Introduction
Cell migration is a fundamental process in cell
biology and refers to the movement of cells from one location to
another. During cell migration, a series of elongated tubular
structures are produced at the trailing edge of the cell. Notably,
cell migration serves a key role in various biological processes,
including embryonic development, wound healing, immune response,
tissue regeneration and tumor metastasis (1). Extracellular vesicles (EVs) are
important mediators of intercellular communication, having notable
roles in various physiological and pathological processes; for
example, cancer cell-derived EVs can modulate the tumor
microenvironment, and EVs from endothelial progenitor cells can
induce a proangiogenic phenotype in terminally differentiated
endothelial cells and promote angiogenesis. Therefore, EVs exhibit
potential as novel biomarkers for diseases, therapeutic agents and
drug delivery vehicles (2,3). Previous studies have revealed that
EVs, including exosomes, microvesicles and apoptotic bodies, serve
important roles in a number of biological processes, including
intercellular communication, tissue homeostasis, cell
differentiation, organ development and remodeling (2–5). In
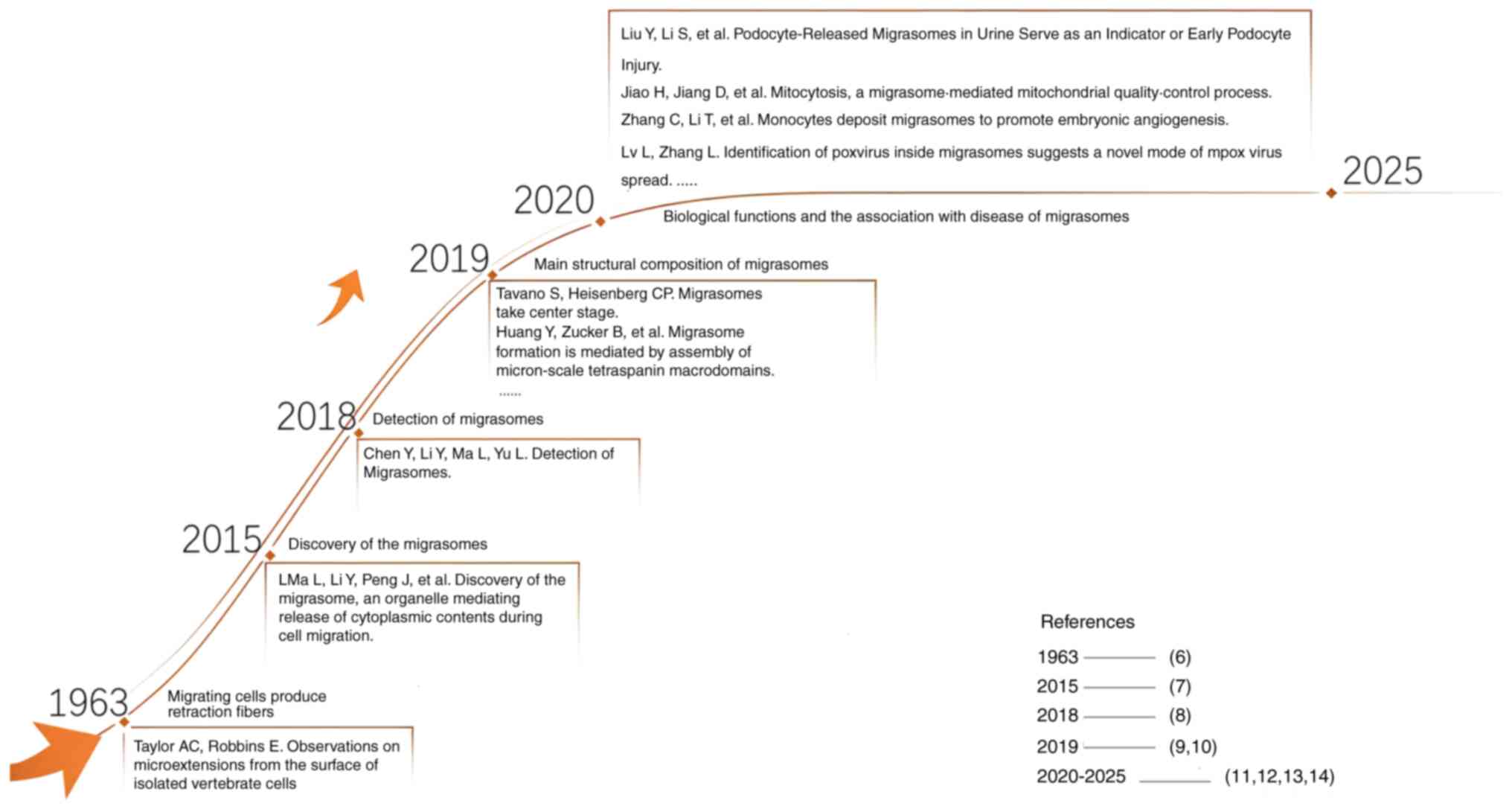
2015, a study by Ma et al (6) discovered a new type EV-like structure
and proposed the concept of migrasomes as novel cellular
organelles. Compared with other EVs, migrasomes exhibit distinct
structural, compositional and functional characteristics. With
diameters typically >1 µm, migrasomes are substantially larger
than exosomes, which have diameters of 30–150 nm. Unlike the
biogenesis of conventional EVs, such as exosomes derived from the
endosomal pathway or microvesicles generated via plasma membrane
budding, migrasome biogenesis is closely associated with cellular
migration (6). This unique
biogenesis mechanism suggests that migrasomes may serve as
migration trail markers, whereas exosomes predominantly facilitate
long-range intercellular communication (Table I). The discovery of migrasomes
provides a new perspective on how cells transport materials and
transmit information through extracellular structures (Fig. 1) (6–14).
 | Table I.Comparison of migrasomes with other
extracellular vesicles. |
Table I.
Comparison of migrasomes with other
extracellular vesicles.
| Feature | Migrasomes | Exosomes | Microvesicles | Apoptotic
bodies | Oncosomes |
|---|
| Structure | Pomegranate-like
with intraluminal vesicles (0.5–3 µm) | Single-membrane
vesicles (30–150 nm) | Single-layered
lipid bilayer vesicles with irregular morphology (100 nm-1 µm) | Irregular large
vesicles (1–5 µm) | Heterogeneous large
vesicles (1–10 µm) |
| Biogenesis | Cell
migration-dependent, formed at retraction fibers | Multivesicular
body-plasma membrane fusion | Plasma membrane
budding | Apoptotic
process | Tumor cell membrane
budding |
| Key components | TSPAN4, TSPAN7,
integrin α5β1 | Tumor
susceptibility gene 101 protein, programmed cell death
6-interacting metalloproteinases protein, CD63, CD81 | Annexin A1, Annexin
A2 | Annexin V, caspase
3 | Tumor-associated
antigens, matrix |
| Lipid
composition | Enriched in
sphingolipids, including sphingomyelin cholesterol |
Sphingolipid-rich |
Phosphatidylserine-rich rich |
Phosphatidylserine-sphingolipids | Cholesterol, and
ceramide, and |
| Functions | Intercellular
communication, migration positioning, mitochondrial quality
control, disease association | Intercellular
communication, immune regulation | Intercellular
communication, inflammatory response | Intercellular
communication, debris clearance | Tumor metastasis,
microenvironment remodeling |
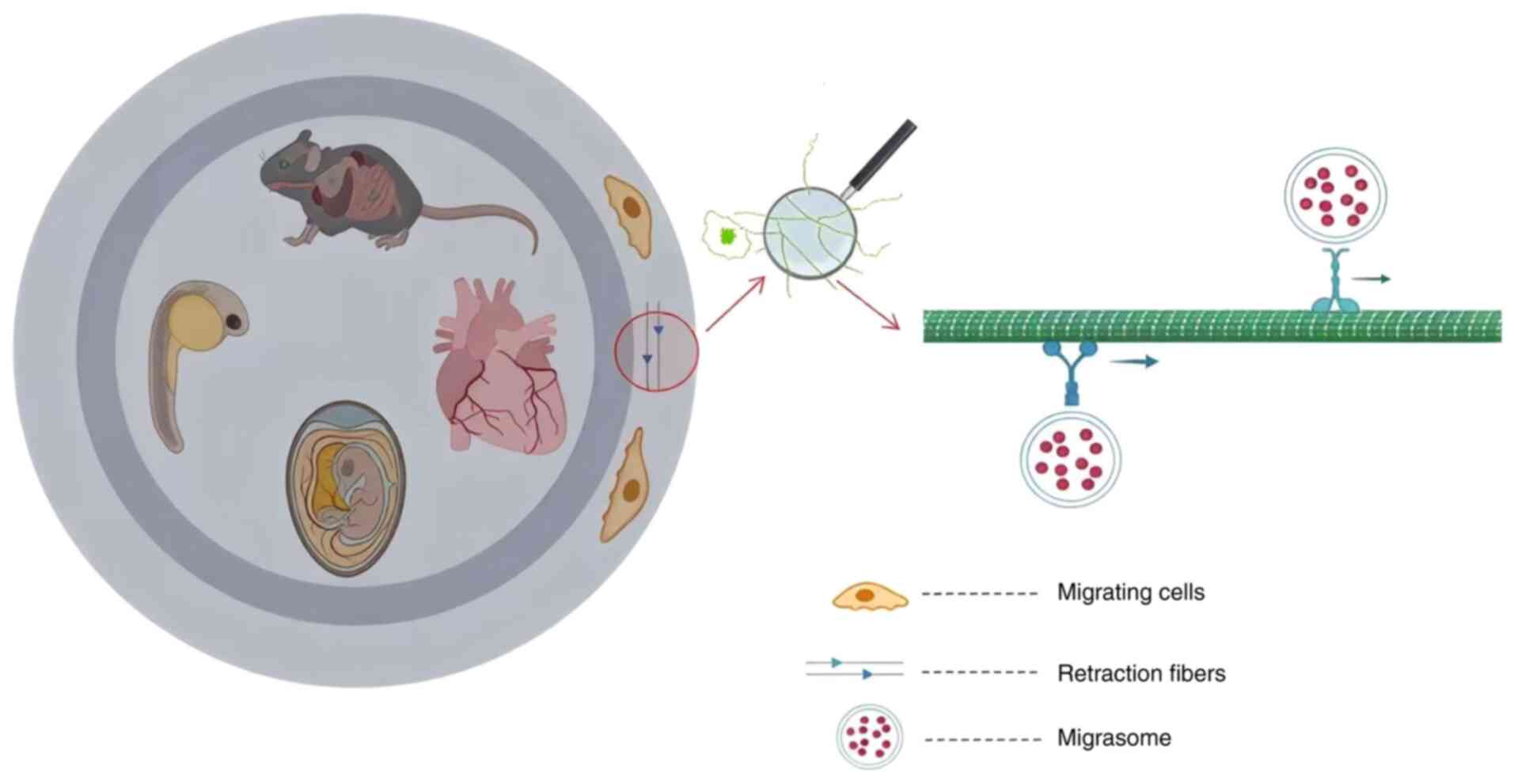
Discovery of migrasomes
The concept of migrasomes was first introduced
through observations of elongated tubular structures located at the
rear of migrating cells. In early studies of migrating cells,
Taylor and Robbins (7) discovered
and documented that elongated tubular structures formed when
migrating cells retracted from a substrate. They designated these
structures ‘retraction fibrils’, which were later named ‘retraction
fibers’ (RFs). Despite the initial lack of interest in these
structures from researchers, in 2012, a study at Tsinghua
University (Beijing, China) led by Yu (15) used transmission electron microscopy
to reveal the migration process of rat kidney cells. The study
revealed that cells leave behind RFs, which, upon further study,
are associated with vesicular structures ranging in diameter from
0.5 to 3 µm (6,15). These structures, situated behind
migrating cells that attach to the RFs left behind during cell
migration (6), were termed
pomegranate-like structures (PLSs) due to their resemblance to
pomegranate seeds. By purifying these structures, tetraspanin
(TSPAN)4, a distinct marker protein for PLS, was identified via
mass spectrometry, which served as a robust reference for future
migrasome research. Through knockdown of the negative regulator
Shank-associated RH domain interacting protein and treatment with
cell migration inhibitors, it was demonstrated that the formation
of PLSs is dependent on cell migration (6). Consequently, PLSs were renamed
‘migrasomes’, being defined as annular organelles formed at the
tips or intersections of RFs, or at the rear edge of migrating
cells (6).
Biological characteristics of
migrasomes
Structural features and main
components
The process of migrasome production can be
visualized dynamically via time-lapse imaging techniques (6,16).
Subsequent studies have confirmed the widespread presence of
migrasomes across a variety of species, tissues, organs and cell
types, including rat eyes, lungs, intestines, zebrafish embryos,
chick chorioallantoic membranes and human coronary artery
endothelial cells (6,11,17–20)
(Fig. 2). As a novel type of
organelle, migrasomes are characterized as membrane-bound vesicles
with an ellipsoidal shape that harbor numerous smaller vesicles.
Their composition primarily comprises proteins, lipids and nucleic
acids (6,21). Notably, migrasomes exhibit a
distinctive protein profile, which includes membrane proteins,
contractile proteins, cytoskeletal proteins, chaperones, vesicular
trafficking proteins and cell adhesion proteins, >50% of which
are membrane-related (6,21,22).
These proteins are engaged predominantly in biological processes,
including cell migration, cell matrix adhesion, lipid degradation,
protein glycosylation and glycoprotein metabolism (21).
In comparison with the cell membrane and overall
lipid composition of the cell, migrasomes are notably enriched in
sphingolipids, such as sphingomyelin (SM), ceramide,
monosialodihexosylgangliosides and glycosphingolipids, including
monoglycosylceramide, diglycosylceramide and triglycosylceramide.
In a study by Liang et al (23), it was demonstrated that ceramide
and SM are essential for the formation and maintenance of
migrasomes. Furthermore, filipin III staining and quantitative
analysis revealed that migrasomes are enriched in cholesterol, an
important component for the physical properties and structural
integrity of the migrasome membrane; notably, TSPAN4, TSPAN7 and
cholesterol assemble into TSPAN-enriched microdomain (TEMs), the
enrichment of which stiffens the plasma membrane, facilitating
migrasome initiation (9). Ongoing
lipidomics analysis of migrasomes anticipates the discovery of
additional lipid components, thereby verifying the growing profile
of known biological characteristics and functions of
migrasomes.
Migrasomes are rich in nucleic acids. A study by Zhu
et al (24) employed
SYTO™ 14 fluorescence staining to demonstrate the
presence of RNA within migrasomes. Sequencing analyses revealed
that migrasomes predominantly contain long-chain mRNAs associated
with cell metabolism, intracellular membrane transport, cell
adhesion, vesicle fusion and the assembly of subcellular membrane
structures. These mRNAs can be translated into proteins within
recipient cells, participating in the biological responses of
recipient cells and regulating their life processes. For example,
the phosphatase and tensin homolog (PTEN) mRNA delivered from
migrasomes to recipient cells is translated into the PTEN protein,
which inhibits the proliferation of the recipient cells (24). Nonetheless, the mechanisms of
migrasome RNA sorting and transport have yet to be elucidated, as
does the presence of DNA within migrasomes, necessitating further
investigation.
Markers of migrasomes
TSPAN4/7, and integrins α1, α3, α5 and β1, which are
expressed on the migrasome membrane, serve as important structural
markers of migrasomes, with TSPAN4 being the most distinguishing
marker, which exhibits the clearest expression when examined by
confocal microscopy (6).
Nonetheless, the detection of migrasomes using fluorescently
labeled marker proteins presents limitations, including complex
procedures, extended experimental duration and difficulty (25). Consequently, Chen et al
(25) investigated
fluorescently-labelled wheat germ agglutinin (WGA), which is a more
rapid, straightforward and less invasive marker than TSPAN4.
However, WGA may non-specifically bind to structures containing
sialic acid and N-acetyl-D-glucosamine, and its fluorescence
intensity can be influenced by various factors; for example, it may
also enter the cell and bind to intracellular components, leading
to enhanced nonspecific signals. In addition, different cell lines
or tissue types naturally exhibit variations in the glycosylation
levels of their cell membrane surfaces, which can also interfere
with the results, potentially limiting its application in
quantitative migrasome analysis (25). Additional specific protein markers
include N-deacetylase/N-sulfotransferase 1,
phosphatidylinositol glycan anchor biosynthesis class K,
carboxypeptidase Q and epidermal growth factor domain-specific
O-linked N-acetylglucosamine transferase; however, due to
marked variations in protein content across different cell types,
these markers are not uniformly present in migrasomes derived from
the same cell source (21,22). Furthermore, migrasome-associated
mRNAs can be labelled with the nucleic acid stain SYTO 14, making
them secondary markers for migrasomes (24,26).
Mechanisms of migrasome formation
During cell migration, RFs are extended from the
posterior end of the cell, with migrasomes situated at the termini
or bifurcations of these fibers (6). When RFs break, migrasomes may leak or
rupture, thereby releasing their contents into the extracellular
space (6). The formation of
migrasomes is a complex process that involves the regulation of
multiple molecules and signaling pathways.
Migrasome formation depends on cell
migration
The formation of migrasomes is contingent upon
cellular migration, a finding initially presented by Ma et
al (6). A study revealed that,
among 563 compounds capable of reducing migrasome numbers in
cultured cells, 507 also decreased RF formation, reinforcing the
association of migrasome production with cell migration (27). Subsequent research conducted by Fan
et al (28) revealed a
lower migrasome count in turning cells compared with those
migrating in a straight line, highlighting the importance of
migration continuity and speed in migrasome formation. Directional
changes during migration lead to fewer RFs and migrasomes, with the
removal of vimentin from cells having been shown to impair
migration and reduce migrasome numbers (29,30).
In conclusion, these findings suggested that the formation of
migrasomes is markedly regulated by cell migration behavior.
Nucleation: SM synthase (SMS)2 foci
are the starting point of migrasome biogenesis
SM, synthesized from ceramide by SMS, is a key
component of the plasma membrane, and is involved in signaling and
membrane transport (23). A
previous study has shown that SM and ceramide are enriched on
migrasomes and are present at the sites of migrasome formation;
furthermore, ceramide is unevenly distributed on different
migrasomes, and as migrasome biogenesis proceeds, SM levels
continuously increase, indicating that ceramide can be converted
into SM on migrasomes (23).
Hydrolyzing SM on migrasomes or knocking out ceramide synthase 5 to
reduce SM synthesis severely impairs the formation of migrasomes,
demonstrating the importance of SM for migrasome formation
(23).
Mammalian cells contain two principal types of SMS:
i) SMS1, which is localized in the Golgi apparatus; and ii) SMS2,
which is located in both the Golgi apparatus and the plasma
membrane (31). SMS2 synthesizes
SM from ceramide in the plasma membrane (23,31).
Consequently, it is plausible that SMS2 modulates migrasome
formation by facilitating SM production. Experimental evidence,
including the knockout of SMS2 and treatment with SMS2 inhibitors
that hinder SM synthesis, has demonstrated that migrasome formation
and growth are impeded in SM-depleted conditions, whereas the
reintroduction of SM restores migrasome production (23). Furthermore, impaired SM synthesis
reduces cholesterol recruitment, thereby affecting TEM assembly,
since TSPAN4, TSPAN7 and cholesterol assemble into TEMs, the
enrichment of which stiffens the plasma membrane, which is an
important condition for migrasome formation (9).
Research has revealed that SMS2 localizes to foci on
the basal membrane at the leading edge of a cell, which predestines
migrasome formation sites (23).
These foci mature into migrasomes during cell migration (23). Intracellular SMS2 foci initially
adhere to the section of the membrane of the migrating cell that is
interacting with a substrate to generate motility, resembling focal
adhesions (FAs), which are the sites where cells are linked to the
ECM in which integrins are highly enriched. However, previous
studies have failed to detect FA markers within migrasomes, and
active forms or markers of FA components such as integrin α5,
integrin β1 or the FA kinase paxillin do not colocalize with
intracellular SMS2 foci, underscoring that SMS2 focus formation is
independent of FAs (23,32). To elucidate the role of SMS2 foci
in migrasome formation, researchers identified an SMS2 mutant,
S217A, that cannot form foci. The formation of migrasomes in cells
expressing this mutation has been shown to be markedly diminished
(23). Notably, the inability to
form SMS2 foci prevents migrasome formation, even when exogenous SM
is added (23). These findings
suggest that SMS2 foci not only regulate migrasome formation by
synthesizing SM, but may also be involved in other important
intracellular signaling processes that are required for migrasome
biosynthesis and function. Future investigations should explore the
precise mechanisms of SMS2 focus assembly, the selection of
assembly sites and the adhesive mechanisms of SMS2 foci to fully
elucidate the contribution of SMS2 foci to migrasome formation and
function.
Expansion: TSPAN4 and cholesterol
mediate migrasome formation
TSPANs constitute a family of small hydrophobic
proteins characterized by four transmembrane domains, comprising a
total of 33 distinct members in mammalian cells. These proteins
facilitate the organization of functional higher-order protein
complexes on the cell membrane through interactions with adhesion
molecules, enzymes and signaling proteins, thereby forming the
structures known as TEMs (33,34).
Initial research identified TSPAN4 as a marker of migrasomes;
however, subsequent investigations have suggested that the roles of
TSPAN family members may extend beyond this initial
characterization. By establishing a stable normal rat kidney (NRK)
cell line that expresses various levels of TSPAN4 and green
fluorescent protein (GFP), a study by Huang et al (9) demonstrated that the overexpression of
14 types of TSPAN family members enhances migrasome formation in a
dose-dependent manner, with TSPAN4 exhibiting the most pronounced
effect. Conversely, knockout of the TSPAN4 gene was shown to
markedly reduce the number of migrasomes, underscoring the
importance of TSPAN4 for migrasome formation. During the migrasome
formation process, the signal from TSPAN4-GFP during the growth
phase of migrasomes rapidly re-emerges on their surface, whereas no
such recovery occurs when migrasomes tend to mature and stabilize
(9). Furthermore, high-speed
imaging revealed that TSPAN4-GFP forms rapidly-moving-discrete
spots on RFs and migrasomes that assemble on the surface of
migrasomes during their growth phase. Taken together, these
findings suggest that TSPAN4 is recruited to migrasomes
specifically during the growth phase (9).
By developing an in vitro system to simulate
migrasome formation, researchers have demonstrated that TSPAN4 and
cholesterol are sufficient to reconstitute migrasome-like
structures, further validating their role in this process (9). To elucidate how the assembly of TEMs
promotes migrasome formation, a theoretical model was proposed,
identifying three key energetic drivers: i) The bending energy of
the migrasome and RF membranes; ii) the membrane tension energy;
and iii) the boundary energy at the migrasome-RF interface
(9). A subsequent study using a
biomimetic system divided migrasome growth into two phases: i) A
local swelling phase, driven by membrane tension and potentially
other cellular factors (such as the cytoskeleton, specific lipids,
ion fluxes, mechanosensitive signaling and adhesion complexes); and
ii) a TSPAN4 migration-enrichment phase mediated by TSPAN family
proteins (35). Notably, the
biomimetic system lacks cytoskeletal components, which may regulate
migrasome formation in vivo; therefore, discrepancies
between artificial and cellular systems may exist (6,17,21,22).
TSPAN4, TSPAN7 and cholesterol assemble into TEMs, whose enrichment
stiffens the plasma membrane, facilitating migrasome initiation.
Changes in TSPAN4 curvature will affect the swelling of the plasma
membrane, the reduced intrinsic curvature of TSPAN4 directs TEMs to
low-curvature membrane swellings, stabilizing these protrusions and
promoting migrasome maturation (9,10,18,35,36).
In summary, TEMs composed of TSPAN4 and cholesterol are important
for migrasome biogenesis.
Maturation
Integrin and extracellular matrix (ECM) protein
pairing determines migrasome formation
Integrins are transmembrane receptors that bind ECM
proteins and link to the cytoskeleton via intracellular adapters.
These heterodimeric proteins, which are composed of α and β chains,
are important for cell adhesion, spreading, migration and matrix
remodeling (37). Migrasomes,
which adhere to ECM sites during cell migration, are enriched with
integrin α5β1, as revealed by mass spectrometry (32). Fluorescence staining confirms that
integrin β1 in migrasomes is in an activated, ligand-bound state,
demonstrating direct ECM binding. Three-dimensional imaging and
total internal reflection fluorescence microscopy have further
revealed the localization of integrins α5 and β1 at the migrasome
base, supporting their role in migrasome anchorage (32).
Although FAs also contain high integrin
concentrations, FA markers are absent in migrasomes, indicating a
distinct adhesion mechanism (32,38).
A functional study has revealed that NRK cells produce markedly
more migrasomes on fibronectin-coated surfaces than on laminin 511-
and collagen I-coated surfaces, with minimal migrasome formation on
uncoated coverslips. This increase in migrasome formation on
fibronectin-coated surfaces compared with other ECM components is
associated with an elevated integrin α5 expression in NRK cells, as
α5 knockout impairs migrasome production on fibronectin but not on
other ECM components (32).
Conversely, integrin α3 overexpression increases the number of
migrasomes on laminin 511. Similarly, integrin α1 overexpression
enhances migrasome formation on collagen IV in Chinese hamster
ovarian cells without affecting formation on other ECM components
(32). These findings demonstrate
that migrasome formation depends on specific integrin-ECM
pairings.
Phosphatidylinositol 4,5-bisphosphate
(PIP2)-Rab35 axis regulates migrasome formation
PIP2, a plasma membrane lipid synthesized by
phosphatidylinositol 4-phosphate 5-kinase type-1α kinases,
regulates key subcellular processes, including the regulation of
ion channels and transporters, clathrin-dependent and -independent
endocytosis, exocytosis regulation, actin polymerization, efficient
FA turnover and the regulation of cell-cell contacts) (39,40).
PIP2 localizes to migrasomes, as confirmed by phospholipase Cγ-PH
domain-GFP fusion protein probes and antibody staining (41). Kinetic experiments have revealed
that PIP2 recruitment precedes TSPAN4 and integrin α5 recruitment,
both of which are important for migrasome biogenesis.
Phosphatidylinositol-4-phosphate 5-kinase type-1α (PIP5K1A)
inhibition disrupts migrasome formation, implicating PIP2 synthesis
in this process (41).
PIP2 likely functions by recruiting binding
partners, such as Rab35, a GTPase involved in organelle biogenesis,
endosomal trafficking and actin regulation (42). Rab35 is recruited to migrasome
formation sites in a PIP2-dependent manner, and its depletion
disrupts RF elongation and migrasome assembly (41). Rab35 also interacts with integrin
α5 via the GFFKR motif, recruiting integrins α5 to migrasome sites,
although the direct binding mechanism remains unconfirmed (41,43).
Thus, the PIP2-Rab35 axis orchestrates migrasome formation by
coupling lipid signaling with integrin trafficking. Future studies
should address PIP5K1A recruitment dynamics, Rab35-integrin α5
interactions and the clinical relevance of the PIP2-Rab35 signaling
axis in cancer and migration-associated diseases.
Roles of Rho-associated protein kinase
(ROCK)1 and programmed death-ligand 1 (PD-L1) in migrasome
formation
Through a chemical genetic screen designed to
identify compounds and protein targets that disrupt migrasome
formation, a study by Lu et al (27) identified SAR407899, an inhibitor of
ROCK1 and ROCK2, which stably suppresses migrasome biogenesis.
ROCK1 and ROCK2 are serine/threonine kinases that act downstream of
the small GTPase Ras homolog family member A (RhoA), regulating
diverse cellular processes, including actin cytoskeleton
organization, cell adhesion and migration, via the ROCK-Rho
signaling pathway (44). Although
ROCK2 knockdown does not affect migrasome formation, ROCK1
depletion markedly reduces the migrasome abundance per cell
(27). Notably, ROCK1 knockdown
also impairs cell migration. To distinguish the effects of RFs and
migration from migrasome formation, researchers have quantified the
number of migrasomes per 100 µm RFs (27). The findings of this experiment
revealed that ROCK1-depleted cells generate notably weaker traction
forces than healthy cells, supporting the premise that migrasome
formation depends on ROCK1-mediated traction and other ECM
components (such as fibronectin) adhesion (27).
PD-L1 is best known as an immune checkpoint molecule
that facilitates cancer cell migration (45). Emerging evidence has indicated that
PD-L1 has an intrinsic capability to facilitate sustained cellular
migration, a process important for migrasome biogenesis.
High-resolution time-lapse microscopy has demonstrated that PD-L1
accumulates at the trailing edge of migrating cancer cells, where
it forms distinct structures that move directionally during
retraction (46). Given that RFs
form at the rear of the cell during migration, a subsequent study
demonstrated that PD-L1 localizes not only in RFs, but also in
migrasomes. Notably, PD-L1 enhances migrasome formation
independently of cell migration (46). PD-L1 closely associates with
integrin β4, co-localizing at the rear of the cell, and later in
RFs and migrasomes. A further investigation revealed that PD-L1
recruits integrin β4 to the trailing edge; this recruitment
stimulates contractility via the dynamics of the cell, a mechanism
by which PD-L1 maintains rear polarity and reduces membrane tension
(46). Additionally, PD-L1
activates RhoA by coupling integrin β4 to the cytoskeleton, further
promoting rear contraction (actin cytoskeleton-mediated
contractility) (46). Taken
together, these findings underscore the dual role of PD-L1 in
facilitating cell migration and migrasome formation. However, the
precise mechanisms of PD-L1 in migrasome biogenesis, and its
broader functional implications, warrant further exploration.
Other factors affecting migrasome
formation
Regarding the formation mechanisms of migrasomes in
different cell types, the universal core mechanism involves the
following sequence of events: i) Cell migration initiation; ii) the
formation of TEMs; and subsequently iii) the recruitment, fusion
and maturation of microvesicles (the precursors of migrasomes)
(6,9). However, migrasome formation is a
complex biological process influenced by multiple factors, and the
underlying mechanisms may vary across cell types. Some examples are
as follows: i) Triggering signals vary, for example monocytes and
macrophages may initiate migrasome generation through tissue damage
and clearance signals, with these migrasomes potentially
participating in angiogenesis regulation or damaged mitochondrial
clearance, whereas cancer cells may trigger migrasome formation via
oncogenic or metastasis-driving signals to promote cancer
progression; and ii) regulatory factors may be cell type-specific.
For example, normal human dermal fibroblasts exhibit modulated
migrasome formation through peptide-modified matrices, which affect
contractile fiber quantity and length (different peptide-modified
substrates influence the strength of cell migration, such as
migration distance and duration, thereby affecting the number and
length of RFs) (47). Mouse
embryonic stem cells exhibit calcium ion and synaptotagmin-1
(Syt1)-regulated migrasome production, where calcium promotes
migrasome formation via Syt1 (48). Furthermore, in H4 human glioma
cells, osmotic regulation may control migrasome formation, as
hypotonic conditions induce the formation of TSPAN4-enriched
migrasome-like vesicles on RFs. These cholesterol-dependent
vesicles exhibit migrasome characteristics but originate from
osmotic stress rather than from cell migration (49). While conclusive identification of
these cholesterol-dependent vesicles as migrasomes requires further
evidence, these observations provide valuable perspectives on
cellular osmoregulation and migrasome biophysical responses in
tissues. A previous report indicated that high-salt diets
exacerbate ischemic brain injury by promoting excessive migrasome
formation in microglia and macrophages, reducing their
post-ischemic populations alongside astrocytes (17).
Migrasomes from different cell types participate in
distinct biological activities. Nevertheless, research on most
cell-type-specific migrasomes remains preliminary, and the
generation mechanisms of these migrasomes have not been fully
elucidated. Currently, evidence confirms only their existence and
functional roles in various biological responses. For example,
migrasomes produced by zebrafish embryonic cells can regulate the
formation of zebrafish embryonic organs; migrasomes derived from
adipose stem cells serve a key role in adipose tissue regeneration;
and migrasomes from neutrophils affect the coagulation function of
the body (18,50). Specialized investigations into
cell-type-specific formation mechanisms remain lacking, preventing
targeted discussion of these mechanisms at present. Exploring these
heterogeneous-origin migrasomes and their impacts on cellular
functions and disease pathogenesis constitute an important future
research direction.
Biological functions of migrasomes
Migrasomes, notable organelles in cell migration,
are increasingly recognized for their roles in cellular physiology
and pathology. Their formation and function are closely associated
not only with cellular-migratory capacity, but also with
intercellular communication, tissue remodeling and immune
regulation. These microvesicles serve as carriers for the
intercellular transfer of biomolecules, such as proteins, mRNAs and
microRNAs, thereby modulating the gene expression and functional
states of recipient cells. The present section elucidates the
biological functions of migrasomes in organ morphogenesis,
angiogenesis, mitochondrial quality control and immune regulation,
highlighting their notable roles in maintaining tissue homeostasis
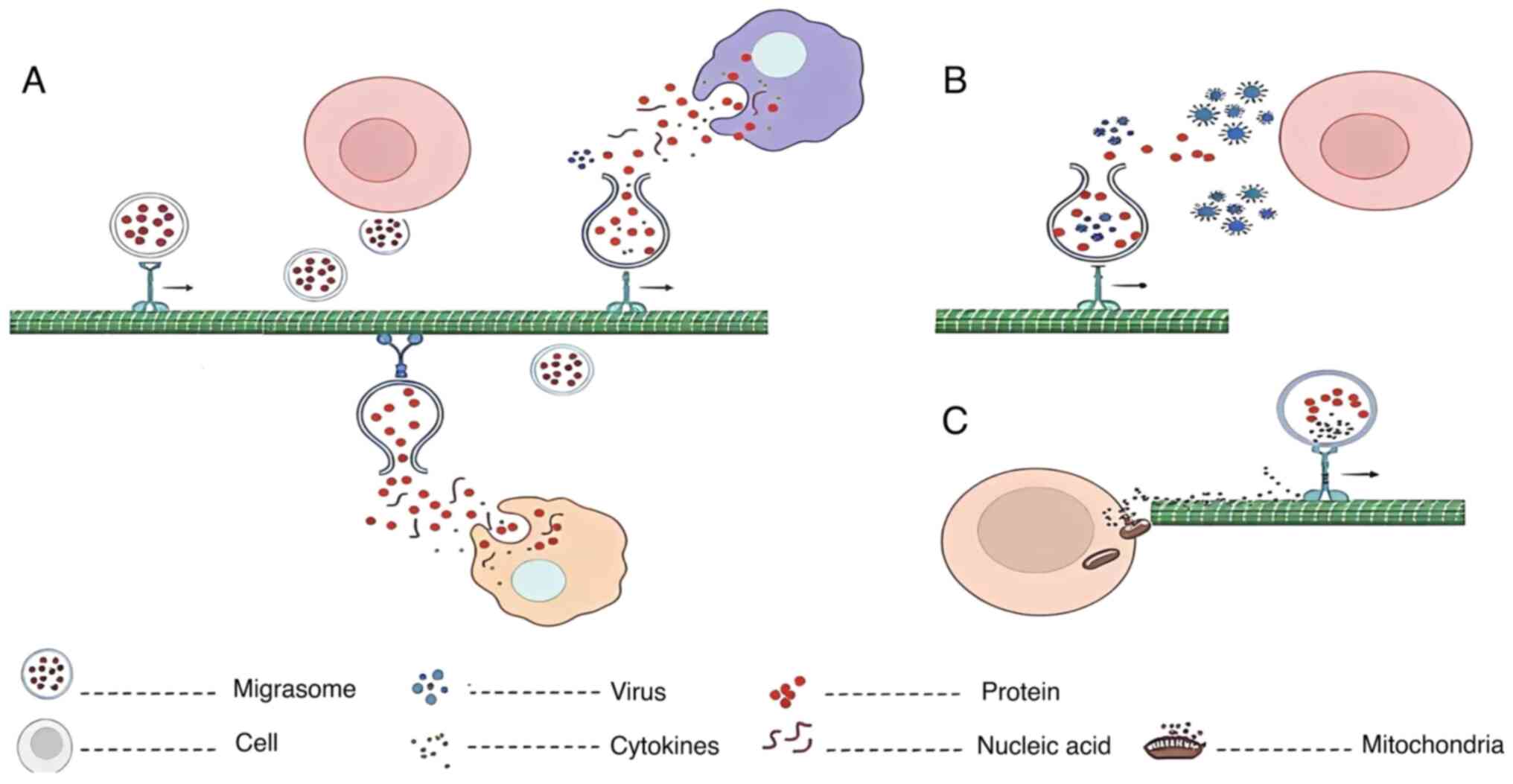
and contributing to disease pathogenesis (Fig. 3). A deeper understanding of these
functions underscores the complexity and diversity of migrasomes in
biological systems and their potential as therapeutic targets in
medical research.
Communication and regulatory
functions
Migrasomes constitute a notable secretory pathway in
migrating cells, encapsulating diverse cytoplasmic contents
(6,51). Migrasomes form and mature on RFs of
migrating cells before they detach, rupture and release their
contents (6) (Fig. 3A). Migrasomes can also be
internalized by neighboring cells, or adhere to cell surfaces or
the ECM without being engulfed. For example, interactions with the
ECM enable migrasomes to attach to specific cell surfaces or
tissues, facilitating intercellular communication (32). These vesicles transport
intracellular biomolecules, including proteins, mRNAs and
microRNAs. Recent studies have indicated that secretory proteins,
such as signaling molecules, are actively transported from
migrating cells into migrasomes via kinesin-mediated carriers, akin
to targeted neurotransmitter release in neuronal systems (51). These molecules can be transferred
to adjacent or distant cells, altering recipient cell-gene
expression and function. For example, Zhu et al (24) added purified migrasomes derived
from L929 cells to U87-MG, MDA-MB-468 and PC3 cells that do not
express the PTEN protein, and demonstrated that migrasomes mediate
the transfer of PTEN mRNA and protein, inhibiting proliferation in
recipient cells.
Migrasomes are enriched with cytokines, including
chemokines and morphogens, which enables them to act as signaling
molecule-carriers, and thereby influence cell behavior. In
zebrafish embryos, gastrula-derived migrasomes contain high levels
of C-X-C motif chemokine ligand (CXCL)12a (18). These migrasomes, produced during
mesoderm and endoderm cell gastrulation, regulate organ
development. Mutations that reduce migrasome numbers lead to severe
developmental defects, which can be rescued by migrasome
supplementation, underscoring their role in organogenesis (18). Further studies have revealed that
CXCL12 in migrasomes interacts with C-X-C chemokine receptor type 4
(CXCR4) on dorsal precursor cells (DFCs), inducing chemotaxis and
ensuring typical organ morphogenesis (35). Migrasomes thus spatially and
temporally distribute signaling molecules, releasing CXCL12 upon
rupture to modulate DFC behavior. Similarly, adipose-derived stem
cells produce CXCL12-enriched migrasomes that promote adipose
tissue regeneration via CXCR4/RhoA signaling (50).
A study by Zhang et al (11) identified migrasome-producing
monocytes in the chorioallantoic membrane of chicken embryos, where
these vesicles are rich in proangiogenic factors, including TGF-β3,
VEGFA and CXCL12. In this context, migrasomes induce endothelial
tube formation; their depletion via monocyte clearance or TSPAN4
knockout inhibits angiogenesis, whereas supplementation restores
monocyte recruitment and capillary formation (11). Monocytes leverage migrasomes to
deliver angiogenic factors, thereby creating a microenvironment
conducive to blood vessel growth. Notably, migrasome-derived CXCL12
recruits additional monocytes, forming a positive feedback loop
(11,52).
Multipotent mesenchymal stem cells (MSCs) are
precursors to various cell types, with the ability to support
tissue homeostasis, promote hematopoiesis and interact with cancer
cells. Previous research on MSCs has focused on MSC-derived EVs
(MSC-EVs), indicating that MSC-EVs have functions similar to those
of MSC (53–58). A recent investigation demonstrated
that MSCs generate migrasomes that attract leukemia KG-1a cells and
CD34+ hematopoietic stem cells via the CXCR4-CXCL12 axis
(59). These migrasomes, enriched
in TSPANs, including CD166 and TSPAN4, and endosomal markers, such
as Rab7 and CD63, are influenced by ECM components such as
fibronectin and laminins. In addition to influencing cell migration
and thereby affecting the formation of retraction fibers, ECM
components can also impact the process of TEM formation (59). Migrasomes produced by MSCs attract
leukemia KG-1a cells and CD34+ hematopoietic stem cells
through the CXCR4-CXCL12 axis, thereby facilitating communication
between the MSCs and these cells, thus highlighting the role of
migrasomes in MSC-hematopoietic cell interactions, offering
insights into their functions in health and disease.
Neutrophil-derived migrasomes adsorb coagulation
factors from the plasma; migrasomes actively bind and enrich
coagulation factors on their surface by virtue of their unique
membrane composition, particularly cholesterol esters, and then
localize to injury sites and modulate coagulation by activating
platelets (60). These migrasomes
also regulate immune responses by delivering cytokines and
chemokines to injury sites, enhancing immune cell recruitment
during inflammation (60).
Conversely, migrasomes have been shown to suppress immunity by
transporting immunosuppressive molecules, thereby maintaining
immune tolerance (46).
Participation in maintaining cellular
homeostasis
Mitochondria, the cellular ‘powerhouses’, maintain
homeostasis through energy production and quality control
mechanisms, such as mitophagy (61,62).
A previous study demonstrated that migrasomes mediate mitocytosis,
a process in which damaged mitochondria are selectively expelled to
sustain cellular health (12)
(Fig. 3C). Under stress
conditions, such as carbonyl cyanide m-chlorophenyl hydrazone
treatment, mitochondria undergo fragmentation via kinesin-5B
(KIF5B)-mediated transport and myosin head domain-containing
protein 1 (MYO19)/density-regulated protein 1-dependent fission,
accumulating at the cell periphery for subsequent migrasome
encapsulation. Knocking out TSPAN9 or taking other measures to
block the formation of migrasomes has been shown to cause migrating
cells to lose their mitocytosis capability, leading to the
accumulation of damaged mitochondria within the cells and affecting
cell viability (12,63,64).
Although both mitocytosis and mitophagy contribute
to cellular homeostasis, their mechanisms differ markedly.
Mitocytosis, which is mediated by migrasomes, selectively removes
mildly damaged mitochondria from migrating cells. In this process,
damaged mitochondria selectively bind to intracellular dynein to
facilitate their transport out of the cell, they are then
transported towards the cell periphery by outwards motor proteins,
such as KIF5B. MYO19 further facilitates this process by anchoring
mitochondria to cortical actin, thereby promoting their
incorporation into migrasomes (12,63,64).
By contrast, mitophagy primarily degrades damaged mitochondria via
the autophagy pathway. Upon mitochondrial damage, PTEN-induced
kinase 1 accumulates on the outer mitochondrial membrane, where it
recruits parkin to ubiquitinate outer membrane proteins. This
ubiquitination marks the mitochondria for autophagosomal
engulfment, and subsequent lysosomal degradation (61,62).
Mitocytosis complements mitophagy by incrementally
clearing mildly damaged mitochondria. Migrasomes may also transfer
mitochondrial components, including mitochondrial DNA and
mitochondrial microRNAs, to recipient cells, influencing their
function.
Migrasomes mediate virus spread
Migrasomes facilitate viral dissemination,
facilitating the evasion of antiviral therapies (Fig. 3B). Vaccinia virus induces
migrasomes containing viral particles, enabling their spread
despite tecovirimat treatment (13,65).
Similarly, herpes simplex virus-2 exploits migrasomes as ‘Trojan
horses’ for intercellular transmission (66). Taken together, these findings
illuminate novel viral spread mechanisms and suggest potential
targets for antiviral strategies.
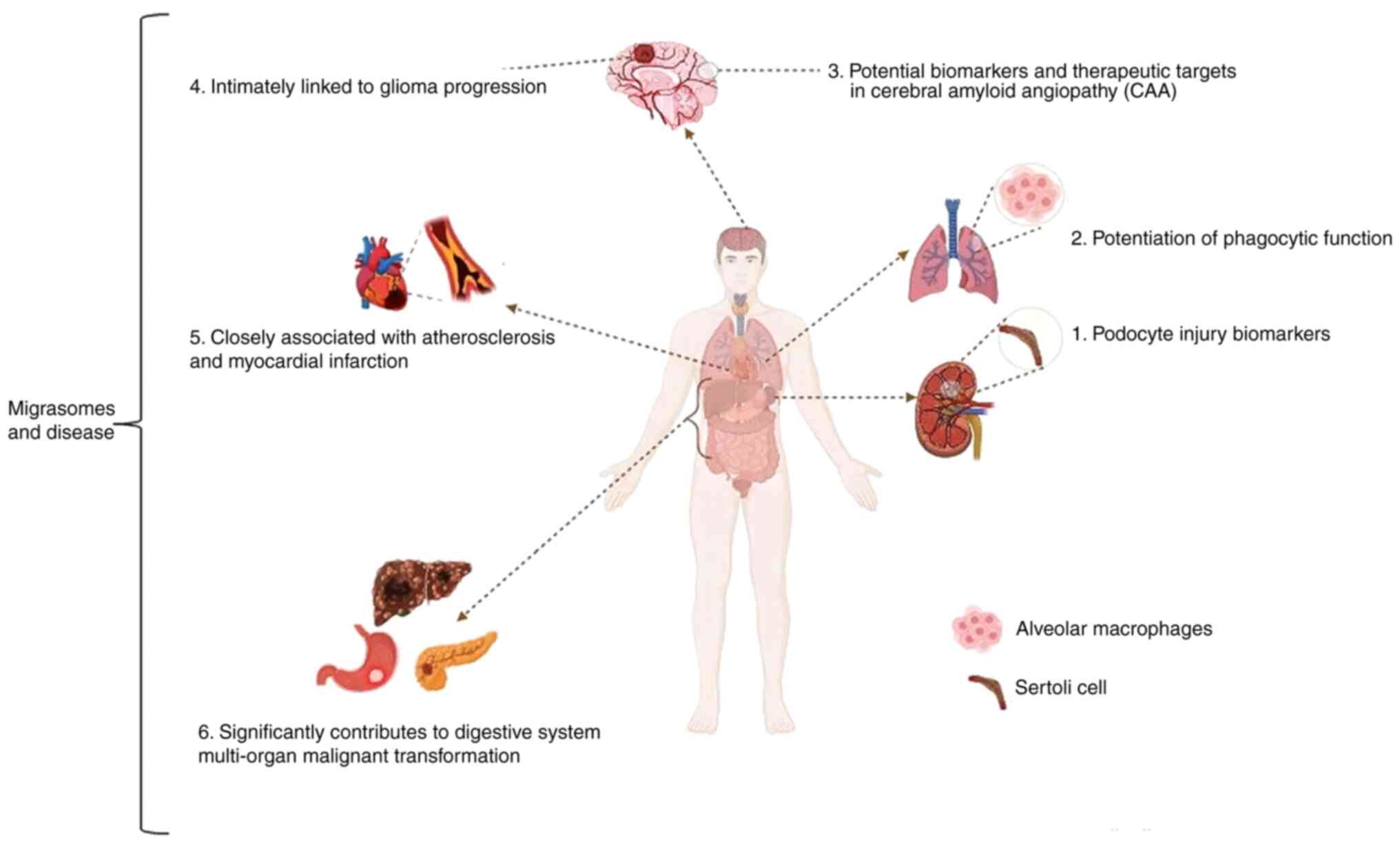
Migrasomes and disease
As research advances, the understating of migrasomes
is improving, with their structure and physiological functions
becoming increasingly elucidated. Studies have demonstrated that
migrasomes perform notable roles in intercellular communication and
signal transduction, as well as in the pathogenesis of various
diseases (Fig. 4). These findings
highlight the potential of migrasomes for clinical applications in
disease diagnosis and treatment.
The association between migrasomes and kidney
disease is particularly notable. Studies have identified
podocyte-derived migrasomes in urine as early biomarkers of
podocyte injury (14,67). Podocytes, which regulate glomerular
permeability, are terminally differentiated cells incapable of
regeneration; therefore, early detection of their injury is
important for managing glomerular diseases (14). Research indicates that podocytes
generate migrasomes during migration, with their numbers markedly
increasing during kidney injury. Furthermore, Rac-1 inhibitors that
target a small Rho family GTPase that is overactivated in podocyte
injury dose-dependently suppress lipopolysaccharide-induced
migrasome release, underscoring the diagnostic potential of the
urinary presence of podocyte migrasomes (14).
Migrasomes also exhibit therapeutic relevance for
post-stroke pneumonia. Bone marrow (BM)-MSC-derived migrasomes,
which are packed with dermcidin, display dual effects that reduce
pulmonary bacteria load and enhance LC3-associated phagocytosis
(LAP) of macrophages (68). These
migrasomes not only exert antibacterial effects, but also augment
LAP in macrophages, facilitating bacterial clearance. Consequently,
BM-MSC-derived migrasomes represent a promising alternative to
conventional antibiotics for preventing and treating post-stroke
pneumonia.
In neurodegenerative disorders, migrasomes have been
implicated in cerebral amyloid angiopathy (CAA). β-amyloid protein
40-induced macrophage-derived migrasomes adhere to the vasculature
in biopsies from patients with CAA and mouse models, delivering the
protein CD5L to vascular walls and triggering complement-dependent
cytotoxicity, thereby compromising the blood-brain barrier
(69). These observations suggest
that macrophage-derived migrasomes and complement activation are
potential biomarkers and therapeutic targets for CAA.
Emerging evidence further links migrasomes to
atherosclerosis, myocardial infarction and malignancies. TSPAN4, a
migrasome-forming protein, is strongly associated with
atherosclerosis regression-associated macrophages according to
single-cell sequencing, Gene Expression Omnibus datasets and The
Cancer Genome Atlas analyses, and is also associated with plaque
hemorrhage and rupture (70). In
another study, low-intensity pulsed ultrasound has been shown to
mitigate myocardial ischemia-reperfusion injury via
migrasome-mediated mitochondrial quality control; the possible
mechanism involves promoting the formation of migrasomes,
potentially by enhancing cell motility, which enables migrasomes to
exert mitocytosis activity, facilitating the removal of damaged
mitochondria from cells (71).
TSPAN4 is also upregulated in hepatocellular carcinoma, gastric
cancer and glioblastoma (GBM), where it influences tumor-associated
macrophages (72). In
hepatocellular carcinoma, migrasomes guide cancer cell invasion
(73); in pancreatic cancer,
pancreatic cancer cell-derived migrasomes modulate
immunosuppressive factors in the tumor microenvironment, thereby
accelerating progression (74). In
GBM, TSPAN4 promotes the progression of GBM by regulating epidermal
growth factor receptor stability, whereas migrasome-autophagosome
crosstalk alleviates endoplasmic reticulum stress and
migrasome-mediated GBM-microenvironment communication may drive
recurrence (75–77). Collectively, these findings
underscore the therapeutic potential of targeting TSPAN4 and
migrasomes in severe diseases.
Summary and outlook
Migrasomes, a novel class of EVs, have attracted
considerable attention in the field of cell biology, with their
important roles in cellular processes gradually being elucidated.
Research on migrasomes has advanced markedly, from structural
characterization to functional exploration. These vesicles not only
serve important roles in normal physiological processes, but also
modulate cell behavior and disease progression under pathological
conditions, highlighting their potential as biomarkers and
therapeutic targets.
The field of migrasome research is rapidly evolving,
but still faces substantial challenges. Firstly, a deeper
mechanistic understanding of migrasome biogenesis and its
regulatory networks, particularly the contributions of lipid and
protein components, is important. Secondly, further experimental
validation is needed to clarify how migrasome-derived biomolecules,
such as RNAs and proteins, influence recipient cell functions and
mediate intercellular communication. Additionally, investigating
migrasome heterogeneity across cell types and tissues, as well as
their functional alterations in disease states, remains a key
research direction. Advances in single-cell sequencing and
high-resolution imaging technologies are expected to provide deeper
insights into the biology and disease-related functions of
migrasomes.
Clinically, migrasome research may offer novel
strategies for early disease diagnosis and therapy (Table II). For example, migrasomes could
serve as biomarkers for monitoring kidney or neurodegenerative
diseases, or their formation and function could be therapeutically
targeted. Furthermore, migrasome involvement in viral transmission
suggests their potential applications in infectious disease
research. As knowledge on the function of migrasomes expands, their
impact on research and clinical therapeutics is expected to grow.
In summary, migrasome research presents both challenges and
opportunities, with future discoveries poised to further elucidate
the complexity of cellular processes while advancing diagnostic and
therapeutic innovations.
 | Table II.Dual roles of migrasomes in
diseases. |
Table II.
Dual roles of migrasomes in
diseases.
| Disease type | Functional
mechanism | Potential
applications |
|---|
| Developmental
defects | Loss of
CXCL12+ migrasomes leads to abnormal organ morphogenesis
in zebrafish embryos (18) | Early intervention
targets for developmental disorders |
| Tumor
microenvironment | MSC-derived
migrasomes recruit leukemia cells; pancreatic cancer migrasomes
enrich immunosuppressive factors (59,74) | Blocking
migrasome-mediated tumor metastasis |
| Ischemic
injury | Adipose stem
cell-derived migrasomes activate Ras homolog family member A via
CXCL12, promoting vascular regeneration (50) | Tissue engineering
and regenerative medicine |
| Inflammation and
infection | Neutrophil-derived
migrasomes enhance coagulation or antibacterial functions, such as
releasing dermcidin in post-stroke pneumonia (68) | Novel
anti-infection strategies |
| Autoimmune
diseases | Migrasomes deliver
immunosuppressive molecules, such as interleukin-10, to maintain
immune tolerance (46) | Immunomodulatory
therapies for autoimmune disorders |
Acknowledgements
Not applicable.
Funding
The present study was supported by the Jiangsu Provincial Key
Medical Discipline (grant no. ZDXK202227) and the Wuxi Taihu Talent
Program (grant no. WX0302B010507200065).
Availability of data and materials
Not applicable.
Authors' contributions
YC and QW wrote the manuscript. YC, XL, BZ and HZ
revised the manuscript. XL supervised the present review and guided
each author who participated in writing the article. Data
authentication is not applicable. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
EV
|
extracellular vesicle
|
|
RF
|
retraction fiber
|
|
PLS
|
pomegranate-like structure
|
|
TSPAN
|
tetraspanin
|
|
SM
|
sphingomyelin
|
|
WGA
|
wheat germ agglutinin
|
|
SMS
|
sphingomyelin synthase
|
|
TEM
|
tetraspanin-enriched microdomain
|
|
FA
|
focal adhesion
|
|
NRK
|
normal rat kidney
|
|
ECM
|
extracellular matrix
|
|
PIP2
|
phosphatidylinositol
4,5-bisphosphate
|
|
GFP
|
green fluorescent protein
|
|
PIP5K1A
|
phosphatidylinositol-4-phosphate
5-kinase type-1α
|
|
PD-L1
|
programmed death-ligand 1
|
|
ROCK
|
Rho-associated protein kinase
|
|
RhoA
|
Ras homolog family member A
|
|
PTEN
|
phosphatase and tensin homolog
|
|
CXCL
|
C-X-C motif chemokine ligand
|
|
CXCR4
|
C-X-C chemokine receptor type 4
|
|
DFC
|
dorsal precursor cell
|
|
MSC
|
mesenchymal stem cell
|
|
KIF5B
|
kinesin-5B
|
|
MYO19
|
myosin head domain-containing protein
1
|
|
BM-MSCs
|
bone marrow mesenchymal stem
cells
|
|
Syt1
|
synaptotagmin-1
|
|
CAA
|
cerebral amyloid angiopathy
|
|
GBM
|
glioblastoma
|
References
|
1
|
Trepat X, Chen Z and Jacobson K: Cell
migration. Compr Physiol. 2:2369–2392. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ratajczak J, Wysoczynski M, Hayek F,
Janowska-Wieczorek A and Ratajczak MZ: Membrane-derived
microvesicles: Important and underappreciated mediators of
cell-to-cell communication. Leukemia. 20:1487–1495. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bruno S, Chiabotto G, Favaro E, Deregibus
MC and Camussi G: Role of extracellular vesicles in stem cell
biology. Am J Physiol Cell Physiol. 317:C303–C313. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hopkin K: Core concept: Extracellular
vesicles garner interest from academia and biotech. Proc Natl Acad
Sci USA. 113:9126–9128. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kang T, Atukorala I and Mathivanan S:
Biogenesis of extracellular vesicles. Subcell Biochem. 97:19–43.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ma L, Li Y, Peng J, Wu D, Zhao X, Cui Y,
Chen L, Yan X, Du Y and Yu L: Discovery of the migrasome, an
organelle mediating release of cytoplasmic contents during cell
migration. Cell Res. 25:24–38. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Taylor AC and Robbins E: Observations on
microextensions from the surface of isolated vertebrate cells. Dev
Biol. 6:660–673. 1963. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen Y, Li Y, Ma L and Yu L: Detection of
migrasomes. Methods Mol Biol. 1749:43–49. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang Y, Zucker B, Zhang S, Elias S, Zhu
Y, Chen H, Ding T, Li Y, Sun Y, Lou J, et al: Migrasome formation
is mediated by assembly of micron-scale tetraspanin macrodomains.
Nat Cell Biol. 21:991–1002. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tavano S and Heisenberg CP: Migrasomes
take center stage. Nat Cell Biol. 21:918–920. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang C, Li T, Yin S, Gao M, He H, Li Y,
Jiang D, Shi M, Wang J and Yu L: Monocytes deposit migrasomes to
promote embryonic angiogenesis. Nat Cell Biol. 24:1726–1738. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jiao H, Jiang D, Hu X, Du W, Ji L, Yang Y,
Li X, Sho T, Wang X, Li Y, et al: Mitocytosis, a migrasome-mediated
mitochondrial quality-control process. Cell. 184:2896–2910.e13.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lv L and Zhang L: Identification of
poxvirus inside migrasomes suggests a novel mode of mpox virus
spread. J Infect. 87:160–162. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu Y, Li S, Rong W, Zeng C, Zhu X, Chen
Q, Li L, Liu ZH and Zen K: Podocyte-released migrasomes in urine
serve as an indicator for early podocyte injury. Kidney Dis
(Basel). 6:422–433. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu L: Migrasomes: The knowns, the known
unknowns and the unknown unknowns: A personal perspective. Sci
China Life Sci. 64:162–166. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu J, Lu Z, Jiang D, Guo Y, Qiao H, Zhang
Y, Zhu T, Cai Y, Zhang X, Zhanghao K, et al: Iterative tomography
with digital adaptive optics permits hour-long intravital
observation of 3D subcellular dynamics at millisecond scale. Cell.
184:3318–3332.e17. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Schmidt-Pogoda A, Strecker JK, Liebmann M,
Massoth C, Beuker C, Hansen U, König S, Albrecht S, Bock S, Breuer
J, et al: Dietary salt promotes ischemic brain injury and is
associated with parenchymal migrasome formation. PLoS One.
13:e02098712018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang D, Jiang Z, Lu D, Wang X, Liang H,
Zhang J, Meng Y, Li Y, Wu D, Huang Y, et al: Migrasomes provide
regional cues for organ morphogenesis during zebrafish
gastrulation. Nat Cell Biol. 21:966–977. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gagat M, Zielińska W, Mikołajczyk K,
Zabrzyński J, Krajewski A, Klimaszewska-Wiśniewska A, Grzanka D and
Grzanka A: CRISPR-based activation of endogenous expression of tpm1
inhibits inflammatory response of primary human coronary artery
endothelial and smooth muscle cells induced by recombinant human
tumor necrosis factor α. Front Cell Dev Biol. 9:6680322021.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ardalan M, Hosseiniyan Khatibi SM, Rahbar
Saadat Y, Bastami M, Nariman-Saleh-Fam Z, Abediazar S, Khalilov R
and Zununi Vahed S: Migrasomes and exosomes; different types of
messaging vesicles in podocytes. Cell Biol Int. 46:52–62. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhao X, Lei Y, Zheng J, Peng J, Li Y, Yu L
and Chen Y: Identification of markers for migrasome detection. Cell
Discov. 5:272019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang Y, Wang J, Ding Y, Zhang J, Xu Y, Xu
J, Zheng S and Yang H: Migrasome and tetraspanins in vascular
homeostasis: Concept, present, and future. Front Cell Dev Biol.
8:4382020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liang H, Ma X, Zhang Y, Liu Y, Liu N,
Zhang W, Chen J, Liu B, Du W, Liu X and Yu L: The formation of
migrasomes is initiated by the assembly of sphingomyelin synthase 2
foci at the leading edge of migrating cells. Nat Cell Biol.
25:1173–1184. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhu M, Zou Q, Huang R, Li Y, Xing X, Fang
J, Ma L, Li L, Yang X and Yu L: Lateral transfer of mRNA and
protein by migrasomes modifies the recipient cells. Cell Res.
31:237–240. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen L, Ma L and Yu L: WGA is a probe for
migrasomes. Cell Discov. 5:132019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gustafson CM, Roffers-Agarwal J and
Gammill LS: Chick cranial neural crest cells release extracellular
vesicles that are critical for their migration. J Cell Sci.
135:jcs2602722022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lu P, Liu R, Lu D, Xu Y, Yang X, Jiang Z,
Yang C, Yu L, Lei X and Chen Y: Chemical screening identifies ROCK1
as a regulator of migrasome formation. Cell Discov. 6:512020.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fan C, Shi X, Zhao K, Wang L, Shi K, Liu
YJ, Li H, Ji B and Jiu Y: Cell migration orchestrates migrasome
formation by shaping retraction fibers. J Cell Biol.
221:e2021091682022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ivaska J, Pallari HM, Nevo J and Eriksson
JE: Novel functions of vimentin in cell adhesion, migration, and
signaling. Exp Cell Res. 313:2050–2062. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jiu Y, Peränen J, Schaible N, Cheng F,
Eriksson JE, Krishnan R and Lappalainen P: Vimentin intermediate
filaments control actin stress fiber assembly through GEF-H1 and
RhoA. J Cell Sci. 130:892–902. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huitema K, van den Dikkenberg J, Brouwers
JF and Holthuis JC: Identification of a family of animal
sphingomyelin synthases. EMBO J. 23:33–44. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu D, Xu Y, Ding T, Zu Y, Yang C and Yu L:
Pairing of integrins with ECM proteins determines migrasome
formation. Cell Res. 27:1397–1400. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zuidscherwoude M, Göttfert F, Dunlock VM,
Figdor CG, van den Bogaart G and van Spriel AB: The tetraspanin web
revisited by super-resolution microscopy. Sci Rep. 5:122012015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hemler ME: Tetraspanin proteins mediate
cellular penetration, invasion, and fusion events and define a
novel type of membrane microdomain. Annu Rev Cell Dev Biol.
19:397–422. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dharan R, Huang Y, Cheppali SK, Goren S,
Shendrik P, Wang W, Qiao J, Kozlov MM, Yu L and Sorkin R:
Tetraspanin 4 stabilizes membrane swellings and facilitates their
maturation into migrasomes. Nat Commun. 14:10372023. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Dharan R, Goren S, Cheppali SK, Shendrik
P, Brand G, Vaknin A, Yu L, Kozlov MM and Sorkin R: Transmembrane
proteins tetraspanin 4 and CD9 sense membrane curvature. Proc Natl
Acad Sci USA. 119:e22089931192022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zaidel-Bar R: Job-splitting among
integrins. Nat Cell Biol. 15:575–577. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wehrle-Haller B: Structure and function of
focal adhesions. Curr Opin Cell Biol. 24:116–124. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kolay S, Basu U and Raghu P: Control of
diverse subcellular processes by a single multi-functional lipid
phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2]. Biochem J.
473:1681–1692. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hammond GRV: Does PtdIns(4,5)P2
concentrate so it can multi-task? Biochem Soc Trans. 44:228–233.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ding T, Ji J, Zhang W, Liu Y, Liu B, Han
Y, Chen C and Yu L: The phosphatidylinositol
(4,5)-bisphosphate-Rab35 axis regulates migrasome formation. Cell
Res. 33:617–627. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Klinkert K and Echard A: Rab35 GTPase: A
central regulator of phosphoinositides and F-actin in endocytic
recycling and beyond. Traffic. 17:1063–1077. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Allaire PD, Seyed Sadr M, Chaineau M,
Seyed Sadr E, Konefal S, Fotouhi M, Maret D, Ritter B, Del Maestro
RF and McPherson PS: Interplay between Rab35 and Arf6 controls
cargo recycling to coordinate cell adhesion and migration. J Cell
Sci. 126:722–731. 2013.PubMed/NCBI
|
|
44
|
Lock FE, Ryan KR, Poulter NS, Parsons M
and Hotchin NA: Differential regulation of adhesion complex
turnover by ROCK1 and ROCK2. PLoS One. 7:e314232012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yu W, Hua Y, Qiu H, Hao J, Zou K, Li Z, Hu
S, Guo P, Chen M, Sui S, et al: PD-L1 promotes tumor growth and
progression by activating WIP and β-catenin signaling pathways and
predicts poor prognosis in lung cancer. Cell Death Dis. 11:5062020.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang M, Xiong C and Mercurio AM: PD-LI
promotes rear retraction during persistent cell migration by
altering integrin β4 dynamics. J Cell Biol. 221:e2021080832022.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Saito S, Tanaka M, Tatematsu S and Okochi
M: Peptide-modified substrate enhances cell migration and migrasome
formation. Mater Sci Eng C Mater Biol Appl. 131:1124952021.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Han Y and Yu L: Calcium ions promote
migrasome formation via Synaptotagmin-1. J Cell Biol.
223:e2024020602024. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yoshikawa K, Saito S, Kadonosono T, Tanaka
M and Okochi M: Osmotic stress induces the formation of
migrasome-like vesicles. FEBS Lett. 598:437–445. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chen Y, Li Y, Li B, Hu D, Dong Z and Lu F:
Migrasomes from adipose derived stem cells enrich CXCL12 to recruit
stem cells via CXCR4/RhoA for a positive feedback loop mediating
soft tissue regeneration. J Nanobiotechnology. 22:2192024.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Jiao H, Li X, Li Y, Guo Y, Hu X, Sho T,
Luo Y, Wang J, Cao H, Du W, et al: Localized, highly efficient
secretion of signaling proteins by migrasomes. Cell Res.
34:572–585. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Strzyz P: Migrasomes promote angiogenesis.
Nat Rev Mol Cell Biol. 24:842023. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Nawaz M, Fatima F, Vallabhaneni KC,
Penfornis P, Valadi H, Ekström K, Kholia S, Whitt JD, Fernandes JD,
Pochampally R, et al: Extracellular vesicles: Evolving factors in
stem cell biology. Stem Cells Int. 2016:10731402016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Rani S, Ryan AE, Griffin MD and Ritter T:
Mesenchymal stem cell-derived extracellular vesicles: Toward
cell-free therapeutic applications. Mol Ther. 23:812–823. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Tan X, Gong YZ, Wu P, Liao DF and Zheng
XL: Mesenchymal stem cell-derived microparticles: A promising
therapeutic strategy. Int J Mol Sci. 15:14348–14363. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Bruno S and Camussi G: Role of mesenchymal
stem cell-derived microvesicles in tissue repair. Pediatr Nephrol.
28:2249–2254. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhang Y, Chopp M, Meng Y, Katakowski M,
Xin H, Mahmood A and Xiong Y: Effect of exosomes derived from
multipluripotent mesenchymal stromal cells on functional recovery
and neurovascular plasticity in rats after traumatic brain injury.
J Neurosurg. 122:856–867. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Xin H, Li Y, Buller B, Katakowski M, Zhang
Y, Wang X, Shang X, Zhang ZG and Chopp M: Exosome-mediated transfer
of miR-133b from multipotent mesenchymal stromal cells to neural
cells contributes to neurite outgrowth. Stem Cells. 30:1556–1564.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Deniz IA, Karbanová J, Wobus M, Bornhäuser
M, Wimberger P, Kuhlmann JD and Corbeil D: Mesenchymal stromal
cell-associated migrasomes: A new source of chemoattractant for
cells of hematopoietic origin. Cell Commun Signal. 21:362023.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Jiang D, Jiao L, Li Q, Xie R, Jia H, Wang
S, Chen Y, Liu S, Huang D, Zheng J, et al: Neutrophil-derived
migrasomes are an essential part of the coagulation system. Nat
Cell Biol. 26:1110–1123. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Sugiura A, McLelland GL, Fon EA and
McBride HM: A new pathway for mitochondrial quality control:
Mitochondrial-derived vesicles. EMBO J. 33:2142–2156. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Poole LP and Macleod KF: Mitophagy in
tumorigenesis and metastasis. Cell Mol Life Sci. 78:3817–3851.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Baumann K: Damaged mitochondria are
discarded via migrasomes. Nat Rev Mol Cell Biol. 22:4422021.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Mehra C and Pernas L: Move it to lose it:
Mitocytosis expels damaged mitochondria. Dev Cell. 56:2014–2015.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Zhao W, Tang X and Zhang L:
Virus-containing migrasomes enable poxviruses to evade
tecovirimat/ST-246 treatment. J Infect. 88:203–205. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Liu Y, Zhu Z, Li Y, Yang M and Hu Q:
Migrasomes released by HSV-2-infected cells serve as a conveyance
for virus spread. Virol Sin. 38:643–645. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Yang R, Zhang H, Chen S, Lou K, Zhou M,
Zhang M, Lu R, Zheng C, Li L, Chen Q, et al: Quantification of
urinary podocyte-derived migrasomes for the diagnosis of kidney
disease. J Extracell Vesicles. 13:e124602024. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Li T, Su X, Lu P, Kang X, Hu M, Li C, Wang
S, Lu D, Shen S, Huang H, et al: Bone marrow mesenchymal stem
cell-derived dermcidin-containing migrasomes enhance LC3-associated
phagocytosis of pulmonary macrophages and protect against
post-stroke pneumonia. Adv Sci (Weinh). 10:e22064322023. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Hu M, Li T, Ma X, Liu S, Li C, Huang Z,
Lin Y, Wu R, Wang S, Lu D, et al: Macrophage lineage cells-derived
migrasomes activate complement-dependent blood-brain barrier damage
in cerebral amyloid angiopathy mouse model. Nat Commun.
14:39452023. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zheng Y, Lang Y, Qi B and Li T: TSPAN4 and
migrasomes in atherosclerosis regression correlated to myocardial
infarction and pan-cancer progression. Cell Adh Migr. 17:14–19.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Sun P, Li Y, Yu W, Chen J, Wan P, Wang Z,
Zhang M, Wang C, Fu S, Mang G, et al: Low-intensity pulsed
ultrasound improves myocardial ischaemia-reperfusion injury via
migrasome-mediated mitocytosis. Clin Transl Med. 14:e17492024.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Zhang Z, Zhang T, Zhang R, Zhang Z and Tan
S: Migrasomes and tetraspanins in hepatocellular carcinoma: Current
status and future prospects. Future Sci OA. 9:FSO8902023.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Zhang K, Zhu Z, Jia R, Wang NA, Shi M,
Wang Y, Xiang S, Zhang Q and Xu L: CD151-enriched migrasomes
mediate hepatocellular carcinoma invasion by conditioning cancer
cells and promoting angiogenesis. J Exp Clin Cancer Res.
43:1602024. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Zhang R, Peng J, Zhang Y, Zheng K, Chen Y,
Liu L, Li T, Liu J, Li Y, Yang S, et al: Pancreatic cancer
cell-derived migrasomes promote cancer progression by fostering an
immunosuppressive tumor microenvironment. Cancer Lett.
605:2172892024. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Dong Y, Tang X, Zhao W, Liu P, Yu W, Ren
J, Chen Y, Cui Y, Chen J and Liu Y: TSPAN4 influences glioblastoma
progression through regulating EGFR stability. iScience.
27:1104172024. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Lee SY, Choi SH, Kim Y, Ahn HS, Ko YG, Kim
K, Chi SW and Kim H: Migrasomal autophagosomes relieve endoplasmic
reticulum stress in glioblastoma cells. BMC Biol. 22:232024.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Köktürk S, Doğan S, Yılmaz CE, Cetinkol Y
and Mutlu O: Expression of brain-derived neurotrophic factor and
formation of migrasome increases in the glioma cells induced by the
adipokinetic hormone. Rev Assoc Med Bras (1992). 70:e202313372024.
View Article : Google Scholar : PubMed/NCBI
|