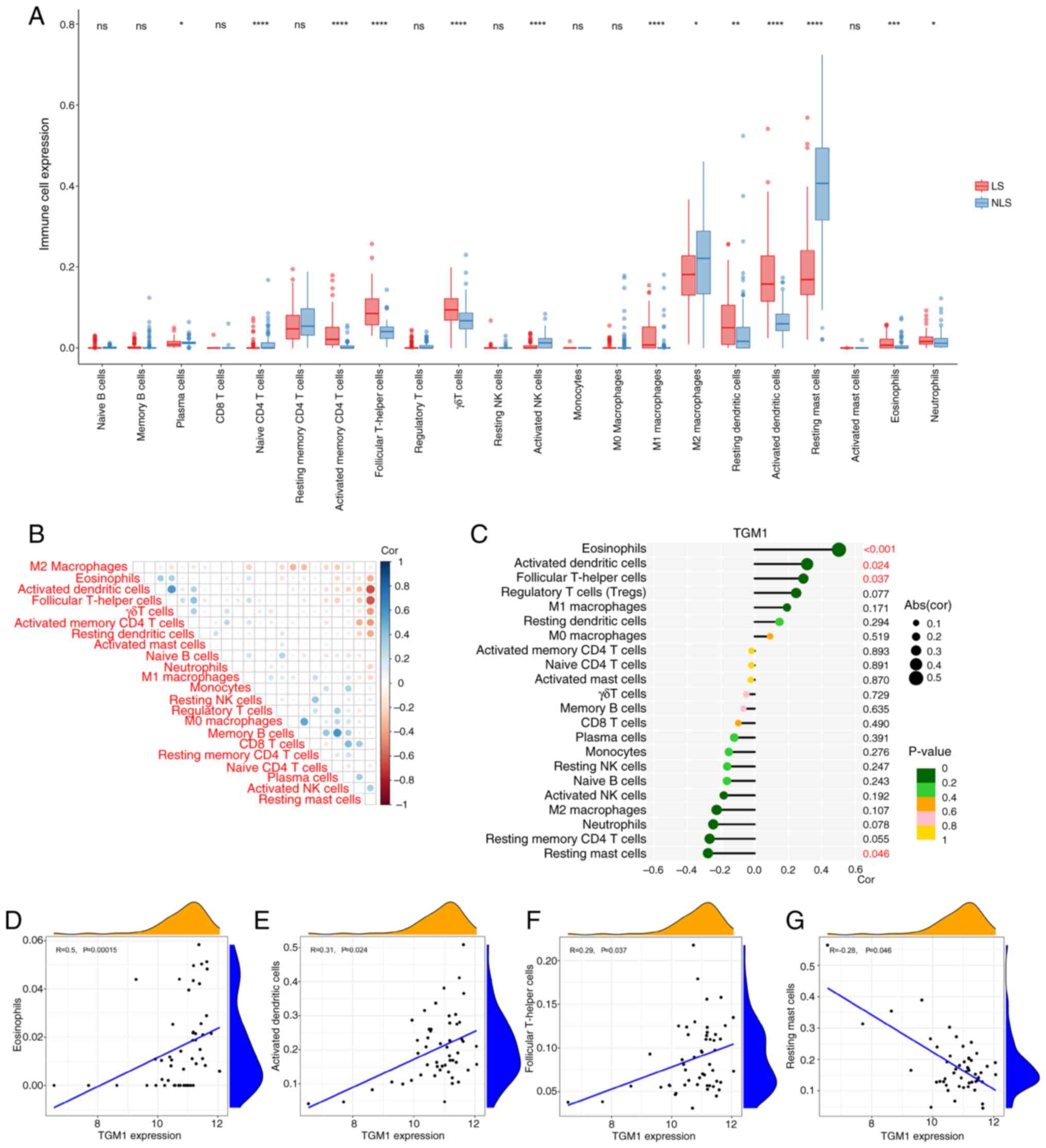
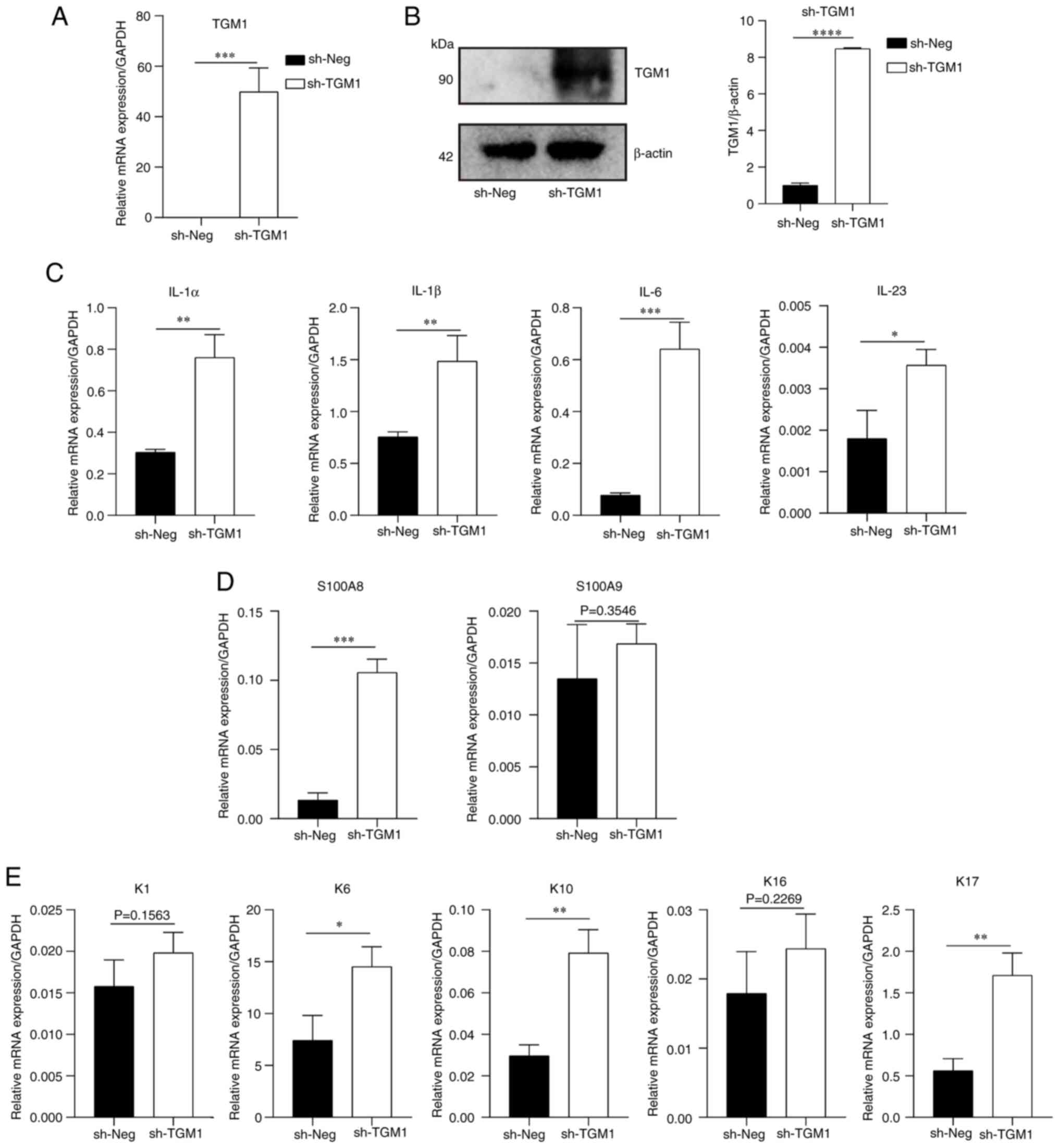
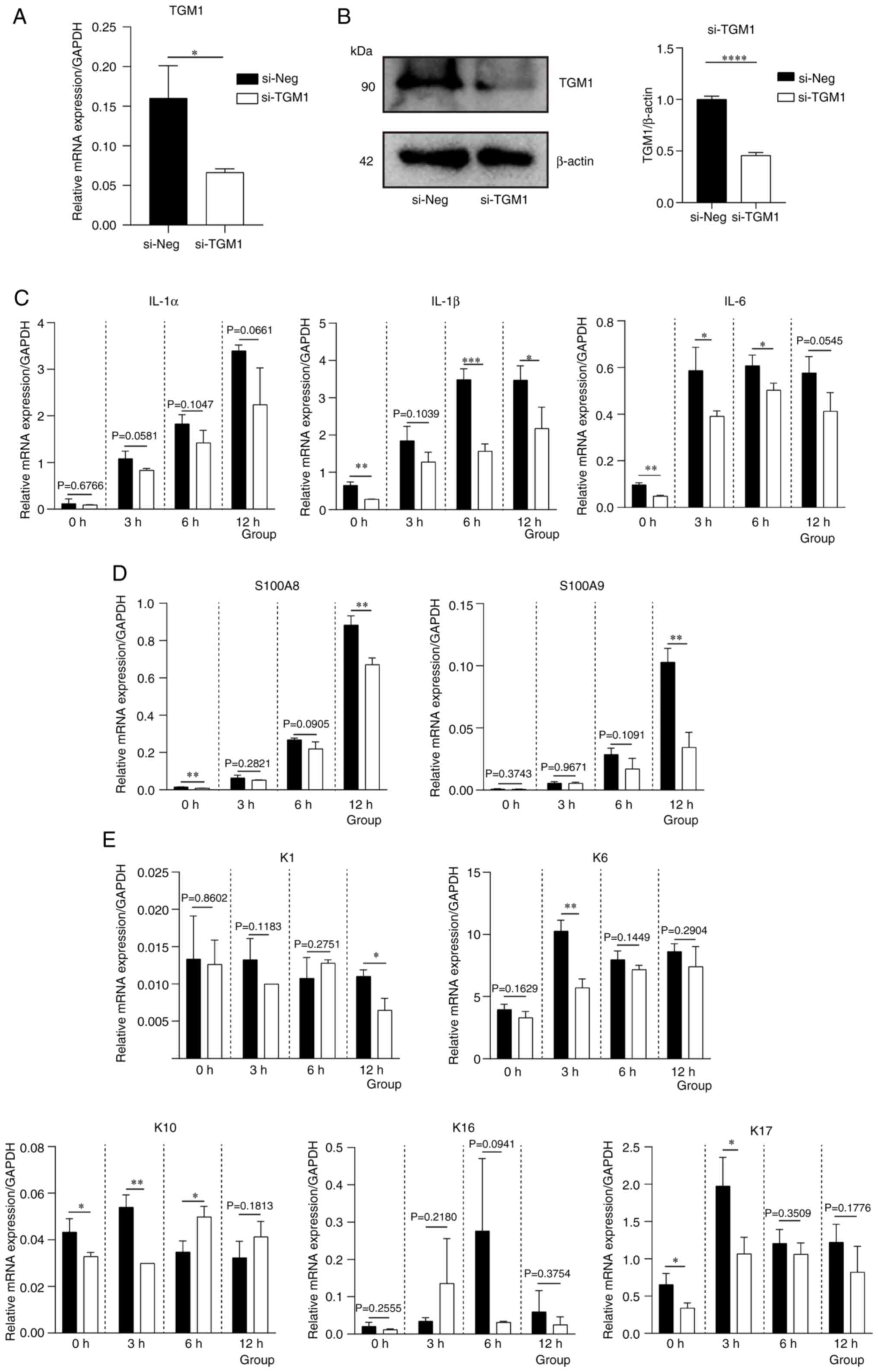
|
1
|
Griffiths CE and Barker JN: Pathogenesis
and clinical features of psoriasis. Lancet. 370:263–271. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Stern RS, Nijsten T, Feldman SR, Margolis
DJ and Rolstad T: Psoriasis is common, carries a substantial burden
even when not extensive, and is associated with widespread
treatment dissatisfaction. J Investig Dermatol Symp Proc.
9:136–139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kurd SK and Gelfand JM: The prevalence of
previously diagnosed and undiagnosed psoriasis in US adults:
Results from NHANES 2003–2004. J Am Acad Dermatol. 60:218–224.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lee EB, Wu KK, Lee MP, Bhutani T and Wu
JJ: Psoriasis risk factors and triggers. Cutis. 102:18–20.
2018.PubMed/NCBI
|
|
5
|
Guo J, Zhang H, Lin W, Lu L, Su J and Chen
X: Signaling pathways and targeted therapies for psoriasis. Signal
Transduct Target Ther. 8:4372023. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Baliwag J, Barnes DH and Johnston A:
Cytokines in psoriasis. Cytokine. 73:342–350. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Michalak-Stoma A, Pietrzak A, Szepietowski
JC, Zalewska-Janowska A, Paszkowski T and Chodorowska G: Cytokine
network in psoriasis revisited. Eur Cytokine Netw. 22:160–168.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Singh R, Koppu S, Perche PO and Feldman
SR: The cytokine mediated molecular pathophysiology of psoriasis
and its clinical implications. Int J Mol Sci. 22:127932021.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kofoed K, Skov L and Zachariae C: New
drugs and treatment targets in psoriasis. Acta Derm Venereol.
95:133–139. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Furtunescu AR, Georgescu SR, Tampa M and
Matei C: Inhibition of the JAK-STAT Pathway in the treatment of
psoriasis: A review of the literature. Int J Mol Sci. 25:46812024.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mavropoulos A, Rigopoulou EI, Liaskos C,
Bogdanos DP and Sakkas LI: The role of p38 MAPK in the
aetiopathogenesis of psoriasis and psoriatic arthritis. Clin Dev
Immunol. 2013:5697512013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang M and Zhang X: The role of
PI3K/AKT/FOXO signaling in psoriasis. Arch Dermatol Res. 311:83–91.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Correa da Rosa J, Kim J, Tian S, Tomalin
LE, Krueger JG and Suárez-Fariñas M: Shrinking the psoriasis
assessment gap: Early Gene-expression profiling accurately predicts
response to Long-Term treatment. J Invest Dermatol. 137:305–312.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ding J, Gudjonsson JE, Liang L, Stuart PE,
Li Y, Chen W, Weichenthal M, Ellinghaus E, Franke A, Cookson W, et
al: Gene expression in skin and lymphoblastoid cells: Refined
statistical method reveals extensive overlap in cis-eQTL signals.
Am J Hum Genet. 87:779–789. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Russell CB, Rand H, Bigler J, Kerkof K,
Timour M, Bautista E, Krueger JG, Salinger DH, Welcher AA and
Martin DA: Gene expression profiles normalized in psoriatic skin by
treatment with brodalumab, a human anti-IL-17 receptor monoclonal
antibody. J Immunol. 192:3828–3836. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Guan S, Xu Z, Yang T, Zhang Y, Zheng Y,
Chen T, Liu H and Zhou J: Identifying potential targets for
preventing cancer progression through the PLA2G1B recombinant
protein using bioinformatics and machine learning methods. Int J
Biol Macromol. 276:1339182024. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wei C, Wei Y, Cheng J, Tan X, Zhou Z, Lin
S and Pang L: Identification and verification of diagnostic
biomarkers in recurrent pregnancy loss via machine learning
algorithm and WGCNA. Front Immunol. 14:12418162023. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z,
Feng T, Zhou L, Tang W, Zhan L, et al: clusterProfiler 4.0: A
universal enrichment tool for interpreting omics data. Innovation
(Camb. 2:1001412021.PubMed/NCBI
|
|
21
|
Ding R, Qu Y, Wu CH and Vijay-Shanker K:
Automatic gene annotation using GO terms from cellular component
domain. BMC Med Inform Decis Mak. 18 (Suppl 5):S1192018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
von Mering C, Huynen M, Jaeggi D, Schmidt
S, Bork P and Snel B: STRING: A database of predicted functional
associations between proteins. Nucleic Acids Res. 31:258–261. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Newman AM, Liu CL, Green MR, Gentles AJ,
Feng W, Xu Y, Hoang CD, Diehn M and Alizadeh AA: Robust enumeration
of cell subsets from tissue expression profiles. Nat Methods.
12:453–457. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Boehncke WH and Schön MP: Psoriasis.
Lancet. 386:983–994. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Langley RG and Ellis CN: Evaluating
psoriasis with psoriasis area and severity index, psoriasis global
assessment, and lattice system Physician's global assessment. J Am
Acad Dermatol. 51:563–569. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang J, Feng X, Zeng J, Zhang S, Zhang J,
Guo P, Yu H, Sun M, Wu J, Li M, et al: Aberrant HO-1/NQO1-Reactive
oxygen Species-ERK signaling pathway contributes to aggravation of
TPA-induced irritant contact dermatitis in Nrf2-deficient mice. J
Immunol. 208:1424–1433. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Banerjee S, Biehl A, Gadina M, Hasni S and
Schwartz DM: JAK-STAT signaling as a target for inflammatory and
autoimmune diseases: Current and future prospects. Drugs.
77:521–546. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kim BH, Na KM, Oh I, Song IH, Lee YS, Shin
J and Kim TY: Kurarinone regulates immune responses through
regulation of the JAK/STAT and TCR-mediated signaling pathways.
Biochem Pharmacol. 85:1134–1144. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Grabarek B, Krzaczyński J, Strzałka-Mrozik
B, Wcisło-Dziadecka D and Gola J: The influence of ustekinumab on
expression of STAT1, STAT3, STAT4, SOCS2, and IL17 in patients with
psoriasis and in a control. Dermatol Ther. 32:e130292019.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Baird L and Yamamoto M: The Molecular
mechanisms regulating the KEAP1-NRF2 pathway. Mol Cell Biol.
40:e00099–20. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Helwa I, Patel R, Karempelis P,
Kaddour-Djebbar I, Choudhary V and Bollag WB: The antipsoriatic
agent monomethylfumarate has antiproliferative, prodifferentiative,
and anti-inflammatory effects on keratinocytes. J Pharmacol Exp
Ther. 352:90–97. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bojanowski K, Ibeji CU, Singh P, Swindell
WR and Chaudhuri RK: A Sensitization-free Dimethyl fumarate
prodrug, isosorbide Di-(Methyl Fumarate), provides a topical
treatment candidate for psoriasis. JID Innov. 1:1000402021.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Afonina IS, Van Nuffel E and Beyaert R:
Immune responses and therapeutic options in psoriasis. Cell Mol
Life Sci. 78:2709–2727. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xu M, Lu H, Lee YH, Wu Y, Liu K, Shi Y, An
H, Zhang J, Wang X, Lai Y and Dong C: An Interleukin-25-Mediated
autoregulatory circuit in keratinocytes plays a pivotal role in
psoriatic skin inflammation. Immunity. 48:787–798.e784. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
McGeachy MJ, Cua DJ and Gaffen SL: The
IL-17 family of cytokines in health and disease. Immunity.
50:892–906. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Baker KJ, Brint E and Houston A:
Transcriptomic and functional analyses reveal a tumour-promoting
role for the IL-36 receptor in colon cancer and crosstalk between
IL-36 signalling and the IL-17/ IL-23 axis. Br J Cancer.
128:735–747. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kirkham BW, Kavanaugh A and Reich K:
Interleukin-17A: A unique pathway in immune-mediated diseases:
Psoriasis, psoriatic arthritis and rheumatoid arthritis.
Immunology. 141:133–142. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Martin DA, Towne JE, Kricorian G, Klekotka
P, Gudjonsson JE, Krueger JG and Russell CB: The emerging role of
IL-17 in the pathogenesis of psoriasis: Preclinical and clinical
findings. J Invest Dermatol. 133:17–26. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ly K, Smith MP, Thibodeaux Q, Reddy V,
Liao W and Bhutani T: Anti IL-17 in psoriasis. Expert Rev Clin
Immunol. 15:1185–1194. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Nemes Z, Marekov LN, Fésüs L and Steinert
PM: A novel function for transglutaminase 1: Attachment of
long-chain omega-hydroxyceramides to involucrin by ester bond
formation. Proc Natl Acad Sci USA. 96:8402–8407. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Baker P, Huang C, Radi R, Moll SB, Jules E
and Arbiser JL: Skin barrier function: The interplay of physical,
chemical, and immunologic properties. Cells. 12:27452023.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Montero-Vilchez T,
Segura-Fernández-Nogueras MV, Pérez-Rodríguez I, Soler-Gongora M,
Martinez-Lopez A, Fernández-González A, Molina-Leyva A and
Arias-Santiago S: Skin barrier function in psoriasis and atopic
dermatitis: Transepidermal water loss and temperature as useful
tools to assess disease severity. J Clin Med. 10:3592021.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Armstrong AW and Read C: Pathophysiology,
clinical presentation, and treatment of psoriasis: A review. JAMA.
323:1945–1960. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Maroto-Morales D, Montero-Vilchez T and
Arias-Santiago S: Study of skin barrier function in psoriasis: The
impact of emollients. Life (Basel). 11:6512021.PubMed/NCBI
|
|
47
|
Farasat S, Wei MH, Herman M, Liewehr DJ,
Steinberg SM, Bale SJ, Fleckman P and Toro JR: Novel
transglutaminase-1 mutations and genotype-phenotype investigations
of 104 patients with autosomal recessive congenital ichthyosis in
the USA. J Med Genet. 46:103–111. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kulski JK, Kenworthy W, Bellgard M, Taplin
R, Okamoto K, Oka A, Mabuchi T, Ozawa A, Tamiya G and Inoko H: Gene
expression profiling of Japanese psoriatic skin reveals an
increased activity in molecular stress and immune response signals.
J Mol Med (Berl). 83:964–975. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wu R, Li D, Zhang S, Wang J, Chen K, Tuo
Z, Miyamoto A, Yoo KH, Wei W, Zhang C, et al: A pan-cancer analysis
of the oncogenic and immunological roles of transglutaminase 1
(TGM1) in human cancer. J Cancer Res Clin Oncol. 150:1232024.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang J, Xiao Y, Wu R and Zhang C: TGM1
could predict overall survival for patients with urinary bladder
cancer. Asian J Surg. 46:5373–5375. 2023. View Article : Google Scholar : PubMed/NCBI
|