Introduction
Cervical cancer is one of the most common types of
gynecological cancer and ranks as the fourth leading cause of
cancer-related mortality among women, and 14th overall among all
types of cancer (1,2). According to the GLOBOCAN 2022 report,
~660,000 new cases of cervical cancer were diagnosed, with ~340,000
mortalities attributed to the disease (3). Extensive evidence suggests that
cervical cancer is primarily caused by human papillomavirus
infection, with additional contributing factors associated with
social factors (such as income, education and region) and
individual behaviors (such as age, sex, smoking status, nutrition,
activity level) (4,5). This is exemplified by the obvious
regional differences in the incidence and mortality of cervical
cancer within China. Surveillance data from the Chinese cancer
registry shows that incidence and mortality rates in rural areas
are consistently higher than those in urban areas. This disparity
is attributed to inequalities in health education, access to HPV
screening, and availability of vaccination programs. Furthermore,
it is estimated that among younger women, this gap in incidence
rates between urban and rural populations is likely to widen
further, highlighting the critical role of socioeconomic factors in
disease outcomes (4). Excessive
cell division and proliferation are central to the development of
cervical cancer (6,7), making the inhibition of cancer cell
proliferation and the induction of apoptosis key therapeutic
strategies.
Endophytic fungi, which reside within the tissues of
medicinal plants and co-evolve with their hosts (8), produce a diverse array of natural
products (9,10). These products exhibit a range of
biological activities, including antioxidant, anti-inflammatory,
antibacterial, antiviral and anticancer properties (11). Anti-tumor natural products have
been classified into several structural groups, such as alkaloids
(12), terpenoids (13), steroids (14), quinones (15) and esters (10).
Taxus wallichiana var. mairei, a rare
and endangered plant native to southern China, is well-known for
producing the anti-cancer drug paclitaxel (16). Fungal endophytes isolated from
Taxus wallichiana var. mairei include species such as
Phoma medicaginis (17),
Diaporthe phaseolorum (18)
and Nigrospora oryzae (19). The genus Alternaria (family
Dematiaceae) consists of globally distributed filamentous fungi and
plant pathogens (20,21), known for producing a variety of
secondary metabolites (22).
Dihydroalterperylenol (DAP) is a perylenequinone
metabolite characterized by a highly conjugated pentacyclic core. A
previous study has reported that DAP does not exhibit cytotoxicity
(23). The present study
demonstrated the potent cytotoxic activity of DAP isolated from the
plant endophytic fungus Alternaria semiverrucosa in HeLa
cells and explored its apoptosis-inducing mechanism through
transcriptome analysis, molecular docking and molecular dynamics
(MD) simulations studies.
Materials and methods
Materials and instruments for chemical
separation
Silica gel (200–300 mesh; Qingdao Haiyang Chemical
Co. Ltd.), Sephadex LH-20 gel (Shanghai Yuanye Bio-Technology Co.
Ltd.), Octadecylsilyl-bonded (ODS) silica gel (cat. no. RP-18;
Merck KGaA) and organic solvents (methanol, CAS No: 67-56-1;
dichloromethane, CAS No: 75-09-2; petroleum ether, CAS No:
8032-32-4; ethyl acetate, CAS No: 141-78-6; All the aforementioned
reagents are analytical grade; Tianjin Fuyu Fine Chemical Co. Ltd.)
were used for column chromatography. Thin-layer chromatography was
monitored using silica gel plates (type H; Qingdao Dingkang
Silicone Co. Ltd.). One-dimensional nuclear magnetic resonance
(NMR) spectra were recorded using Bruker Avance-500 and 600
spectrometers (Beijing Obel Scientific Instrument Co., Ltd.).
Fungal material and fermentation
All specimens of Taxus chinensis var.
mairei were obtained through artificial propagation and
exclusively sourced from the Wanshun Seedling Farm Guiyang, China.
Voucher specimens of the host plant have been deposited at the
Engineering Research Center for Utilization of Characteristic
Bio-Pharmaceutical Resources in Southwest China, Ministry of
Education, Guizhou University, with the accession number ZZ-HJ-13.
The endophytic fungus Alternaria semiverrucosa was isolated
from the healthy branches of Taxus chinensis var.
mairei. The fungal isolate Alternaria semiverrucosa
was preserved at the Guizhou Academy of Agricultural Sciences under
strain code (dried culture; holotype no. GZAAS 22-2029; ex-type
living culture, no. GZCC 22-2029). The fungus was cultured on
potato dextrose agar plates for 10 days. The mycelia were then
sliced and incubated in 300 ml Yeast extract-Peptone-Starch (YPS)
medium (glucose 20.0 g, peptone 10.0 g, yeast extract 5.0 g,
KH2PO4 1.0 g, MgSO4 1.0 g and 1 l
distilled water) for 3 days at 28°C. A 15 ml aliquot of the seed
culture was transferred to 400 × 1 l Erlenmeyer flasks, each
containing 300 ml of YPS culture medium. The flasks were shaken on
a rotary shaker (140 rpm) at 28°C for 3 days, followed by static
cultivation for 30 days.
Extraction and isolation
After cultivation, the broth and mycelia were
separated by gauze. The broth was extracted with ethyl acetate
(EtOAc) three times. The EtOAc extract was collected and
concentrated under vacuum to yield 120.0 g of extract, while the
mycelia were extracted with methanol to obtain a 250.0 g extract
after evaporation under reduced pressure. Both the EtOAc and
methanol extracts were subjected to silica gel column
chromatography with a step gradient of
CH2Cl2-MeOH (1:0 → 0:1 v/v), yielding 28
fractions (Fr.1-Fr.28) from the EtOAc extract and 22 fractions
(Fr.A-Fr.V) from the methanol extract. Fr.11 was separated on an
ODS column, eluting with a CH3OH/H2O
gradient, yielding five subfractions (Fr.11.1-Fr.11.5). Fr.11.2 was
further purified using a silica gel column, resulting in compound 1
(11.9 mg). Fr.11.3 was purified by Sephadex LH-20 and silica gel
column chromatography to yield compound 4 (27.9 mg). Fr.16 was
separated on an ODS column, eluted with a methanol-water gradient
(10–100%), yielding six subfractions (Fr.16.1-Fr.16.6). Fr.16.3 was
further purified on silica gel and Sephadex LH-20 to obtain
compound 2 (9.8 mg) and Fr.16.6 was chromatographed on an ODS
column to yield compound 3 (7.2 mg). Fr.13 was purified by ODS
column chromatography, followed by repeated silica gel column
purification, yielding compounds 5 (27.9 mg) and 6 (18.4 mg).
Compound 7 (21.8 mg) was obtained by repeated Sephadex LH-20 and
silica gel column chromatography. Fr.8 was separated on an RP-C18
column using a methanol-water gradient to yield six subfractions
(Fr.8.1-Fr.8.6). Fr.8.1 was purified by repeated Sephadex LH-20
chromatography to yield compound 8 (18.9 mg). Fr.I was separated by
RP-C18 column chromatography with a methanol-water gradient
(20–80%), yielding five subfractions (Fr.I.1-Fr.I.5). Fr.I.2 was
further purified by Sephadex LH-20, resulting in compound 9 (18.8
mg). Fr.I.3 was purified by silica gel column chromatography to
yield compound 10 (31.5 mg).
Cell culture and cytotoxicity
assay
A total of six human cancer cell lines (PC3 cat. no.
CRL-1435; LNCaP, cat. no. CRL-1740; HeLa, cat. no. CCL-2; SiHa,
cat. no. HTB-35; K562, cat. no. CCL-243; and HEL, cat. no. TIB-180)
were purchased from Typical Cultures Depository of the United
States of America and are currently maintained in the Key
Laboratory of Chemistry for Natural Products of Guizhou, Chinese
Academy of Sciences in Guiyang, China. Human gingival fibroblasts
(HGF-1) were provided by Guiyang Stomatological Hospital in
Guiyang, China. The inhibitory effects of the compounds on tumor
cells were assessed using the MTT assay, as described in a previous
study (24). A volume of 20 µl 5
mg/ml 3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide
(MTT; M158055, Aladdin) solution was added to each well and
incubated for 4 h at 37°C. After centrifugation at 20 × g for 10
min at room temperature to remove the supernatant, 150 µl DMSO was
added, followed by shaking on a shaker for 15 min. Absorbance at
490 nm was measured using a microplate reader to calculate the cell
proliferation inhibition rate. Inhibition rate of the
proliferation=1-(Average optical density of experimental
group/Average optical density of control group) ×100%. The cells
were cultured in RPMI 1640 medium (cat. no. A-CSH807-500ml;
Biogradetech, Inc.) supplemented with 10% fetal bovine serum (cat.
no. C0230; Beyotime Institute of Biotechnology) and 1%
penicillin-streptomycin solution, in an incubator for 24 h at 37°C
under 5% CO2 atmosphere. Cell viability was calculated
as a percentage relative to control wells. The half-maximal
inhibitory concentration (IC50) values were determined
using MTT viability curves, and the data were analyzed with
GraphPad Prism 8.0 (Dotmatics). Doxorubicin (cat. no. D1515; Merck
KGaA) was used as a positive control and was applied at
concentrations of 0.0, 0.5, 1.0, 2.0, and 4.0 µM. Cells were
treated for 24 h at 37°C under 5% CO2 atmosphere,
consistent with standard protocols for cytotoxicity assessment.
Flow cytometric analysis of cell cycle
and apoptosis
Cell cycle progression was analyzed using the BD
Cycletest Plus DNA Reagent Kit (cat. no. 340242; Becton, Dickinson
and Company), and apoptosis was assessed with the BD Annexin V-FITC
Apoptosis Detection Kit I (cat. no. 556547; Becton, Dickinson and
Company). Cells were seeded into 6-well plates at a density of
3×105/well. After 24 h, the cells were treated with 0.1%
DMSO (negative control), compound 1 (3.0, 6.0 and 9.0 µM) for 48 h
at 37°C. The cells were washed twice with cold PBS and then
resuspended in 100 µl of binding buffer. Annexin V PE (5 µl) and
7-AAD (5 µl) were added, and the mixture was incubated in the dark
at 37°C for 15 min. Apoptosis and cell cycle distribution were
analyzed using flow cytometry (NovoCyte 2040R; ACEA Biosciences,
Inc.) (24).
RNA-Sequencing (RNA-seq) and data
analysis
HeLa cells were pretreated with compound 1 at 6.0 µM
for 24 h at 37°C and then cells were washed with PBS and
subsequently lysed directly in the culture dish using 1.5 ml of
Trizol™ reagent (cat. no. 15596026CN; Invitrogen; Thermo Fisher
Scientific, Inc.). The lysis process was allowed to proceed for 10
min at room temperature. Following lysis, the lysate was
transferred to a 2.0 ml sterile centrifuge tube and stored at −80°C
for subsequent RNA extraction. Total RNA was extracted using
Trizol™ reagent following the manufacturer's protocol. RNA
concentration was measured using a NanoDrop 2000 spectrophotometer
(Thermo Fisher Scientific, Inc.) and RNA integrity was assessed
using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.). RNA
libraries were constructed and the sequencing was performed using
the NovaSeq 6000 S4 Reagent Kit (300 cycles; cat. no. 20028315;
Illumina, Inc.) and sequences using the Illumina HiSeq platform
(New England Biolabs, Inc.). The library loading concentration was
0.5 nM, and quantification was conducted using qPCR. Paired-end
sequencing with a read length of 150 bp (PE150) was carried out.
The mRNA reads were mapped to the reference genome using HISAT2
software (version 2.2.1; http://ccb.jhu.edu/software/hisat/index.shtml). Gene
expression was quantified as fragments per kilobase of exon model
per million mapped reads, based on the number of uniquely mapped
reads. Differential expression analysis was carried out using edgeR
software (version 3.32.1; parameters: P<0.05; fold change ≥1.5).
The results were visualized using a volcano plot, revealing
differentially expressed metabolism-related genes and lncRNAs in
cervical cancer and adjacent samples. Gene ontology (GO) enrichment
and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway
enrichment analyses were conducted using the clusterProfiler R
package and the iDEP 1.1 web tool (https://bioinformatics.sdstate.edu/idep96/) (25). Heatmap analysis was performed using
TBtools 2.200 (26), red
represents upregulated gene expression.
Reverse transcription-quantitative PCR
(RT-qPCR) for transcriptomics
HeLa cells were seeded in 12-well plates overnight
and pretreated with compound 1 at different concentrations (3.0,
6.0 and 9.0 µM) at 37°C for 24 h. Total RNA was extracted from the
cells using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) and cDNA was synthesized using the HiScript cDNA
Synthesis Mix kit (Jiangsu CoWin Biotech Co., Ltd.). RT-qPCR was
carried out using the UltraSYBR Mixture (With ROX) kit (Jiangsu
CoWin Biotech Co., Ltd.). GAPDH was used as the internal reference
gene. The primer sequences for TGIF2, ID1 and BMP2 were obtained
from previous publications (Table
SI). The PCR reaction mixture contained 2.0 µl cDNA template,
21.0 µl DNase-free ddH2O, 1.0 µl forward and reverse
primers, and 25.0 µl Ultra SYBR Mixture. The relative mRNA
expression levels were measured using the Real-Time PCR Detection
System (CFX Connect; Bio-Rad Laboratories, Inc.). qPCR was
performed using the following thermocycling protocol: Initial
pre-denaturation at 95°C for 5 min; 35 cycles consisting of
denaturation at 95°C for 45 sec, annealing at 56°C for 50 sec, and
extension at 72°C for 40 sec; followed by a final extension step at
72°C for 5 min. The relative expression ratio was using the
2−ΔΔCq method (27).
Molecular docking studies
Crystal structures of TGIF2 [Protein Data Bank (PDB)
code, 2dmn], ID1 (PDB code, 6mgn) and BMP2 (PDB code, 1rew) were
obtained from the Research Collaboratory for Structural
Bioinformatics Protein Data Bank (https://www.rcsb.org/). The docking parameters were
calculated using PyMOL 2.2.0 (https://github.com/schrodinger/pymol-open-source;
center and size for the docking box were defined as follows, TGIF2:
Center_x, −3.094; center_y, −3.281; center_z, 3.948 with size_x,
65.45; size_y, 65.45; size_z, 65.45; ID1: Center_x, 18.573;
center_y, 14.631; center_z, 12.395 with size_x, 46.55; size_y,
46.55; size_z, 46.55; BMP2: Center_x, −28.157; center_y, 67.095;
center_z, 21.448 with size_x, 65.45; size_y, 65.45; size_z, 65.45).
Molecular docking studies were carried out to explore the binding
modes of compound 1 with TGIF2, ID1 and BMP2 using AutoDock Vina
1.2.3 (https://github.com/ccsb-scripps/AutoDock-Vina/releases/tag/v1.2.3).
The best-scoring docking poses were selected based on the Vina
docking scores and further analyzed visually using PyMOL 2.2.0 and
Discovery Studio 2021 software (Version 21.1.0.20298; https://www.3ds.com/products-services/biovia/products/molecular-modeling-simulation/biovia-discovery-studio/).
Molecular dynamics simulation
MD simulations were carried out using the Amber24
software suite (version 2024; http://ambermd.org/) (28), applying the ff19SB force field
(29,30) and the OPC water model (31). The complex was placed in a cubic
water box, with electrostatic and van der Waals interactions having
cut off distances set to 1.0 nm. A time step of 2 fs was used for
the integration and long-range electrostatic interactions were
handled using the Particle Mesh Ewald method (32). The system was maintained at a
temperature of 300 K and a pressure of 1 bar. Initial energy
minimization was carried out (33), followed by 200 ps of constant
number of particles, volume, and energy equilibration and 100 ps of
constant number of particles, pressure, and temperature
equilibration dynamics. Temperature control was achieved using the
V-rescale method (34), while
pressure control was applied using the Parrinello-Rahman approach
(35). Following the
equilibration, 200 ns of production dynamics were carried out. Key
metrics such as root mean square deviation (RMSD), root mean square
fluctuation (RMSF), radius of gyration (Rg) and hydrogen bond
counts were calculated using Amber's built-in Cpptraj tool.
Statistical analysis
Data analysis was carried out using SPSS Statistics
21.0 (IBM Corp.) and GraphPad Prism 7.0 (Dotmatics). Results are
presented as the mean ± SD of three independent experiments.
Statistical analysis was conducted using the unpaired Student's
t-test or the one-way or two-way ANOVA, followed by Šídák's
multiple comparisons test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Compounds from Alternaria
semiverrucosa and cytotoxic activity assay
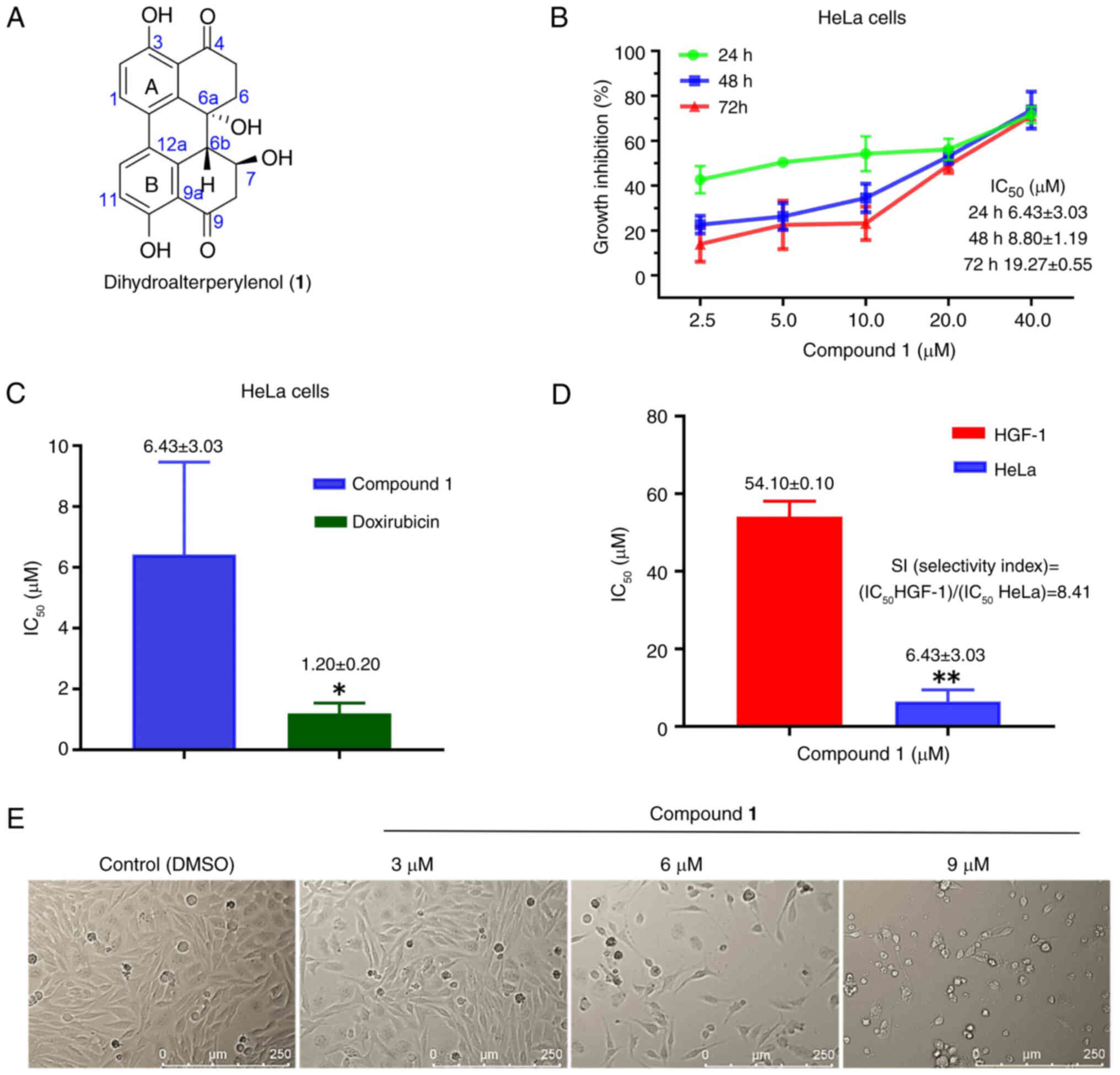
Structures of the ten known compounds were
identified (Fig. S1) as i) DAP
(Figs. 1A, S2 and B) (36), ii) Fonsecinone A (Fig. S3A and iii) (37), Aurasperone A (Fig. S4A-B) (38), iv)
3β,5α,9α-trihydroxy-(22E,24R)-ergosta-7,22-diene-6-one (Fig. S5A and B) (30), v) Gargalol B (Fig. S6A and B) (39), vi)
(22E,24R)-ergosta-7,22-dien-3β,5α,6β,9α-tetraol (Fig. S7A and B) (40), vii)
11-[(6-Deoxy-α-L-mannopyranosyl)oxy]-3-hydroxyhexadecanoic acid
(Fig. S8A and B), viii)
Penisochroman A (Fig. S9A and B)
(41), viiii) β-adenosine
(Fig. S10A and B) (42) and x) uridine (Fig. S11A and B) (43), by comparing their 1H, 13C NMR
(Data S1) and mass spectrometry
data with those in the literature.
The aforementioned compounds were tested for
cytotoxicity in vitro against human tumor cell lines (PC3,
LNCaP, HeLa, SiHa, K562, and HEL). As shown in Table SII, compound 1 exhibited an
inhibition rate of 49.48% against HeLa cells, and demonstrated the
most potent inhibitory effect. HeLa cells were exposed to varying
concentrations of 2.5, 5.0, 10.0, 20.0 and 40.0 µmol/l for 24, 48
and 72 h. As illustrated in Fig.
1B, within the same time frame, the inhibitory effect on cell
proliferation became progressively more pronounced with increasing
treatment concentration. However, when considering the duration of
exposure, the inhibitory effect diminished over time. Statistical
analysis indicated IC50 values of 6.43±3.03 µmol/l at 24
h, 8.80±1.19 µmol/l at 48 h and 19.27±0.55 µmol/l at 72 h (Fig. 1B). This was significantly higher
when compared with the positive control (IC50=1.20±0.20
µM; Fig. 1C) at 24 h
(P=0.0310).
Compound 1 was also tested in normal HGF-1 cells as
a reference, and exhibited a markedly reduced inhibitory effect
when compared with the HeLa cells (P=0.0055), with an
IC50 value of 54.10±0.10 µmol/l (Fig. 1D), suggesting limited cytotoxic
activity. The safety profile of a drug is typically assessed using
the selectivity index (SI), which is defined as: SI=IC50
(normal cells)/IC50 (tumor cells), with a SI value of
8.41. An increased SI value indicates a broader therapeutic window
and reduced cytotoxicity towards normal cells. Morphological
changes in the cells became apparent after 24 h of treatment
compared with the control group (Fig.
1E). As drug concentration increased, intercellular spaces
widen, cellular morphology becomes more rounded, and the presence
of cell debris becomes increasingly evident.
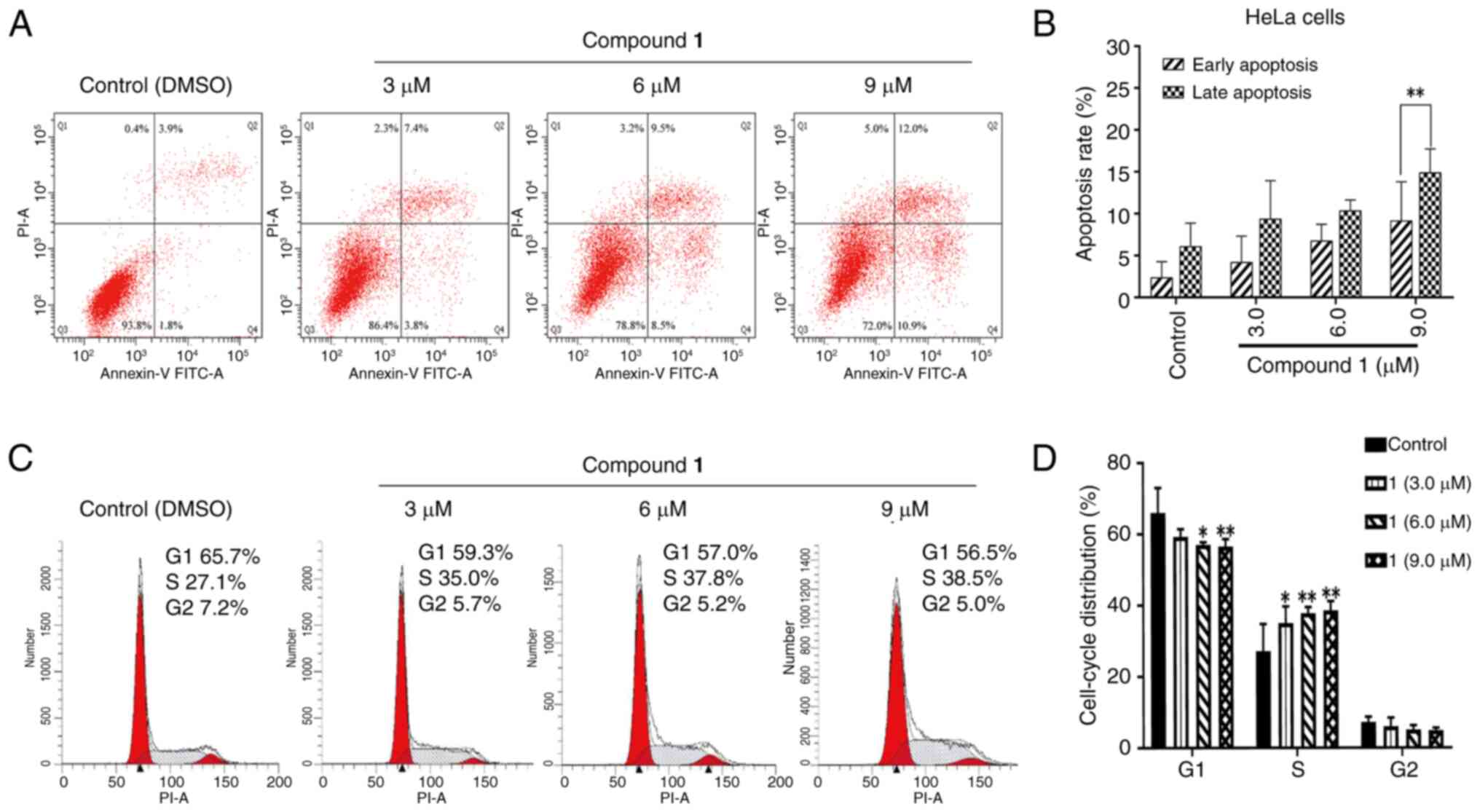
DAP induced apoptosis and cell cycle
G1/S arrest in HeLa cancer cells
Compound 1 was subsequently evaluated for its
effects on the cell cycle and apoptosis. As shown in Fig. 2A and B, these findings suggest that
compound 1 induces apoptosis in HeLa cells. Notably, the proportion
of early apoptotic cells was reduced compared with that of late
apoptotic cells after a 24 h treatment with compound 1 (P=0.0065),
indicating that the majority of the cytotoxic effects of compound 1
on HeLa cells are due to the induction of late-stage apoptotic cell
death. The impact of compound 1 on cell cycle distribution was
evaluated using flow cytometry. As illustrated in Fig. 2C, compared with the control group,
alterations in the G1 and S phases were observed with increasing
concentrations of compound 1. Statistical analysis (Fig. 2D) revealed that when the
concentration of compound 1 reached 6.0 µmol/l, the proportion of
HeLa cells in the G1 phase was significantly reduced compared with
the control group (P=0.0126), with a significant decrease observed
at 9.0 µmol/l (P=0.0090). By contrast, cell accumulation was
evident in the S phase. A significant increase in the S phase
population was detected at a concentration of 3.0 µmol/l
(P=0.0280), which further increased markedly at 6.0 µmol/l
(P=0.0043). No notable changes were observed in the G2/M phase.
These findings suggest that the growth inhibitory effect of
compound 1 on HeLa cells is associated with cell cycle arrest in
the S phase.
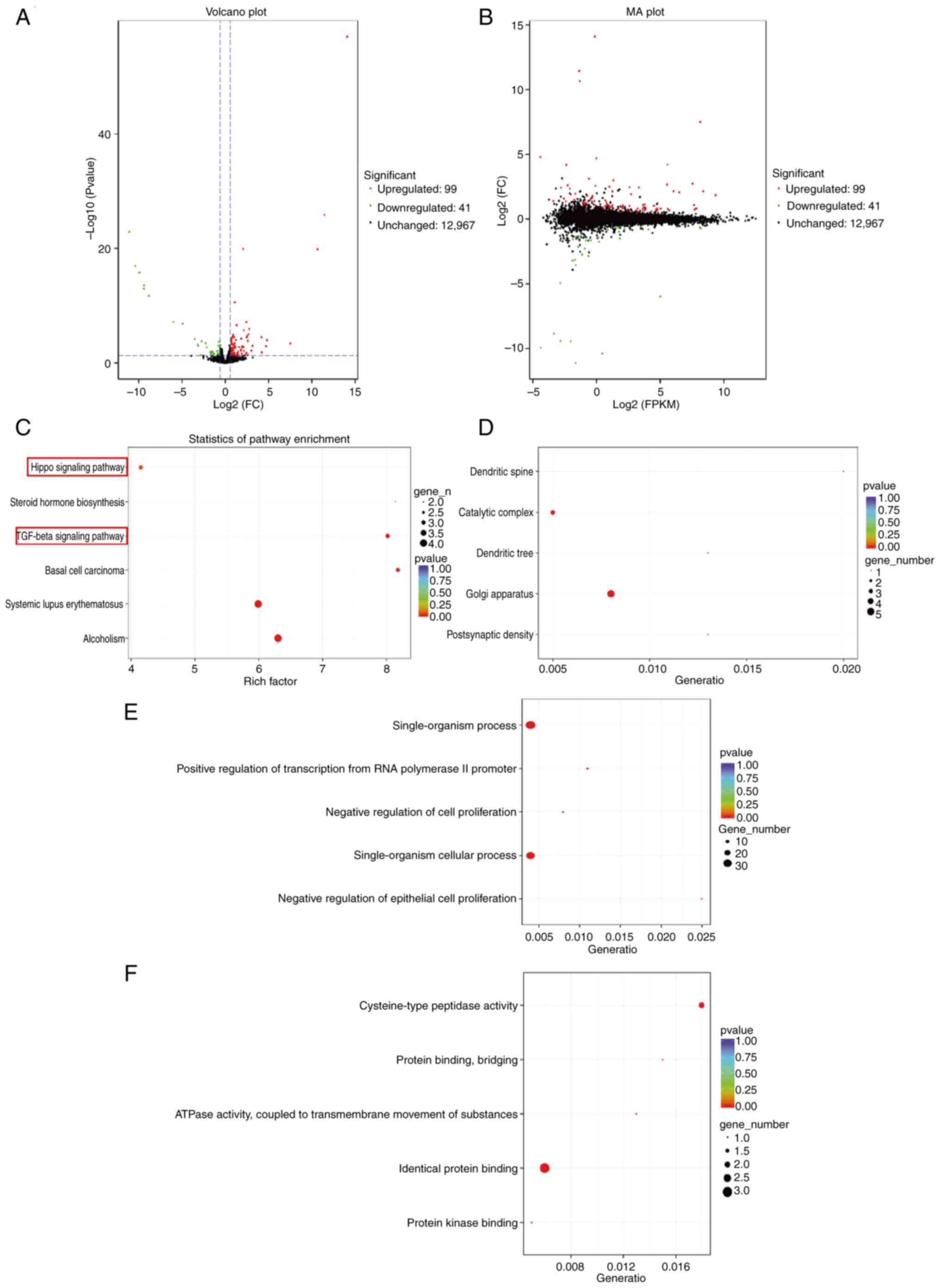
Transcriptome analysis
To further investigate the molecular mechanisms
underlying the effects of DAP, transcriptome sequencing (RNA-seq)
was carried out in HeLa cells. This analysis identified 140
differentially expressed genes (DEGs), including 99 upregulated and
41 downregulated genes (Fig. 3A and
B). GO and KEGG enrichment analyses revealed significant
enrichment in pathways associated with tumor cell proliferation,
particularly the Hippo signaling and TGFβ signaling pathways
(Fig. 3C) have been implicated in
the regulation of cell apoptosis. Among the pathways enriched with
a P<0.05 (Fig. 3C), two
apoptosis-related signaling pathways, specifically the TGF-β
signaling pathway and the Hippo signaling pathway, were identified.
Within the TGF-β signaling pathway, the differentially expressed
genes were TGIF2, BMP2, and ID1; in contrast, the Hippo signaling
pathway exhibited differential expression of TGIF2 and BMP2.
Collectively, three distinct differentially expressed genes (TGIF2,
BMP2, and ID1) were observed across both signaling pathways. GO
analysis was conducted on DEGs to elucidate the molecular
mechanisms underlying their regulation. In the cellular component
category (Fig. 3D), significant
enrichment was observed for postsynaptic density (P=0.00052), Golgi
apparatus (P=0.00099), dendritic tree (P=0.00162), catalytic
complex (P=0.00294) and dendritic spine (P=0.00392). In the
biological process category (Fig.
3E), notable terms included ‘negative regulation of epithelial
cell proliferation’ (P=7.20×−06), ‘single-organism
cellular process’ (P=1.90×−05), ‘negative regulation of
cell proliferation’ (P=2.50×−05), ‘positive regulation
of transcription from RNA polymerase II promoter’
(P=3.30×−05) and ‘single-organism process’ (P=0.00012).
In the molecular function category (Fig. 3F), enriched terms were ‘protein
kinase binding’ (P=0.00032), ‘identical protein binding’
(P=0.00125), ‘ATPase activity coupled to transmembrane movement of
substances’ (P=0.00131), ‘protein binding, bridging’ (P=0.0042) and
‘cysteine-type peptidase activity’ (P=0.0043). The GO analysis
indicated that the inhibitory effect of compound 1 on HeLa cell
proliferation was primarily associated with the regulation of
cellular proliferation processes.
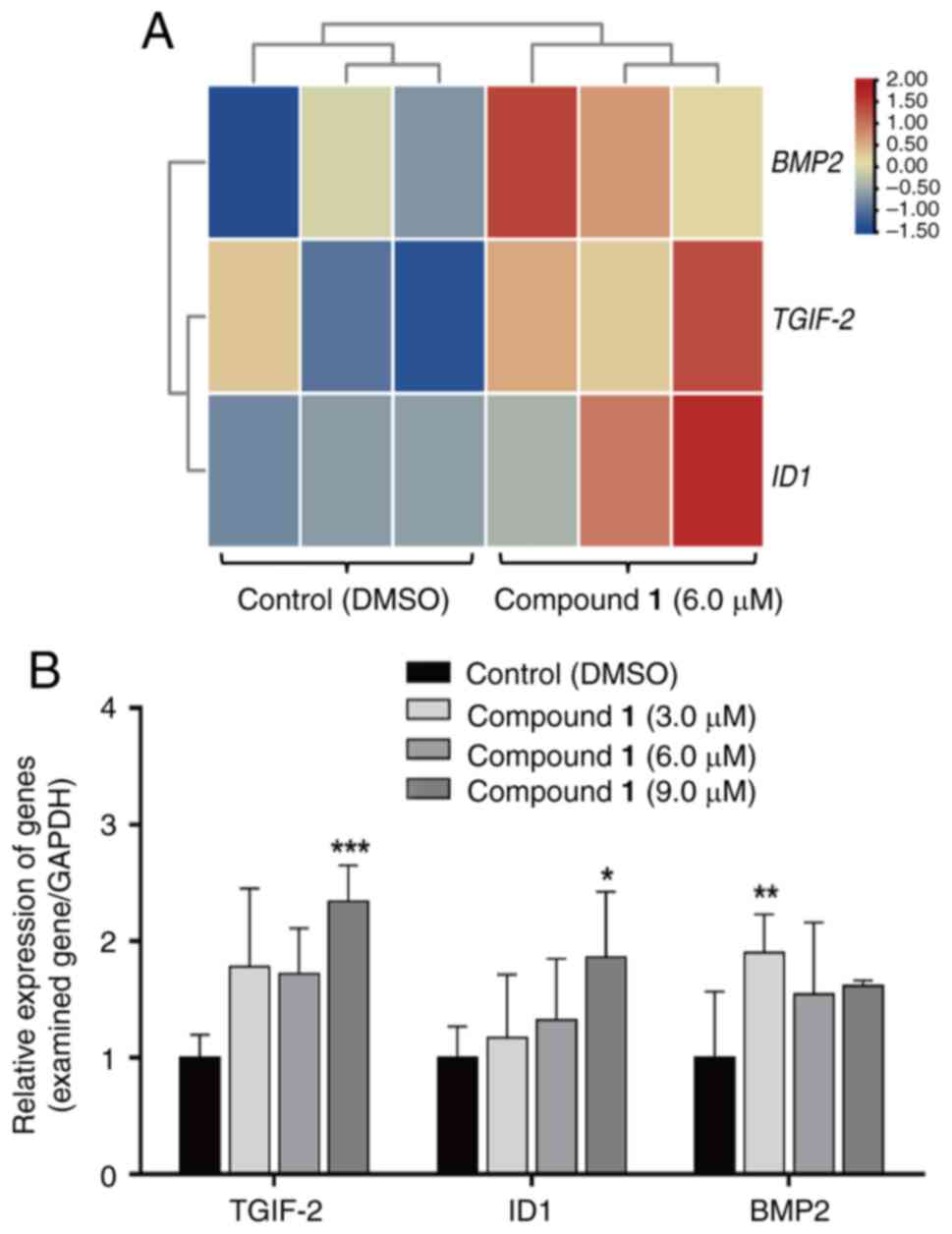
mRNA expression of DAP in HeLa
cells
To investigate the expression patterns of key genes
in the TGFβ signaling pathway and Hippo signaling pathway, TGIF2,
ID1 and BMP2 were selected. Heatmaps were generated using TBtool
(https://gitee.com/an–jiaxin/TBtools), revealing that
all three genes exhibited significant upregulation compared with
the control group (Fig. 4A).
Additionally, RT-qPCR analysis was conducted to evaluate the
expression levels of these genes under treatment with different
concentrations of compound 1. Compared with the control group, the
expression levels of all three genes were increased. Specifically,
the expression of TGIF2 and ID1 showed a significant increase at
9.0 µmol/l compound 1 (P=0.0006 and P=0.0215), while BMP2
expression significantly increased at 3.0 µmol/l Compound 1
(P=0.0035) (Fig. 4B). These
results indicate that the gene expression trends observed in
transcriptome sequencing are consistent with those obtained from
RT-qPCR analysis, verifying that the expression levels of TGIF2,
ID1 and BMP2 are upregulated following compound 1 treatment.
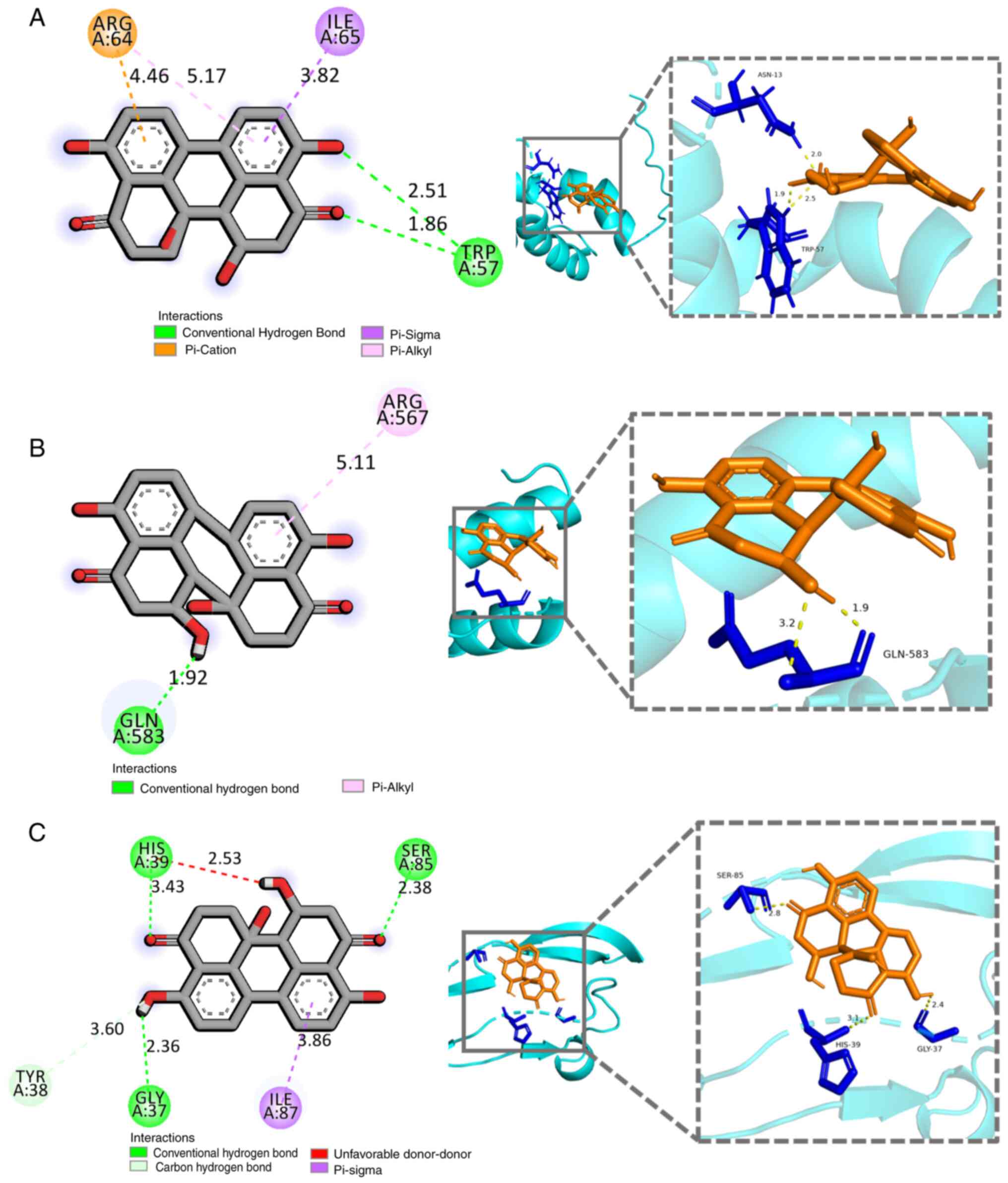
Docking studies of DAP with TGIF2, ID1
and BMP2
Molecular docking studies were conducted to explore
the potential binding sites of compound 1 with TGIF2, ID1 and BMP2.
The docking results revealed that the binding energies between
compound 1 and TGIF2, ID1 and BMP2 were all <-5.0 kcal/mol
(Table SIII). Specifically, a
hydrogen bond was formed between the 7-OH group of compound 1 and
Trp57 of TGIF2 (Fig. 5A).
Additionally, π-Cation and π-Sigma interactions were observed
between the two benzene rings of compound 1 and Arg64 and Ile65.
For ID1, a hydrogen bond was formed between the 7-OH group of
compound 1 and Gln583, while the A-ring of compound 1 interacted
with Arg567 of ID1, with distances of 1.92 Å and 5.11 Å,
respectively (Fig. 5B). As shown
in Fig. 5C, a hydrogen bond
interaction was observed between 7-OH of compound 1 and the
carbonyl group (C-4) of BMP2, with distances of 3.43 Å and 2.53 Å,
respectively. Furthermore, the benzene B-ring of compound 1
interacted with Ile87 of BMP2 via a π-σ bond.
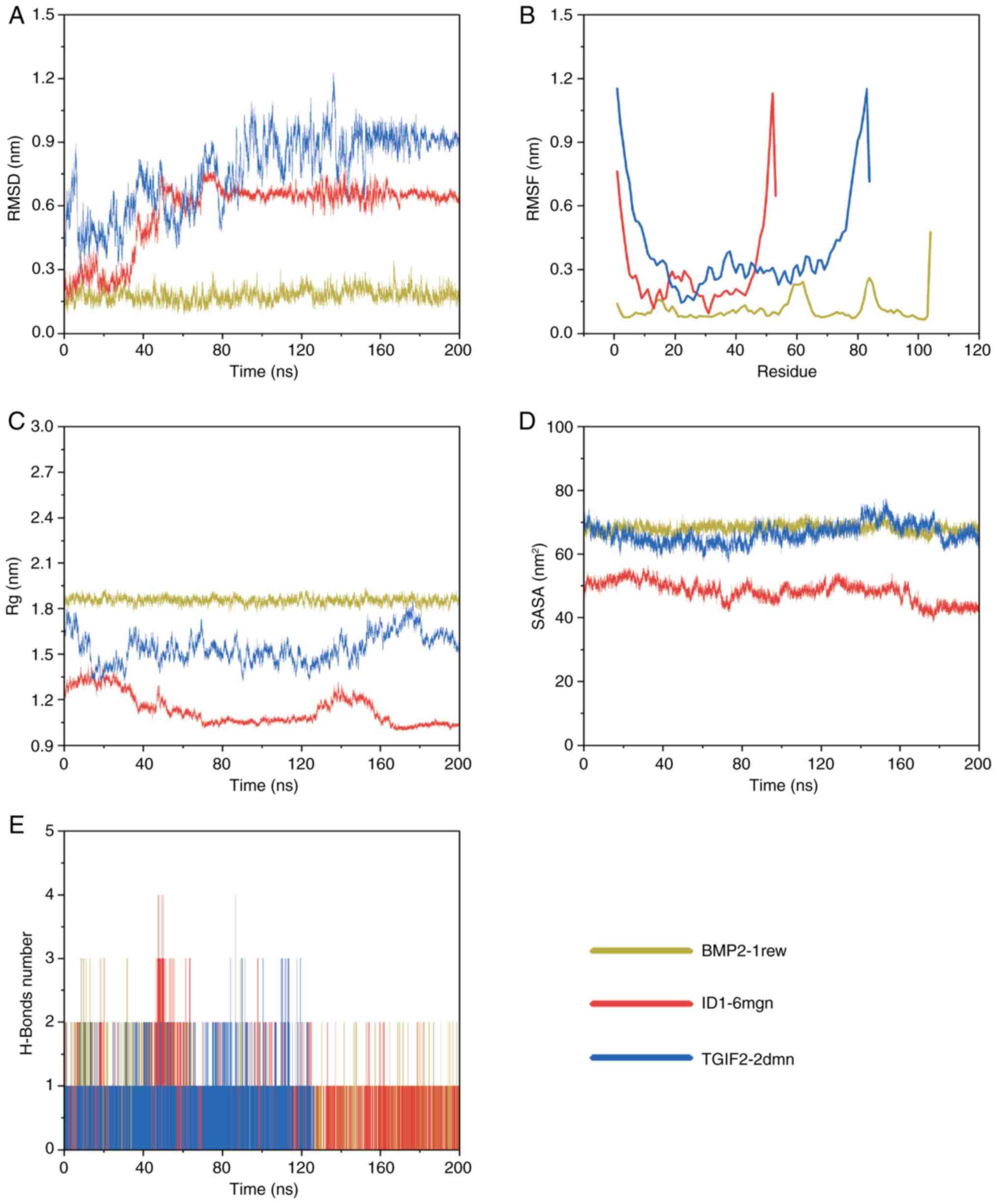
RMSD
RMSD is a key indicator of protein conformational
variability, providing insights into the stability of ligand-target
protein interactions. Higher RMSD values indicate greater
structural alterations within the complex, while lower values
suggest more stable binding configurations (44). As shown in Fig. 6A, the BMP2-1rew complex exhibited
notable fluctuations in RMSD during the initial phase of the
simulation (0–10 ns) before stabilizing between 10 and 200 ns.
Similarly, the ID1-6mgn complex displayed considerable RMSD
variations from 0 to 80 ns, followed by stabilization from 80 to
200 ns. The TGIF2-2dmn complex also experienced considerable RMSD
fluctuations during the first 155 ns, ultimately stabilizing from
155 to 200 ns. These results indicate that all three complexes,
BMP2-1rew, ID1-6mgn and TGIF2-2dmn underwent substantial
conformational changes early in the simulation before achieving
stability at different time intervals.
RMSF
RMSF measures the flexibility of individual amino
acid residues during MD simulations. Higher RMSF values indicate
increased flexibility at specific residues (45). As depicted in Fig. 6B, the BMP2-1rew complex exhibited
fluctuations at residues 58–63, 83–85 and 104. The ID1-6mgn complex
revealed marked mobility in residues 1–4 and 48–53, while the
TGIF2-2dmn complex demonstrated considerable movement in residues
1–4 and 79–84. Despite these localized fluctuations, the overall
structural stability of all three complexes, BMP2-1rew, ID1-6mgn
and TGIF2-2dmn, remained relatively intact.
Radius of gyration (Rg)
Rg radius is an important metric for evaluating the
compactness of molecular structures. Higher Rg values suggest a
more extended conformation, while lower values indicate a more
condensed structure (46). As
shown in Fig. 6C, after
equilibration, the BMP2-1rew complex exhibited minimal fluctuations
and stabilized at ~1.86 nm. The ID1-6mgn complex revealed limited
variation, stabilizing at ~1.08 nm. Similarly, the TGIF2-2dmn
complex demonstrated minor fluctuations, stabilizing at ~1.52 nm.
These results suggest that the BMP2-1rew complex adopts a
relatively extended conformation, indicative of weaker
intermolecular interactions. By contrast, the ID1-6mgn complex
adopts a more compact structure, implying stronger intermolecular
interactions. The TGIF2-2dmn complex falls in between, revealing
moderate intermolecular interactions.
Solvent-accessible surface area
(SASA)
SASA quantifies the exposure of protein molecules to
their surrounding water environment, reflecting changes in
hydrophobic and hydrophilic properties (47). As shown in Fig. 6D, SASA values for the entire system
remained relatively stable throughout the simulation. Among the
three complexes, the ID1-6mgn complex exhibited the most compact
structure, followed by TGIF2-2dmn, with BMP2-1rew displaying the
least compact conformation. These findings suggest that the
ID1-6mgn complex likely has the strongest intermolecular
interactions, while the BMP2-1rew complex likely exhibits the
weakest.
Hydrogen bonds
Hydrogen bonds are among the strongest non-covalent
interactions and serve as important indicators of binding strength
during molecular simulations (48). The frequency and number of hydrogen
bonds observed in the simulations reflect the dynamic nature of
protein-ligand interactions. A higher number of hydrogen bonds
typically correlates with greater binding stability (48). As shown in Fig. 6E, the BMP2-1rew complex formed
between 1 and 3 hydrogen bonds, whereas both the ID1-6mgn and
TGIF2-2dmn complexes formed between 1 and 4 hydrogen bonds. These
findings suggest that the interactions within the ID1-6mgn and
TGIF2-2dmn complexes are likely more robust compared with those
observed in the BMP2-1rew complex.
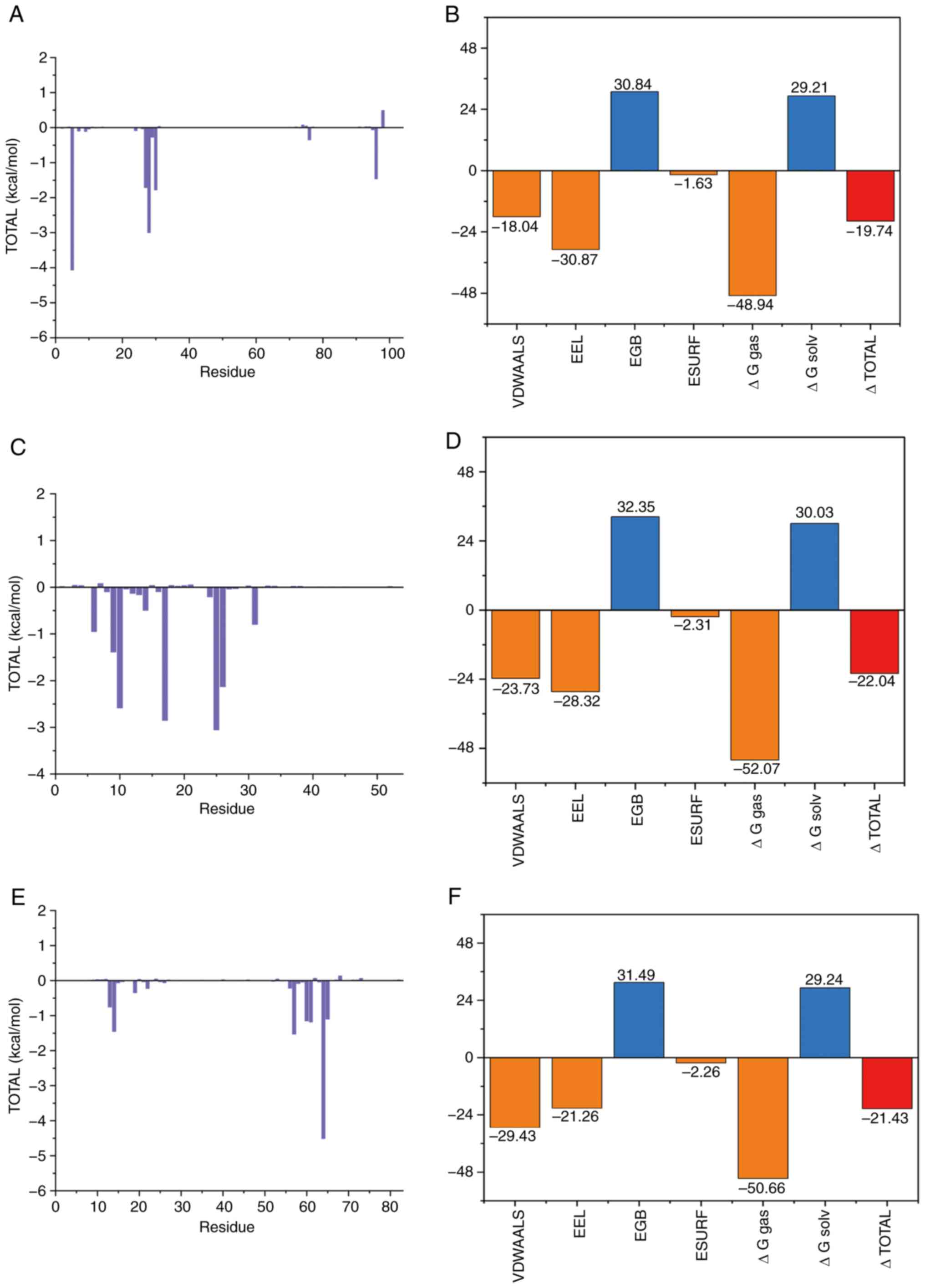
Combination free energy
Molecular mechanics generalized born surface area is
a method (49) that is widely
employed post-simulation to estimate the binding free energy of
molecular complexes, providing insights into their stability. A
lower binding free energy value suggests a stronger binding
affinity (49). As shown in
Fig. 7A, the key residues
contributing to the binding free energy of the BMP2-1rew complex
include ARG-5, HIE-28, PHE-30, TYR-27, VAL-96, ILE-76, ALA-29,
TYR-9, PRO-7 and PRO-24 from the protein. The binding free energy
for this complex is calculated at −19.74 kcal/mol, with
contributions from van der Waals interactions (−18.04 kcal/mol),
electrostatic potential energy (−30.87 kcal/mol) and non-polar
solvation effects (−1.63 kcal/mol; Fig. 7B).
For the ID1-6mgn complex (Fig. 7C), the binding free energy
primarily involves residues ALA-25, GLN-17, ARG-10, GLN-26, PHE-9,
ASN-6, ILE-31, ARG-14, LYS-24 and GLY-13. The calculated binding
free energy is −22.04 kcal/mol, with van der Waals forces
contributing −23.73 kcal/mol, electrostatic potential energy
contributing −28.32 kcal/mol and non-polar solvation energy
contributing −2.31 kcal/mol (Fig.
7D).
For the TGIF2-2dmn complex (Fig. 7E), the binding free energy involves
residues ARG-64, TRP-57, LEU-14, ALA-61, ASN-60, ILE-65, ASN-13,
VAL-19, LEU-22 and ASN-56, with the overall binding free energy
calculated as −21.43 kcal/mol (Fig.
7F). The contributions include van der Waals forces (−29.43
kcal/mol), electrostatic potential energy (−21.26 kcal/mol) and
non-polar solvation energy (−2.26 kcal/mol).
These results indicate that DAP exhibits strong
binding affinities with TGIF2, ID1 and BMP2, with van der Waals and
electrostatic interactions carrying out the most significant roles,
while non-polar solvation energy contributes to a lesser extent.
Polar solvation energy appears to have a less favorable effect on
these interactions.
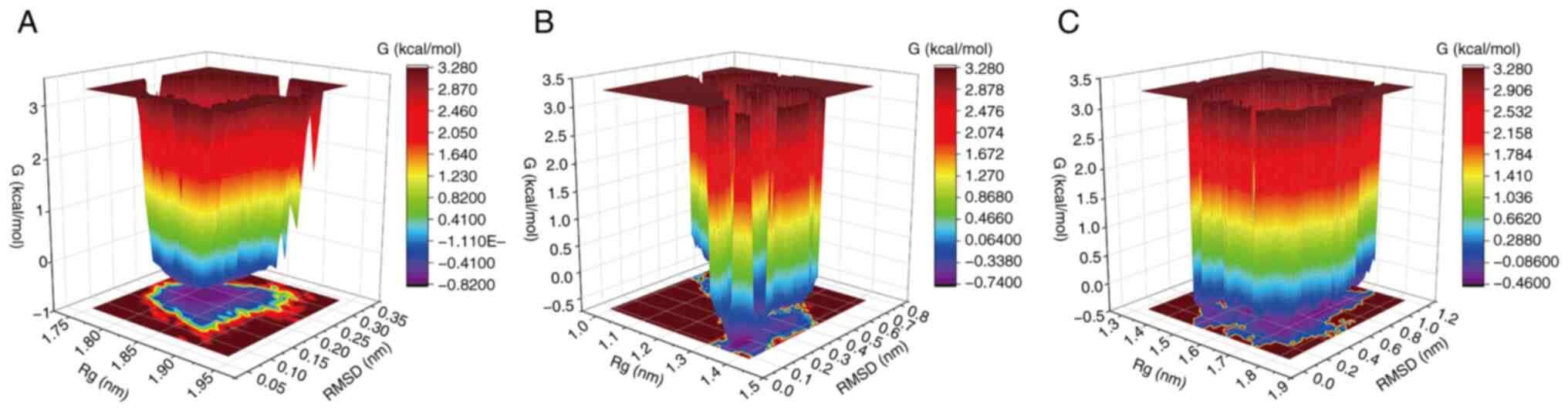
Free energy landscape (FEL)
FEL provides a detailed characterization of the free
energy changes experienced by a molecular system during
simulations. By analyzing the FEL, the characteristic conformations
of a complex can be identified and examined, which serves as an
indicator of conformational stability during the simulation
(50,51). As illustrated in Fig. 8, the BMP2-1rew complex
predominantly adopts a stable conformation within an RMSD range of
0.10–0.30 Å and an Rg range of 1.77–1.90 nm (Fig. 8A). For the ID1-6mgn complex, the
most stable conformation is observed within an RMSD range of
0.0–0.7 Å and an Rg range of 1.0–1.4 nm (Fig. 8B). The TGIF2-2dmn complex
demonstrates two distinct regions of dominant stability: One with
an RMSD of 0.0–1.2 Å and Rg of 1.3–1.8 nm, and another with an RMSD
of 0.8–1.0 Å and Rg of 1.45–1.55 nm (Fig. 8C). These findings indicate that the
BMP2-1rew, ID1-6mgn and TGIF2-2dmn complexes each exhibit stable
conformations within specific RMSD and Rg ranges, which can be
utilized as a basis for understanding the characteristic
conformations and stability of these complexes.
Discussion
Treatment of cervical cancer currently relies on a
combination of surgery, radiotherapy, chemotherapy and
immunotherapy (52). Early-stage
cervical cancer can often be managed with either radical surgery or
radiation therapy (53). However,
both methods suffer from a lack of specificity for cancer cells,
leading to notable side effects despite their clinical efficacy.
Traditional chemotherapy agents for cervical cancer, such as
nedaplatin, paclitaxel and bleomycin, are associated with harmful
side effects (54,55). Consequently, there is a need for
more effective and safer anticancer drugs. Natural products, known
for their low toxicity and ability to target multiple biological
pathways, have gained attention as promising alternatives to
chemical drugs due to their superior biocompatibility (56). As a result, natural products have
become a key source of novel antitumor agents (57).
DAP also known as Altertoxin I, is a quinone-type
mycotoxin first isolated from Alternaria species. A previous
study reveal that DAP has an ID50 value of 20 µg/ml
against HeLa cells (58). DAP has
been evaluated for cytotoxicity in A549, HeLa, U2OS and HepG2 cell
lines, and the results revealed that DAP exhibited minimal
cytotoxicity at 20 µM in these cell lines (43). DAP has demonstrated selective
cytotoxicity against human colon cancer cells (HCT-8) with an
IC50 of 1.78 µM (59),
highlighting its potential in cancer therapy. Chemotherapy, a
mainstay treatment, is frequently hampered by drug resistance and
dose-limiting toxicities (60).
Cisplatin remains the preferred drug for the chemotherapeutic
treatment of cervical cancer, however, several factors, including
its low selectivity, high toxicity, adverse side effects, tumor
multidrug resistance and propensity for recurrence, ultimately
contribute to treatment failure (61). Chemotherapy failure is primarily
caused by drug resistance mechanisms, such as
P-glycoprotein-mediated efflux of chemotherapeutic agents from
tumor cells. Furthermore, chemotherapy is constrained by
dose-limiting toxicities that adversely affect healthy tissues and
compromise the quality of life of patients (62). Additionally, targeted therapies
encounter obstacles due to tumor heterogeneity and the development
of resistance through compensatory signaling pathways (63). Natural products such as DAP offer a
promising strategy to address these issues. Its structural
complexity enables multi-target mechanisms, potentially overcoming
resistance while minimizing off-target effects. In the present
study, DAP exhibited potent activity against HeLa cells
(IC50±6.43 µM) and transcriptome analysis revealed that
DAP induces apoptosis via the Hippo and TGFβ pathways. This dual
effect may circumvent common drug resistance mechanisms. Notably,
DAP exhibited minimal inhibitory activity against HGF-1 after 24 h
of exposure, with an SI value of 8.41. The safety profile of the
drug was evaluated using the SI value, where a higher SI value
indicates a broader therapeutic window and reduced cytotoxicity
towards normal cells. A previous study has demonstrated that an SI
value >1 is indicative of selective antitumor activity (64).
The findings of the present study demonstrated that
the IC50 of DAP exhibits a time-dependent increase,
indicating that prolonged exposure to drug treatment may enhance
the fitness of a cell population. According to existing research
findings (65), the fitness of a
cell population can be improved through gradual exposure to stress,
as cells epigenetically adapt and fine-tune their stress regulatory
networks, thereby establishing stable adaptive states. Initially,
drug-sensitive cells accumulate transcriptional and epigenetic
changes in response to stress, resulting in more robust responses
upon subsequent stimulation. We hypothesize that a comparable
process occurs when DAP is applied to HeLa cells spanning an
extended period, leading to an increase in IC50 from
6.43 µM at 24 h to 19.27 µM at 72 h.
DAP was applied to HeLa cells at concentrations of
3.0, 6.0 and 9.0 µmol/l for 24 h, inducing apoptosis and cell cycle
arrest in a concentration-dependent manner. At the highest
concentration of 9.0 µmol/l, the apoptosis rate reached 24.13%,
with cell cycle arrest occurring predominantly in the S phase.
These findings suggest that the inhibitory effects of DAP on HeLa
cells are associated with apoptosis induction and cell cycle
arrest. To further elucidate the molecular mechanisms underlying
DAP-induced apoptosis, transcriptome sequencing was conducted on
HeLa cells treated with 6.0 µmol/l DAP for 24 h, identifying 140
DEGs. Bioinformatics analysis using KEGG and GO revealed that
significantly enriched pathways were associated with apoptosis,
while GO annotations highlighted terms associated with the
regulation of cell proliferation. Specifically, the TGFβ signaling
pathway and Hippo signaling pathway, both implicated in apoptosis
and enriched in KEGG, were analyzed in detail. Three genes (TGIF2,
ID1 and BMP2) were identified as key mediates in these pathways.
Subsequent RT-qPCR analysis confirmed that the expression trends of
these three genes were consistent with the results from
transcriptome sequencing. This consistency indicates that the
inhibitory effects of DAP on HeLa cells are likely mediated through
the upregulation of TGIF2, ID1 and BMP2.
TGIF2, an important member of the TGIF family,
encodes a DNA-binding transcription factor that carries out a key
role in tumor regulation through multiple mechanisms. The study has
demonstrated that long non-coding RNA SNHG7 upregulates TGIF2
expression by modulating miR-449a, thereby promoting proliferation,
migration, invasion and epithelial-mesenchymal transition in
non-small cell lung cancer (66).
Additionally, non-coding RNA MALAT1 upregulates TGIF2 expression by
negatively regulating miR-129, leading to enhanced proliferation,
migration and invasion in human osteosarcoma MG63 cells (67). In osteosarcoma, TGIF2 serves as a
target gene of miR-34; its upregulation inhibits osteosarcoma tumor
growth in nude mice and promotes apoptosis (68), these findings are consistent with
the upregulation of TGIF2 expression observed in cervical cancer
HeLa cells in the present study.
ID1 is a member of the helix-loop-helix (HLH) family
of transcription factors. ID1 exerts its oncogenic effects by
inhibiting the activity of basic HLH transcription factors, thereby
participating in tumor initiation, progression, cell cycle
regulation and metastasis. It is closely associated with
angiogenesis, tumor differentiation and drug resistance in various
types of cancer (69,70). As a negative regulator of cell
differentiation, ID1 promotes tumorigenesis by activating the MAPK
signaling pathway and inactivating the p16(INK4a)/pRB tumor
suppressor pathway, thus facilitating the growth and proliferation
of prostate cancer cells (71,72).
A study has confirmed that ID1 is highly expressed in ovarian
cancer tissues and its expression associates with the degree of
tumor differentiation, carrying out a key role in ovarian cancer
development (73). Compared with
control cells, SKOV3 cells depleted of ID1 exhibited a notable
reduction in proliferation and invasion capabilities, while
apoptosis was markedly increased (74). The increase in ROS levels following
ID1 overexpression aligns with prior studies revealing ROS inhibits
dendritic cell differentiation and myeloid-derived suppressor cell
expansion in tumor-bearing mice (75), These findings are consistent with
the results of the present study. We hypothesize that DAP
upregulates ID1 expression, increasing ROS levels and inhibiting
cervical cancer cells.
BMP2 is a highly expressed secreted protein in
various types of cancer (76). As
a member of the TGFβ superfamily, BMP2 participates in multiple
biological processes, including normal cell growth and development,
apoptosis, migration, invasion, bone formation and cancer
progression (77). Research
(78) indicates that BMP2 carries
out a key role in the proliferation, migration and differentiation
of neural crest cells. Compared with normal tissues, BMP2 is
considerably upregulated in various cancer tissues, suggesting its
potential as a key biomarker for targeted cancer therapy.
Literature studies have revealed that BMP2 exhibits both pro-cancer
and anti-cancer functions in cancer tissues. For instance, Du et
al (79) demonstrated that
promoter methylation of the BMP2 gene in the breast cancer cell
line MCF-7 downregulates BMP2 expression, enhancing drug resistance
and promoting cancer progression. Conversely, other studies have
revealed that treating the human and mouse breast cancer cell,
MCF-7, with BMP2 inhibits cell migration and proliferation,
revealing its anti-cancer effects (80,81).
In nasopharyngeal carcinoma, the BMP2 promoter
enhances its expression by binding to SOX9, thereby activating the
BMP2-induced mTOR signaling pathway and promoting the
proliferation, migration and invasion of nasopharyngeal carcinoma
cells (77,82). Research reveals that BMP2
suppresses gastric cancer cell proliferation by arresting the cell
cycle via a non-SMAD BMP pathway that downregulates EZH2 (83), consistent with the findings of the
present study. The present study indicates that DAP induces the
upregulation of BMP2 expression, which subsequently influences cell
proliferation. This effect may be attributed to the ability of BMP2
upregulation to activate AMPK, leading to the inhibition of the
mTOR signaling pathway. Specifically, the activation of AMPK
suppresses the activity of the mTORC1 complex via phosphorylation,
thereby disrupting protein synthesis and metabolic processes
essential for cell proliferation (84).
The limited number of DEGs identified at 24 h
post-treatment suggests an early drug response phase rather than
transcriptomic reprogramming. Similar results were seen in other
studies: Only 357 and 455 DEGs were found in two sweet corn
varieties under low-temperature stress (85) and 551 DEGs were detected after
vascular hepatocyte differentiation in pigs (86). The limited number of DEGs observed
in this study may be attributed to three potential factors:
Inadequate drug exposure or low concentrations preventing a
significant transcriptional response, cellular heterogeneity
causing varied drug sensitivities and diluting signals, and the
phenotypic lag effect (87) where
drugs act through early gene activity and protein interactions.
Future work will assess DAP's effects on cervical cancer HeLa cells
by extending treatment time and testing more concentration
levels.
Following DAP treatment, cells exhibited signs of
apoptosis. Transcriptome analysis and RT-qPCR validation confirmed
that this apoptotic response was associated with the upregulation
of three genes: TGIF2, ID1 and BMP2. This finding raises the
question of whether DAP interacts directly with the corresponding
proteins encoded by these genes. To investigate potential
protein-level interactions between DAP and the TGIF2, ID1 and BMP2
proteins, the present study employed molecular docking technology
to evaluate their binding affinities. Molecular docking is based on
the lock-and-key principle, examining how the spatial structure and
electrostatic characteristics of small molecules match the active
sites of their target proteins. The lower the binding energy
between a small molecule and its target protein, the stronger their
interaction. Typically, a binding energy <-4.25 kcal/mol
indicates some degree of binding activity; <-5.0 kcal/mol
suggests good binding activity; and <-7.0 kcal/mol indicates
strong binding activity (88).
While this study elucidated the potential binding
modes of DAP with TGIF2, ID1 and BMP2 proteins using molecular
docking, it is important to acknowledge the intrinsic limitations
of static docking models. Molecular docking generally relies on
rigid or semi-flexible receptor models, which may fail to
adequately capture the conformational dynamics of protein-ligand
complexes under physiological conditions (89). To address these limitations, the
present study incorporated MD simulations for dynamic validation.
To further elucidate the molecular interactions underlying the
anticancer potential of DAP, MD simulations were carried out to
evaluate its binding stability with key regulatory proteins
involved in cervical cancer progression. The simulations analyzed
ligand-protein interactions with TGIF2, ID1 and BMP2 over a
200-nanosecond trajectory. RMSD analysis revealed that all three
complexes exhibited initial structural fluctuations before
achieving stable conformations, suggesting successful ligand
binding. Similarly, RMSF analysis identified key residues
exhibiting flexibility within each protein-ligand complex, further
supporting their dynamic interactions. These computational findings
provide valuable insights into the structural basis of the
biological activity of DAP, reinforcing its potential as a novel
therapeutic candidate for cervical cancer.
The present study provides insights into the
inhibitory mechanisms of DAP on cervical cancer cells but is
limited by using only the HeLa cell line. Initial screening
(Table SII) revealed DAP has
notable activity against HeLa and SiHa cells, however, only 11.9 mg
of purified DAP was available. Due to this limitation, mechanistic
studies focused on HeLa cells. Using a single cell line restricts
the generalizability of the findings, as cervical cancer is
heterogeneous and responses vary across cell lines. Thus, results
obtained with HeLa cells may not fully represent other subtypes.
The proposed mechanisms require further validation for broader
applicability.
Future work will validate key experiments in
additional cell lines such as CaSki and SiHa and, where feasible,
use patient-derived cells or organoids for clinically relevant
models. Simultaneously, this study will establish subcutaneous
xenograft cervical cancer models in immunodeficient nude mice
through the injection of cervical cancer cells. DAP will be
administered subcutaneously at low, medium and high doses to these
models. This study will utilize the untreated models as the control
group, and paclitaxel-treated models as the positive control. In
vivo experiments will evaluate the efficacy of DAP by analyzing
tumor growth, final tumor size, pathological changes and marker
protein expression across all groups. Acute or subchronic toxicity
studies using healthy mice will assess behavioral changes, body
weight, food/water intake, blood profiles, biochemical parameters
and organ histopathology (liver, kidney, spleen, lung, heart and
intestine). These analyses will aim to identify toxic target
organs, determine the maximum tolerated dose and establish a safety
margin. Together, these experiments will provide preclinical data
to assess the clinical potential of DAP and guide its development
as a therapy.
Collectively, a novel endophytic fungal species,
Alternaria semiverrucosa, was isolated from Taxus
chinensis var. marei collected in Guiyang, China. DAP
was subsequently obtained from the fermentation metabolites of
Alternaria semiverrucosa. The inhibitory effects of DAP on
cervical cancer HeLa cells suggest that it exhibits high efficacy
and low toxicity, making it a promising candidate for further
investigation, and warranting further investigation into its
underlying molecular mechanisms and therapeutic potential. Future
studies integrating experimental validation of these molecular
interactions will be key to confirming efficacy and clinical
applicability of DAP.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank Dr Heng Luo for
their assistance and guidance with the in vitro experiments
which were conducted at the Key Laboratory of Chemistry for Natural
Products of Guizhou Province, Chinese Academy of Sciences, Guiyang,
China. The authors would also like to thank Guiyang Stomatological
Hospital (Guiyang, China) for donating HGF-1 human gingival
fibroblasts.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant nos. 32460007, 32170019 and
31670027).
Availability of data and materials
The data generated in the present study may be
found in the National Center for Biotechnology Information under
accession number PRJNA1272013, or at the following URL: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1272013/.
Authors' contributions
JK and MY conceptualized the present study. MY and
YL developed the methodology. LH and ZH were responsible for the
application and implementation of the Molecular dynamics simulation
software. MY, YL and JK conducted the analysis of the experimental
data. MY, YL and ZH carried out cell validation experiments. ZH and
YL managed the transcriptome data curation and analysis. MY and LH
wrote the original draft. JK, MY and YL contributed to reviewing
and editing. LH, ZH and YL handled the microscopy images. JK
supervised the project and administered it. MY, YL, ZH and JK
confirm the authenticity of all the raw data. All authors have read
and agreed to the published version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cohen PA, Jhingran A, Oaknin A and Denny
L: Cervical cancer. Lancet. 393:169–182. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kokhdan EP, Khodavandi P, Ataeyan MH,
Alizadeh F, Khodavandi A and Zaheri A: Anti-cancer activity of
secreted aspartyl proteinase protein from Candida tropicalis on
human cervical cancer HeLa cells. Toxicon. 249:1080732024.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bray F, Jemal A, Soerjomataram I, Siegel
RL, Ferlay J, Sung H and Laversanne M: Global cancer statistics
2022: Globocan estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263.
2024.PubMed/NCBI
|
|
4
|
Luan H: Human papilloma virus infection
and its associated risk for cervical lesions: A cross-sectional
study in Putuo area of Shanghai, China. BMC Womens Health.
23:282023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang R, Pan W, Jin L, Huang W, Li Y, Wu D,
Gao C, Ma D and Liao S: Human papillomavirus vaccine against
cervical cancer: Opportunity and challenge. Cancer Lett.
471:88–102. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xu J, Tan ZC, Shen ZY, Shen XJ and Tang
SM: Cordyceps cicadae polysaccharides inhibit human cervical cancer
hela cells proliferation via apoptosis and cell cycle arrest. Food
Chem Toxicol. 148:1119712021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang X, Song Z, Li Y, Wang H, Zhang S,
Reid AM, Lall N, Zhang J, Wang C, Lee D, et al: Cytotoxic and
antiangiogenetic xanthones inhibiting tumor proliferation and
metastasis from garcinia xipshuanbannaensis. J Nat Prod.
84:1515–1523. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kusari S, Hertweck C and Spiteller M:
Chemical ecology of endophytic fungi: Origins of secondary
metabolites. Chem Biol. 19:792–798. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang HW, Song YC and Tan RX: Biology and
chemistry of endophytes. Nat Prod Rep. 23:753–771. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bashyal BP, Wijeratne EM, Tillotson J,
Arnold AE, Chapman E and Gunatilaka AA: Chlorinated
dehydrocurvularins and alterperylenepoxide A from Alternaria sp.
AST0039, a fungal endophyte of Astragalus lentiginosus. J Nat Prod.
80:427–433. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li SJ, Zhang X, Wang XH and Zhao CQ: Novel
natural compounds from endophytic fungi with anticancer activity.
Eur J Med Chem. 156:316–343. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhu M, Zhang X, Feng H, Dai J, Li J, Che
Q, Gu Q, Zhu T and Li D: Penicisulfuranols A-F, alkaloids from the
mangrove endophytic fungus Penicillium janthinellum HDN13-309. J
Nat Prod. 80:71–75. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chakravarthi BV, Sujay R, Kuriakose GC,
Karande AA and Jayabaskaran C: Inhibition of cancer cell
proliferation and apoptosis-inducing activity of fungal taxol and
its precursor baccatin III purified from endophytic Fusarium
solani. Cancer Cell Int. 13:1052013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu LS, Hu CL, Han T, Zheng CJ, Ma XQ,
Rahman K and Qin LP: Cytotoxic metabolites from Perenniporia
tephropora, an endophytic fungus from Taxus chinensis var. mairei.
Appl Microbiol Biotechnol. 97:305–315. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Uzor PF, Ebrahim W, Osadebe PO, Nwodo JN,
Okoye FB, Müller WE, Lin W, Liu Z and Proksch P: Metabolites from
Combretum dolichopetalum and its associated endophytic fungus
Nigrospora oryzae-evidence for a metabolic partnership.
Fitoterapia. 105:147–150. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wani MC, Taylor HL, Wall ME, Coggon P and
McPhail AT: Plant antitumor agents. VI. The isolation and structure
of taxol, a novel antileukemic and antitumor agent from Taxus
brevifolia. J Am Chem Soc. 93:2325–2327. 1971. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zaiyou J, Li M, Xu G and Zhou X: Isolation
of an endophytic fungus producing baccatin III from Taxus
wallichiana var. mairei. J Ind Microbiol Biotechnol. 40:1297–1302.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zaiyou J, Li M and Xiqiao H: An endophytic
fungus efficiently producing paclitaxel isolated from Taxus
wallichiana var. mairei. Medicine (Baltimore). 96:e74062017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fan ZY, Peng J, Lou JQ, Chen Y, Wu XM, Tan
R and Tan RX: Neuroprotective α-pyrones from Nigrospora oryzae, an
endophytic fungus residing in Taxus chinensis var. mairei.
Phytochemistry. 216:1138732023. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hu D, Fan Y, Tan Y, Tian Y, Liu N, Wang L,
Zhao D, Wang C and Wu A: Metabolic profiling on alternaria toxins
and components of Xinjiang Jujubes incubated with pathogenic
alternaria Alternata and Alternaria tenuissima via orbitrap
high-resolution mass spectrometry. J Agri Food Chem. 65:8466–8474.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Andersen B, Nielsen KF, Fernández Pinto V
and Patriarca A: Characterization of Alternaria strains from
Argentinean blueberry, tomato, walnut and wheat. Int J Food
Microbiol. 196:1–10. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang CL, Wu HM, Liu CL, Zhang X, Guo ZK,
Chen Y, Liu F, Liang Y, Jiao RH, Tan RX and Ge HM: Bialternacins
A-F, aromatic polyketide dimers from an endophytic Alternaria sp. J
Nat Prod. 82:792–797. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xi J, Tian LL, Xi J, Girimpuhwe D, Huang
C, Ma R, Yao X, Shi D, Bai Z, Wu QX and Fang J: Alterperylenol as a
novel thioredoxin reductase inhibitor induces liver cancer cell
apoptosis and ferroptosis. J Agric Food Chem. 70:15763–15775. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li L, Yang M, Yu J, Cheng S, Ahmad M, Wu
C, Wan X, Xu B, Ben-David Y and Luo H: A novel L-phenylalanine
dipeptide inhibits the growth and metastasis of prostate cancer
cells via targeting DUSP1 and TNFSF9. Int J Mol Sci. 23:109162022.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu Y, Xin ZZ, Song J, Zhu XY, Liu QN,
Zhang DZ, Tang BP, Zhou CL and Dai LS: Transcriptome analysis
reveals potential antioxidant defense mechanisms in Antheraea
pernyi in response to zinc stress. J Agric Food Chem. 66:8132–8141.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen C, Chen H, Zhang Y, Thomas HR, Frank
MH, He Y and Xia R: TBtools: An integrative toolkit developed for
interactive analyses of big biological data. Mol Plant.
13:1194–1202. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Case DA, Aktulga HM, Belfon K, Cerutti DS,
Cisneros GA, Cruzeiro VWD, Forouzesh N, Giese TJ, Götz AW, Gohlke
H, et al: Amber Tools. J Chem Inf Model. 63:6183–6191. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shaw DE, Maragakis P, Lindorff-Larsen K,
Piana S, Dror RO, Eastwood MP, Bank JA, Jumper JM, Salmon JK, Shan
Y and Wriggers W: Atomic-level characterization of the structural
dynamics of proteins. Science. 330:341–346. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tian C, Kasavajhala K, Belfon KAA,
Raguette L, Huang H, Migues AN, Bickel J, Wang Y, Pincay J, Wu Q
and Simmerling C: ff19SB: Amino-acid-specific protein backbone
parameters trained against quantum mechanics energy surfaces in
solution. J Chem Theory Comput. 16:528–552. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Izadi S, Anandakrishnan R and Onufriev AV:
Building water models: A different approach. J Phys Chem Lett.
5:3863–3871. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang H, Gao X and Fang J: Multiple
staggered mesh ewald: Boosting the accuracy of the smooth particle
mesh ewald method. J Chem Theory Comput. 12:5596–5608. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Donnelly SM, Lopez NA and Dodin IY:
Steepest-descent algorithm for simulating plasma-wave caustics via
metaplectic geometrical optics. Phys Rev E. 104:0253042021.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bussi G, Donadio D and Parrinello M:
Canonical sampling through velocity rescaling. J Chem Phys.
126:0141012007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Nosé S and Klein ML: Constant pressure
molecular dynamics for molecular systems. Mol Phys. 50:1055–1076.
1983. View Article : Google Scholar
|
|
36
|
Zhang SY, Li ZL, Bai J, Wang Y, Zhang LM,
Wu X and Hua HM: A new perylenequinone from a halotolerant fungus,
Alternaria sp. M6. Chin J Nat Med. 10:68–71. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Campos FR, Barison A, Daolio C, Ferreira
AG and Rodrigues-Fo E: Complete 1H and 13C NMR assignments of
aurasperone A and fonsecinone A, two bis-naphthopyrones produced by
Aspergillus aculeatus. Magn Reson Chem. 43:962–965. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xiong HY, Fei DQ, Zhou JS, Yang CJ and Ma
GL: Steroids and other constituents from the mushroom Armillaria
lueo-virens. Chem Nat Compd. 45:759–761. 2009. View Article : Google Scholar
|
|
39
|
Wu J, Choi JH, Yoshida M, Hirai H, Harada
E, Masuda K, Koyama T, Yazawa K, Noguchi K, Nagasawa K and
Kawagishi H: Osteoclast-forming suppressing compounds, gargalols A,
B, and C, from the edible mushroom Grifola gargal. Tetrahedron.
67:6576–6581. 2011. View Article : Google Scholar
|
|
40
|
Yue JM, Chen SN, Lin ZW and Sun HD:
Sterols from the fungus lactarium volemus. Phytochemistry.
56:801–806. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ding T, Zhou Y, Qin JJ, Yang LJ, Zhang WD
and Shen YH: Chemical constituents from wetland soil fungus
penicillium oxalicum GY1. Fitoterapia. 142:1045302020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Liu S, Sun C, Ha Y, Ma M, Wang N, Zhou Y
and Zhang Z: Novel antibacterial alkaloids from the mariana
trench-derived actinomycete streptomyces sp. SY2255. Tetrahedron
Lett. 137:1549352024. View Article : Google Scholar
|
|
43
|
Wu B, Lin WH, Gao HY, Zheng L, Wu LJ and
Kim CS: Four new antibacterial constituents from Senecio
cannabifolius. Pharm Biol. 44:440–444. 2006. View Article : Google Scholar
|
|
44
|
Pitera JW: Expected distributions of
root-mean-square positional deviations in proteins. J Phys Chem B.
118:6526–6530. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Trott O and Olson AJ: AutoDock vina:
Improving the speed and accuracy of docking with a new scoring
function, efficient optimization, and multithreading. J Comput
Chem. 31:455–461. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Fu Y, Wu F, Huang JH, Chen YC and Luo MB:
Simulation study on the extension of semi-flexible polymer chains
in cylindrical channel. Chin J Polym Sci. 37:1053–1060. 2019.
View Article : Google Scholar
|
|
47
|
Cao X, Hummel MH, Wang Y, Simmerling C and
Coutsias EA: Exact analytical algorithm for the solvent-accessible
surface area and derivatives in implicit solvent molecular
simulations on GPUs. J Chem Theory Comput. 20:4456–4468. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wang Y and Wang Y: HBCalculator: A tool
for hydrogen bond distribution calculations in molecular dynamics
simulations. J Chem Inf Model. 64:1772–1777. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Jiang D, Du H, Zhao H, Deng Y, Wu Z, Wang
J, Zeng Y, Zhang H, Wang X, Wang E, et al: Assessing the
performance of MM/PBSA and MM/GBSA methods. 10. Prediction
reliability of binding affinities and binding poses for RNA-ligand
complexes. Phys Chem Chem Phys. 26:10323–10335. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liao Q: Enhanced sampling and free energy
calculations for protein simulations. Prog Mol Biol Transl Sci.
170:177–213. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Bai F, Xu Y, Chen J, Liu Q, Gu J, Wang X,
Ma J, Li H, Onuchic JN and Jiang H: Free energy landscape for the
binding process of Huperzine A to acetylcholinesterase. Proc Natl
Acad Sci USA. 110:4273–4278. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhang Z, Liu M, An Y, Gao C, Wang T, Zhang
Z, Zhang G, Li S, Li W, Li M and Wang G: Targeting immune
microenvironment in cervical cancer: Current research and advances.
J Transl Med. 23:8882025. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Sharma S, Deep A and Sharma AK: Current
treatment for cervical cancer: An update. Anticancer Agents Med
Chem. 20:1768–1779. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Barra F, Lorusso D, Leone Roberti Maggiore
U, Ditto A, Bogani G, Raspagliesi F and Ferrero S: Investigational
drugs for the treatment of cervical cancer. Expert Opin Investig
Drugs. 26:389–402. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Giudice E, Mirza MR and Lorusso D:
Advances in the management of recurrent cervical cancer: State of
the art and future perspectives. Curr Oncol Rep. 25:1307–1326.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Newman DJ and Giddings LA: Natural
products as leads to antitumor drugs. Phytochem Rev. 13:123–137.
2014. View Article : Google Scholar
|
|
57
|
Baek SH: Editorial for the special issue
‘Anticancer activity and metabolic pathways of natural products
2.0’. Biomedicines. 13:20832025. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Pero RW, Posner H, Blois M, Harvan D and
Spalding JW: Toxicity of metabolites produced by the ‘Alternaria’.
Environ Health Perspect. 4:87–94. 1973. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Fang ZF, Yu SS, Zhou WQ, Chen XG, Ma SG,
Li Y and Qu J: A new isocoumarin from metabolites of the endophytic
fungus Alternaria tenuissima (Nees & T. Nees: Fr.)
Wiltshire. Chinese Chem Lett. 23:317–320. 2012. View Article : Google Scholar
|
|
60
|
Ma Y, Lin Q, Yang Y, Liang W, Salamone SJ,
Li Y, Lin Y, Zhao H, Zhao Y, Fang W, et al: Clinical
pharmacokinetics and drug exposure-toxicity correlation study of
docetaxel based chemotherapy in Chinese head and neck cancer
patients. Ann Transl Med. 8:2362020. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Yang Z, Liu Z, Ablise M, Jia J, Maimaiti
A, Lv ZY, Mutalipu Z, Yan T, Wang Y, Aihaiti A, et al: Design,
synthesis, and in vitro and in vivo anti-drug resistant cervical
cancer activity of novel licochalcone A derivatives based on dual
targeting of VEGFR-2/P-gp. Bioorg Chem. 163:1086392025. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zafar A, Khatoon S, Khan MJ, Abu J and
Naeem A: Advancements and limitations in traditional anti-cancer
therapies: A comprehensive review of surgery, chemotherapy,
radiation therapy, and hormonal therapy. Discov Oncol. 16:6072025.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Raman R, Debata S, Govindarajan T and
Kumar P: Targeting triple-negative breast cancer: Resistance
mechanisms and therapeutic advancements. Cancer Med. 14:e708032025.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Li W, Xu F, Shuai W, Sun H, Yao H, Ma C,
Xu S, Yao H, Zhu Z, Yang DH, et al: Discovery of novel
quinoline-chalcone derivatives as potent antitumor agents with
microtubule polymerization inhibitory activity. J Med Chem.
62:993–1013. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
França GS, Baron M, King BR, Bossowski JP,
Bjornberg A, Pour M, Rao A, Patel AS, Misirlioglu S, Barkley D, et
al: Cellular adaptation to cancer therapy along a resistance
continuum. Nature. 631:876–883. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Pang L, Cheng Y, Zou S and Song J: Long
noncoding RNA SNHG7 contributes to cell proliferation, migration,
invasion and epithelial to mesenchymal transition in non-small cell
lung cancer by regulating miR-449a/TGIF2 axis. Thorac Cancer.
11:264–276. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Liu K, Zhang Y, Liu L and Yuan Q:
MALAT1 promotes proliferation, migration, and invasion of
MG63 cells by upregulation of TGIF2 via negatively
regulating miR-129. Onco Targets Ther. 11:8729–8740. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Xi L, Zhang Y, Kong S and Liang W:
miR-34 inhibits growth and promotes apoptosis of
osteosarcoma in nude mice through targetly regulating TGIF2
expression. Biosci Rep. 38:BSR201800782018. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Weiler S, Ademokun JA and Norton JD: ID
helix-loop-helix proteins as determinants of cell survival in
B-cell chronic lymphocytic leukemia cells in vitro. Mol Cancer.
14:302015. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Ruzinova MB and Benezra R: Id proteins in
development, cell cycle and cancer. Trends Cell Biol. 13:410–418.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Ouyang XS, Wang X, Ling MT, Wong HL, Tsao
SW and Wong YC: Id-1 stimu-lates serum independent prostate cancer
cell proliferation through inactivation of p16(INK4a)/pRB pathway.
Carcinogenesis. 23:721–725. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Ling MT, Wang X, Ouyang XS, Lee TK, Fan
TY, Xu K, Tsao SW and Wong YC: Activation of MAPK signaling pathway
is essential for Id-1 induced serum independent prostate cancer
cell growth. Oncogene. 21:8498–8505. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Su Y, Zheng L, Wang Q, Bao J, Cai Z and
Liu A: The PI3K/Akt pathway upregulates Id1 and integrin α4 to
enhance recruitment of human ovarian cancer endothelial progenitor
cells. BMC Cancer. 10:4592010. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Sun WZ, Li MH, Chu M, Wei LL, Bi MY, He Y
and Yu LB: Id1 knockdown induces the apoptosis and inhibits the
proliferation and invasion of ovarian cancer cells. Eur Rev Med
Pharmacol Sci. 20:2812–2818. 2016.PubMed/NCBI
|
|
75
|
Papaspyridonos M, Matei I, Huang Y, do
Rosario Andre M, Brazier-Mitouart H, Waite JC, Chan AS, Kalter J,
Ramos I, Wu Q, et al: Id1 suppresses anti-tumour immune responses
and promotes tumour progression by impairing myeloid cell
maturation. Nat Commun. 6:68402015. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Wang MH, Zhou XM, Zhang MY, Shi L, Xiao
RW, Zeng LS, Yang XZ, Zheng XFS, Wang HY and Mai SJ: BMP2 promotes
proliferation and invasion of nasopharyngeal carcinoma cells via
mTORC1 pathway. Aging (Albany NY). 9:1326–1340. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Lan L, Evan T, Li H, Hussain A, Ruiz EJ,
Zaw Thin M, Ferreira RMM, Ps H, Riising EM, Zen Y, et al: GREM1 is
required to maintain cellular heterogeneity in pancreatic cancer.
Nature. 607:163–168. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Huang S, Wang Y, Luo L, Li X, Jin X, Li S,
Yu X, Yang M and Guo Z: BMP2 is related to Hirschsprung's disease
and required for enteric nervous system development. Front Cell
Neurosci. 13:5232019. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Du M, Su XM, Zhang T and Xing YJ: Aberrant
promoter DNA methylation inhibits bone morphogenetic protein 2
expression and contributes to drug resistance in breast cancer. Mol
Med Rep. 10:1051–1055. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Waite KA and Eng C: BMP2 exposure results
in decreased PTEN protein degradation and increased PTEN levels.
Hum Mol Genet. 12:679–684. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Buijs JT, van der Horst G, van den Hoogen
C, Cheung H, de Rooij B, Kroon J, Petersen M, van Overveld PG,
Pelger RC and van der Pluijm G: The BMP2/7 heterodimer inhibits the
human breast cancer stem cell subpopulation and bone metastases
formation. Oncogene. 31:2164–2174. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Xiao B, Zhang W, Kuang Z, Lu J, Li W, Deng
C, He Y, Lei T, Hao W, Sun Z and Li L: SOX9 promotes nasopharyngeal
carcinoma cell proliferation, migration and invasion through BMP2
and mTOR signaling. Gene. 715:1440172019. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Chen Z, Yuan L, Li X, Yu J and Xu Z: BMP2
inhibits cell proliferation by downregulating EZH2 in gastric
cancer. Cell Cycle. 21:2298–2308. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Vora M, Mondal A, Jia D, Gaddipati P, Akel
M, Gilleran J, Roberge J, Rongo C and Langenfeld J: Bone
morphogenetic protein signaling regulation of AMPK and PI3K in lung
cancer cells and C. elegans. Cell Biosci. 12:762022. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Mao J, Yu Y, Yang J, Li G, Li C, Qi X, Wen
T and Hu J: Comparative transcriptome analysis of sweet corn
seedlings under low-temperature stress. Crop J. 5:396–406. 2017.
View Article : Google Scholar
|
|
86
|
Chen L, Zhang Y, Chen H, Zhang X, Liu X,
He Z, Cong P, Chen Y and Mo D: Comparative transcriptome analysis
reveals a more complicated adipogenic process in intramuscular stem
cells than that of subcutaneous vascular stem cells. J Agric Food
Chem. 67:4700–4708. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Zhao W, Li J, Chen MM, Luo Y, Ju Z, Nesser
NK, Johnson-Camacho K, Boniface CT, Lawrence Y, Pande NT, et al:
Large-scale characterization of drug responses of clinically
relevant proteins in cancer cell lines. Cancer Cell. 38:829–843.e4.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Istyastono EP, Radifar M, Yuniarti N,
Prasasty VD and Mungkasi S: PyPLIF HIPPOS: A molecular interaction
fingerprinting tool for docking results of autoDock vina and
PLANTS. J Chem Inf Model. 60:3697–3702. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Errington D, Schneider C, Bouysset C and
Dreyer FA: Assessing interaction recovery of predicted
protein-ligand poses. J Cheminform. 17:762025. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Wang L, Zhang J, Zhang W, Zheng M, Guo H,
Pan X, Li W, Yang B and Ding L: The inhibitory effect of adenosine
on tumor adaptive immunity and intervention strategies. Acta Pharm
Sin B. 14:1951–1964. 2024. View Article : Google Scholar : PubMed/NCBI
|