosclerosis.
Numerous studies have demonstrated that, in both
patients and animal models of atherosclerosis, miRNAs, serving as
key epigenetic molecules within exosomes, can significantly
influence the pathological progression of AS through changes in
their expression levels (116,118). In patients with unstable angina,
28 miRNAs were upregulated, among which miR-19b showed the most
pronounced increase. This miRNA may inhibit the transcriptional
activity of the STAT3 signaling pathway, thereby suppressing the
proliferation, migration and angiogenesis of ECs, ultimately
delaying the formation and progression of unstable plaques
(41). Dysfunction of ECs can lead
to atherosclerosis by causing EC senescence, inflammation,
hyperpermeability, oxidative stress and vasodilation abnormalities
(119). Previous studies have
shown that exosomal ncRNAs play a major role in this process. Human
smooth muscle cells in the aorta produce miR-221/222, which
inhibits autophagy in these cells by regulating the PTEN/Akt axis
(120,121). Thrombin-stimulated platelet
exosomes showed enhanced miR-21, miR-339 and miR-223 levels, and
miR-223 could inhibit the expression of intercellular adhesion
molecule-1 (ICAM-1) in inflammatory processes by regulating the
MAPK/NF-κB axis (122). In
addition, exosomal miR-146a modulates inflammatory responses in ECs
by inhibiting interleukin-1 receptor-associated kinase 1 (123), whereas miR-4532 exacerbates EC
injury through its interaction with SP1 (124). A previous study showed that
upregulation of miR-155 can regulate the interaction between ECs
and VSMCs, inhibiting EC migration, proliferation and
reendothelialization (113).
This, in turn, weakens the endothelial barrier, increases vascular
wall permeability and promotes the progression of atherosclerotic
plaque (125).
In addition, the pathophysiology of atherosclerosis
is regulated by exosomal miRNAs derived from non-cardiomyocyte cell
types, such as endothelial cell-derived exosomal miR-92a that
promotes vascular inflammation (126), macrophage-derived exosomal
miR-146a that modulates inflammatory signaling (127), and vascular smooth muscle
cell-derived exosomal miR-221/222 that enhances cell proliferation
and migration (128). Macrophages
in particular possess the unique ability to engulf aggregated and
oxidized low-density lipoprotein via micropinocytosis and scavenger
receptor-mediated uptake, subsequently transforming into
lipid-laden foam cells that constitute the core of atherosclerotic
plaques. Exosomal miR-33a-5p, secreted by these cells, can regulate
ABCA1 expression and promote apoA1-mediated cholesterol efflux,
thereby affecting foam cell formation (129). miR-16-5p accelerates inflammation
and oxidative stress in atherosclerosis by downregulating SMAD7,
and promotes apoptosis (130),
thereby facilitating the progression of atherosclerotic lesions.
Endothelial dysfunction is one of the key initiating factors in the
early stages of atherosclerosis. Previous in vitro studies
have shown that circ_0124644 promotes endothelial injury by
modulating the miR-149-5p/PAPP-A axis, highlighting its pathogenic
role in the progression of AS and its potential as a target for
clinical intervention (131). In
addition, the miR-203-3p/cathepsin S pathway, which functions along
the p38/MAPK axis, has been shown to reverse atherosclerosis
(132). Neutrophil extracellular
traps (NETs) have also been identified as contributors to the
amplification of immune-inflammatory responses in AS (133,134). NETs are a network of densified
chromatin, nuclear histones and granular antimicrobial proteins
that are released by neutrophils in response to microbial antigens
(135). Exosomal miR-146a
released from macrophages upon oxidized low-density lipoprotein
(oxLDL) stimulation can induce the formation of NETs and promote
the progression of atherosclerosis (136). Following oxLDL treatment, human
umbilical vein ECs (HUVECs) exhibit enhanced NF-κB pathway
activation, which upregulates miR-505 expression. MiR-505 then
targets SIRT3, leading to increased reactive oxygen species
production and elevated NET formation (88). VSMCs participate in plaque
formation during the early stages of AS and contribute to plaque
stability in the later stages.
In addition to the aforementioned miRNAs, lncRNAs
are also important epigenetic regulators, and have been shown to be
significantly upregulated in both animal models and patients with
atherosclerosis. Previous studies have shown that exosomal lncRNA
LIPCAR regulates CDK2 and participates in the phenotypic
transformation of VSMCs (137,138). MiR-106a-3p modulates apoptosis by
targeting CASP9 (139), and the
LINC01005/miR-128-3p/KLF4 axis influences VSMC migration and
proliferation (140). These
lncRNAs mediate intercellular communication via exosomes, and serve
as key regulatory nodes in the evolution and stability of
atherosclerotic plaques. In THP-1 cells, lncRNA GAS5 can induce and
regulate apoptosis in both THP-1 monocytes and ECs (141). By contrast, EC-derived lncRNA
MALAT1 exhibits bidirectional effects: On one hand, it promotes M2
macrophage polarization and protects against atherosclerosis
(142), while, on the other hand,
it may also facilitate the formation of NETs, thereby accelerating
the progression of atherosclerosis (143). In addition, several studies have
found that exosomal circRNA-0006896 is closely associated with the
pathogenesis of atherosclerosis (144,145). It is significantly upregulated in
patients with unstable atherosclerotic plaques, and its levels are
positively correlated with triglycerides, low-density lipoprotein
and C-reactive protein. Mechanistically, circRNA-0006896 may
promote EC proliferation and migration by activating the NF-κB
signaling pathway (144,146). Previous in vitro studies
have shown that exosome levels are significantly elevated in the
serum of patients with coronary artery disease, which can
exacerbate inflammatory responses and apoptosis in HUVECs, thereby
promoting endothelial injury and atherosclerosis progression
(147,148). Conversely, exosomal circ_0001785
can alleviate endothelial damage through the miR-513a-5p/TGFBR3
axis, thus exhibiting a protective effect (149).
In summary, exosome-derived miRNAs, lncRNAs and
circRNAs in atherosclerosis profoundly participate in immune
inflammation, apoptosis, lipid metabolism and plaque stability by
targeting multiple key signaling pathways, including MAPK, NF-κB,
STAT3 and p38. These epigenetic factors not only hold promise as
diagnostic biomarkers for AS, but also offer new strategies for
intervening in plaque progression and enhancing therapeutic
precision.
AMI is typically caused by sudden coronary artery
occlusion and is characterized by myocardial cell necrosis and loss
of function, which can have severe clinical consequences (150). Previous studies have shown that
exosomal miRNAs play a pivotal regulatory role in the onset and
progression of myocardial infraction (MI) by modulating a variety
of pathological processes, including autophagy, inflammation,
angiogenesis and apoptosis (151,152), as shown in Table III.
Multiple studies have found that the exosomal miRNA
expression profile derived from cardiomyocytes is significantly
altered in patients with MI. Notably, exosomal miR-125b, miR-499,
miR-133, miR-22, miR-21 and miR-301, which originate from
cardiomyocytes, are markedly upregulated following MI (153,154). These miRNAs are encapsulated
within cardiomyocyte-derived exosomes and are broadly expressed in
damaged myocardial regions. By inhibiting cardiomyocyte death,
modulating local inflammatory responses and promoting ischemic
tissue repair, they help alleviate pathological injury following MI
and may partially contribute to the prevention of complications
such as atherosclerosis (155,156). Furthermore,
inflammation-associated miRNAs, such as miR-146a and miR-25-3p, are
upregulated in the immune environment induced by MI. Notably,
exosomal miR-301 is also elevated and has been shown to suppress
autophagy in cardiomyocytes (109,157). At the translational level,
exosomal miR-499 and miR-133a are significantly elevated in
patients with MI compared to those with stable coronary artery
disease or acute coronary syndrome, demonstrating their potential
utility for clinical diagnosis and disease stratification (94). In addition, miR-26b-5p promotes
ferroptosis by targeting SLC7A11, and its downregulation in
exosomes from patients with AMI helps attenuate myocardial injury
(158), thus offering new
perspectives for anti-ferroptosis therapies.
In addition to those derived from cardiomyocytes,
exosomal ncRNAs from other cell types also play important roles in
the pathology of MI. For example, during ischemic episodes, the
expression of miR-22 in mesenchymal stem cell-derived exosomes is
reduced (159); however, in
vivo supplementation of this miRNA can significantly attenuate
cardiac fibrosis (160),
highlighting its therapeutic potential after AMI. High expression
of miR-21 in mesenchymal stem cell-derived exosomes can
significantly inhibit EC apoptosis and promote angiogenesis,
thereby exerting a protective effect on cardiomyocytes (161). In addition, lncRNAs are involved
in the epigenetic regulation of MI. LncRNA BC002059 has been shown
to reduce infarct size in mice with coronary artery ligation
(162). Mechanistically, BC002059
exerts its cytoprotective effects by influencing the downstream
target gene ABHD10 and subsequently regulating the expression of
miR-19b-3p (163). The roles of
other lncRNAs in the process of MI should not be overlooked, such
as MALAT and H19 (164,165). After AMI, the levels of MALAT1 in
exosomes released from cardiomyocytes are significantly increased
under hyperoxic conditions. MALAT1 enhances neovascularization by
suppressing miR-92a expression (117), thereby contributing to ischemic
tissue repair. Atorvastatin treatment has been shown to induce an
increase in lncRNA H19 release from mesenchymal stem cells
(118). By contrast, under
hypoxic conditions, cardiomyocyte-derived exosomal lncRNA AK139128
regulates cardiac fibrosis by affecting the proliferation,
migration and apoptosis of cardiac fibroblasts (165). These findings suggest that
exosomal lncRNAs not only play functional roles in response to
ischemic injury, but may also serve as important molecular targets
for the treatment of MI.
Overall, exosomal ncRNAs play critical roles in
myocardial infarction as well as other CVDs, including
hypertension, atherosclerosis and HF. Previous research in both
animal models and clinical samples has identified numerous
potential therapeutic targets, such as exosomal miR-92b-5p
regulating cardiomyocyte apoptosis (166), lncRNA H19 enhancing the efficacy
of mesenchymal stem cell-derived exosomes in myocardial infarction
(167). However, there is
considerable heterogeneity in the mechanisms, target pathways and
functional roles of ncRNAs among different types of CVDs. In HF,
exosomal miRNAs primarily regulate fibroblast activity and
influence myocardial remodeling, demonstrating potential for
anti-fibrotic effects and cardiac structural modulation. By
contrast, exosomal ncRNAs associated with atherosclerosis are more
focused on regulating endothelial function and lipid metabolism,
reflecting microenvironment-specific regulatory characteristics
within atherosclerotic lesions. By comparison, those involved in
hypertension predominantly participate in the regulation of
vascular tone and remodeling.
Although these findings reveal distinct mechanisms
by which exosomal ncRNAs act in different CVDs, as shown in
Table III, their clinical
translation still faces important challenges. The majority of
previous studies rely on animal models and in vitro
experiments, with a lack of validation in large-scale human
samples. Future research urgently requires multicenter clinical
data to further clarify the specific mechanisms of exosomal ncRNAs
in cardiovascular pathology.
Prognostic biomarkers may be employed for
identification of diseases and/or prognostic indicators. The most
crucial and essential qualities of effective biomarkers are their
sensitivity, specificity, stability and relative noninvasive
detection. Notably, recent studies (166,168) are focusing on exosomes as
possible biomarkers due to their easy and affordable detection in
multiple bodily fluids, and their ability to function as a
protective transport system against enzymatic degradation. Exosomes
possess the capacity to serve as helpful biomarkers for the early
detection of CVDs, since their cargo for epigenetic pathways varies
depending on the type of CVD (169,170). Therefore, exosomes may be used as
biomarkers to assist diagnosis according to their abnormal
manifestations in different CVDs.
Previous studies have shown that exosomal ncRNAs
exhibit diverse biological functions within the cardiovascular
system. Dicer's heart-specific downregulation in young mice
degrades miRNAs, particularly miR-1, miR-133a, miR-208 and miR-499
(171,172), resulting in atrial expansion,
ventricular remodeling and enlargement, reduced cardiac function,
and early mortality (173,174).
By contrast, overexpression of certain specific miRNAs in
vitro can induce cardiac hypertrophy and HF (44,45),
indicating that the balance of miRNA expression is essential for
maintaining cardiac homeostasis (173). Therefore, exosomal miRNAs and
other epigenetic cargos not only reflect disease states but may
also serve as potential targets for therapeutic intervention.
As therapeutic agents, exosomes hold promise as
viable delivery vehicles due to a range of advantages, including
minimal immunogenicity and toxicity, high biocompatibility,
efficient bio-barrier permeability and natural availability
(174). It has been attempted to
alter exosomes through genetic engineering for medicinal uses
(175,176). The potential to use exosomes as
carriers to deliver certain cargo to particular tissues or organs
is made possible by the capacity to change isolated and purified
exosomes with target molecules and surface-loaded mimics or
inhibitors of epigenetic payload (172,177). This gene regulation points to
exosome epigenetic cargo as a potentially effective treatment
strategy for a range of human diseases.
Previous animal studies have demonstrated the
therapeutic potential of exosomes in various CVD models (168,178,179). Engineered exosomes enriched with
low-density lipoprotein receptor mRNA have been shown to
effectively alleviate hypercholesterolemia in mice (180). Additionally, mesenchymal
stem/stromal cell (MSC)-derived exosomes can reduce cardiomyocyte
apoptosis after myocardial infarction by suppressing SOCS2
signaling via miRNA-185 (181–183). A previous study further confirmed
that pharmacological agents can modulate exosomes to intervene in
the development and progression of CVDs (165). Furthermore, a previous study
found that phenol can attenuate inflammatory responses in HUVECs,
which deaccelerates the progression of atherosclerosis, and
decreases the expression levels of STAT3 and phosphorylated STAT3
in HUVECs by increasing the quantity of exosome miR-223 in HUVECs
treated with THP-1 and exosomes (184). In an I/R injury model in mice,
the injection of cortical diaphysial-derived exosomes decreased the
size of the infarct (185). On
the contrary, chrysin reduced miR-92a expression in HCAEC cells and
accelerated the progression of atherosclerosis (186). Furthermore, exosomal miR-21-3p
derived from nicotine-treated macrophages could bind to PTEN,
thereby promoting VSMC migration and proliferation, and
exacerbating the progression of atherosclerosis (187). In addition, exosomes have shown a
promising potential in promoting cardiac regeneration. Multiple
studies using models of ischemic heart disease have demonstrated
that injection of mesenchymal stem cell-derived exosomes can
effectively improve myocardial remodeling, enhance cardiomyocyte
survival (188,189), and promote EC proliferation,
thereby partially restoring cardiac function (190,191). This ‘cell-free therapy’ offers a
safer and more controllable alternative to traditional cell
transplantation.
In recent years, exosome-based therapeutic
strategies have attracted increasing attention for the treatment of
CVDs (192). Exosomes are
nanoscale, membrane-bound vesicles secreted by cells, and their
unique structural and functional properties make them highly
promising as therapeutic carriers (176,193). The bilayer lipid membrane of
exosomes effectively protects encapsulated bioactive molecules,
such as miRNAs and proteins, enhancing their stability and
biological activity in vivo (194). For example, compared with free
curcumin, curcumin encapsulated in exosomes exhibits higher plasma
concentrations and greater anti-inflammatory effects (195). Furthermore, as non-living
particulate structures, exosomes offer greater controllability
compared to viral vectors or live cell systems, and lack
tumorigenic potential (196). In
multiple studies, exosomes have demonstrated a favorable safety
profile (197,198).
However, despite the numerous advantages of
exosomes as therapeutic tools, their clinical application still
faces notable challenges. First, safety concerns cannot be ignored.
While exosomes derived from autologous cells are generally
considered to have low immunogenicity, artificially engineered
exosome delivery systems may still pose immunogenic risks, follow
unintended pathways away from target recipient cell types, and
result in unexpected off-target effects (199). More importantly, exosomes lack
specific biodistribution in vivo. Once cleared from the
bloodstream, exosomes tend to accumulate in organs such as the
lungs, spleen and liver (200,201), which can result in off-target
effects, reduced therapeutic efficacy and even potential toxicity.
Due to the tendency of RNA to aggregate or degrade, loading
specific RNA cargos into exosomes using conventional
electroporation or sonication techniques presents remarkable
technical challenges, and the efficacy of these approaches remains
uncertain (202). In addition,
there may be a risk of infection when these particles enter the
human bloodstream. More challenging is the substantial variability
in exosomal drug loading capacity and the lack of uniformity,
making it difficult to optimize exosome dosing (140). Furthermore, the optimal
therapeutic time window for exosome administration in vivo
has not yet been determined, which further increases the
uncertainty of clinical translation (203). Therefore, before advancing
exosomes as RNA delivery vehicles for clinical use, it is essential
to ensure their safety in vivo and to minimize immune
responses or off-target effects. In addition, optimization of
loading techniques is needed to improve RNA stability and achieve a
uniform cargo distribution.
Furthermore, the industrialization and clinical
translation of exosomes face important technical bottlenecks.
Currently, ultracentrifugation is the most commonly used isolation
method for exosomes; however, it is labor-intensive, inefficient,
and has limited yield, thus making it difficult to meet the demands
of large-scale clinical applications. On one hand, there is still a
lack of unified and standardized Good Manufacturing Practice-grade
manufacturing processes (204).
On the other hand, exosomes derived from different cell sources
show significant variations in composition, function and efficacy,
leading to poor batch-to-batch reproducibility and substantial
challenges in quality control. Furthermore, delivery efficiency
remains one of the most critical issues. Exosomes are commonly
administered for the treatment of CVDs via intravenous,
intracoronary or intramyocardial injection; however, these
approaches are often associated with procedural complexity or low
cardiac retention rates in clinical practice. Although surface
engineering modifications can enhance targeted delivery (205), exosomes still face challenges
such as off-target effects, rapid clearance and the need for
rigorous safety validation in the complex human physiological
environment.
In summary, exosomes hold great therapeutic promise
for CVDs, particularly due to their advantages in targeting
specificity, delivery efficiency and epigenetic regulation.
However, several key issues must be addressed before exosome-based
therapies can be translated into clinical practice, including
ensuring safety, improving manufacturing processes and enhancing
consistency in therapeutic efficacy.
The present study aimed to review in detail the
exosome-mediated epigenetic mechanisms, such as exosomal ncRNAs and
exosome-mediated DNA methylation, and their role in CVDs.
Concurrently, the present study examined the function of exosomal
phylogenetic pathways in CVD detection and treatment. The miRNA
cargo of exosomes is of particular importance, as these molecules
have the capacity to control the expression of proteins. At
present, research on the mechanisms by which exosomes influence
cardiovascular epigenetics is still ongoing, with current studies
mainly focusing on exosomal ncRNAs. Targeted approaches for the
treatment of CVDs are feasible, as exosomes are capable of
delivering therapeutic agents directly to damaged cells and
tissues. The application of exosomes as delivery vehicles for CVD
therapy is highly promising. Both animal studies and preliminary
clinical trials have shown encouraging results, suggesting that
exosome-based therapies could represent a breakthrough in CVD
treatment. Given that CVD remains a leading cause of mortality
worldwide, this strategy is particularly meaningful for high-risk
populations, including those with comorbidities such as diabetes
and hypertension, or individuals with a history of angina or
myocardial infarction.
However, the clinical application of exosomes still
faces multiple challenges. First, there is a lack of standardized
protocols for exosome isolation, characterization and cargo
detection, resulting in poor reproducibility of experimental
results and efficacy validation across studies, which limits
translational progress. Second, the low loading efficiency and
instability of functional RNAs compromise the stability and
controllability of exosome-based delivery systems. In addition, the
non-specific biodistribution of exosomes in vivo leads to
risks of rapid clearance and off-target effects, which highlights
the need for improved delivery strategies such as surface
modification and magnetic guidance to enhance targeting.
Particularly in the cardiovascular field, achieving safe, stable
and effective therapeutic delivery in complex microenvironments
requires further systematic research and validation.
However, as research on exosomes continues to
advance, their prospects in precision medicine remain highly
promising. In particular, exosomes offer novel insights into
epigenetic regulation and intercellular communication underlying
the pathogenesis and progression of CVDs. Future studies should
focus on both mechanistic exploration and the establishment of
technical standards to facilitate the critical transition from
laboratory research to clinical application.
Not applicable.
The present study was supported by the Jilin Province Science
and Technology Development Plan Project (grant no.
YDZJ202201ZYTS209); the Regional Joint Fund Project of National
Natural Science Foundation of China (grant no. U24A20779); and the
Science and Technology Development Plan of Jilin Province (grant
no. YDZJ202301ZYTS337).
Not applicable.
Conceptualization and study design: LZ and JL.
Writing of the first manuscript draft: YZ. Literature search,
figure reparation (created with BioRender) and manuscript editing:
WJ and YL. Data curation, writing, reviewing and editing: YZ and
SZ. Supervision and project administration: LZ and JL. All authors
contributed to the critical revision of the manuscript, read and
approved the final version of the manuscript. Data authentication
is not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
GBD 2019 Viewpoint Collaborators, . Five
insights from the global burden of disease study 2019. Lancet.
396:1135–1159. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shah R, Patel T and Freedman JE:
Circulating extracellular vesicles in human disease. N Engl J Med.
379:958–966. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Théry C, Witwer KW, Aikawa E, Alcaraz JM,
Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F,
Atkin-Smith GK, et al: Minimal information for studies of
extracellular vesicles 2018 (MISEV2018): A position statement of
the international society for extracellular vesicles and update of
the MISEV2014 guidelines. J Extracell Vesicles. 7:15357502018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sahoo S, Adamiak M, Mathiyalagan P,
Kenneweg F, Kafert-Kasting S and Thum T: Therapeutic and diagnostic
translation of extracellular vesicles in cardiovascular diseases.
Circulation. 143:1426–1449. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Martin-Ventura JL, Roncal C, Orbe J and
Blanco-Colio LM: Role of extracellular vesicles as potential
diagnostic and/or therapeutic biomarkers in chronic cardiovascular
diseases. Front Cell Dev Biol. 10:8138852022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shi Y, Zhang H, Huang S, Yin L, Wang F,
Luo P and Huang H: Epigenetic regulation in cardiovascular disease:
Mechanisms and advances in clinical trials. Signal Transduct Target
Ther. 7:2002022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Khan MI, Alsayed RKME, Choudhry H and
Ahmad A: Exosome-mediated response to cancer therapy: Modulation of
epigenetic machinery. Int J Mol Sci. 23:62222022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nasir A, Bullo MMH, Ahmed Z, Imtiaz A,
Yaqoob E, Jadoon M, Ahmed H, Afreen A and Yaqoob S: Nutrigenomics:
Epigenetics and cancer prevention: A comprehensive review. Crit Rev
Food Sci Nutr. 60:1375–1387. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Schiano C, Benincasa G, Franzese M, Della
Mura N, Pane K, Salvatore M and Napoli C: Epigenetic-sensitive
pathways in personalized therapy of major cardiovascular diseases.
Pharmacol Ther. 210:1075142020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hulshoff MS, Xu X, Krenning G and Zeisberg
EM: Epigenetic regulation of Endothelial-to-mesenchymal transition
in chronic heart disease. Arterioscler Thromb Vasc Biol.
38:1986–1996. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chang W and Wang J: Exosomes and their
noncoding RNA cargo are emerging as new modulators for diabetes
mellitus. Cells. 8:8532019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mathieu M, Martin-Jaular L, Lavieu G and
Théry C: Specificities of secretion and uptake of exosomes and
other extracellular vesicles for cell-to-cell communication. Nat
Cell Biol. 21:9–17. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shao H, Im H, Castro CM, Breakefield X,
Weissleder R and Lee H: New technologies for analysis of
extracellular vesicles. Chem Rev. 118:1917–1950. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Raiborg C and Stenmark H: The ESCRT
machinery in endosomal sorting of ubiquitylated membrane proteins.
Nature. 458:445–452. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang J, Li S, Li L, Li M, Guo C, Yao J
and Mi S: Exosome and exosomal microRNA: Trafficking, sorting, and
function. Genomics Proteomics Bioinformatics. 13:17–24. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kalluri R: The biology and function of
exosomes in cancer. J Clin Invest. 126:1208–1215. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Poddar S, Kesharwani D and Datta M:
Interplay between the miRNome and the epigenetic machinery:
Implications in health and disease. J Cell Physiol. 232:2938–2945.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Petrovic N and Ergun S: miRNAs as
potential treatment targets and treatment options in cancer. Mol
Diagn Ther. 22:157–168. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Quinn JJ and Chang HY: Unique features of
long non-coding RNA biogenesis and function. Nat Rev Genet.
17:47–62. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lou Z, Zhou R, Su Y, Liu C, Ruan W, Jeon
CO, Han X, Lin C and Jia B: Minor and major circRNAs in virus and
host genomes. J Microbiol. 59:324–331. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang T, Long T, Du T, Chen Y, Dong Y and
Huang ZP: Circle the cardiac remodeling with circRNAs. Front
Cardiovasc Med. 8:7025862021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hall IF, Climent M, Viviani Anselmi C,
Papa L, Tragante V, Lambroia L, Farina FM, Kleber ME, März W,
Biguori C, et al: rs41291957 controls miR-143 and miR-145
expression and impacts coronary artery disease risk. EMBO Mol Med.
13:e140602021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jiang F, Chen Q, Wang W, Ling Y, Yan Y and
Xia P: Hepatocyte-derived extracellular vesicles promote
endothelial inflammation and atherogenesis via microRNA-1. J
Hepatol. 72:156–166. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hou Z, Qin X, Hu Y, Zhang X, Li G, Wu J,
Li J, Sha J, Chen J, Xia J, et al: Longterm Exercise-derived
exosomal miR-342-5p: A novel exerkine for cardioprotection. Circ
Res. 124:1386–1400. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chang YJ, Li YS, Wu CC, Wang KC, Huang TC,
Chen Z and Chien S: Extracellular MicroRNA-92a mediates endothelial
Cell-macrophage communication. Arterioscler Thromb Vasc Biol.
39:2492–2504. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sindi HA, Russomanno G, Satta S,
Abdul-Salam VB, Jo KB, Qazi-Chaudhry B, Ainscough AJ, Szulcek R,
Jan Bogaard H, Morgan CC, et al: Therapeutic potential of
KLF2-induced exosomal microRNAs in pulmonary hypertension. Nat
Commun. 11:11852020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Miao Y, Ajami NE, Huang TS, Lin FM, Lou
CH, Wang YT, Li S, Kang J, Munkacsi H, Maurya MR, et al:
Enhancer-associated long non-coding RNA LEENE regulates endothelial
nitric oxide synthase and endothelial function. Nat Commun.
9:2922018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang K, Liu CY, Zhou LY, Wang JX, Wang M,
Zhao B, Zhao WK, Xu S, Fan LH, Zhang XJ, et al: APF lncRNA
regulates autophagy and myocardial infarction by targeting
miR-188-3p. Nat Commun. 6:67792015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liang H, Su X, Wu Q, Shan H, Lv L, Yu T,
Zhao X, Sun J, Yang R, Zhang L, et al: LncRNA 2810403D21Rik/Mirf
promotes ischemic myocardial injury by regulating autophagy through
targeting Mir26a. Autophagy. 16:1077–1091. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Viereck J, Bührke A, Foinquinos A,
Chatterjee S, Kleeberger JA, Xiao K, Janssen-Peters H, Batkai S,
Ramanujam D, Kraft T, et al: Targeting muscle-enriched long
non-coding RNA H19 reverses pathological cardiac hypertrophy. Eur
Heart J. 41:3462–3474. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Omura J, Habbout K, Shimauchi T, Wu WH,
Breuils-Bonnet S, Tremblay E, Martineau S, Nadeau V, Gagnon K,
Mazoyer F, et al: Identification of long noncoding RNA H19 as a new
biomarker and therapeutic target in right ventricular failure in
pulmonary arterial hypertension. Circulation. 142:1464–1484. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hua X, Wang YY, Jia P, Xiong Q, Hu Y,
Chang Y, Lai S, Xu Y, Zhao Z, Song J, et al: Multi-level
transcriptome sequencing identifies COL1A1 as a candidate marker in
human heart failure progression. BMC Med. 18:22020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mao YY, Wang JQ, Guo XX, Bi Y and Wang CX:
Circ-SATB2 upregulates STIM1 expression and regulates vascular
smooth muscle cell proliferation and differentiation through
miR-939. Biochem Biophys Res Commun. 505:119–125. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shen L, Hu Y, Lou J, Yin S, Wang W, Wang
Y, Xia Y and Wu W: CircRNA-0044073 is upregulated in
atherosclerosis and increases the proliferation and invasion of
cells by targeting miR-107. Mol Med Rep. 19:3923–3932.
2019.PubMed/NCBI
|
|
35
|
Sygitowicz G and Sitkiewicz D: Involvement
of circRNAs in the development of heart failure. Int J Mol Sci.
23:141292022. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wu N, Li C, Xu B, Xiang Y, Jia X, Yuan Z,
Wu L, Zhong L and Li Y: Circular RNA mmu_circ_0005019 inhibits
fibrosis of cardiac fibroblasts and reverses electrical remodeling
of cardiomyocytes. BMC Cardiovasc Disord. 21:3082021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang M, Wang Z, Cheng Q, Wang Z, Lv X,
Wang Z and Li N: Circular RNA (circRNA) CDYL induces myocardial
regeneration by ceRNA after myocardial infarction. Med Sci Monit.
26:e9231882020.PubMed/NCBI
|
|
38
|
Leimena C and Qiu H: Non-Coding RNA in the
pathogenesis, progression and treatment of hypertension. Int J Mol
Sci. 19:9272018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sun Y, Yang Z, Zheng B, Zhang XH, Zhang
ML, Zhao XS, Zhao HY, Suzuki T and Wen JK: A novel regulatory
mechanism of smooth muscle α-Actin expression by
NRG-1/circACTA2/miR-548f-5p Axis. Circ Res. 121:628–635. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yang K, Zhou Q, Qiao B, Shao B, Hu S, Wang
G, Yuan W and Sun Z: Exosome-derived noncoding RNAs: Function,
mechanism, and application in tumor angiogenesis. Mol Ther Nucleic
Acids. 27:983–997. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Qian Z, Shen Q, Yang X, Qiu Y and Zhang W:
The role of extracellular vesicles: An epigenetic view of the
cancer microenvironment. Biomed Res Int. 2015:6491612015.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Radhakrishna U, Albayrak S, Zafra R, Baraa
A, Vishweswaraiah S, Veerappa AM, Mahishi D, Saiyed N, Mishra NK,
Guda C, et al: Placental epigenetics for evaluation of fetal
congenital heart defects: Ventricular Septal Defect (VSD). PLoS
One. 14:e02002292019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li S, Geng Q, Chen H, Zhang J, Cao C,
Zhang F, Song J, Liu C and Liang W: The potential inhibitory
effects of miR-19b on vulnerable plaque formation via the
suppression of STAT3 transcriptional activity. Int J Mol Med.
41:859–867. 2018.PubMed/NCBI
|
|
44
|
Small EM and Olson EN: Pervasive roles of
microRNAs in cardiovascular biology. Nature. 469:336–342. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li Q, Song XW, Zou J, Wang GK, Kremneva E,
Li XQ, Zhu N, Sun T, Lappalainen P, Yuan WJ, et al: Attenuation of
microRNA-1 derepresses the cytoskeleton regulatory protein
twinfilin-1 to provoke cardiac hypertrophy. J Cell Sci.
123:2444–2452. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Heidersbach A, Saxby C, Carver-Moore K,
Huang Y, Ang YS, de Jong PJ, Ivey KN and Srivastava D: microRNA-1
regulates sarcomere formation and suppresses smooth muscle gene
expression in the mammalian heart. Elife. 2:e013232013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Van Rooij E, Sutherland LB, Liu N,
Williams AH, McAnally J, Gerard RD, Richardson JA and Olson EN: A
signature pattern of stress-responsive microRNAs that can evoke
cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA.
103:18255–18260. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Benzoni P, Nava L, Giannetti F, Guerini G,
Gualdoni A, Bazzini C, Milanesi R, Bucchi A, Baruscotti M and
Barbuti A: Dual role of miR-1 in the development and function of
sinoatrial cells. J Mol Cell Cardiol. 157:104–112. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ji R, Cheng Y, Yue J, Yang J, Liu X, Chen
H, Dean DB and Zhang C: MicroRNA expression signature and
antisense-mediated depletion reveal an essential role of MicroRNA
in vascular neointimal lesion formation. Circ Res. 100:1579–1588.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Nazari-Jahantigh M, Wei Y, Noels H, Akhtar
S, Zhou Z, Koenen RR, Heyll K, Gremse F, Kiessling F, Grommes J, et
al: MicroRNA-155 promotes atherosclerosis by repressing Bcl6 in
macrophages. J Clin Invest. 122:4190–202. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zernecke A, Bidzhekov K, Noels H,
Shagdarsuren E, Gan L, Denecke B, Hristov M, Köppel T, Jahantigh
MN, Lutgens E, et al: Delivery of microRNA-126 by apoptotic bodies
induces CXCL12-dependent vascular protection. Sci Signal.
2:ra812009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Mattei AL, Bailly N and Meissner A: DNA
methylation: A historical perspective. Trends Genet. 38:676–707.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhu L, Zhu C, Wang J, Yang R and Zhao X:
The association between DNA methylation of 6p21.33 and AHRR in
blood and coronary heart disease in Chinese population. BMC
Cardiovasc Disord. 22:3702022. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhang Y, Yang R, Burwinkel B, Breitling
LP, Holleczek B, Schöttker B and Brenner H: F2RL3 methylation in
blood DNA is a strong predictor of mortality. Int J Epidemiol.
43:1215–1225. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Guarrera S, Fiorito G, Onland-Moret NC,
Russo A, Agnoli C, Allione A, Di Gaetano C, Mattiello A, Ricceri F,
Chiodini P, et al: Gene-specific DNA methylation profiles and
LINE-1 hypomethylation are associated with myocardial infarction
risk. Clin Epigenetics. 7:1332015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhang W, Song M, Qu J and Liu GH:
Epigenetic modifications in cardiovascular aging and diseases. Circ
Res. 123:773–786. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Nakamura TY, Iwata Y, Arai Y, Komamura K
and Wakabayashi S: Activation of Na+/H+ exchanger 1 is sufficient
to generate Ca2+ signals that induce cardiac hypertrophy and heart
failure. Circ Res. 103:891–899. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Chen S, Ma J, Chi J, Zhang B, Zheng X,
Chen J and Liu J: Roles and potential clinical implications of
tissue transglutaminase in cardiovascular diseases. Pharmacol Res.
177:1060852022. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Westerman K, Sebastiani P, Jacques P, Liu
S, DeMeo D and Ordovás JM: DNA methylation modules associate with
incident cardiovascular disease and cumulative risk factor
exposure. Clin Epigenetics. 11:1422019. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
GTEx Consortium; Laboratory, Data Analysis
& Coordinating Center (LDACC)-Analysis Working Group;
Statistical Methods groups-Analysis Working Group; Enhancing GTEx
(eGTEx) groups; NIH Common Fund; NIH/NCI; NIH/NHGRI; NIH/NIMH;
NIH/NIDA; Biospecimen Collection Source Site-NDRI et al., . Genetic
effects on gene expression across human tissues. Nature.
550:204–213. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Walaszczyk E, Luijten M, Spijkerman AMW,
Bonder MJ, Lutgers HL, Snieder H, Wolffenbuttel BHR and van
Vliet-Ostaptchouk JV: DNA methylation markers associated with type
2 diabetes, fasting glucose and HbA1c levels: A systematic review
and replication in a case-control sample of the Lifelines study.
Diabetol. 61:354–368. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Richard MA, Huan T, Ligthart S, Gondalia
R, Jhun MA, Brody JA, Irvin MR, Marioni R, Shen J, Tsai PC, et al:
DNA methylation analysis identifies loci for blood pressure
regulation. American. J Human Genet. 101:888–902. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Kennel PJ, Liao X, Saha A, Ji R, Zhang X,
Castillero E, Brunjes D, Takayama H, Naka Y, Thomas T, et al:
Impairment of myocardial glutamine homeostasis induced by
suppression of the amino acid carrier SLC1A5 in failing myocardium.
Circ Heart Fail. 12:e0063362019. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Fernández-Sanlés A, Sayols-Baixeras S,
Curcio S, Subirana I, Marrugat J and Elosua R: DNA Methylation and
Age-independent cardiovascular risk, an Epigenome-wide approach:
The REGICOR study (REgistre GIroní del COR). Arterioscler Thromb
Vasc Biol. 38:645–652. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Guarrera S, Fiorito G, Onland-Moret NC,
Russo A, Agnoli C, Allione A, Di Gaetano C, Mattiello A, Ricceri F,
Chiodini P, et al: Gene-specific DNA methylation profiles and
LINE-1 hypomethylation are associated with myocardial infarction
risk. Clin Epigenetics. 7:1332015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Shenker NS, Polidoro S, van Veldhoven K,
Sacerdote C, Ricceri F, Birrell MA, Belvisi MG, Brown R, Vineis P
and Flanagan JM: Epigenome-wide association study in the European
Prospective Investigation into Cancer and Nutrition (EPIC-Turin)
identifies novel genetic loci associated with smoking. Hum Mol
Genet. 22:843–851. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Zaina S, Heyn H, Carmona FJ, Varol N,
Sayols S, Condom E, Ramírez-Ruz J, Gomez A, Gonçalves I, Moran S
and Esteller M: DNA methylation map of human atherosclerosis. Circ
Cardiovasc Genet. 7:692–700. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Ligthart S, Marzi C, Aslibekyan S,
Mendelson MM, Conneely KN, Tanaka T, Colicino E, Waite LL, Joehanes
R, Guan W, et al: DNA methylation signatures of chronic low-grade
inflammation are associated with complex diseases. Genome Biol.
17:2552016. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Navas-Acien A, Domingo-Relloso A, Subedi
P, Riffo-Campos AL, Xia R, Gomez L, Haack K, Goldsmith J, Howard
BV, Best LG, et al: Blood DNA methylation and incident coronary
heart disease: Evidence from the strong heart study. JAMA Cardiol.
6:1237–1246. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Luo X, Hu Y, Shen J, Liu X, Wang T, Li L
and Li J: Integrative analysis of DNA methylation and gene
expression reveals key molecular signatures in acute myocardial
infarction. Clin Epigenetics. 14:462022. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Yoshioka J, Imahashi K, Gabel SA, Chutkow
WA, Burds AA, Gannon J, Schulze PC, MacGillivray C, London RE,
Murphy E and Lee RT: Targeted deletion of thioredoxin-interacting
protein regulates cardiac dysfunction in response to pressure
overload. Circ Res. 101:1328–1338. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Dorn LE, Lasman L, Chen J, Xu X, Hund TJ,
Medvedovic M, Hanna JH, van Berlo JH and Accornero F: The
N6-methyladenosine mRNA methylase METTL3 controls cardiac
homeostasis and hypertrophy. Circulation. 139:533–545. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Yin T, Wang N, Jia F, Wu Y, Gao L, Zhang J
and Hou R: Exosome-based WTAP siRNA delivery ameliorates myocardial
ischemia-reperfusion injury. Eur J Pharm Biopharm. 197:1142182024.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Madsen A, Krause J, Höppner G, Hirt MN,
Tan WLW, Lim I, Hansen A, Nikolaev VO, Foo RSY, Eschenhagen T and
Stenzig J: Hypertrophic signaling compensates for contractile and
metabolic consequences of DNA methyltransferase 3A loss in human
cardiomyocytes. J Mol Cell Cardiol. 154:115–123. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Madsen A, Höppner G, Krause J, Hirt MN,
Laufer SD, Schweizer M, Tan WLW, Mosqueira D, Anene-Nzelu CG, Lim
I, et al: An important role for DNMT3A-Mediated DNA methylation in
cardiomyocyte metabolism and contractility. Circulation.
142:1562–1578. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Schiano C, Balbi C, Burrello J, Ruocco A,
Infante T, Fiorito C, Panella S, Barile L, Mauro C, Vassalli G and
Napoli C: De novo DNA methylation induced by circulating
extracellular vesicles from acute coronary syndrome patients.
Atherosclerosis. 354:41–52. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Fang X, Poulsen RR, Wang-Hu J, Shi O,
Calvo NS, Simmons CS, Rivkees SA and Wendler CC: Knockdown of DNA
methyltransferase 3a alters gene expression and inhibits function
of embryonic cardiomyocytes. FASEB J. 30:3238–3255. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Stenzig J, Schneeberger Y, Löser A, Peters
BS, Schaefer A, Zhao RR, Ng SL, Höppner G, Geertz B, Hirt MN, et
al: Pharmacological inhibition of DNA methylation attenuates
pressure overload-induced cardiac hypertrophy in rats. J Mol Cell
Cardiol. 120:53–63. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Bomer N, Grote Beverborg N, Hoes MF,
Streng KW, Vermeer M, Dokter MM, IJmker J, Anker SD, Cleland JGF,
Hillege HL, et al: Selenium and outcome in heart failure. Eur J
Heart Fail. 22:1415–1423. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Zhu H, Wang X, Meng X, Kong Y, Li Y, Yang
C, Guo Y, Wang X, Yang H, Liu Z and Wang F: Selenium
supplementation improved cardiac functions by suppressing
DNMT2-Mediated GPX1 Promoter DNA methylation in AGE-induced heart
failure. Oxid Med Cell Longev. 2022:54029972022. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Ramachandran B, Stabley JN, Cheng SL,
Behrmann AS, Gay A, Li L, Mead M, Kozlitina J, Lemoff A, Mirzaei H,
et al: A GTPase-activating protein-binding protein
(G3BP1)/antiviral protein relay conveys arteriosclerotic Wnt
signals in aortic smooth muscle cells. J Biol Chem. 293:7942–7968.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Putra SED, Reichetzeder C, von Websky K,
Neuber C, Halle H, Kleuser B, Krämer BK and Hocher B: Association
between placental global DNA methylation and blood pressure during
human pregnancy. J Hypertens. 40:1002–1009. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Shao H, Chung J, Lee K, Balaj L, Min C,
Carter BS, Hochberg FH, Breakefield XO, Lee H, Weissleder R, et al:
Chip-based analysis of exosomal mRNA mediating drug resistance in
glioblastoma. Nat Commun. 6:69992015. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Yu W, Zhang L, Wei Q and Shao A:
O6-Methylguanine-DNA methyltransferase (MGMT): Challenges and new
opportunities in glioma chemotherapy. Front Oncol. 9:15472019.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Cao YL, Zhuang T, Xing BH, Li N and Li Q:
Exosomal DNMT1 mediates cisplatin resistance in ovarian cancer.
Cell Biochem Funct. 35:296–303. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Chen M and Wong CM: The emerging roles of
N6-methyladenosine (m6A) deregulation in liver carcinogenesis. Mol
Cancer. 19:442020. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Li T, Hu PS, Zuo Z, Lin JF, Li X, Wu QN,
Chen ZH, Zeng ZL, Wang F, Zheng J, et al: METTL3 facilitates tumor
progression via an mA-IGF2BP2-dependent mechanism in colorectal
carcinoma. Mol Cancer. 18:1122019. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Vu LP, Pickering BF, Cheng Y, Zaccara S,
Nguyen D, Minuesa G, Chou T, Chow A, Saletore Y, MacKay M, et al:
The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid
differentiation of normal hematopoietic and leukemia cells. Nat
Med. 23:1369–1376. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Han M, Liu Z, Xu Y, Liu X, Wang D, Li F,
Wang Y and Bi J: Abnormality of m6A mRNA methylation is involved in
Alzheimer's disease. Front Neurosci. 14:982020. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Fang J, Wu X, He J, Zhang H, Chen X, Zhang
H, Novakovic B, Qi H and Yu X: RBM15 suppresses hepatic insulin
sensitivity of offspring of gestational diabetes mellitus mice via
m6A-mediated regulation of CLDN4. Mol Med. 29:232023. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Liu Y, Wang X, Huang M, Luo A, Liu S, Cai
M, Li W, Yuan S, Zheng Z, Liu X and Tang C: METTL3 facilitates
kidney injury through promoting IRF4-mediated plasma cell
infiltration via an m6A-dependent manner in systemic lupus
erythematosus. BMC Med. 22:5112024. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Henning RJ: Cardiovascular exosomes and
MicroRNAs in cardiovascular physiology and pathophysiology. J
Cardiovasc Transl Res. 14:195–212. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Boulanger CM, Loyer X, Rautou PE and
Amabile N: Extracellular vesicles in coronary artery disease. Nat
Rev Cardiol. 14:259–272. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Waldenström A and Ronquist G: Role of
exosomes in myocardial remodeling. Circ Res. 114:315–324. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Lao KH, Zeng L and Xu Q: Endothelial and
smooth muscle cell transformation in atherosclerosis. Curr Opin
Lipidol. 26:449–456. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Wang Z, Gao D, Wang S, Lin H, Wang Y and
Xu W: Exosomal microRNA-1246 from human umbilical cord mesenchymal
stem cells potentiates myocardial angiogenesis in chronic heart
failure. Cell Biol Int. 45:2211–2225. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Wu T, Chen Y, Du Y, Tao J, Li W, Zhou Z
and Yang Z: Circulating exosomal miR-92b-5p is a promising
diagnostic biomarker of heart failure with reduced ejection
fraction patients hospitalized for acute heart failure. J Thorac
Dis. 10:6211–6220. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Wu T, Chen Y, Du Y, Tao J, Zhou Z and Yang
Z: Serum exosomal MiR-92b-5p as a potential biomarker for acute
heart failure caused by dilated cardiomyopathy. Cell Physiol
Biochem. 46:1939–1950. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Wang L, Liu J, Xu B, Liu YL and Liu Z:
Reduced exosome miR-425 and miR-744 in the plasma represents the
progression of fibrosis and heart failure. Kaohsiung J Med Sci.
34:626–633. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Xie Y, Hang JZ, Zhang N and Liu G:
Clinical significance of MiR-27a expression in serum exosomes in
patients with heart failure. Cell Mol Biol (Noisy-le-grand).
67:324–331. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Natrus L, Labudzynskyi D, Muzychenko P,
Chernovol P and Klys Y: Plasma-derived exosomes implement
miR-126-associated regulation of cytokines secretion in PBMCs of
CHF patients in vitro. Acta Biomed. 93:e20220662022.PubMed/NCBI
|
|
102
|
Fuchs FD and Whelton PK: High blood
pressure and cardiovascular disease. Hypertension. 75:285–292.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Bress AP, Colantonio LD, Cooper RS, Kramer
H, Booth JN III, Odden MC, Bibbins-Domingo K, Shimbo D, Whelton PK,
Levitan EB, et al: Potential cardiovascular disease events
prevented with adoption of the 2017 American college of
Cardiology/American heart association blood pressure guideline.
Circulation. 139:24–36. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Gan L, Guo X, Dong S and Sun C: The
biology of exosomes and exosomal non-coding RNAs in cardiovascular
diseases. Front Pharmacol. 16:15293752025. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Bhaskara M, Anjorin O and Wang M:
Mesenchymal stem Cell-derived exosomal microRNAs in cardiac
regeneration. Cells. 12:28152023. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Jayaseelan VP and Arumugam P: Exosomal
microRNAs as a promising theragnostic tool for essential
hypertension. Hypertens Res. 43:74–75. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Ren XS, Tong Y, Qiu Y, Ye C, Wu N, Xiong
XQ, Wang JJ, Han Y, Zhou YB, Zhang F, et al: MiR155-5p in
adventitial fibroblasts-derived extracellular vesicles inhibits
vascular smooth muscle cell proliferation via suppressing
angiotensin-converting enzyme expression. J Extracell Vesicles.
9:16987952019. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Wu N, Ye C, Zheng F, Wan GW, Wu LL, Chen
Q, Li YH, Kang YM and Zhu GQ: MiR155-5p inhibits cell migration and
oxidative stress in vascular smooth muscle cells of spontaneously
hypertensive rats. Antioxidants (Basel). 9:2042020. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Zou X, Wang J, Chen C, Tan X, Huang Y,
Jose PA, Yang J and Zeng C: Secreted monocyte miR-27a, via
mesenteric arterial Mas receptor-eNOS pathway, causes hypertension.
Am J Hypertens. 33:31–42. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Osada-Oka M, Shiota M, Izumi Y, Nishiyama
M, Tanaka M, Yamaguchi T, Sakurai E, Miura K and Iwao H:
Macrophage-derived exosomes induce inflammatory factors in
endothelial cells under hypertensive conditions. Hypertens Res.
40:353–360. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Oh J, Matkovich SJ, Riek AE, Bindom SM,
Shao JS, Head RD, Barve RA, Sands MS, Carmeliet G, Osei-Owusu P, et
al: Macrophage secretion of miR-106b-5p causes renin-dependent
hypertension. Nat Commun. 11:47982020. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Rodosthenous RS, Kloog I, Colicino E,
Zhong J, Herrera LA, Vokonas P, Schwartz J, Baccarelli AA and Prada
D: Extracellular vesicle-enriched microRNAs interact in the
association between long-term particulate matter and blood pressure
in elderly men. Environ Res. 167:640–649. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Ren Y and Zhang H: Emerging role of
exosomes in vascular diseases. Front Cardiovasc Med.
10:10909092023. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Chen L, Hu L, Li Q, Ma J and Li H:
Exosome-encapsulated miR-505 from ox-LDL-treated vascular
endothelial cells aggravates atherosclerosis by inducing NET
formation. Acta Biochim Biophys Sin (Shanghai). 51:1233–1241. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Shaheen N, Shaheen A, Diab RA and Desouki
MT: MicroRNAs (miRNAs) role in hypertension: Pathogenesis and
promising therapeutics. Ann Med Surg (Lond). 86:319–328. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Han J, Kang X, Su Y, Wang J, Cui X, Bian Y
and Wu C: Plasma exosomes from patients with coronary artery
disease promote atherosclerosis via impairing vascular endothelial
junctions. Sci Rep. 14:298132024. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Guo B, Zhuang TT, Li CC, Li F, Shan SK,
Zheng MH, Xu QS, Wang Y, Lei LM, Tang KX, et al: MiRNA-132/212
encapsulated by adipose tissue-derived exosomes Worsen
atherosclerosis progression. Cardiovasc Diabetol. 23:3312024.
View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Wehbe Z, Wehbe M, Al Khatib A, Dakroub AH,
Pintus G, Kobeissy F and Eid AH: Emerging understandings of the
role of exosomes in atherosclerosis. J Cell Physiol.
240:e314542025. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Xu S, Ilyas I, Little PJ, Li H, Kamato D,
Zheng X, Luo S, Li Z, Liu P, Han J, et al: Endothelial dysfunction
in atherosclerotic cardiovascular diseases and beyond: From
mechanism to pharmacotherapies. Pharmacol Rev. 73:924–967. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Alidadi M, Hjazi A, Ahmad I, Mahmoudi R,
Sarrafha M, Reza Hosseini-Fard S and Ebrahimzade M: Exosomal
non-coding RNAs: Emerging therapeutic targets in atherosclerosis.
Biochem Pharmacol. 212:1155722023. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Li L, Wang Z, Hu X, Wan T, Wu H, Jiang W
and Hu R: Human aortic smooth muscle cell-derived exosomal
miR-221/222 inhibits autophagy via a PTEN/Akt signaling pathway in
human umbilical vein endothelial cells. Biochem Biophys Res Commun.
479:343–350. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Li J, Tan M, Xiang Q, Zhou Z and Yan H:
Thrombin-activated platelet-derived exosomes regulate endothelial
cell expression of ICAM-1 via microRNA-223 during the
thrombosis-inflammation response. Thromb Res. 154:96–105. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Zhong X, Gao W, Wu R, Liu H and Ge J:
Dendritic cell exosome-shuttled miRNA146a regulates exosome-induced
endothelial cell inflammation by inhibiting IRAK-1: A feedback
control mechanism. Mol Med Rep. 20:5315–5323. 2019.PubMed/NCBI
|
|
124
|
Liu P, Wang S, Wang G, Zhao M, Du F, Li K,
Wang L, Wu H, Chen J, Yang Y and Su G: Macrophage-derived exosomal
miR-4532 promotes endothelial cells injury by targeting SP1 and
NF-κB P65 signalling activation. J Cell Mol Med. 26:5165–5180.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Zheng B, Yin WN, Suzuki T, Zhang XH, Zhang
Y, Song LL, Jin LS, Zhan H, Zhang H, Li JS and Wen JK:
Exosome-Mediated miR-155 transfer from smooth muscle cells to
endothelial cells induces endothelial injury and promotes
atherosclerosis. Mol Ther. 25:1279–1294. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Loyer X, Potteaux S, Vion AC, Guérin CL,
Boulkroun S, Rautou PE, Ramkhelawon B, Esposito B, Dalloz M, Paul
JL, et al: Inhibition of microRNA-92a prevents endothelial
dysfunction and atherosclerosis in mice. Circ Res. 114:434–443.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Masson JJR, Cherry CL, Murphy NM,
Sada-Ovalle I, Hussain T, Palchaudhuri R, Martinson J, Landay AL,
Billah B, Crowe SM and Palmer CS: Polymorphism rs1385129 Within
Glut1 Gene SLC2A1 is linked to poor CD4+ T cell recovery in
Antiretroviral-Treated HIV+ individuals. Front Immunol. 9:9002018.
View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Lee MA, Raad N, Song MH, Yoo J, Lee M,
Jang SP, Kwak TH, Kook H, Choi EK, Cha TJ, et al: The matricellular
protein CCN5 prevents adverse atrial structural and electrical
remodelling. J Cell Mol Med. 24:11768–11778. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Stamatikos A, Knight E, Vojtech L, Bi L,
Wacker BK, Tang C and Dichek DA: Exosome-mediated transfer of
Anti-miR-33a-5p from transduced endothelial cells enhances
macrophage and vascular smooth muscle cell cholesterol efflux. Hum
Gene Ther. 31:219–232. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Chen F, Li J, She J, Chen T and Yuan Z:
Exosomal microRNA-16-5p from macrophage exacerbates atherosclerosis
via modulating mothers against decapentaplegic homolog 7. Microvasc
Res. 142:1043682022. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Wang G, Li Y, Liu Z, Ma X, Li M, Lu Q, Li
Y, Lu Z, Niu L, Fan Z and Lei Z: Circular RNA circ_0124644
exacerbates the ox-LDL-induced endothelial injury in human vascular
endothelial cells through regulating PAPP-A by acting as a sponge
of miR-149-5p. Mol Cell Biochem. 471:51–61. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Lin B, Xie W, Zeng C, Wu X, Chen A, Li H,
Jiang R and Li P: Transfer of exosomal microRNA-203-3p from
dendritic cells to bone marrow-derived macrophages reduces
development of atherosclerosis by downregulating Ctss in mice.
Aging (Albany NY). 13:15638–15658. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Adamidis PS, Pantazi D, Moschonas IC,
Liberopoulos E and Tselepis AD: Neutrophil extracellular traps
(NETs) and atherosclerosis: Does hypolipidemic treatment have an
effect? J Cardiovasc Dev Dis. 11:722024.PubMed/NCBI
|
|
134
|
Sano M, Maejima Y, Nakagama S,
Shiheido-Watanabe Y, Tamura N, Hirao K, Isobe M and Sasano T:
Neutrophil extracellular traps-mediated Beclin-1 suppression
aggravates atherosclerosis by inhibiting macrophage autophagy.
Front Cell Dev Biol. 10:8761472022. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Brinkmann V, Reichard U, Goosmann C,
Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y and Zychlinsky A:
Neutrophil extracellular traps kill bacteria. Science.
303:1532–1535. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Zhang YG, Song Y, Guo XL, Miao RY, Fu YQ,
Miao CF and Zhang C: Exosomes derived from oxLDL-stimulated
macrophages induce neutrophil extracellular traps to drive
atherosclerosis. Cell Cycle. 18:2674–268. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Wang X, Li D, Chen H, Wei X and Xu X:
Expression of Long Noncoding RNA LIPCAR promotes cell
proliferation, cell migration, and change in phenotype of vascular
smooth muscle cells. Med Sci Monit. 25:7645–7651. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Hu N, Zeng X, Tang F and Xiong S: Exosomal
long non-coding RNA LIPCAR derived from oxLDL-treated THP-1 cells
regulates the proliferation of human umbilical vein endothelial
cells and human vascular smooth muscle cells. Biochem Biophys Res
Commun. 575:65–72. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Liu Y, Zhang WL, Gu JJ, Sun YQ, Cui HZ, Bu
JQ and Chen ZY: Exosome-mediated miR-106a-3p derived from ox-LDL
exposed macrophages accelerated cell proliferation and repressed
cell apoptosis of human vascular smooth muscle cells. Eur Rev Med
Pharmacol Sci. 24:7039–7050. 2020.PubMed/NCBI
|
|
140
|
Zhang Z, Yi D, Zhou J, Zheng Y, Gao Z, Hu
X, Ying G, Peng X and Wen T: Exosomal LINC01005 derived from
oxidized low-density lipoprotein-treated endothelial cells
regulates vascular smooth muscle cell phenotypic switch.
Biofactors. 46:743–753. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Chen L, Yang W, Guo Y, Chen W, Zheng P,
Zeng J and Tong W: Exosomal lncRNA GAS5 regulates the apoptosis of
macrophages and vascular endothelial cells in atherosclerosis. PLoS
One. 12:e01854062017. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Huang C, Han J, Wu Y, Li S, Wang Q, Lin W
and Zhu J: Exosomal MALAT1 derived from oxidized low-density
lipoprotein-treated endothelial cells promotes M2 macrophage
polarization. Mol Med Rep. 18:509–515. 2018.PubMed/NCBI
|
|
143
|
Gao H, Wang X, Lin C, An Z, Yu J, Cao H,
Fan Y and Liang X: Exosomal MALAT1 derived from ox-LDL-treated
endothelial cells induce neutrophil extracellular traps to
aggravate atherosclerosis. Biol Chem. 401:367–376. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Wen Y, Chun Y, Lian ZQ, Yong ZW, Lan YM,
Huan L, Xi CY, Juan LS, Qing ZW, Jia C and Ji ZH:
circRNA-0006896-miR1264-DNMT1 axis plays an important role in
carotid plaque destabilization by regulating the behavior of
endothelial cells in atherosclerosis. Mol Med Rep. 23:3112021.
View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Lu Z, Tang H, Li S, Zhu S, Li S and Huang
Q: Role of circulating exosomes in cerebrovascular diseases: A
comprehensive review. Curr Neuropharmacol. 21:1575–1593. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Yu Z, Huang Q, Zhang Q, Wu H and Zhong Z:
CircRNAs open a new era in the study of cardiovascular disease
(review). Int J Mol Med. 47:49–64. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Zhang P, Liang T, Wang X, Wu T, Xie Z, Yu
Y and Yu H: Serum-derived exosomes from patients with coronary
artery disease induce endothelial injury and inflammation in human
umbilical vein endothelial cells. Int Heart J. 62:396–406. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Zarà M, Campodonico J, Cosentino N, Biondi
ML, Amadio P, Milanesi G, Assanelli E, Cerri S, Biggiogera M,
Sandrini L, et al: Plasma exosome profile in ST-Elevation
myocardial infarction patients with and without Out-of-Hospital
cardiac arrest. Int J Mol Sci. 22:80652021. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Tong X, Dang X, Liu D, Wang N, Li M, Han
J, Zhao J, Wang Y, Huang M, Yang Y, et al: Exosome-derived
circ_0001785 delays atherogenesis through the ceRNA network
mechanism of miR-513a-5p/TGFBR3. J Nanobiotechnology. 21:3622023.
View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Byrne RA, Rossello X, Coughlan JJ, Barbato
E, Berry C, Chieffo A, Claeys MJ, Dan GA, Dweck MR, Galbraith M, et
al: 2023 ESC Guidelines for the management of acute coronary
syndromes. Eur Heart J. 44:3720–382. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Ong SG, Lee WH, Huang M, Dey D, Kodo K,
Sanchez-Freire V, Gold JD and Wu JC: Cross talk of combined gene
and cell therapy in ischemic heart disease: role of exosomal
microRNA transfer. Circulation. 130 (Suppl 1):S60–S69. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Saparov A, Ogay V, Nurgozhin T, Chen WCW,
Mansurov N, Issabekova A and Zhakupova J: Role of the immune system
in cardiac tissue damage and repair following myocardial
infarction. Inflamm Res. 66:739–751. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Li Y, Yang R, Guo B, Zhang H, Zhang H, Liu
S and Li Y: Exosomal miR-301 derived from mesenchymal stem cells
protects myocardial infarction by inhibiting myocardial autophagy.
Biochem Biophys Res Commun. 514:323–332. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
De Rosa S, Fichtlscherer S, Lehmann R,
Assmus B, Dimmeler S and Zeiher AM: Transcoronary concentration
gradients of circulating microRNAs. Circulation. 124:1936–1944.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Peng Y, Zhao JL, Peng ZY, Xu WF and Yu GL:
Exosomal miR-25-3p from mesenchymal stem cells alleviates
myocardial infarction by targeting pro-apoptotic proteins and EZH2.
Cell Death Dis. 11:3172020. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Wang W, Zheng Y, Wang M, Yan M, Jiang J
and Li Z: Exosomes derived miR-126 attenuates oxidative stress and
apoptosis from ischemia and reperfusion injury by targeting ERRFI1.
Gene. 690:75–80. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Pan J, Alimujiang M, Chen Q, Shi H and Luo
X: Exosomes derived from miR-146a-modified adipose-derived stem
cells attenuate acute myocardial infarction-induced myocardial
damage via downregulation of early growth response factor 1. J Cell
Biochem. 120:4433–4443. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Li H, Ding J, Liu W, Wang X, Feng Y, Guan
H and Chen Z: Plasma exosomes from patients with acute myocardial
infarction alleviate myocardial injury by inhibiting ferroptosis
through miR-26b-5p/SLC7A11 axis. Life Sci. 322:1216492023.
View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Feng Y, Huang W, Wani M, Yu X and Ashraf
M: Ischemic preconditioning potentiates the protective effect of
stem cells through secretion of exosomes by targeting Mecp2 via
miR-22. PLoS One. 9:e886852014. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Wang J, Huang W, Xu R, Nie Y, Cao X, Meng
J, Xu X, Hu S and Zheng Z: MicroRNA-24 regulates cardiac fibrosis
after myocardial infarction. J Cell Mol Med. 16:2150–2160. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Wang K, Jiang Z, Webster KA, Chen J, Hu H,
Zhou Y, Zhao J, Wang L, Wang Y, Zhong Z, et al: Enhanced
Cardioprotection by human endometrium mesenchymal stem cells driven
by exosomal MicroRNA-21. Stem Cells Transl Med. 6:209–222. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Liao B, Dong S, Xu Z, Gao F, Zhang S and
Liang R: MiR-19b-3p regulated by BC002059/ABHD10 axis promotes cell
apoptosis in myocardial infarction. Biol Direct. 17:202022.
View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Shyu KG, Wang BW, Fang WJ, Pan CM and Lin
CM: Hyperbaric oxygen-induced long non-coding RNA MALAT1 exosomes
suppress MicroRNA-92a expression in a rat model of acute myocardial
infarction. J Cell Mol Med. 24:12945–12954. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Huang P, Wang L, Li Q, Tian X, Xu J, Xu J,
Xiong Y, Chen G, Qian H, Jin C, et al: Atorvastatin enhances the
therapeutic efficacy of mesenchymal stem cells-derived exosomes in
acute myocardial infarction via up-regulating long non-coding RNA
H19. Cardiovasc Res. 116:353–367. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Wang L and Zhang J: Exosomal lncRNA
AK139128 Derived from hypoxic cardiomyocytes promotes apoptosis and
inhibits cell proliferation in cardiac fibroblasts. Int J
Nanomedicine. 15:3363–3376. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Cheng M, Yang J, Zhao X, Zhang E, Zeng Q,
Yu Y, Yang L, Wu B, Yi G, Mao X, et al: Circulating myocardial
microRNAs from infarcted hearts are carried in exosomes and
mobilise bone marrow progenitor cells. Nat Commun. 10:9592019.
View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Huang P, Wang L, Li Q, Tian X, Xu J, Xu J,
Xiong Y, Chen G, Qian H, Jin C, et al: Atorvastatin enhances the
therapeutic efficacy of mesenchymal stem cells-derived exosomes in
acute myocardial infarction via up-regulating long non-coding RNA
H19. Cardiovasc Res. 116:353–367. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Zhang Y, Liu Y, Liu H and Tang WH:
Exosomes: Biogenesis, biologic function and clinical potential.
Cell Biosci. 9:192019. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Xu H, Du X, Xu J, Zhang Y, Tian Y, Liu G,
Wang X, Ma M, Du W, Liu Y, et al: Pancreatic β cell microRNA-26a
alleviates type 2 diabetes by improving peripheral insulin
sensitivity and preserving β cell function. PLoS Biol.
18:e30006032020. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Fu Q, Jiang H, Wang Z, Wang X, Chen H,
Shen Z, Xiao L, Guo X and Yang T: Injury factors alter miRNAs
profiles of exosomes derived from islets and circulation. Aging
(Albany NY). 10:3986–3999. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
da Costa Martins PA, Bourajjaj M, Gladka
M, Kortland M, van Oort RJ, Pinto YM, Molkentin JD and De Windt LJ:
Conditional dicer gene deletion in the postnatal myocardium
provokes spontaneous cardiac remodeling. Circulation.
118:1567–1576. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Chen JF, Murchison EP, Tang R, Callis TE,
Tatsuguchi M, Deng Z, Rojas M, Hammond SM, Schneider MD and Selzman
CH: Targeted deletion of Dicer in the heart leads to dilated
cardiomyopathy and heart failure. Proc Natl Acad Sci USA.
105:2111–2116. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Landgraf P, Rusu M, Sheridan R, Sewer A,
Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M,
et al: A mammalian microRNA expression atlas based on small RNA
library sequencing. Cell. 129:1401–1414. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Kalluri R and LeBleu VS: The biology,
function, and biomedical applications of exosomes. Science.
367:eaau69772020. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Liang Y, Duan L, Lu J and Xia J:
Engineering exosomes for targeted drug delivery. Theranostics.
11:3183–3195. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Vader P, Mol EA, Pasterkamp G and
Schiffelers RM: Extracellular vesicles for drug delivery. Adv Drug
Deliv Rev. 106:148–156. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
177
|
O'Brien K, Breyne K, Ughetto S, Laurent LC
and Breakefield XO: RNA delivery by extracellular vesicles in
mammalian cells and its applications. Nat Rev Mol Cell Biol.
21:585–606. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
178
|
Liu Y, Wang M, Yu Y, Li C and Zhang C:
Advances in the study of exosomes derived from mesenchymal stem
cells and cardiac cells for the treatment of myocardial infarction.
Cell Commun Signal. 21:2022023. View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Das D, Jothimani G, Banerjee A, Dey A,
Duttaroy AK and Pathak S: A brief review on recent advances in
diagnostic and therapeutic applications of extracellular vesicles
in cardiovascular disease. Int J Biochem Cell Biol. 173:1066162024.
View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Li Z, Zhao P, Zhang Y, Wang J, Wang C, Liu
Y, Yang G and Yuan L: Exosome-based Ldlr gene therapy for familial
hypercholesterolemia in a mouse model. Theranostics. 11:2953–2965.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Huang L, Yang L, Ding Y, Jiang X, Xia Z
and You Z: Human umbilical cord mesenchymal stem cells-derived
exosomes transfers microRNA-19a to protect cardiomyocytes from
acute myocardial infarction by targeting SOX6. Cell Cycle.
19:339–353. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Li Y, Zhou J, Zhang O, Wu X, Guan X, Xue
Y, Li S, Zhuang X, Zhou B, Miao G and Zhang L: Bone marrow
mesenchymal stem cells-derived exosomal microRNA-185 represses
ventricular remolding of mice with myocardial infarction by
inhibiting SOCS2. Int Immunopharmacol. 80:1061562020. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Zhao J, Li X, Hu J, Chen F, Qiao S, Sun X,
Gao L, Xie J and Xu B: Mesenchymal stromal cell-derived exosomes
attenuate myocardial ischaemia-reperfusion injury through
miR-182-regulated macrophage polarization. Cardiovasc Res.
115:1205–1216. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Liu Y, Li C, Wu H, Xie X, Sun Y and Dai M:
Paeonol attenuated inflammatory response of endothelial cells via
stimulating Monocytes-Derived exosomal MicroRNA-223. Front
Pharmacol. 9:11052018. View Article : Google Scholar : PubMed/NCBI
|
|
185
|
Schena GJ, Murray EK, Hildebrand AN,
Headrick AL, Yang Y, Koch KA, Kubo H, Eaton D, Johnson J, Berretta
R, et al: Cortical bone stem cell-derived exosomes' therapeutic
effect on myocardial ischemia-reperfusion and cardiac remodeling.
Am J Physiol Heart Circ Physiol. 321:H1014–H1029. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Lin CM, Wang BW, Pan CM, Fang WJ, Chua SK,
Cheng WP and Shyu KG: Chrysin boosts KLF2 expression through
suppression of endothelial cell-derived exosomal microRNA-92a in
the model of atheroprotection. Eur J Nutr. 60:4345–4355. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Zhu J, Liu B, Wang Z, Wang D, Ni H, Zhang
L and Wang Y: Exosomes from nicotine-stimulated macrophages
accelerate atherosclerosis through miR-21-3p/PTEN-mediated VSMC
migration and proliferation. Theranostics. 9:6901–6919. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
188
|
Bergmann O, Bhardwaj RD, Bernard S, Zdunek
S, Barnabé-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA,
Druid H, et al: Evidence for cardiomyocyte renewal in humans.
Science. 324:98–102. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Smith RR, Barile L, Cho HC, Leppo MK, Hare
JM, Messina E, Giacomello A, Abraham MR and Marbán E: Regenerative
potential of cardiosphere-derived cells expanded from percutaneous
endomyocardial biopsy specimens. Circulation. 115:896–908. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
190
|
Teerlink JR, Metra M, Filippatos GS,
Davison BA, Bartunek J, Terzic A, Gersh BJ, Povsic TJ, Henry TD,
Alexandre B, et al: Benefit of cardiopoietic mesenchymal stem cell
therapy on left ventricular remodelling: Results from the
congestive heart failure cardiopoietic regenerative therapy
(CHART-1) study. Eur J Heart Fail. 19:1520–1529. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
191
|
Bartunek J, Terzic A, Behfar A and Wijns
W: Clinical experience with regenerative therapy in heart failure:
Advancing care with cardiopoietic stem cell interventions. Circ
Res. 122:1344–1346. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
192
|
Wang Y, Zhao M, Liu S, Guo J, Lu Y, Cheng
J and Liu J: Macrophage-derived extracellular vesicles: Diverse
mediators of pathology and therapeutics in multiple diseases. Cell
Death Dis. 11:9242020. View Article : Google Scholar : PubMed/NCBI
|
|
193
|
EL Andaloussi S, Mäger I, Breakefield XO
and Wood MJ: Extracellular vesicles: Biology and emerging
therapeutic opportunities. Nat Rev Drug Discov. 12:347–357. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
194
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
195
|
Sun D, Zhuang X, Xiang X, Liu Y, Zhang S,
Liu C, Barnes S, Grizzle W, Miller D and Zhang HG: A Novel
nanoparticle drug delivery system: The Anti-inflammatory activity
of curcumin is enhanced when encapsulated in exosomes. Mol Ther.
18:1606–1614. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
196
|
Sherman LS, Shaker M, Mariotti V and
Rameshwar P: Mesenchymal stromal/stem cells in drug therapy: New
perspective. Cytotherapy. 19:19–27. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
197
|
Ubanako P, Mirza S, Ruff P and Penny C:
Exosome-mediated delivery of siRNA molecules in cancer therapy:
Triumphs and challenges. Front Mol Biosci. 11:14479532024.
View Article : Google Scholar : PubMed/NCBI
|
|
198
|
Sun L, Xu R, Sun X, Duan Y, Han Y, Zhao Y,
Qian H, Zhu W and Xu W: Safety evaluation of exosomes derived from
human umbilical cord mesenchymal stromal cell. Cytotherapy.
18:413–422. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
199
|
Fu W, Li T, Chen H, Zhu S and Zhou C:
Research progress in Exosome-based nanoscale drug carriers in tumor
therapies. Front Oncol. 12:9192792022. View Article : Google Scholar : PubMed/NCBI
|
|
200
|
Smyth T, Kullberg M, Malik N, Smith-Jones
P, Graner MW and Anchordoquy TJ: Biodistribution and delivery
efficiency of unmodified tumor-derived exosomes. J Control Release.
199:145–155. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
201
|
Frolova L and Li ITS: Targeting
capabilities of native and bioengineered extracellular vesicles for
drug delivery. Bioengineering (Basel). 9:4962022. View Article : Google Scholar : PubMed/NCBI
|
|
202
|
Jeyaram A, Lamichhane TN, Wang S, Zou L,
Dahal E, Kronstadt SM, Levy D, Parajuli B, Knudsen DR, Chao W and
Jay SM: Enhanced loading of functional miRNA cargo via pH gradient
modification of extracellular vesicles. Mol Ther. 28:975–985. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
203
|
Perocheau D, Touramanidou L, Gurung S,
Gissen P and Baruteau J: Clinical applications for exosomes: Are we
there yet? Br J Pharmacol. 178:2375–2392. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
204
|
Lamparski HG, Methadamani A, Yao JY, Patel
S, Hsu DH, Ruegg C and Le PJ: Production and characterization of
clinical grade exosomes derived from dendritic cells. J Immunol
Methods. 270:211–226. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
205
|
Liu Y, Li D, Liu Z, Zhou Y, Chu D, Li X,
Jiang X, Hou D, Chen X and Chen Y: Targeted exosome-mediated
delivery of opioid receptor Mu siRNA for the treatment of morphine
relapse. Sci Rep. 5:175432015. View Article : Google Scholar : PubMed/NCBI
|