Inflammatory bowel disease (IBD) is a type of
noninfectious chronic gastrointestinal inflammation disease that
includes Crohn's disease (CD) and ulcerative colitis (UC) and is
characterized by chronic or intermittent inflammation of the
intestinal mucosa (1). Globally,
in 2019, 405,000 (95% UI 361,000 to 457,000) new cases of IBD and
41,000 (95% UI 35,000 to 45,000) deaths from IBD have been reported
(2). It is projected that the
age-standardized mortality rate from IBD may continue to decline
from 2020 to 2050 (2). The results
of clinical trials have indicated primary non-response rates of up
to 40%, and a significant challenge is the inability to predict
which treatment will benefit individual patients (3). The investigation of preclinical human
IBD mechanisms is one of the key concerns (4).
The mucus layer of the intestines is the first
interface to insulate the complex microbial environment, and the
integrity of the mucus layer directly affects intestinal barrier
function and the development of IBD (5). Mucus is a dilute, aqueous and
viscoelastic secretion that contains 90–95% water, 1–2% lipids,
electrolytes, 29 core proteins and other substances; mucins are the
primary structural and functional components in mucus and are
present at concentrations of 1–5% (6). Mucus separates the epithelium and
millions of antigens from food, the environment and the microbiome
while permitting nutrient absorption and gas exchange (7).
Goblet cells secrete large polymers of gel-forming
mucin 2 (MUC2), which compose the structural backbone of the mucus
that coats the epithelial surface (8). The MUC2 mucus barrier plays important
roles in the response to changes in dietary patterns, MUC2 mucus
barrier dysfunction, contact stimulation with colonic epithelial
cells, and mucosal and submucosal inflammation during the
occurrence and development of IBD (9,10).
MUC2 gene expression is reduced or absent in patients with CD,
whereas MUC2 protein expression and secretion are decreased in
active UC, resulting in a thin mucus layer and increased intestinal
absorption (11). The absence of
MUC2 expression in the mucus layer renders animals vulnerable to
intestinal inflammation, which leads to the development of
spontaneous colitis and predisposes them to the development of
colorectal cancer (12).
MUC2-knockout mice present clinical signs of colitis along with
mucosal thickening, increased proliferation, and superficial
erosion (13).
The present review aimed to explore the importance
of MUC2 in the mucus barrier and the novel UPR pathway to provide
new ideas for future research on IBD.
The human MUC gene family encodes 19 typical
mucin-type glycoproteins, which are recognized by the Human Genome
Organization Gene Nomenclature Committee (http://www.genenames.org) and can be divided into
three subgroups, membrane-bound (including MUC1, MUC3A and MUC3B),
secreted gel-forming (including MUC2, MUC5AC and MUC5B) and
secreted non-gel-forming (MUC7 and MUC8) (Table I). Among these, MUC2 is the most
important secreted and gel-forming component of the intestinal
mucus in the human intestine and provides a first line of defense
against pathogens in the gut immune system (20).
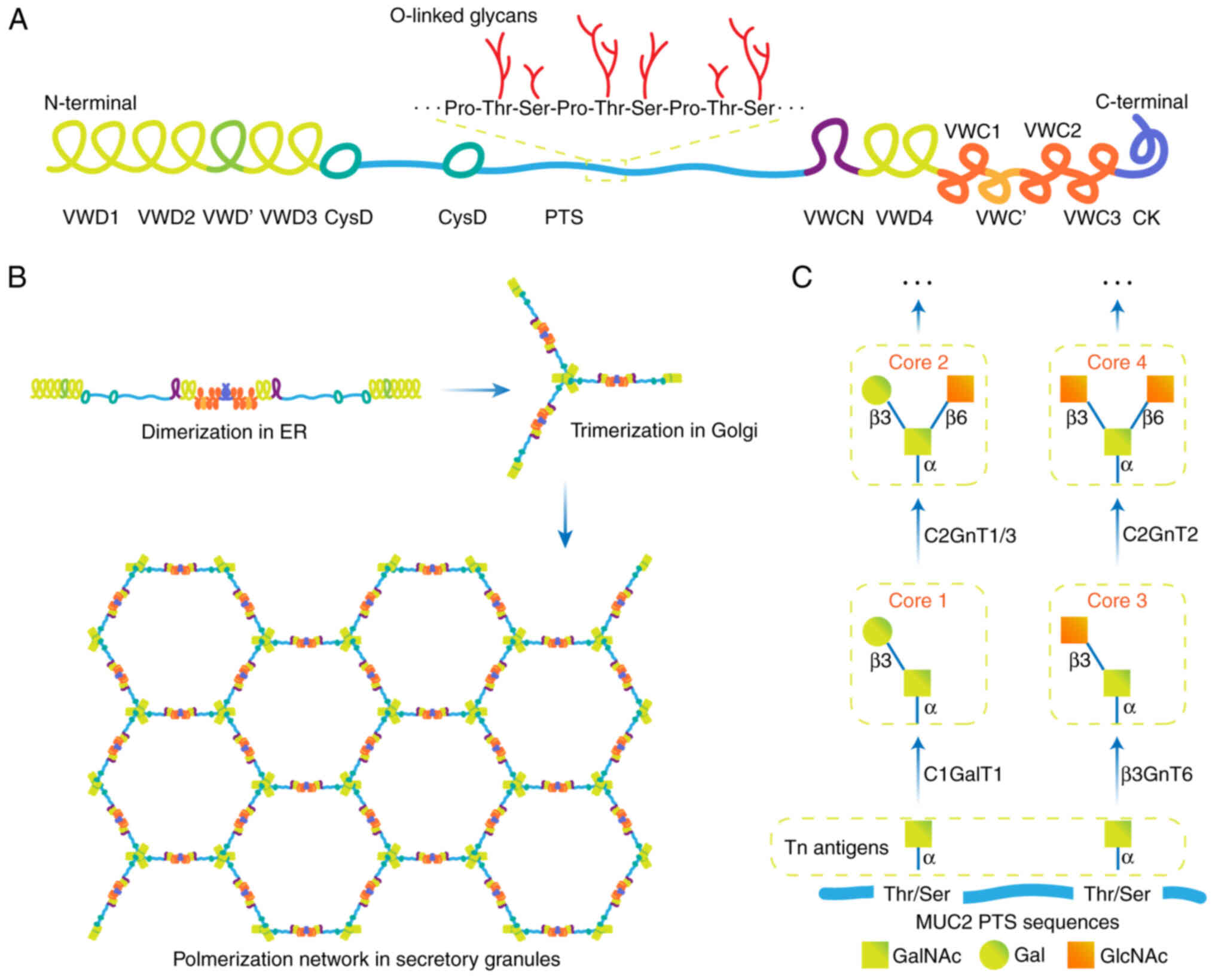
Cysteine residues located at the N- and C-termini of
MUC2 are highly glycosylated, which increases the hydrophilicity of
MUC2 and allows lubrication of the intestinal mucosa (25). MUC2 contains 215 cysteine amino
acids that make up >10% of the total amino acids outside the PTS
mucin domains (26). As all
cysteines need to be interlinked (oxidized) to exit the ER, the
correct assembly of MUC2 is a formidable challenge (26). A MUC2 monomer contains up to 1,600
O-glycans and 30 N-glycans, resulting in more than 3,300 terminal
sugar residues (27).
MUC2 monomers are N-glycosylated within the ER, a
process that enables correct MUC2 folding, stable dimer formation
and maturation (22). The CK
domain in the C-terminus of MUC2 forms a dimer via disulfide bonds
between three cysteine amino acids via the ER machinery (Fig. 1B) (28). The correct folding of MUC2 is
assisted by various molecular chaperones that prevent the
misfolding and aggregation of proteins in the ER, such as binding
immunoglobulin protein (BiP, GRP78), anterior gradient 2 (AGR2),
calnexin and calreticulin (29).
Next, MUC2 dimers are transported to the Golgi, and
the hydroxy amino acids (Ser and Thr) of the PTS domains become
O-glycosylated, in which >80% of all Ser and Thr residues carry
O-linked glycans (30). The
typically O-glycosylated compounds are largely concentrated in the
PTS domains, which resemble bottle brushes with protein cores at
their centers and oligosaccharides as their bristles (31). In the trans-Golgi network, the
glycosylated dimers are then trimerized via disulfide bonding in
VWD3, and a net-like structure is generated (Fig. 1B) (32). The glycosylated MUC2 monomer has a
mass of ~2.5 MDa and the polymer may have a mass of >100 MDa
(33). An important function of
O-glycans is to cover the protein backbone and thus protect the
mucin from proteolytic enzymes while simultaneously binding water
to generate a gel (33). Glycans
also act as attachment sites for bacteria, as they are important
food sources (34). An increase in
the number of small glycans is found in most patients with active
UC, and a decrease in the number of more complex compounds has been
detected (31).
The distribution of glycosyltransferases (GalNAc-Ts)
throughout the Golgi apparatus is not homogenous, and each
compartment has a distinct composition of GalNAc-Ts (35). To achieve controlled, sequential
extension, the GalNAc-Ts are spatially arranged according to the
order of monosaccharide addition (27). First, O-glycosylation is a covalent
linkage initiated by 1 of the 20 GalNAc-Ts that add
N-acetylgalactosamine (GalNAc) to the Ser or Thr amino acid
residues in the MUC2 PTS sequence (36). The Pro amino acids in the PTS
ensure a non-folded structure allowing the GalNAc-Ts of the Golgi
apparatus to access the protein core (37). The structures formed during this
initial stage of modification are known as Tn antigens. Next, the
glucan chains are further extended to form four core structures
known as Core 1–4 (Fig. 1C)
(38). The addition of galactose
(Gal) by Core 1 β1-3-galactosyltransferase (C1GalT1) is known as
core 1 (39). The addition of
N-acetylglucosamine (GlcNAc) by Core 3
β1-3-N-acetylglucosaminyltransferase (β3GnT6) to peptide-bound
GalNAc is known as Core 3 (40).
Core 2 is formed by the addition of β1-6GlcNAc to the GalNAcs of
Core 1 through Core 2 β1,6-N-acetylglucosaminyltransferase 1/3
(C2GnT1/3) (41). Core 4 is formed
by the addition of β1-6GlcNAc to the GalNAc of Core 3 through Core
2 β1,6-N-acetylglucosaminyltransferase-2 (C2GnT2) (38). Groups that can be attached to core
structures include galactose, sialic acid, sulfate and fucose
(42). The order in which the
glycans interact with different GalNAc-Ts plays a critical role in
O-glycosylation since specific monosaccharides, such as fucose or
N-acetylneuraminic acid, can limit further elongation (27). There are large differences in MUC2
characteristics between animal species, such as mouse colonic MUC2,
which largely contains Core 2 and minor amounts of Core 3 and Core
4, whereas human MUC2 predominantly contains Core 3 structures,
Core 4 to a lesser extent, and essentially lacks Core 1 (42).
MUC2 forms large net-like structures that are
densely packed in the secretory granules of goblet cells (43). Fully synthesized MUC2 is released
from goblet cells through baseline secretion or via
Ca2+−dependent stimulated secretion (44). The pH along the secretory pathway
shifts gradually from 7.2 in the ER to 6.0 in the trans-Golgi
network, and to 5.2 in the secretory granule (45). Moreover, the intragranular
Ca2+ concentration increases, suggesting that the
packing of MUC2 may be pH- and Ca2+-dependent (45). The N-terminal trimers spontaneously
form concatenated rings with long-extended mucin domains standing
perpendicular to this structure and joining to other MUC2 molecules
end-to-end through their C-terminus (46).
In addition, the hydrolytic activities of some
proteases also play a role in the process of MUC2 volume expansion
(50). Calcium-activated chloride
channel regulator 1 is highly abundant in intestinal mucus, with an
N-terminal metalloprotease domain, a central von Willebrand A
domain and a C-terminal fibronectin type III domain that likely has
both metallohydrolase activity and structural roles and can process
N-terminal MUC2 via proteolysis (51). Transglutaminase 3, the dominant
cross-linking enzyme in the mouse colon, catalyzes cross-linking of
the MUC2 protein and increases the resistance of MUC2 to
degradation (52). Under the
synergistic effects of numerous factors, MUC2 forms a net-like
structure and establishes a sturdy mucus barrier.
In response to environmental factors, such as
pathogenic bacterial infection, there is an increased demand for
MUC2 synthesis, resulting in a significant protein folding and
modification burden on the ER of goblet cells (53). Additionally, owing to the large
size (~2.5 MDa of a monomer) and complex structure of MUC2, it is
prone to misfolding, which may result in the accumulation of
misfolded proteins within the ER lumen, thereby triggering ER
stress (54). Abnormal synthesis
of MUC2 induces a unique model of IBD caused by ER stress (55). In the Winnie and Eeyore mouse
models (which carry single missense MUC2 mutations), misfolded MUC2
proteins heavily accumulate in membranous structures, and
quantitative PCR reveals a two- to threefold increase in the
expression of BiP mRNA (an ER stress marker) in the proximal and
distal colon (16). Winnie and
Eeyore mice develop mild spontaneous inflammation of the colon
accompanied by goblet cell ER stress (16). Winnie mice exhibit altered mucus
production as early as 4 weeks of age, after which colonic
inflammation begins (56).
ER stress aggravates MUC2 synthesis abnormalities
and causes a series of chain reactions (57). The synthesis of MUC2 begins in the
ER, where it must be properly folded and bound to a molecular
chaperone before it can be transported to the Golgi apparatus
(58). Once abnormal MUC2 folding
occurs, it is difficult to perform glycosylation, sulfation and
other modifications, which are performed in the Golgi apparatus,
leading to further functional defects in MUC2 (31). For example, the O-glycosylation
patterns of MUC2 are significantly altered in patients with IBD,
especially in patients with UC; these patterns include
O-glycosylation reduction, a reduction in the length of the
O-glycan chains of MUC2 and genetic defects in GalNAc-Ts enzymes
(59). Sulfation is catalyzed
mainly by sulfotransferases located in the Golgi apparatus, and
sulfotransferase adds sulfuric acid groups to the sugar chains of
MUC2, resulting in the formation of negatively charged molecules
whose hydration and viscosity properties are increased (11). Since the presence of a sugar chain
is necessary for sulfation, the obstruction of MUC2 glycosylation
reduces the number of sulfation sites, thus affecting the addition
and distribution of sulfuric acid groups and resulting in changes
in the charge and viscosity properties of MUC2 (60). Moreover, delayed transport or
misfolding of MUC2 due to ER stress can affect the recognition
sites of proteolytic enzymes, resulting in the failure of MUC2 to
undergo proper hydrolysis and extracellular secretion of MUC2,
further impairing intestinal barrier function (61).
UPR signaling pathways are activated to stop
improper translation, refold unfolded proteins, and degrade
irreversible unfolded proteins via the ER-associated degradation
(ERAD) pathway, in which unfolded/misfolded proteins that have
accumulated in the ER are transported to the cytosol for
degradation by the ubiquitin-proteasome system (62). UPR sensors monitor ER protein
folding capacity, and their effectors balance protein synthesis,
folding, trafficking, and degradation to alleviate ER stress
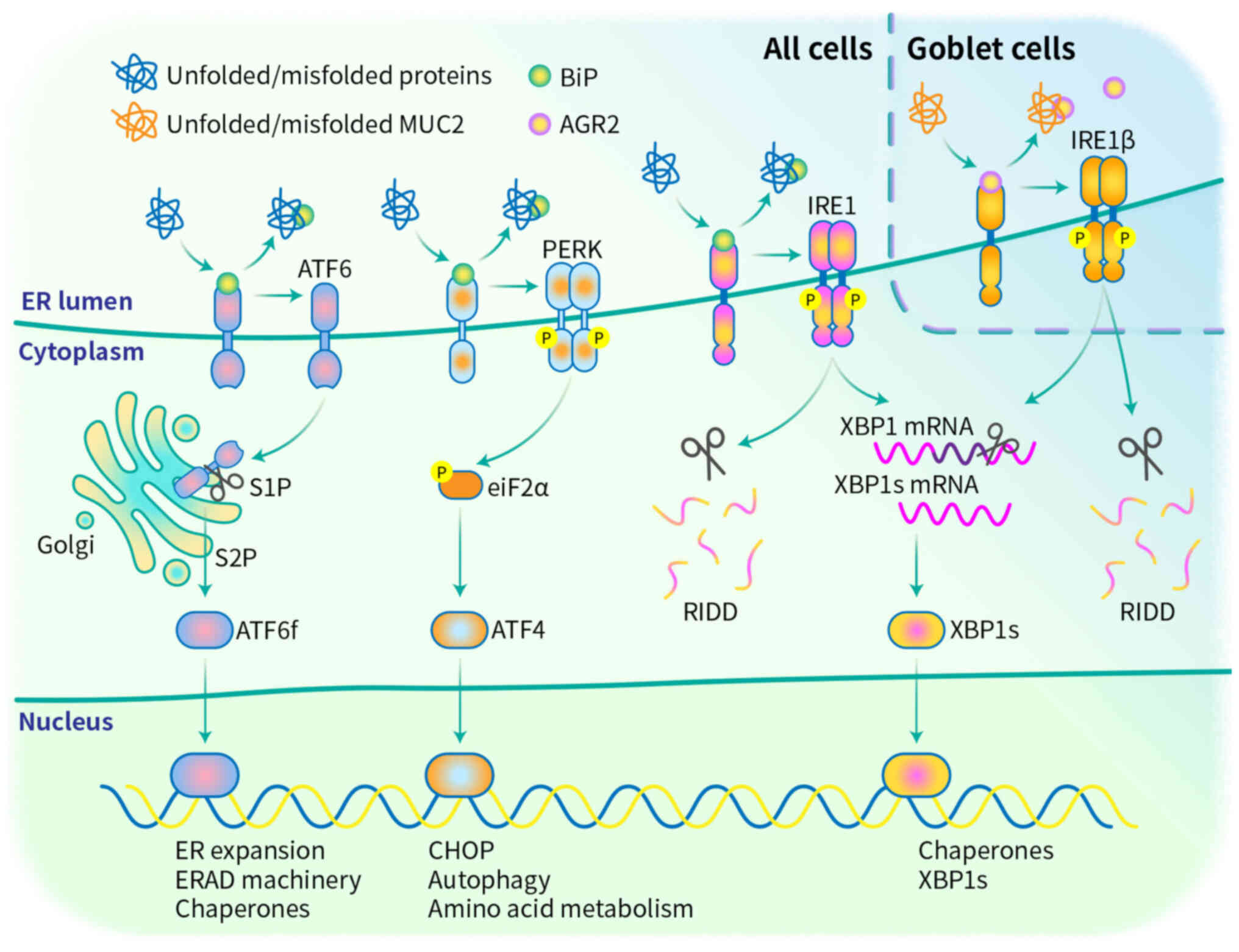
(62). The mammalian UPR has three
known signaling branches: Pancreatic ER kinase (PERK), ER
transmembrane inositol-requiring enzymes 1α and β (IRE1α and IRE1β)
and activating transcription factor 6 (ATF6) (Fig. 2). These transmembrane sensor
proteins have an ER luminal sensor domain and a cytosolic effector
domain, thereby transmitting the protein folding status inside the
ER to other cellular compartments via intracellular signaling
pathways (63). In the absence of
misfolded proteins in the ER lumen, these cascades are usually
maintained in an inactivated state by the chaperone BiP (64). However, under elevated ER stress,
unfolded proteins compete for BiP binding, thereby detaching BiP
from the sensing molecules, which activates the signaling cascade
to overcome the stress environment and sustain homeostasis
(64). Moreover, under protracted
and acute ER stress, the UPR initiates cascades that can trigger
apoptosis in goblet cells, which are essential for the production
of MUC2 (65). Thus, maintaining
the balance of the UPR is very important.
IRE1 proteins, which include IRE1α and IRE1β,
possess both an ER luminal sensor domain and a cytosolic
endoribonuclease (RNase) domain and show kinase activity (66). Kinase activity enables
trans-autophosphorylation, which activates RNase activity (67). Activated IRE1 splices X-Box Binding
Protein 1 (XBP1) mRNA and removes an inhibitory 26-nucleotide
intron from the XBP1 transcript (68). The mRNA is subsequently re-ligated
by RNA terminal phosphorylase B, generating a functionally active
isoform of XBP1 (XBP1s) (69).
XBP1s bind to ER stress response element (ERSE), ER stress response
element II (ERSE II) and UPR element sequences (70). This process upregulates the
expression of genes that encode proteins that promote protein
refolding (ER chaperone proteins), aid in the destruction of
proteins that are beyond repair (components of the ERAD machinery)
and encode proteins that facilitate the expansion of the ER, thus
increasing protein folding capacity (71). In addition to splicing XBP1, IRE1
RNase activity can cleave other RNA targets in a process known as
regulated IRE1-dependent decay (72).
PERK is a transmembrane protein located in the ER,
and its N-terminal regulatory motif is located in the lumen and
adjoins the cytosolic eIF2α kinase domain (73). PERK is maintained in an inactive
state by the interaction of its ER luminal domain with BiP
(74). Once a misfolded protein is
produced, BiP dissociates from PERK due to its preferential binding
to hydrophobic residues of misfolded proteins (75). Once disassociated, following
phosphorylation at Thr980 by autophosphorylation, PERK dimerizes to
form an active homodimer (76).
The activation of PERK leads to the phosphorylation of eukaryotic
initiation factor 2 (eIF2α) at Ser51 (77). Phosphorylated eIF2α perturbs 80S
ribosome assembly inhibiting protein translation, thus blocking the
production of an additional influx of nascent polypeptides that
could worsen the ER stress (78).
Moreover, activating transcription factor 4 (ATF4)
escapes translational attenuation by eIF2α phosphorylation because
ATF4 has upstream open reading frames (ORFs) in its 5′-untranslated
region (79). These upstream ORFs
prevent translation of ATF4 under normal conditions and are
bypassed only when eIF2α is phosphorylated; therefore, ATF4
translation occurs (80). ATF4
translation activates the expression of the transcription factor
CCAAT/enhancer binding protein-homologous protein, a master
regulator of ER stress-induced apoptosis and plays pro-apoptotic
roles in the stress response (81). Chronic or prolonged PERK signaling
can lead to goblet cell apoptosis in IBD (82).
ATF6 proteins are type II transmembrane proteins
that contain basic leucine zipper (bZIP) motifs in their cytosolic
domains and a C-terminus protrudes that into the ER lumen (83). After disassociation from BiP under
ER/oxidative stress conditions, ATF6 translocates from the ER to
the Golgi apparatus, where it is cleaved by site-1 protease (S1P)
and site-2 protease (S2P) to remove the luminal and transmembrane
domains (84). This process
results in the generation of a cytosolic fragment with
transcription factor activity which is designated as ATF6f, the
released ATF6f then translocate to the nucleus and regulates the
expression of genes encoding BiP, XBP1s and ERAD components
(85).
Unlike IRE1α, which plays a broad role in the UPR by
splicing XBP1 mRNA and degrading misfolded proteins, IRE1β has
specialized functions in secretory epithelial cells, especially in
goblet cells (86). Vertebrates
express two IRE1 paralogs, IRE1α, which is universally expressed
and IRE1β, which is specifically expressed within mucus-secreting
cells in the respiratory and gastrointestinal tracts (87). Single-cell RNA sequencing revealed
that the abundance of IRE1β mRNAs is ~50-fold greater than that of
IRE1α mRNAs in the goblet cells of the small intestinal epithelium
(88). Loss of IRE1β expression
results in the accumulation of misfolded MUC2 precursor proteins in
the ER of immature goblet cells, and defects in MUC2 maturation and
its accumulation in the ER lead to marked ER abnormalities and
signs of ER stress (89).
The role of IRE1β in intestinal mucus barrier
homeostasis can't be replaced by IRE1α (90). The transplantation of microbiota
from conventionally raised mice to germ-free mice restored goblet
cell numbers; this restoration was completely abolished in
IRE1β-deficient mice, despite normal IRE1α expression, but it
failed to compensate for IRE1β function (91). Compared with normal colonic
tissues, the tumor tissues exhibited a significant increase in
XBP1s mRNA expression levels, no significant difference in IRE1α
mRNA expression, and a significant decrease in both IRE1β and MUC2
mRNA and protein levels (92).
Moreover, the expression levels of 23 genes associated with the
mechanistic target of rapamycin complex 1 signaling pathway were
increased in the IRE1β-knockout intestinal epithelial cells (IECs)
compared with IRE1α knockout IECs in both an ER stress mouse model
treated with tunicamycin, and a colitis mouse model induced by 2.5%
DSS (93). These findings
indicated that, unlike IRE1α, IRE1β exerts a novel non-canonical
splicing target gene unique in IECs.
AGR2 selectively binds to IRE1β, but not IRE1α.
AGR2, a member of the protein disulfide isomerase (PDI) family with
redox and molecular chaperone functions that process and modify
immature mucins into mature mucins, is highly expressed in goblet
cells (94). AGR2 forms disulfide
bonds with the N- and C-terminal cysteine-rich sections of MUC2
(94). Impaired AGR2 function,
such as H117Y mutation and S-glutathiolation modification, directly
suppresses MUC2 biosynthesis, inducing ER stress and driving IBD
pathogenesis (58,95). Bio-Layer Interferometry revealed a
half-maximal binding concentration of 19 µM for binding of the
IRE1β luminal domain (LD) to AGR2, whereas an immobilized IRE1α LD
resulted in significantly weaker binding signals with AGR2
(96). The interaction between
IRE1β and AGR2 does not involve disulfide bonds, and AGR2 binding
favors the formation of IRE1β LD monomers (96). Both the C81S and H117Y mutations in
AGR2 abrogate its ability to bind and inhibit IRE1β activity
(97). Importantly, both the
N-terminus (residues 21–1,397) and the C-terminus (residues
4,198-6,178) of MUC2 have a derepression effect on the IRE1β-AGR2
pair, and the derepression effect on IRE1α is relatively weak under
the same conditions (96).
These results suggest that the IRE1β-mediated UPR
likely works by binding with AGR2 in a manner such as that of BiP
and IRE1α, by antagonizing IRE1β LD dimerization (Fig. 2) (98). The IRE1β-AGR2 pathway may be an
important UPR pathway in goblet cells.
MUC2 dysfunction disrupts intestinal host-commensal
homeostasis. An imbalance in host-microbiota interactions leading
to a thinner mucus layer may be an early sign of IBD (99). The exposed terminal glycans on the
outer surface of MUC2 can be recognized by bacteria and lectins,
providing adhesion binding sites for intestinal commensal bacteria
and utilizing mucin O-glycans as an energy source (27). Escherichia coli secretes the
metalloprotease SslE, Vibrio cholerae secretes the
metalloprotease TagA, and Candida albicans secretes the
aspartyl protease Sap2, which can all degrade mucins, potentially
allowing microbial penetration of the mucosal barrier (100). Decreased MUC2 expression induced
by REGγ gene deficiency in goblet cells leads to dysregulation of
the intestinal flora (101).
ST6GalNAc1 (ST6)-mediated sialylation protects MUC2 from bacterial
proteolytic (such as protease of C1 esterase inhibitor and
O-glycoprotease) degradation to maintain mucus integrity, and ST6
R319Q mutation mice harbor a different microbiome with less
diversity (102).
MUC2 dysfunction leads to an imbalance in the
extracellular matrix (ECM), resulting in ECM remodeling, and
further exacerbating intestinal fibrosis in IBD. The ECM forms a
complex network of multidomain macromolecules composed of collagen,
elastin, fibronectin, laminin, aminoglycans and proteoglycans
(103). ECM provides structural
support to IECs while cooperatively establishing intestinal barrier
function with the mucus layer (103). Upon disruption of the mucus
barrier, bacterial products such as lipopolysaccharides recruit
neutrophils, which release MMPs and elastase, leading to excessive
degradation of ECM components including elastin, collagen,
fibronectin, and proteoglycans (104). Although probiotics generally have
beneficial effects, there is a risk that probiotics (such as
Bacteroides fragilis and Lactobacillus gasseri) may
degrade the ECM components abundant in either the mucosa or
submucosa (collagen I and IV, laminin, fibronectin and hyaluronic
acid) and exacerbate inflammation (105).
Although appropriate UPR have beneficial effects,
chronic and aberrant UPR aggravates ECM remodeling, which in turn
exacerbates MUC2 dysfunction. The IRE1α/XBP1 arm of the UPR
upregulates the expression of integrins, laminins, collagens, MMP1,
MMP10 and MMP9 in epithelial cells, which further leads to the
remodeling of the basement membrane (106). Excessive degradation of the ECM
may inhibit the differentiation of intestinal stem cells into
goblet cells by increasing yes-associated protein 1 (YAP)-dependent
signaling, thereby reducing the MUC2 synthesis (107,108). Moreover, excessive
proinflammatory cytokines such as tumor necrosis factor α and
interleukin 13 produced in the ECM upregulate MUC2 mRNA synthesis,
which increases the metabolic load on intestinal cells and causes
ER stress in IECs (109,110).
The development of artificial mucus layer and
small-molecule drugs targeting the UPR signaling pathway have shown
efficacy in IBD. An artificial mucus layer formed from a
thiol-substituted hyaluronic acid derivative excellent protection
against the penetration of Escherichia coli (111). Epithelial cell ER-targeted
protein, a recombinant variant of the cholera toxin B subunit,
induces colon epithelial mucosal healing in colitis by activating
the IRE1α/XBP1 arm of the UPR in colon epithelial cells (112,113). Integrated stress response
inhibitor, a small molecule inhibitor of the UPR, blocks eIF2α
phosphorylation and PERK pathway activation, thereby preserving
intestinal epithelial integrity in IBD by mitigating aberrant
apoptosis (114). The development
of small-molecule drugs targeting the IRE1β-AGR2-MUC2 axis of the
UPR in goblet cells and further elucidating the functional
distinctions between IRE1α and IRE1β may provide new strategies for
mucosal healing in IBD.
The effects of UPR signaling pathways are intricate
and interconnected and involve both positive and negative effects.
The present review emphasizes the positive impact of the UPR on the
synthesis of MUC2 in early-IBD. But its negative effects, such as
the induction of apoptosis and ECM remodeling, cannot be ignored.
Focusing solely on isolated aspects of mucus layer function yields
limited understanding, research and therapeutic strategies should
adopt a holistic approach.
Not applicable.
The present study was supported by Three-year Action Plan for
the Construction of the Public Health System of Songjiang
(Shanghai, China; 2023-2025) (grant no. SJGW6-23).
Not applicable.
ZWY and FZ designed and supervised the review. ZX,
LYL and JZX conducted literature organization and revised the
review. ZWY contributed to draft the manuscript. ZWY and FZ
reviewed this manuscript. All authors contributed to the manuscript
and approved all aspects of the work. All authors read and approved
the final version of the manuscript. Data authentication is not
applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Gilliland A, Chan JJ, De Wolfe TJ, Yang H
and Vallance BA: Pathobionts in inflammatory bowel disease:
Origins, underlying mechanisms, and implications for clinical care.
Gastroenterology. 166:44–58. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhou JL, Bao JC, Liao XY, Chen YJ, Wang
LW, Fan YY, Xu QY, Hao LX, Li KJ, Liang MX, et al: Trends and
projections of inflammatory bowel disease at the global, regional
and national levels, 1990–2050: a bayesian age-period-cohort
modeling study. BMC Public Health. 23:25072023. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wyatt NJ, Watson H, Anderson CA, Kennedy
NA, Raine T, Ahmad T, Allerton D, Bardgett M, Clark E, Clewes D, et
al: Defining predictors of responsiveness to advanced therapies in
Crohn's disease and ulcerative colitis: protocol for the
IBD-RESPONSE and nested CD-metaRESPONSE prospective, multicentre,
observational cohort study in precision medicine. BMJ Open.
14:e0736392024. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Heller C, Moss AC and Rubin DT: Overview
to challenges in IBD 2024–2029. Inflamm Bowel Dis. 30 (Suppl
2):S1–S4. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gasaly N, Hermoso MA and Gotteland M:
Butyrate and the fine-tuning of colonic homeostasis: Implication
for inflammatory bowel diseases. Int J Mol Sci. 22:30612021.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
van der Post S, Jabbar KS, Birchenough G,
Arike L, Akhtar N, Sjovall H, Johansson MEV and Hansson GC:
Structural weakening of the colonic mucus barrier is an early event
in ulcerative colitis pathogenesis. Gut. 68:2142–2151. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Reznik N, Gallo AD, Rush KW, Javitt G,
Fridmann-Sirkis Y, Ilani T, Nairner NA, Fishilevich S, Gokhman D,
Chacón KN, et al: Intestinal mucin is a chaperone of multivalent
copper. Cell. 185:4206–4215.e11. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tonetti FR, Eguileor A and Llorente C:
Goblet cells: Guardians of gut immunity and their role in
gastrointestinal diseases. eGastroenterology. 2:e1000982024.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Leoncini G, Cari L, Ronchetti S, Donato F,
Caruso L, Calafà C and Villanacci V: Mucin expression profiles in
ulcerative colitis: New Insights on the histological mucosal
healing. Int J Mol Sci. 25:18582024. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yao D, Dai W, Dong M, Dai C and Wu S: MUC2
and related bacterial factors: Therapeutic targets for ulcerative
colitis. EBioMedicine. 74:1037512021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kang Y, Park H, Choe BH and Kang B: The
role and function of mucins and its relationship to inflammatory
bowel disease. Front Med (Lausanne). 9:8483442022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu M, Wu Y, Li J, Bao Y, Guo Y and Yang W:
The dynamic changes of gut microbiota in Muc2 deficient mice. Int J
Mol Sci. 19:28092018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Van der Sluis M, De Koning BA, De Bruijn
AC, Velcich A, Meijerink JP, Van Goudoever JB, Büller HA, Dekker J,
Van Seuningen I, Renes IB and Einerhand AW: Muc2-deficient mice
spontaneously develop colitis, indicating that MUC2 is critical for
colonic protection. Gastroenterology. 131:117–129. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Engevik MA, Herrmann B, Ruan W, Engevik
AC, Engevik KA, Ihekweazu F, Shi Z, Luck B, Chang-Graham AL,
Esparza M, et al: Bifidobacterium dentium-derived
y-glutamylcysteine suppresses ER-mediated goblet cell stress and
reduces TNBS-driven colonic inflammation. Gut Microbes. 13:1–21.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Das I, Png CW, Oancea I, Hasnain SZ,
Lourie R, Proctor M, Eri RD, Sheng Y, Crane DI, Florin TH and
McGuckin MA: Glucocorticoids alleviate intestinal ER stress by
enhancing protein folding and degradation of misfolded proteins. J
Exp Med. 210:1201–1216. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Heazlewood CK, Cook MC, Eri R, Price GR,
Tauro SB, Taupin D, Thornton DJ, Png CW, Crockford TL, Cornall RJ,
et al: Aberrant mucin assembly in mice causes endoplasmic reticulum
stress and spontaneous inflammation resembling ulcerative colitis.
PLoS Med. 5:e542008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
López-Cauce B, Puerto M, García JJ,
Ponce-Alonso M, Becerra-Aparicio F, Del Campo R, Peligros I,
Fernández-Aceñero MJ, Gómez-Navarro Y, Lara JM, et al: Akkermansia
deficiency and mucin depletion are implicated in intestinal barrier
dysfunction as earlier event in the development of inflammation in
interleukin-10-deficient mice. Front Microbiol. 13:10838842023.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wiseman RL, Mesgarzadeh JS and Hendershot
LM: Reshaping endoplasmic reticulum quality control through the
unfolded protein response. Mol Cell. 82:1477–1491. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gao H, He C, Hua R, Guo Y, Wang B, Liang
C, Gao L, Shang H and Xu JD: Endoplasmic reticulum stress of gut
enterocyte and intestinal diseases. Front Mol Biosci. 9:8173922022.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fekete E and Buret AG: The role of mucin
O-glycans in microbiota dysbiosis, intestinal homeostasis, and
host-pathogen interactions. Am J Physiol Gastrointest Liver
Physiol. 324:G452–G465. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gum JR Jr, Hicks JW, Toribara NW, Siddiki
B and Kim YS: Molecular cloning of human intestinal mucin (MUC2)
cDNA. Identification of the amino terminus and overall sequence
similarity to prepro-von Willebrand factor. J Biol Chem.
269:2440–2446. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu Y, Yu Z, Zhu L, Ma S, Luo Y, Liang H,
Liu Q, Chen J, Guli S and Chen X: Orchestration of MUC2-The key
regulatory target of gut barrier and homeostasis: A review. Int J
Biol Macromol. 236:1238622023. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gallego P, Garcia-Bonete MJ, Trillo-Muyo
S, Recktenwald CV, Johansson MEV and Hansson GC: The intestinal
MUC2 mucin C-terminus is stabilized by an extra disulfide bond in
comparison to von Willebrand factor and other gel-forming mucins.
Nat Commun. 14:19692023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Stanforth KJ, Zakhour MI, Chater PI,
Wilcox MD, Adamson B, Robson NA and Pearson JP: The MUC2 gene
product: Polymerisation and post-secretory organisation-current
models. Polymers (Basel). 16:16632024. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu Y, Yu X, Zhao J, Zhang H, Zhai Q and
Chen W: The role of MUC2 mucin in intestinal homeostasis and the
impact of dietary components on MUC2 expression. Int J Biol
Macromol. 164:884–891. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pelaseyed T, Bergström JH, Gustafsson JK,
Ermund A, Birchenough GM, Schütte A, van der Post S, Svensson F,
Rodríguez-Piñeiro AM, Nyström EE, et al: The mucus and mucins of
the goblet cells and enterocytes provide the first defense line of
the gastrointestinal tract and interact with the immune system.
Immunol Rev. 260:8–20. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Luis AS and Hansson GC: Intestinal mucus
and their glycans: A habitat for thriving microbiota. Cell Host
Microbe. 31:1087–1100. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Arike L and Hansson GC: The densely
o-glycosylated MUC2 mucin protects the intestine and provides food
for the commensal bacteria. J Mol Biol. 428:3221–3229. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
McCool DJ, Okada Y, Forstner JF and
Forstner GG: Roles of calreticulin and calnexin during mucin
synthesis in LS180 and HT29/A1 human colonic adenocarcinoma cells.
Biochem J. 341((Pt 3)): 593–600. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Johansson ME, Larsson JM and Hansson GC:
The two mucus layers of colon are organized by the MUC2 mucin,
whereas the outer layer is a legislator of host-microbial
interactions. Proc Natl Acad Sci USA. 108 (Suppl 1):S4659–S4665.
2011. View Article : Google Scholar
|
|
31
|
Larsson JM, Karlsson H, Crespo JG,
Johansson ME, Eklund L, Sjövall H and Hansson GC: Altered
O-glycosylation profile of MUC2 mucin occurs in active ulcerative
colitis and is associated with increased inflammation. Inflamm
Bowel Dis. 17:2299–2307. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Javitt G, Khmelnitsky L, Albert L, Bigman
LS, Elad N, Morgenstern D, Ilani T, Levy Y, Diskin R and Fass D:
Assembly mechanism of mucin and von willebrand factor polymers.
Cell. 183:717–729.e16. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bergstrom KS and Xia L: Mucin-type
O-glycans and their roles in intestinal homeostasis. Glycobiology.
23:1026–1037. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Paone P and Cani PD: Mucus barrier, mucins
and gut microbiota: The expected slimy partners? Gut. 69:2232–2243.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kouka T, Akase S, Sogabe I, Jin C,
Karlsson NG and Aoki-Kinoshita KF: Computational modeling of
o-linked glycan biosynthesis in CHO Cells. Molecules. 27:17662022.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nielsen MI, de Haan N, Kightlinger W, Ye
Z, Dabelsteen S, Li M, Jewett MC, Bagdonaite I, Vakhrushev SY and
Wandall HH: Global mapping of GalNAc-T isoform-specificities and
O-glycosylation site-occupancy in a tissue-forming human cell line.
Nat Commun. 13:62572022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bennett EP, Mandel U, Clausen H, Gerken
TA, Fritz TA and Tabak LA: Control of mucin-type O-glycosylation: A
classification of the polypeptide GalNAc-transferase gene family.
Glycobiology. 22:736–756. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Brockhausen I, Wandall HH, Hagen KGT and
Stanley P: O-GalNAc Glycans. Essentials of Glycobiology. Varki A,
Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Mohnen D,
Kinoshita T, Packer NH, Prestegard JH, Schnaar RL and Seeberger PH:
Cold Spring Harbor Laboratory Press; New York, USA: pp. 117–128.
2022
|
|
39
|
Xia L, Ju T, Westmuckett A, An G, Ivanciu
L, McDaniel JM, Lupu F, Cummings RD and McEver RP: Defective
angiogenesis and fatal embryonic hemorrhage in mice lacking core
1-derived O-glycans. J Cell Biol. 164:451–459. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bergstrom K, Fu J, Johansson ME, Liu X,
Gao N, Wu Q, Song J, McDaniel JM, McGee S, Chen W, et al: Core 1-
and 3-derived O-glycans collectively maintain the colonic mucus
barrier and protect against spontaneous colitis in mice. Mucosal
Immunol. 10:91–103. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Schwientek T, Yeh JC, Levery SB, Keck B,
Merkx G, van Kessel AG, Fukuda M and Clausen H: Control of O-glycan
branch formation. Molecular cloning and characterization of a novel
thymus-associated core 2 beta1, 6-n-acetylglucosaminyltransferase.
J Biol Chem. 275:11106–11113. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hansson GC: Mucins and the Microbiome.
Annu Rev Biochem. 89:769–793. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Song C, Chai Z, Chen S, Zhang H, Zhang X
and Zhou Y: Intestinal mucus components and secretion mechanisms:
what we do and do not know. Exp Mol Med. 55:681–691. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang Z and Shen J: The role of goblet
cells in Crohn's disease. Cell Biosci. 14:432024. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Birchenough GM, Johansson ME, Gustafsson
JK, Bergström JH and Hansson GC: New developments in goblet cell
mucus secretion and function. Mucosal Immunol. 8:712–719. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ambort D, Johansson ME, Gustafsson JK,
Nilsson HE, Ermund A, Johansson BR, Koeck PJ, Hebert H and Hansson
GC: Calcium and pH-dependent packing and release of the gel-forming
MUC2 mucin. Proc Natl Acad Sci USA. 109:5645–5650. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Harrison CA, Laubitz D, Ohland CL,
Midura-Kiela MT, Patil K, Besselsen DG, Jamwal DR, Jobin C, Ghishan
FK and Kiela PR: Microbial dysbiosis associated with impaired
intestinal Na(+)/H(+) exchange accelerates and exacerbates colitis
in ex-germ free mice. Mucosal Immunol. 11:1329–1341. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ridley C, Kouvatsos N, Raynal BD, Howard
M, Collins RF, Desseyn JL, Jowitt TA, Baldock C, Davis CW,
Hardingham TE and Thornton DJ: Assembly of the respiratory mucin
MUC5B: A new model for a gel-forming mucin. J Biol Chem.
289:16409–16420. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Johansson ME, Phillipson M, Petersson J,
Velcich A, Holm L and Hansson GC: The inner of the two Muc2
mucin-dependent mucus layers in colon is devoid of bacteria. Proc
Natl Acad Sci USA. 105:15064–15069. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Nyström EEL, Birchenough GMH, van der Post
S, Arike L, Gruber AD, Hansson GC and Johansson MEV:
Calcium-activated chloride channel regulator 1 (CLCA1) controls
mucus expansion in colon by proteolytic activity. EBioMedicine.
33:134–143. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Nyström EEL, Arike L, Ehrencrona E,
Hansson GC and Johansson MEV: Calcium-activated chloride channel
regulator 1 (CLCA1) forms non-covalent oligomers in colonic mucus
and has mucin 2-processing properties. J Biol Chem.
294:17075–17089. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Sharpen JDA, Dolan B, Nyström EEL,
Birchenough GMH, Arike L, Martinez-Abad B, Johansson MEV, Hansson
GC and Recktenwald CV: Transglutaminase 3 crosslinks the secreted
gel-forming mucus component Mucin-2 and stabilizes the colonic
mucus layer. Nat Commun. 13:452022. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ma X, Dai Z, Sun K, Zhang Y, Chen J, Yang
Y, Tso P, Wu G and Wu Z: Intestinal epithelial cell endoplasmic
reticulum stress and inflammatory bowel disease pathogenesis: An
update review. Front Immunol. 8:12712017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Verjan Garcia N, Hong KU and Matoba N: The
unfolded protein response and its implications for novel
therapeutic strategies in inflammatory bowel disease. Biomedicines.
11:20662023. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Chieppa M, De Santis S and Verna G: Winnie
mice: A chronic and progressive model of ulcerative colitis.
Inflamm Bowel Dis. 31:1158–1167. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Liso M, De Santis S, Verna G, Dicarlo M,
Calasso M, Santino A, Gigante I, Eri R, Raveenthiraraj S,
Sobolewski A, et al: A specific mutation in muc2 determines early
dysbiosis in colitis-prone winnie mice. Inflamm Bowel Dis.
26:546–556. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Kaser A and Blumberg RS: Endoplasmic
reticulum stress in the intestinal epithelium and inflammatory
bowel disease. Semin Immunol. 21:156–163. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Al-Shaibi AA, Abdel-Motal UM, Hubrack SZ,
Bullock AN, Al-Marri AA, Agrebi N, Al-Subaiey AA, Ibrahim NA,
Charles AK; COLORS in IBD-Qatar Study Group, ; et al: Human AGR2
deficiency causes mucus barrier dysfunction and infantile
inflammatory bowel disease. Cell Mol Gastroenterol Hepatol.
12:1809–1830. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kudelka MR, Stowell SR, Cummings RD and
Neish AS: Intestinal epithelial glycosylation in homeostasis and
gut microbiota interactions in IBD. Nat Rev Gastroenterol Hepatol.
17:597–617. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Grondin JA, Kwon YH, Far PM, Haq S and
Khan WI: Mucins in intestinal mucosal defense and inflammation:
Learning from clinical and experimental studies. Front Immunol.
11:20542020. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Pelaseyed T and Hansson GC: Membrane
mucins of the intestine at a glance. J Cell Sci. 133:jcs2409292020.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Hosomi S, Kaser A and Blumberg RS: Role of
endoplasmic reticulum stress and autophagy as interlinking pathways
in the pathogenesis of inflammatory bowel disease. Curr Opin
Gastroenterol. 31:81–88. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Li H, Wen W and Luo J: Targeting
endoplasmic reticulum stress as an effective treatment for
alcoholic pancreatitis. Biomedicines. 10:1082022. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Oikawa D, Kimata Y, Kohno K and Iwawaki T:
Activation of mammalian IRE1alpha upon ER stress depends on
dissociation of BiP rather than on direct interaction with unfolded
proteins. Exp Cell Res. 315:2496–2504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Deka D, D'Incà R, Sturniolo GC, Das A,
Pathak S and Banerjee A: Role of ER stress mediated unfolded
protein responses and ER stress inhibitors in the pathogenesis of
inflammatory bowel disease. Dig Dis Sci. 67:5392–5406. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Le Goupil S, Laprade H, Aubry M and Chevet
E: Exploring the IRE1 interactome: From canonical signaling
functions to unexpected roles. J Biol Chem. 300:1071692024.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Ferri E, Le Thomas A, Wallweber HA, Day
ES, Walters BT, Kaufman SE, Braun MG, Clark KR, Beresini MH,
Mortara K, et al: Activation of the IRE1 RNase through remodeling
of the kinase front pocket by ATP-competitive ligands. Nat Commun.
11:63872020. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Hetz C, Zhang K and Kaufman RJ:
Mechanisms, regulation and functions of the unfolded protein
response. Nat Rev Mol Cell Biol. 21:421–438. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Lu Y, Liang FX and Wang X: A synthetic
biology approach identifies the mammalian UPR RNA ligase RtcB. Mol
Cell. 55:758–770. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Yamamoto K, Yoshida H, Kokame K, Kaufman
RJ and Mori K: Differential contributions of ATF6 and XBP1 to the
activation of endoplasmic reticulum stress-responsive cis-acting
elements ERSE, UPRE and ERSE-II. J Biochem. 136:343–350. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Ong G and Logue SE: Unfolding the
interactions between endoplasmic reticulum stress and oxidative
stress. Antioxidants (Basel). 12:9812023. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Lu M, Lawrence DA, Marsters S,
Acosta-Alvear D, Kimmig P, Mendez AS, Paton AW, Paton JC, Walter P
and Ashkenazi A: Opposing unfolded-protein-response signals
converge on death receptor 5 to control apoptosis. Science.
345:98–101. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Wek RC, Anthony TG and Staschke KA:
Surviving and adapting to stress: Translational control and the
integrated stress response. Antioxid Redox Signal. 39:351–373.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Urra H and Hetz C: Fine-tuning PERK
signaling to control cell fate under stress. Nat Struct Mol Biol.
24:789–790. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Kopp MC, Larburu N, Durairaj V, Adams CJ
and Ali MMU: UPR proteins IRE1 and PERK switch BiP from chaperone
to ER stress sensor. Nat Struct Mol Biol. 26:1053–1062. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Elvira R, Cha SJ, Noh GM, Kim K and Han J:
PERK-Mediated eIF2α phosphorylation contributes to the protection
of dopaminergic neurons from chronic heat stress in drosophila. Int
J Mol Sci. 21:8452020. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Gorbatyuk MS, Starr CR and Gorbatyuk OS:
Endoplasmic reticulum stress: New insights into the pathogenesis
and treatment of retinal degenerative diseases. Prog Retin Eye Res.
79:1008602020. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Ajoolabady A, Lindholm D, Ren J and
Pratico D: ER stress and UPR in Alzheimer's disease: Mechanisms,
pathogenesis, treatments. Cell Death Dis. 13:7062022. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Saito A, Ochiai K, Kondo S, Tsumagari K,
Murakami T, Cavener DR and Imaizumi K: Endoplasmic reticulum stress
response mediated by the PERK-eIF2(alpha)-ATF4 pathway is involved
in osteoblast differentiation induced by BMP2. J Biol Chem.
286:4809–4818. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Vattem KM and Wek RC: Reinitiation
involving upstream ORFs regulates ATF4 mRNA translation in
mammalian cells. Proc Natl Acad Sci USA. 101:11269–11274. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Hooper KM, Barlow PG, Henderson P and
Stevens C: Interactions between autophagy and the unfolded protein
response: Implications for inflammatory bowel disease. Inflamm
Bowel Dis. 25:661–671. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Yin S, Li L, Tao Y, Yu J, Wei S, Liu M and
Li J: The inhibitory effect of artesunate on excessive endoplasmic
reticulum stress alleviates experimental colitis in mice. Front
Pharmacol. 12:6297982021. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Lei Y, Yu H, Ding S, Liu H, Liu C and Fu
R: Molecular mechanism of ATF6 in unfolded protein response and its
role in disease. Heliyon. 10:e259372024. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Ye J, Rawson RB, Komuro R, Chen X, Davé
UP, Prywes R, Brown MS and Goldstein JL: ER stress induces cleavage
of membrane-bound ATF6 by the same proteases that process SREBPs.
Mol Cell. 6:1355–1364. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Jin JK, Blackwood EA, Azizi K, Thuerauf
DJ, Fahem AG, Hofmann C, Kaufman RJ, Doroudgar S and Glembotski CC:
ATF6 decreases myocardial ischemia/reperfusion damage and links ER
stress and oxidative stress signaling pathways in the heart. Circ
Res. 120:862–875. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Grey MJ, Cloots E, Simpson MS, LeDuc N,
Serebrenik YV, De Luca H, De Sutter D, Luong P, Thiagarajah JR,
Paton AW, et al: IRE1β negatively regulates IRE1α signaling in
response to endoplasmic reticulum stress. J Cell Biol.
219:e2019040482020. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Luo H, Gong WY, Zhang YY, Liu YY, Chen Z,
Feng XL, Jiao QB and Zhang XW: IRE1β evolves to be a guardian of
respiratory and gastrointestinal mucosa. Heliyon. 10:e390112024.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Haber AL, Biton M, Rogel N, Herbst RH,
Shekhar K, Smillie C, Burgin G, Delorey TM, Howitt MR, Katz Y, et
al: A single-cell survey of the small intestinal epithelium.
Nature. 551:333–339. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Cloots E, Simpson MS, De Nolf C, Lencer
WI, Janssens S and Grey MJ: Evolution and function of the
epithelial cell-specific ER stress sensor IRE1β. Mucosal Immunol.
14:1235–1246. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Johansson ME and Hansson GC: Goblet cells
need some stress. J Clin Invest. 132:e1620302022. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Grey MJ, De Luca H, Ward DV, Kreulen IA,
Bugda Gwilt K, Foley SE, Thiagarajah JR, McCormick BA, Turner JR
and Lencer WI: The epithelial-specific ER stress sensor ERN2/IRE1β
enables host-microbiota crosstalk to affect colon goblet cell
development. J Clin Invest. 132:e1535192022. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Dai F, Dong S, Rong Z, Xuan Q, Chen P,
Chen M, Fan Y and Gao Q: Expression of inositol-requiring enzyme 1β
is downregulated in azoxymethane/dextran sulfate sodium-induced
mouse colonic tumors. Exp Ther Med. 17:3181–3188. 2019.PubMed/NCBI
|
|
93
|
Deng R, Wang M, Promlek T, Druelle-Cedano
C, Murad R, Davidson NO and Kaufman RJ: IRE1α and IRE1β protect
intestinal epithelium and suppress colorectal tumorigenesis through
distinct mechanisms. Preprint. bioRxiv [Preprint].
2025.05.01.651751. 2025.
|
|
94
|
Ye X, Wu J, Li J and Wang H: anterior
gradient protein 2 promotes mucosal repair in pediatric ulcerative
colitis. Biomed Res Int. 2021:64838602021. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Wu D, Su S, Zha X, Wei Y, Yang G, Huang Q,
Yang Y, Xia L, Fan S and Peng X: Glutamine promotes O-GlcNAcylation
of G6PD and inhibits AGR2 S-glutathionylation to maintain the
intestinal mucus barrier in burned septic mice. Redox Biol.
59:1025812023. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Neidhardt L, Cloots E, Friemel N, Weiss
CAM, Harding HP, McLaughlin SH, Janssens S and Ron D: The
IRE1β-mediated unfolded protein response is repressed by the
chaperone AGR2 in mucin producing cells. EMBO J. 43:719–753. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Cloots E, Guilbert P, Provost M, Neidhardt
L, Van de Velde E, Fayazpour F, De Sutter D, Savvides SN, Eyckerman
S and Janssens S: Activation of goblet-cell stress sensor IRE1β is
controlled by the mucin chaperone AGR2. EMBO J. 43:695–718. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Bertolotti A: Keeping goblet cells
unstressed: new insights into a general principle. EMBO J.
43:663–665. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Cadwell K and Loke P: Gene-environment
interactions shape the host-microbial interface in inflammatory
bowel disease. Nat Immunol. 26:1023–1035. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Valle Arevalo A and Nobile CJ:
Interactions of microorganisms with host mucins: A focus on Candida
albicans. FEMS Microbiol Rev. 44:645–654. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Zhu X, Li Y, Tian X, Jing Y, Wang Z, Yue
L, Li J, Wu L, Zhou X, Yu Z, et al: REGγ mitigates
radiation-induced enteritis by preserving mucin secretion and
sustaining microbiome homeostasis. Am J Pathol. 194:975–988. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Yao Y, Kim G, Shafer S, Chen Z, Kubo S, Ji
Y, Luo J, Yang W, Perner SP, Kanellopoulou C, et al: Mucus
sialylation determines intestinal host-commensal homeostasis. Cell.
185:1172–1188.e28. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Vilardi A, Przyborski S, Mobbs C, Rufini A
and Tufarelli C: Current understanding of the interplay between
extracellular matrix remodelling and gut permeability in health and
disease. Cell Death Discov. 10:2582024. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Zhu Y, Huang Y, Ji Q, Fu S, Gu J, Tai N
and Wang X: Interplay between extracellular matrix and neutrophils
in diseases. J Immunol Res. 2021:82433782021. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Porras AM, Zhou H, Shi Q, Xiao X; JRI Live
Cell Bank, ; Longman R and Brito IL: Inflammatory bowel
disease-associated gut commensals degrade components of the
extracellular matrix. mBio. 13:e02201222022. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Zhao Y, Qiao D, Skibba M and Brasier AR:
The IRE1α-XBP1s Arm of the unfolded protein response activates
N-glycosylation to remodel the subepithelial basement membrane in
paramyxovirus infection. Int J Mol Sci. 23:90002022. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Onfroy-Roy L, Hamel D, Foncy J, Malaquin L
and Ferrand A: Extracellular matrix mechanical properties and
regulation of the intestinal stem cells: When mechanics control
fate. Cells. 9:26292020. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Gjorevski N, Sachs N, Manfrin A, Giger S,
Bragina ME, Ordóñez-Morán P, Clevers H and Lutolf MP: Designer
matrices for intestinal stem cell and organoid culture. Nature.
539:560–564. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Fionda C and Sciumè G: A little ER stress
isn't bad: The IRE1α/XBP1 pathway shapes ILC3 functions during
intestinal inflammation. J Clin Invest. 134:e1822042024. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Ren C, Dokter-Fokkens J, Figueroa Lozano
S, Zhang Q, de Haan BJ, Zhang H, Faas MM and de Vos P: Fibroblasts
impact goblet cell responses to lactic acid bacteria after exposure
to inflammatory cytokines and mucus disruptors. Mol Nutr Food Res.
63:e18014272019. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Zhang G, Song D, Ma R, Li M, Liu B, He Z
and Fu Q: Artificial mucus layer formed in response to ROS for the
oral treatment of inflammatory bowel disease. Sci Adv.
10:eado82222024. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Royal JM, Oh YJ, Grey MJ, Lencer WI,
Ronquillo N, Galandiuk S and Matoba N: A modified cholera toxin B
subunit containing an ER retention motif enhances colon epithelial
repair via an unfolded protein response. FASEB J. 33:13527–13545.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Kittle WM, Reeves MA, Fulkerson AE,
Hamorsky KT, Morris DA, Kitterman KT, Merchant ML and Matoba N:
Preclinical long-term stability and forced degradation assessment
of EPICERTIN, a mucosal healing biotherapeutic for inflammatory
bowel disease. Pharmaceutics. 17:2592025. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Zheng T, Huang KY, Tang XD, Wang FY and Lv
L: Endoplasmic reticulum stress in gut inflammation: Implications
for ulcerative colitis and Crohn's disease. World J Gastroenterol.
31:1046712025. View Article : Google Scholar : PubMed/NCBI
|