Introduction
Psoriasis is an immune-mediated dermatological
condition characterized by erythematous and scaly plaques affecting
the entire integumentary system (1). Empirical observations indicate that
it severely impacts 2–4% of the population, leading to serious
psychological anguish and diminished quality of life (2,3).
Numerous contributing factors have been identified in this context
(4). Genetic underpinnings and
external influences, including smoking cessation, anxiety and
alcohol consumption, have been shown to contribute to the
development of psoriasis (4).
Systematic exploration has revealed that psoriasis
is predominantly regulated by interactions among numerous cytokines
and multifaceted signaling pathways (5). Notably, studies have demonstrated
that several cytokines, including IL-2, IL-6, IL-17, IL-23, TNF-α
and IFN-γ, are important for the advancement of psoriasis (6,7).
Consequently, biological therapies targeting these cytokines,
particularly the TNF-α, IL-12/IL-23, IL-17 and IL-23/IL-39
pathways, have been recognized as promising treatments for
psoriasis (8,9). The Janus kinase (JAK)/STAT pathway is
a primary inflammatory mechanism involved in the development of
psoriasis. Specific cytokines, particularly IL-17 and IL-23, are
important in this process by relaying signals and regulating the
transcriptional expression of targets within the JAK/STAT pathway.
This process is important for the advancement of psoriatic disease
(10). Signaling pathways,
including MAPK and PI3K/AKT signaling, also contribute to the
development of psoriasis by influencing the disease state (11,12).
Notably, while existing research has yielded some insights into
psoriasis mechanisms, current findings remain insufficient.
Therefore, it is key to identify novel diagnostic markers to
clarify the pathogenesis of psoriasis.
The present study utilized the GSE30999, GSE53552
and GSE13355 datasets from the Gene Expression Omnibus (GEO)
database containing lesional skin (LS) and non-lesional skin (NLS)
tissues from individuals with psoriasis. The datasets were divided
into a training set, GSE30999, and two validation sets, GSE53552
and GSE13355. The analysis of these datasets employed the following
methods: Variable selection, model training, protein-protein
interaction (PPI) analysis, Gene Set Enrichment Analysis (GSEA) and
single-gene immune infiltration analysis. To specifically address
the course of psoriatic disease, 163 distinct differentially
expressed genes (DEGs) were identified. The DEGs were examined
utilizing the following machine learning algorithms: Least absolute
shrinkage and selection operator (LASSO) regression, support vector
machine-recursive feature elimination (SVM-RFE) and random forest
(RF). The findings from these investigations yielded notable
insights into the mechanism of transglutaminase 1 (TGM1). To
clarify the role of TGM1 during psoriasis, an investigation of PPIs
was conducted. Furthermore, single-gene GSEA and single-gene immune
infiltration analysis were implemented to elucidate the underlying
activities and biological pathways of TGM1.
Materials and methods
Data collection
Gene expression profiling data of psoriasis were
retrieved from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). The GSE30999,
GSE13355 and GSE53552 datasets were used in the present study.
GSE30999 (13) includes 85
patients with psoriasis including 85 biopsy samples of LS and 85
matched biopsy samples of NLS. GSE13355 (14) includes 58 patients with psoriasis
including 58 biopsy samples of LS and 58 biopsy samples of NLS.
GSE53552 (15) includes 25
patients with psoriasis including 25 biopsy samples of LS and 24
biopsy samples of NLS. GSE30999 was selected as training dataset
and both the GSE13355 and GSE53552 datasets were selected as
validation datasets.
Identification of DEGs
‘Limma (version 3.58.1)’, a package of R software
(version 4.3.0, http://www.r-project.org/), was used to identify DEGs
in LS and NLS of patients with psoriasis (16). The present study used
|log2[fold change (FC)]|>1 and P<0.05 as cutoff
thresholds to extract statistically significant DEGs. However, when
looking for signature genes, not all DEGs were included in later
calculations. For example, Guan et al (17) created three algorithms to choose
potential genes for lung cancer from a total of 51 PPI-related
DEGs. Wei et al (18)
adopted three methods to analyze the DEGs of the most important
modules in the results of a weighted gene co-expression network
analysis to screen significant variables. Therefore, the present
study only used the significant DEGs for subsequent analysis. DEGs
with |FC|>2 were initially isolated, but the top DEGs with
|log2FC|>3 were then screened for the following
analysis in order to prevent interference of genes with little
variation and to reduce the workload of the calculations.
Eventually, the R packages ‘pheatmap’ and ‘ggplot2’ were used to
depict the DEGs in a heatmap and a volcano plot.
GSEA
GSEA is a computer tool that assesses the type of
gene expression in a certain functional gene set and reveals the
underlying biochemical pathways driving complex disease (19). In the present study, the
‘clusterProfiler’ package (4.10.0) was employed to perform GSEA
(20). The reference gene set was
‘c2.cp.kegg_medicus.v2024.1.Hs.symbols.gmt’
(gsea-msigdb.org/gsea/msigdb/human/collections.jsp#C2), downloaded
from the MsigDB database (https://www.gsea-msigdb.org/gsea/msigdb). To avoid
interference from unchanged genes, the top 10,000 genes from all
probes in the GSE30999 dataset were selected for GSEA based on
|logFC| descending order. Kyoto Encyclopedia of Genes and Genomes
(KEGG, genome.jp/kegg/) pathway terms with a P-value <0.05 were
selected as significant.
In addition, the present study investigated the
potential function of signature genes in psoriasis via single-gene
GSEA. The ‘clusterProfiler’ R package was utilized to calculate the
correlation between signature and other genes and the genes were
then ordered according to their correlation scores. The ranking
genes were chosen as the test gene set for KEGG enrichment
analysis. P<0.05 was used as a criterion for significant
items.
Functional and pathway enrichment
analysis
The present study used Gene Ontology (GO) and KEGG
analyses to unravel the functional implications and potential
biological processes (BPs) of the significant DEGs with
|log2FC|>3. GO analysis, a widely recognized
bioinformatics tool, makes it possible to systematically classify
and annotate genes according to BP, cellular component (CC) and
molecular function (MF) (21). By
subjecting genes to GO and KEGG pathway analysis, the present study
identified the notable BPs and pathways that were substantially
enriched in these genes. The present study used the bioinformatics
website (bioinformatics.com.cn/) to perform GO and KEGG pathway
analysis. Only GO and KEGG terms with P<0.05 were kept as
significantly enriched. The top 10 results in GO and KEGG pathway
analyses were selected and plotted as bubble plots and gene-pathway
association network diagrams.
PPI network analysis
The identified genes were imported into the STRING
database (version 12.0) (https://cn.string-db.org/) (22) to obtain the PPI network, with a
minimum required interaction score of ≥0.4. The PPI network was
visualized using Cytoscape v3.10.1 (23).
Screening of signature genes using
machine learning methods
To further identify the signature genes for
psoriasis, the present study adopted LASSO, SVM-RFE and RF
analyses. A 10-fold cross-verification of LASSO was performed using
the ‘glmnet’ package (version 4.1–8,
cran.r-project.org/web/packages/glmnet/index.html) of R software to
identify significant genes, with the minimal λ value considered
optimal. The RF algorithm was executed by the ‘randomForest’
package (version 4.7–1.1, http://cran.r-project.org/web/packages/randomForest/index.html)
of R software, and the top 15 genes were selected as potential
candidates. The ‘e1071’ R package (version 1.7–16, http://cran.r-project.org/web/packages/e1071/index.html)
was used to implement the SVM-RFE algorithm to detect the
classifier with the least possible cross-validation error. The
genes identified by all three methods were considered to be the
signature genes for psoriasis, as shown in a Venn diagram. Finally,
the area under the curve (AUC) of the receiver operating
characteristic curve (ROC) was analyzed using the R package ‘pROC’
(version 1.18.5,
search.r-project.org/CRAN/refmans/pROC/html/pROC-package.html) to
assess the diagnostic efficacy of signature genes in the GSE30999,
GSE13355 and GSE53552 datasets.
Immune infiltration analysis
To explore the distribution of immune cells in LS
and NLS of patients with psoriasis in the GSE30999 dataset, the
present study employed the CIBERSORT algorithm of the R software
based on the ‘CIBERSORT’ R package (version 0.1.0, http://github.com/Moonerss/CIBERSORT/blob/main/R/CIBERSORT.R)
(24), which contains a gene
expression matrix of 22 immune cell types. The difference in
infiltrated immune cells between the LS and NLS groups was analyzed
using the Wilcoxon rank-sum test and visualized with a boxplot
using the ‘ggpubr’ R package (version 0.6.0,
cran.r-project.org/web/packages/ggpubr/index.html). P<0.05 was
considered to indicate a statistically significant difference. The
Spearman correlation between immune cell types was calculated and
illustrated using the ‘corrplot’ tool (version 0.92,
cran.r-project.org/web/packages/corrplot/index.html) in R.
For single-gene immune infiltration analysis, the
Spearman correlation between the expression of signature genes and
the gene expression matrix of 22 infiltrating immune cell types was
calculated and displayed in a lollipop graph visualized using the
‘ggpubr’ R package (version 0.6.0,
cran.r-project.org/web/packages/ggpubr/index.html). The immune
cells significantly associated with signature genes (P<0.05)
were extracted and scatter plots were generated.
Patients and tissue samples
The present study included 10 patients with
psoriasis (4 female and 6 male patients; age, 20–49; mean age,
36.3±6.3 years) and 10 healthy controls (5 female and 5 male
individuals; age, 21–50; mean age, 34.3±6.7 years). The control
samples were obtained from the Plastic Surgery Department of
Tianjin Academy of Traditional Chinese Medicine Affiliated Hospital
(Tianjin, China). Patients with psoriasis were recruited from the
Department of Dermatology of Tianjin Academy of Traditional Chinese
Medicine Affiliated Hospital (Tianjin, China) between October 2022
and March 2023, and were selected based on objective criteria such
as age, sex and health status to avoid self-selection bias. The
inclusion criteria for patients with psoriasis were: i) Clinical
presentation consistent with typical features of plaque psoriasis;
ii) histological evidence of abnormal epidermal proliferation and
keratinocyte differentiation (25); iii) disease duration ≥2 years, with
severity during the active phase assessed by experienced
dermatologists using the Psoriasis Area and Severity Index
(26); and iv) no systemic
treatment within 4 weeks prior to sampling and no topical treatment
within 2 weeks. A total of four doctors were involved in the
diagnosis of psoriasis and samples with disagreements were excluded
from the study. Exclusion criteria included comorbid diabetes,
renal insufficiency, history of malignancy, severe cardiovascular
or cerebrovascular disease, and other autoimmune or
immunodeficiency disorders. Patient samples were obtained from skin
lesions on the upper arm or leg (1.0×1.0 cm), and control samples
were collected from normal skin during plastic or reconstructive
surgery. All samples were fixed in 4% paraformaldehyde at 4°C for
12 h and subsequently processed into 5 µm paraffin sections.
H&E staining and
immunohistochemistry (IHC)
H&E staining and IHC were conducted as described
previously (27). Sections were
incubated with a primary antibody against TGM1 (1: 100 cat. no.
12912-3-AP; Proteintech Group, Inc.) overnight at 4°C. The slides
were rinsed with PBS with 0.1% Tween-20 and treated with the
secondary goat anti-rabbit HRP antibody (1:200; cat. no. HS101-01;
TransGen Biotech Co., Ltd.) for 1 h at room temperature. Images
were captured using a light microscope (Leica DM2000; Leica
Microsystems GmbH).
TGM1 overexpression
HaCaT cells (immortalized human keratinocyte cell
line; cat. no. CLS300493; Cell Line Service) were cultured in DMEM
(high glucose) (cat. no. PM150210; Pricella®;
Elabscience Bionovation Inc.) supplemented with 10%
TransSerum® EQ Fetal Bovine Serum (cat. no. FS201-02;
TransGen Biotech Co., Ltd.) and 1% penicillin-streptomycin (cat.
no. FG101-01; TransGen Biotech Co., Ltd.) at 37°C. The
overexpression plasmid pLV3-CMV-TGM1-human and the control plasmid
pLV3-CMV-Empty-human (2 µg/ml, MiaoLing Plasmid Platform,) were
transfected into HaCaT cells using Lipofectamine® 3000
reagent (cat. no. L3000015; Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol at 37°C for 72 h.
Reverse transcription-quantitative PCR (RT-qPCR) was used to assess
the mRNA expression levels of TGM1 in the HaCaT cells 72 h after
transfection.
TGM1 knockdown
HaCaT cells were transfected with small interfering
RNA (siRNA) targeting TGM1 (si-TGM1) using
Lipofectamine® 2000 (cat. no. 11668030; Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. The sequence for si-TGM1 was as follows: Sense (5′-3′),
GGUGAAUAGUGACAAGGUGUAdTdT; and antisense (5′-3′),
UACACCUUGUCACUAUUCACCdTdT (75 nM, Ruilai Biotechnology (Tianjin)
Co., Ltd.). A non-targeting siRNA was used as the negative control
[sense, 5′-UUCUCCGAACGUGUCACGUdTdT-3′ and antisense
[5′-ACGUGACACGUUCGGAGAAdTdT-3′; 75 nM, Ruilai Biotechnology
(Tianjin) Co., Ltd.). Following 48 h of transfection at 37°C, HaCaT
cells were stimulated with a combination of recombinant human
IL-17A (10 ng/ml; PeproTech, Inc.; Thermo Fisher Scientific, Inc.),
IL-22 (10 ng/ml; PeproTech, Inc.; Thermo Fisher Scientific, Inc.),
Oncostatin M (10 ng/ml; PeproTech, Inc.; Thermo Fisher Scientific,
Inc.), TNF-α (10 ng/ml; PeproTech, Inc.; Thermo Fisher Scientific,
Inc.) and IL-1α (10 ng/ml; PeproTech, Inc.; Thermo Fisher
Scientific, Inc.), collectively referred to as M5. The cells were
exposed to M5 for 0, 3, 6 or 12 h at 37°C and harvested for
subsequent analysis.
RT-qPCR
Total RNA collected from skin tissue and HaCat cells
was extracted using TransZol-Up reagent (cat. no. ET111-01;
TransGen Biotech Co., Ltd.) according to the manufacturer's
instructions. cDNA was synthesized using the All-in-One FirstStrand
cDNA Synthesis SuperMix for RT-PCR (cat. no. AE341-02; TransGen
Biotech Co., Ltd.). The RT-PCR reaction procedure is as follows:
37°C 15 min, 85°C 1 min, 4°C holding. qPCR was performed on a 7500
Fast Real-Time PCR System (Applied Biosystems; Thermo Fisher
Scientific, Inc.) with PerfectStart Green qPCR SuperMix (cat. no.
AQ602-24; TransGen Biotech Co., Ltd.). The RT-qPCR reaction
procedure is as follows: (95°C 30 sec, 95°C 5 sec, 60°C 30 sec) for
40 cycles, 95°C 15 sec, 60°C 1 min, 95°C 15 sec. Relative mRNA
expression levels were calculated using the 2−ΔΔCq
method (28), with GAPDH as the
internal control. The primer sequences used for RT-qPCR are listed
in Table I.
 | Table I.Primer sequences for reverse
transcription-quantitative PCR. |
Table I.
Primer sequences for reverse
transcription-quantitative PCR.
| Gene | Forward
(5′-3′) | Reverse
(5′-3′) |
|---|
| Human-GAPDH |
GGAGCGAGATCCCTCCAAAAT |
GGCTGTTGTCATACTTCTCATGG |
| Human-IL-1α |
TGGTAGTAGCAACCAACGGGA |
ACTTTGATTGAGGGCGTCATTC |
| Human-IL-1β |
CAGAAGTACCTGAGCTCGCC |
GGTCGGAGATTCGTAGCTGG |
| Human-IL-6 |
CCTGAACCTTCCAAAGATGGC |
TTCACCAGGCAAGTCTCCTCA |
| Human-IL-23 |
ATGTTCCCCATATCCAGTGTG |
GCTCCCCTGTGAAAATATCCG |
| Human-S100A8 |
AGTGTCCTCAGTATATCA |
CATCTTTATCACCAGAATG |
| Human-S100A9 |
CAACACCTTCCACCAATAC |
TCATTCTTATTCTCCTTCTTGAG |
| Human-K1 |
AGAGTGGACCAACTGAAGAGT |
ATTCTCTGCATTTGTCCGCTT |
| Human-K6 |
CTGGAGGCATCCAAGAGGTCA |
GCAGGGTCCACTTTGTTTCCA |
| Human-K10 |
TCCTACTTGGACAAAGTTCGGG |
CCCCTGATGTGAGTTGCCA |
| Human-K16 |
CAGCGAACTGGTACAGAGCA |
GTTCTCCAGGGATGCTTTCA |
| Human-K17 |
GGTGGGTGGTGAGATCAATGT |
CGCGGTTCAGTTCCTCTGTC |
| Human-TGM1 |
GCACCACACAGACGAGTATGA |
GGTGATGCGATCAGAGGATTC |
Western blot analysis
HaCaT Cells were lysed using RIPA lysis buffer
(TransGen Biotech Co., Ltd.) containing phenylmethylsulfonyl
fluoride as a protease inhibitor. Protein concentrations were
quantified with the Omni-Easy™ Ready-to-Use BCA Protein Assay kit
(Epizyme; Ipsen Pharma) according to the manufacturer's protocol.
Equal amounts of total protein (15–20 µg/lane) were separated by
10–15% SDS-PAGE and transferred to PVDF membranes. The membranes
were blocked with blocking buffer (5% skimmed milk powder in TBST)
for 1 h at room temperature and subsequently incubated overnight at
4°C with primary antibodies, including anti-TGM1 (1:1,000; cat. no.
12912-3-AP; Proteintech Group, Inc.) and anti-β-actin (1:1,000,
cat. no. YT0099; ImmunoWay Biotechnology Company).
Following three 10-min washes with TBS with 0.1%
Tween-20, membranes were incubated with HRP-conjugated secondary
antibodies (1:2,000) at room temperature for 1 h. Protein bands
were then visualized with ECL reagent (cat. no. SQ201; EpiZyme,
using an enhanced chemiluminescence detection system (Bioworld
Technology, Inc.), and images were acquired using a Tanon 5200
chemiluminescence imaging system (Tanon Science and Technology Co.,
Ltd.). The gray values were analyzed using ImageJ (version 1.54,
imagej.net/ij/).
Statistical analysis
Statistical analyses were performed using GraphPad
Prism software 8.0 (Dotmatics). Data are presented as the mean ± SD
for at least three individual experiments. The statistical
significance of differences was determined using the unpaired,
two-tailed Student's t-test. Other statistical analyses in the
present study were performed using R software 4.3.0
(r-project.org/). P<0.05 was considered to indicate a
statistically significant difference.
Results
Identification of DEGs
The ‘limma’ R package retrieved 1,845 DEGs from the
GSE30999 dataset based on P<0.05 and FC>2 or FC<-2,
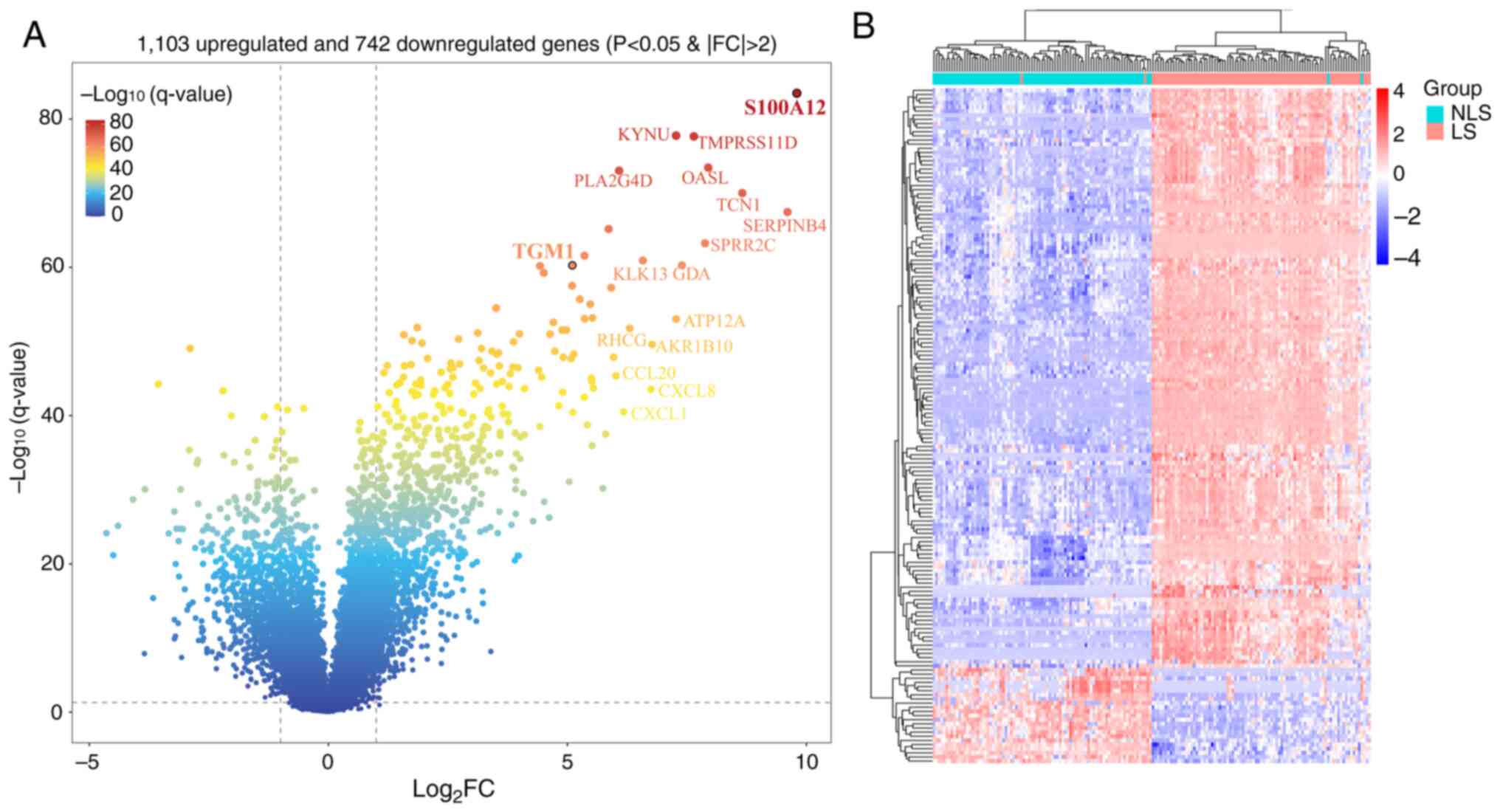
including 1,103 upregulated and 742 downregulated genes (Fig. 1A). Most of the significant DEGs
were upregulated in LS of psoriasis compared with NLS. In addition,
163 significant DEGs were selected based on
|log2FC|>3 and used for subsequent analysis (Fig. 1B).
Functional enrichment analysis of the
gene set in the GSE30999 dataset
To identify pathways enriched in the GSE30999
dataset within the context of psoriasis, GSEA was conducted. To
avoid disturbance from unperturbed genes, the present study
selected the top 10,000 genes from a total of 19,099 probes in the
GSE30999 dataset based on descending order of |log(FC)| values for
GSEA. GSEA showed that a number of upregulated pathways, including
‘Cytokine-JAK-STAT signaling pathway’, ‘DNA replication licensing’,
‘Origin unwinding and elongation’ and ‘Pre-IC formation’, and
downregulated pathways, including ‘ACTH/cortisol signaling
pathway’, ‘Keap1-Nrf2 signaling pathway’, ‘RTK-PLCG-ITPR signaling
pathway’ and ‘TSH-TG signaling pathway’, were enriched in the
GSE30999 dataset (Fig. 2).
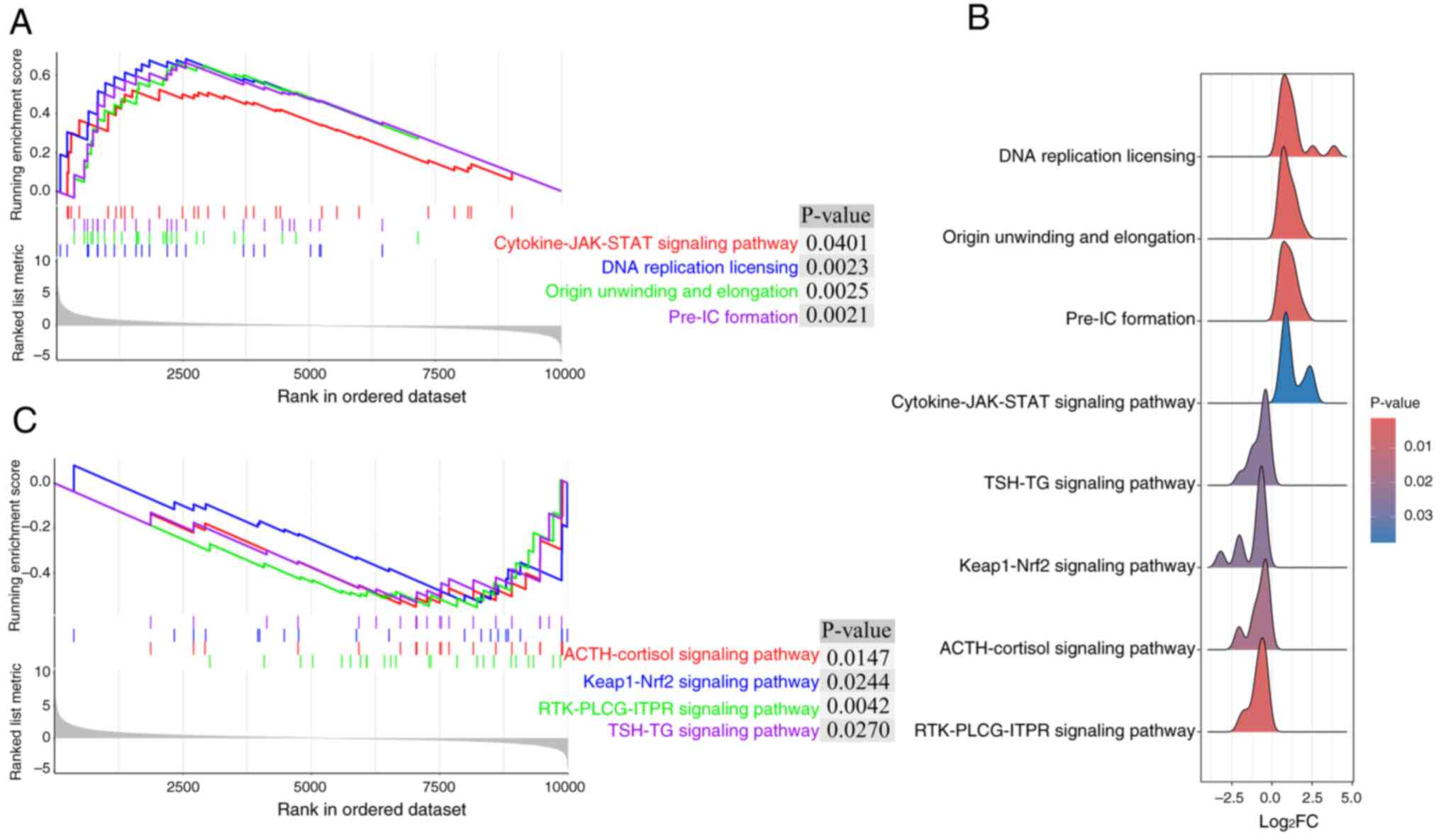 | Figure 2.Gene Set Enrichment Analysis of the
top 10,000 differentially expressed genes in the GSE30999 dataset.
(A) A total of four upregulated pathways were enriched in the
GSE30999 dataset. (B) A total of four significant downregulated
pathways were enriched in the GSE30999 dataset. (C) Ridgeline plots
of the four upregulated pathways and four downregulated pathways
enriched in the GSE30999 dataset. JAK, Janus kinase; ACTH,
adrenocorticotropic hormone; Keap1, Kelch-like ECH-associated
protein 1; Nrf2, nuclear factor erythroid 2-related factor 2; PLCG,
phospholipase Cγ; ITPR, inositol 1,4,5-trisphosphate receptor; TSH,
thyroid-stimulating hormone; TG, thyroglobulin. RTK, receptor
tyrosine kinase; Pre-IC: Pre-Initiation Complex. |
Functional annotation of 163 DEGs
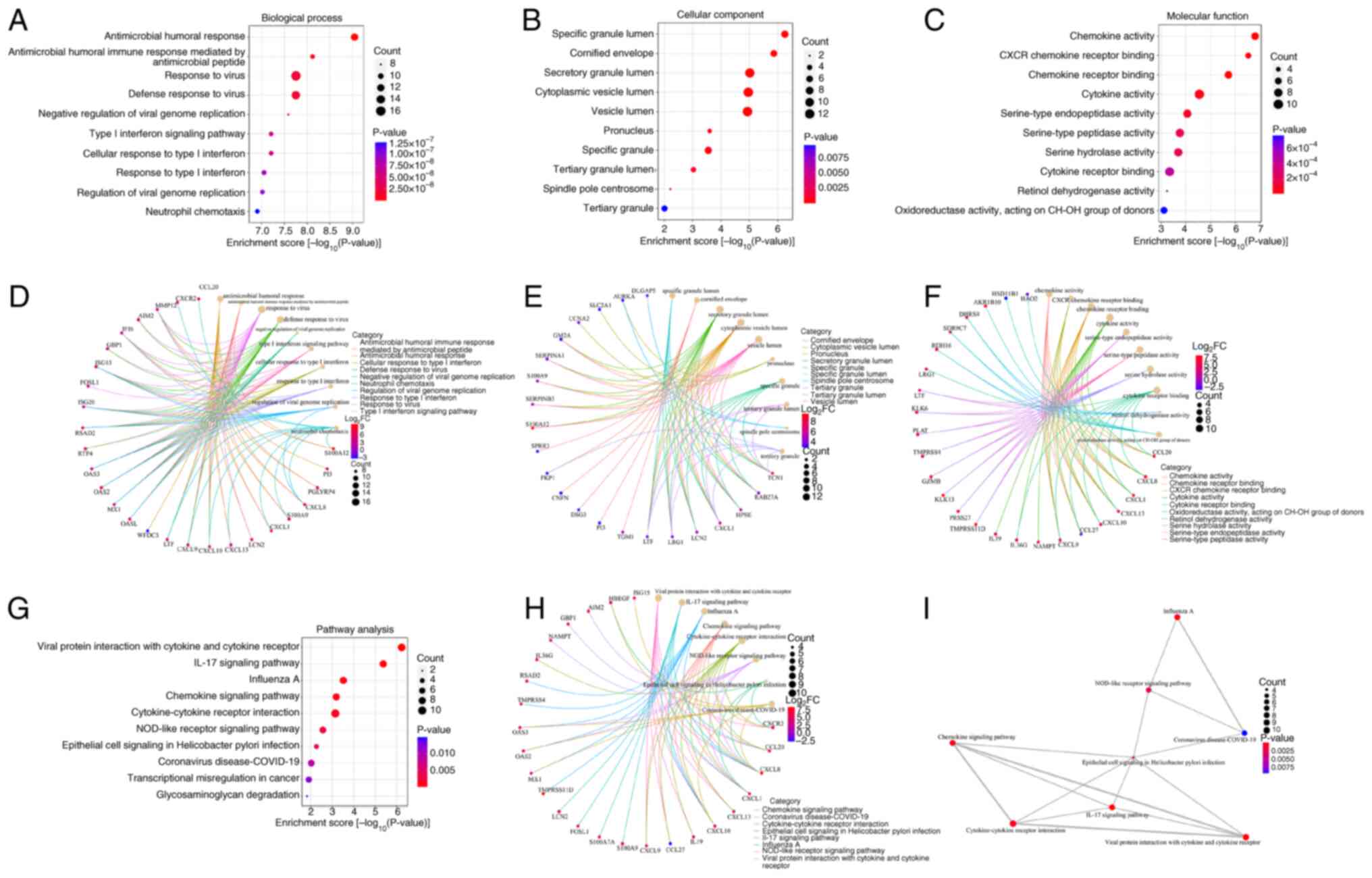
The present study used GO enrichment analysis to
obtain a more precise functional overview of the top 163 DEGs. A
total of 284 BP terms, 17 CC terms and 50 MF terms were identified,
and the top 10 GO terms of each category are shown in Fig. 3. PBH was obtained via
Benjamini and Hochberg (BH) correction. The top 10 GO BP terms were
‘antimicrobial humoral response’
(PBH=2.11×10−6), ‘antimicrobial humoral
immune response mediated by antimicrobial peptide’
(PBH=8.87×10−6), ‘response to virus’
(PBH=1.04×10−5), ‘defense response to
virus’(PBH=1.04×10−5), ‘negative regulation
of viral genome replication’ (PBH=1.22×10−5),
‘type I interferon signaling pathway’
(PBH=2.07×10−5), ‘cellular response to type I
interferon’ (PBH=2.07×10−5), ‘response to
type I interferon’ (PBH=2.52×10−5),
‘regulation of viral genome replication’
(PBH=2.52×10−5) and ‘neutrophil chemotaxis’
(PBH=2.94×10−5; Fig. 3A and D). The most significant GO CC
and MF terms were ‘specific granule lumen’
(PBH=1.29×10−4) and ‘chemokine activity’
(PBH=5.97×10−5), respectively (Fig. 3B, C, E and -F).
A total of 21 pathways were identified using KEGG
analysis and the top 10 pathways are shown in Fig. 3. These were ‘Viral protein
interaction with cytokine and cytokine receptor’
(PBH=9.67×10−5), ‘IL-17 signaling pathway’
(PBH=3.53×10−4), ‘Influenza A’
(PBH=1.67×10−2), ‘Chemokine signaling
pathway’ (PBH=2.41×10−2), ‘Cytokine-cytokine
receptor interaction’ (PBH=2.41×10−2),
‘NOD-like receptor signaling pathway’
(PBH=7.56×10−2), ‘Epithelial cell signaling
in Helicobacter pylori infection’
(PBH=1.29×10−1), ‘Coronavirus
disease-COVID-19’ (PBH=1.97×10−1),
‘Transcriptional misregulation in cancer’
(PBH=2.31×10−1) and ‘Glycosaminoglycan
degradation’ (PBH=2.53×10−1) (Fig. 3G-I).
Selection of signature DEGs using
machine learning techniques
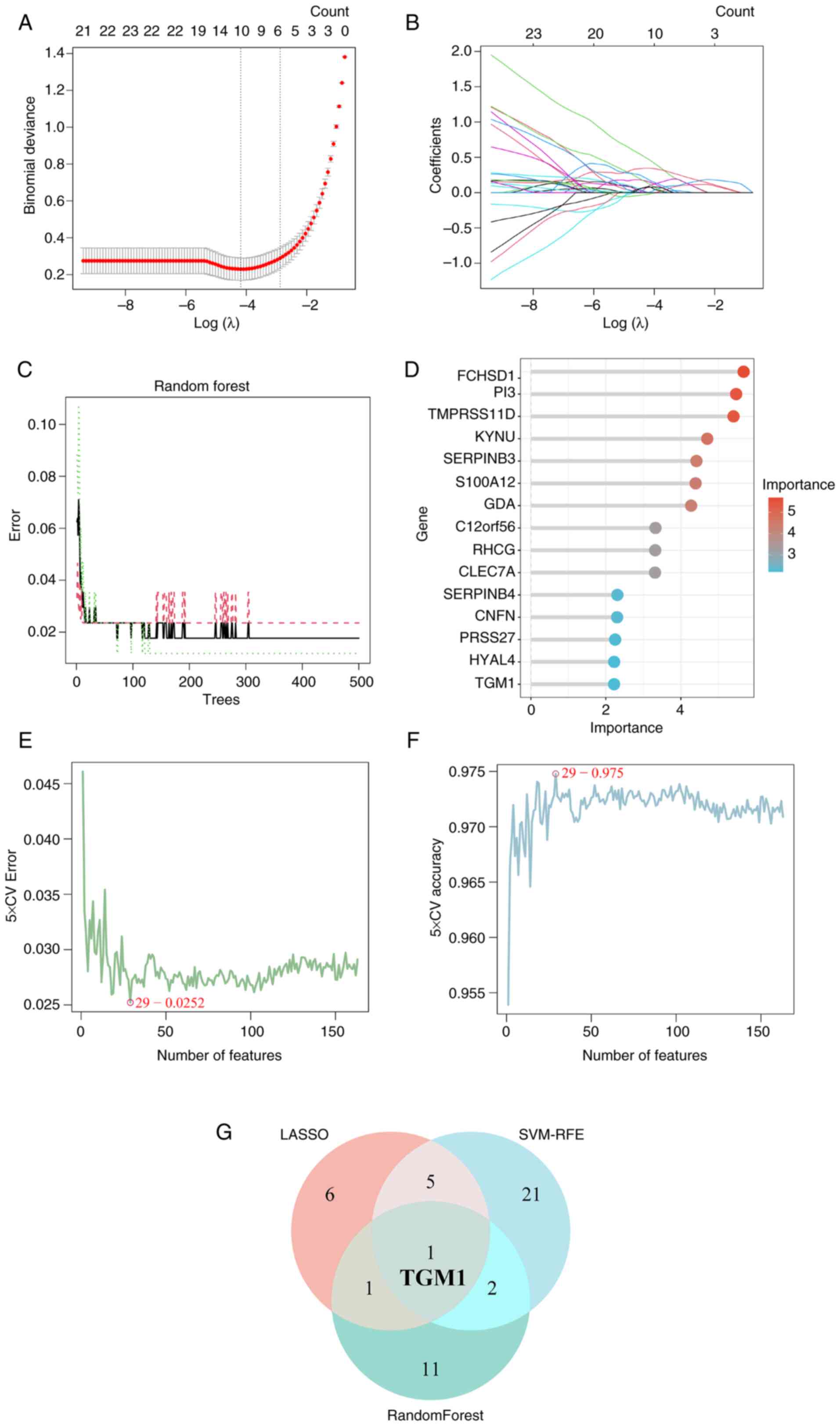
The present study utilized three machine learning
methods (LASSO, RF and SVM-RFE) to extract the possible diagnostic
DEGs from a total of 163 DEGs. The LASSO regression algorithm
identified 13 core genes with optimal λ values (λ.min, 0.01264872)
in the GSE30999 dataset (Fig. 4A and
B), which were SYNCRIP, VNN1, GBP1, TGM1, TMPRSS11D,
CYP2C18, PRKCQ, RSAD2, FUT3, HAO2, NLRP2, TMEM86A and
LYPD5. Using a RF algorithm, 35 genes with an importance
score >1.0 were identified, and the top 15 in importance were
chosen as signature DEGs (Fig. 4C and
D). Through SVM-RFE analysis, a total of 29 DEGs were
identified with an accuracy of 0.975 and an error rate of 0.0252
(Fig. 4E and F). Ultimately, these
prediction methods identified TGM1. Therefore, TGM1
was employed as a diagnostic marker of psoriasis in later
investigations (Fig. 4G).
Identification of diagnostic markers
in psoriasis
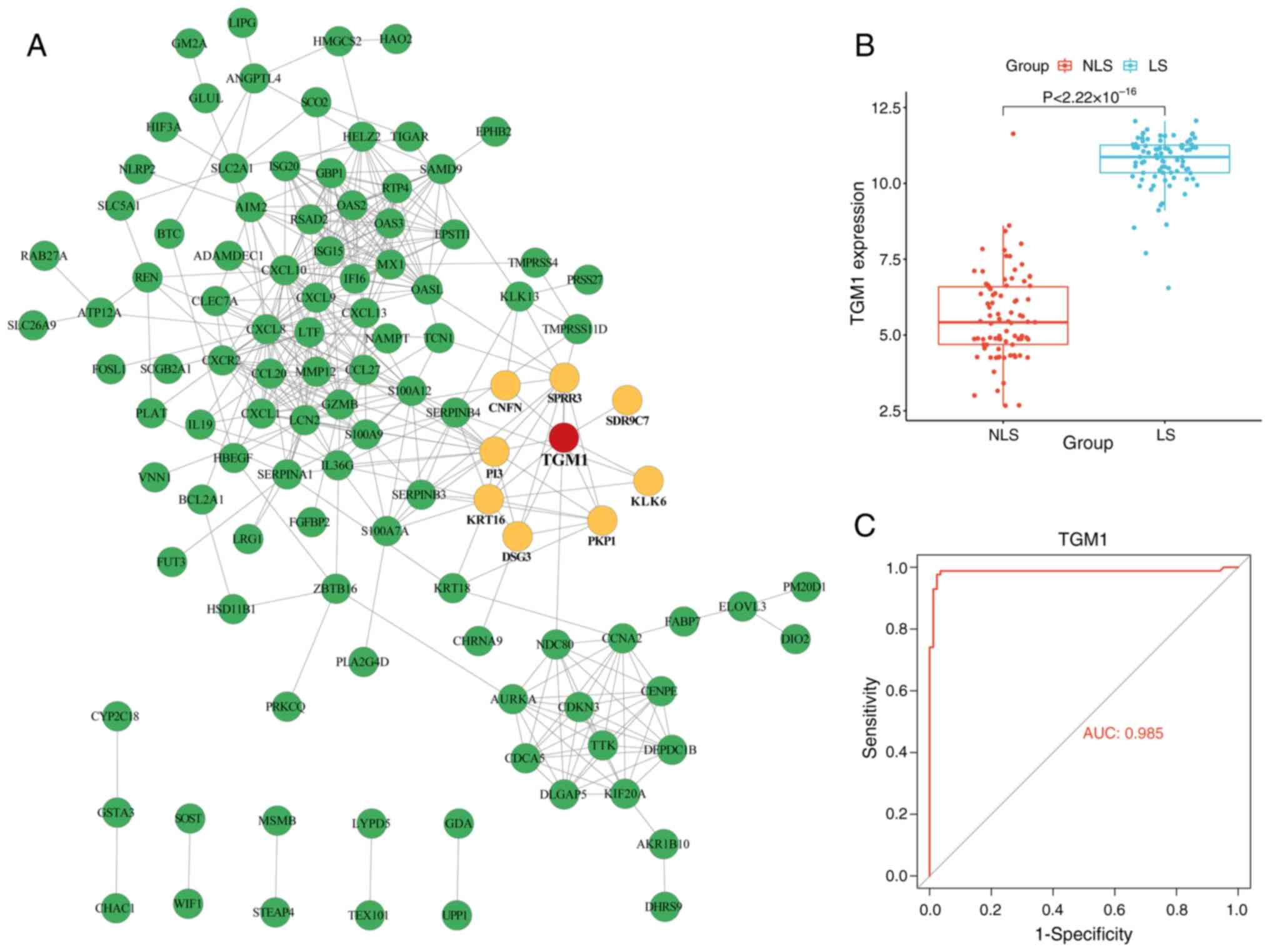
PPIs are important in numerous biological pathways.
The majority of proteins serve their functions through interactions
with a large number of other proteins. Therefore, the 163
identified DEGs were submitted to the STRING database to acquire
their interaction data with TGM1. Of the 163 DEGs, a total of 52
genes were disconnected, and thus, removed from the PPI network.
After removing the 52 disconnected genes, the final PPI network
contained 111 nodes and 388 edges. Among the 111 connected
proteins, CNFN, SPRR3, SDR9C7, PI3, KRT16, DSG3, PKP1 and KLK6 were
directly connected with TGM1 (Fig.
5A). Additionally, in comparison with the NLS tissue, the LS
tissues of psoriasis exhibited a significant increase in TGM1
expression (Fig. 5B). ROC curves
highlighted the robust diagnostic potential of TGM1 as biomarkers
for psoriasis (Fig. 5C).
Identification of potential pathways
of TGM1 by single-gene GSEA
Single-gene GSEA is a bioinformatics tool used to
investigate the biological pathways enriched by genes associated
with a specific gene, and thus, to reveal its particular biological
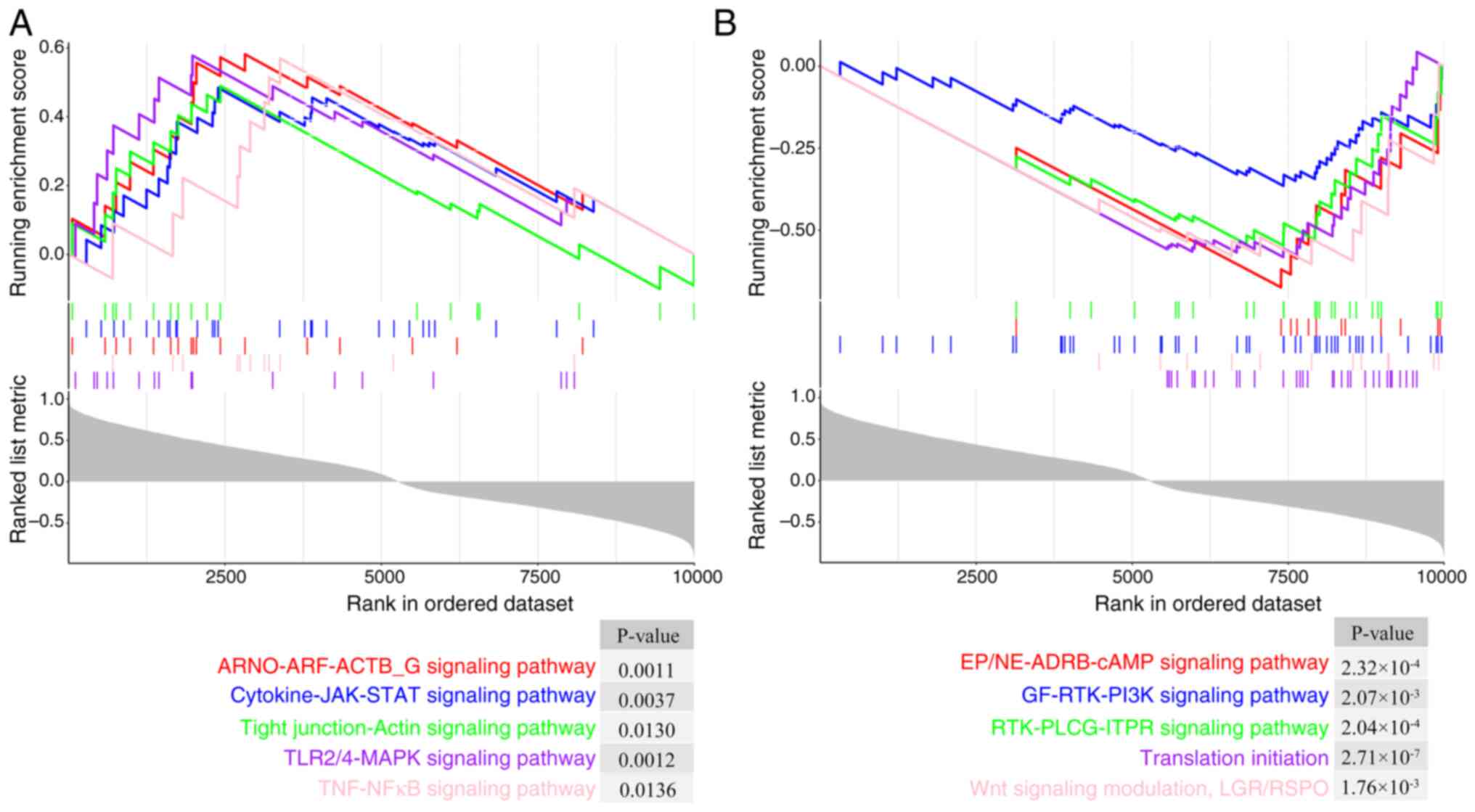
activity. Single-gene GSEA of TGM1 was conducted on LS samples, and
revealed that TGM1 was primarily enriched in upregulated pathways,
including the ‘ARNO-ARF-ACTB_G signaling pathway’ [false discovery
rate (FDR), 0.0062], ‘Cytokine-JAK-STAT signaling pathway’ (FDR,
0.0140), ‘Tight junction-Actin signaling pathway’ (FDR, 0.0320),
‘TLR2/4-MAPK signaling pathway’ (FDR, 0.0064) and ‘TNF-NFκB
signaling pathway’ (FDR, 0.0325) (Fig.
6A). Downregulated pathways included the ‘EP/NE-ADRB-cAMP
signaling pathway’ (FDR, 1.71×10−3), ‘GF-RTK-PI3K
signaling pathway’ (FDR, 9.21×10−3), ‘RTK-PLCG-ITPR
signaling pathway’ (FDR, 1.57×10−3), ‘Translation
initiation’ (FDR, 2.29×10−5) and ‘Wnt signaling
modulation, LGR/RSPO’ (FDR, 8.13×10−3) (Fig. 6B).
Validation of signature genes
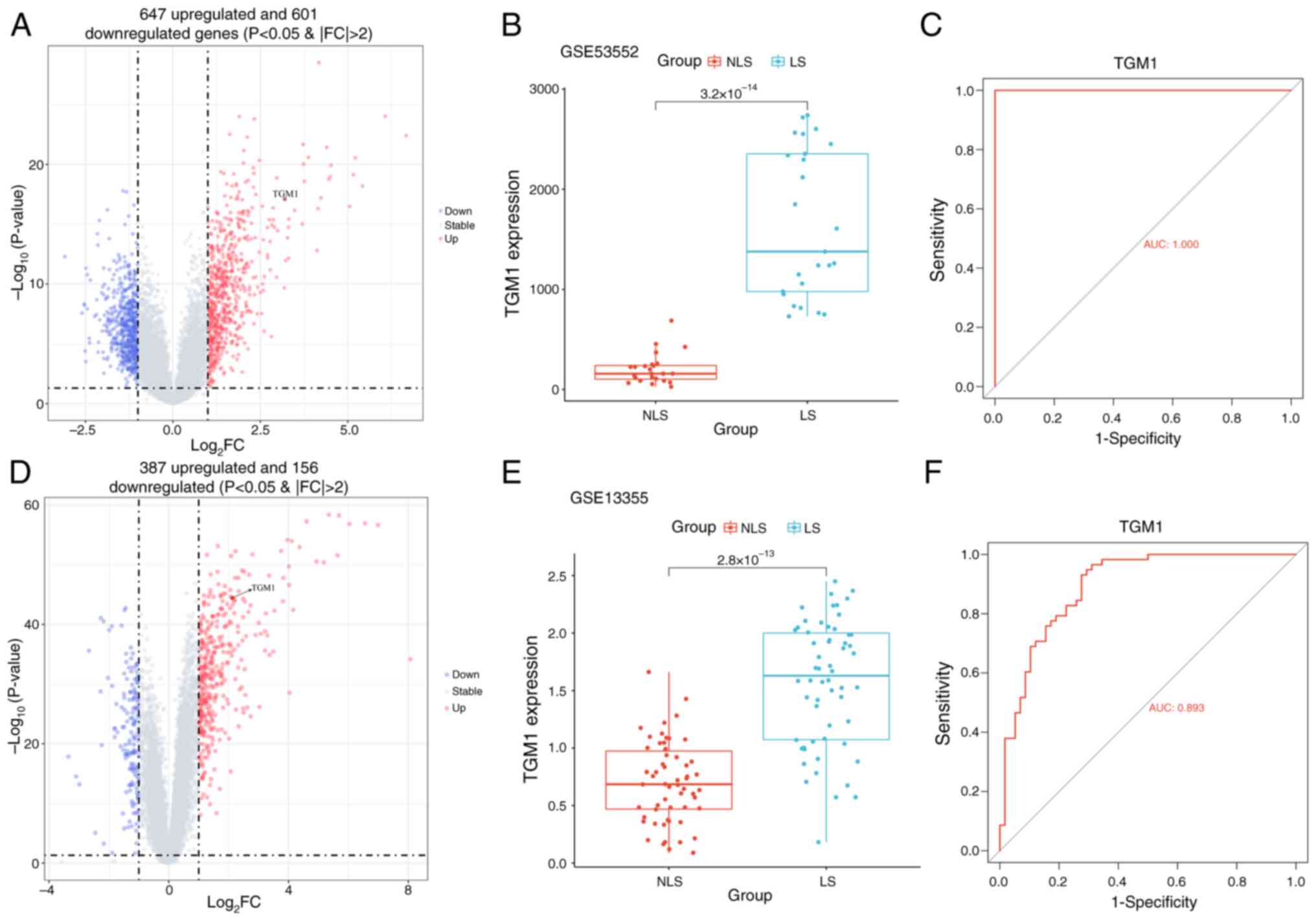
The present study first examined TGM1 expression in
the GSE53552 and GSE13355 datasets. The results showed that TGM1
expression was significantly increased in LS tissues of psoriasis
compared with the NLS tissues in the GSE53552 (Fig. 7A and B) and GSE13355 (Fig. 7D and E) datasets. Additionally, the
diagnostic efficacy of TGM1 was also validated using the GSE53552
and GSE13355 datasets, which showed a high associated value with
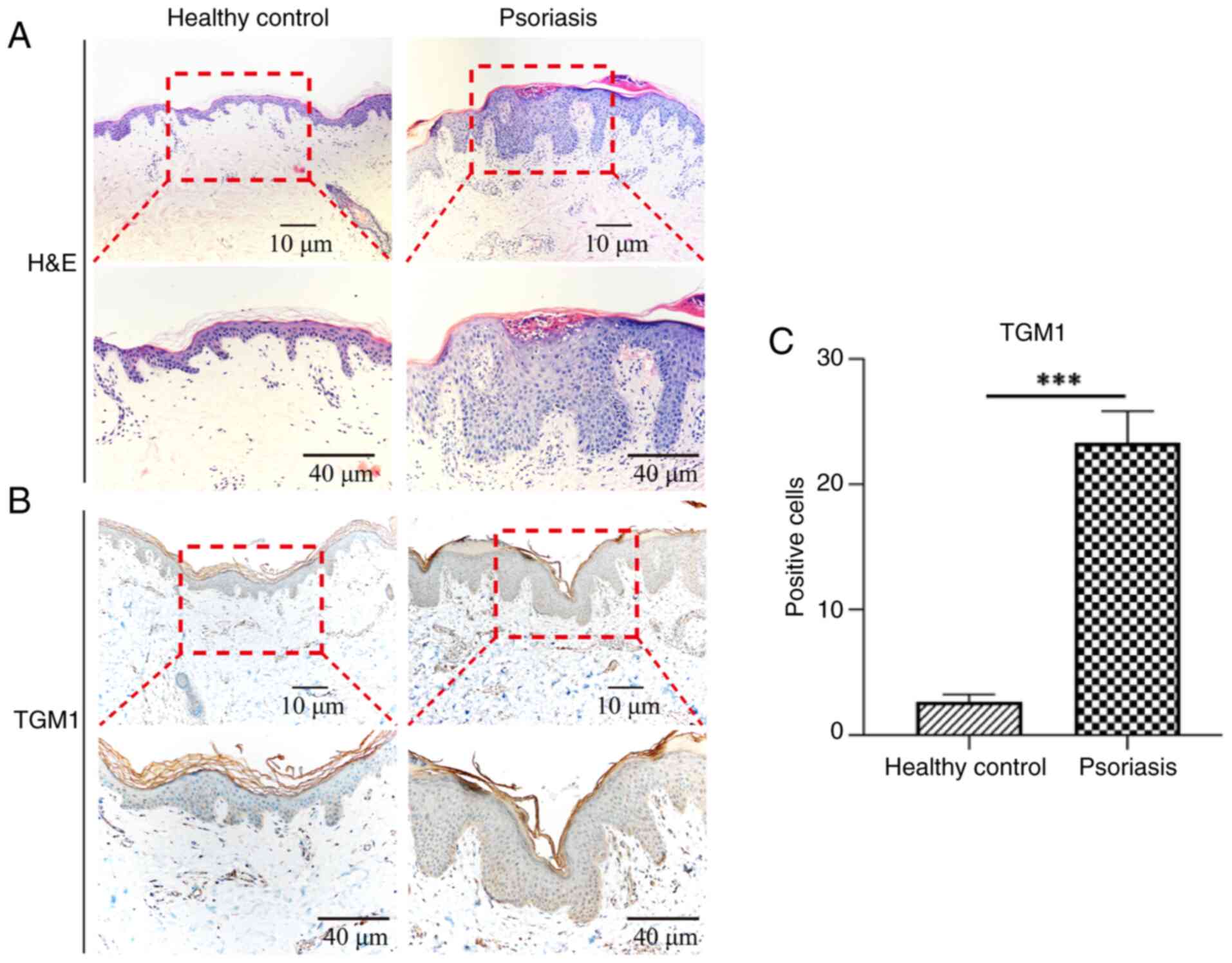
AUC values of 1.000 and 0.893, respectively (Fig. 7C and F). Finally, IHC was performed
to evaluate TGM1 protein expression levels in affected areas of
patients with psoriasis. IHC showed that TGM1 expression exceeded
normal levels in psoriatic lesions (Fig. 8).
Immune cell infiltration in
psoriasis
The immune cell infiltration results based on
GSE30999 showed that plasma, naïve CD4 T cells, activated memory
CD4 T cells, follicular T helper cells, γδT cells, activated NK
cells, M1 macrophages, M2 macrophages, resting dendritic cells,
activated dendritic cells, resting Mast cells, eosinophils and
neutrophils were significantly increased in the LS group compared
with the NLS group, while activated natural killer cells and
resting mast cells were significantly decreased (Fig. 9A). The correlation study revealed a
significant positive correlation between activated dendritic and
follicular helper T cells as well as between memory B cells and
naïve CD4 T cells (Fig. 9B). A
significant negative correlation was observed between activated
dendritic cells and resting mast cells as well as between
follicular T helper cells and resting mast cells (Fig. 9B). In order to further validate the
association between TGM1 expression and the immune component, the
present study employed single-gene immune infiltration analysis.
The results showed association between TGM1 expression and
eosinophils, activated dendritic cells and follicular T-helper
cells (Fig. 9C-F). There was an
association between TGM1 expression and resting mast cells
(Fig. 9C and G).
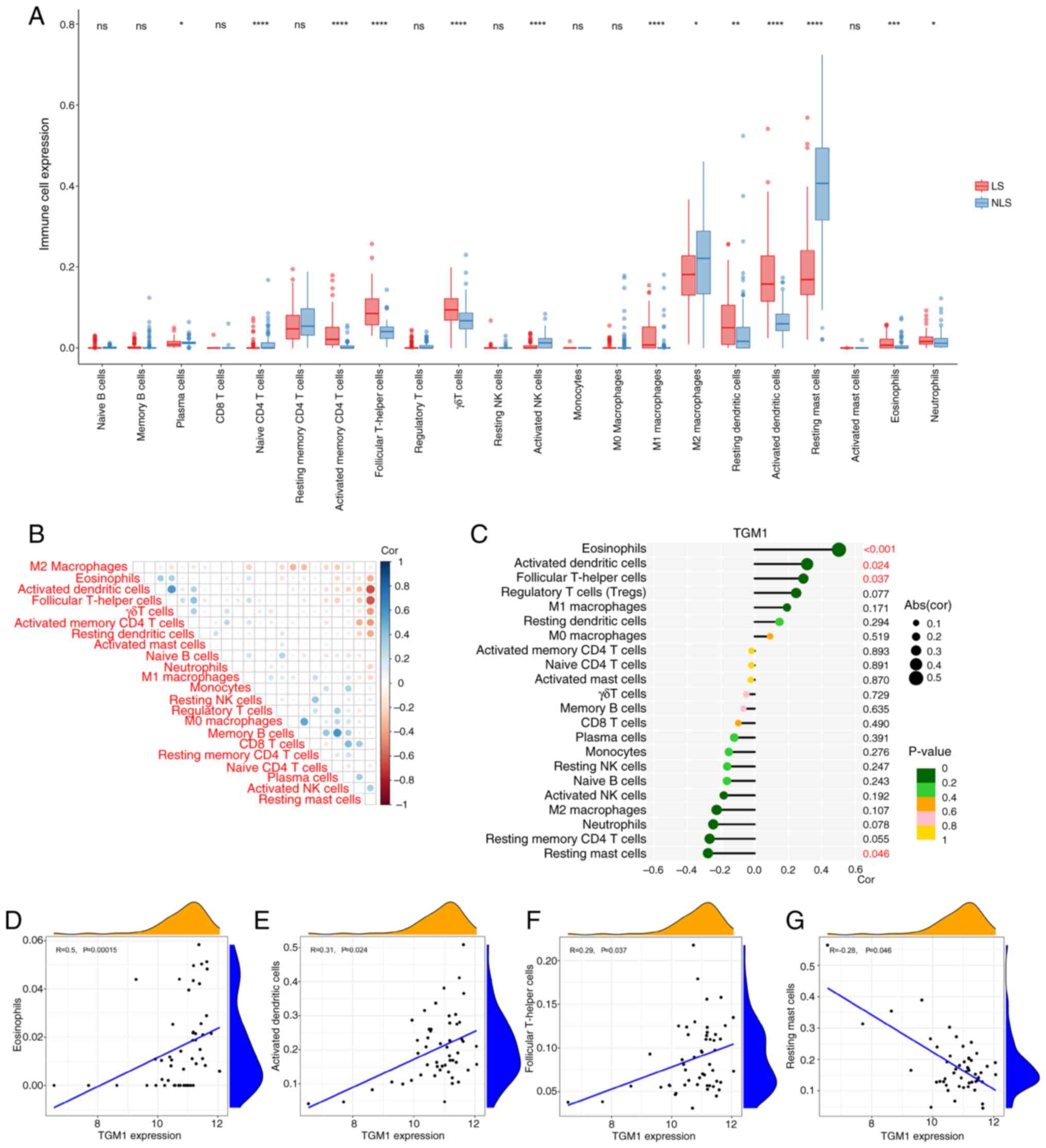 | Figure 9.Correlation of TGM1 expression with
infiltrating immune cells in psoriasis. (A) Expression of immune
cells between LS and NLS. (B) Correlation between 22 types of
infiltrating immune cells between LS and NLS. (C) Lollipop graph
showing the correlation between TGM1 expression and 22 types of
infiltrating immune cells in psoriasis. Scatter plots showing that
(D) eosinophils, (E) activated dendritic cells, (F) follicular T
helper cells and (G) resting mast cells were significantly
correlated with TGM1 expression. *P<0.05, **P<0.01,
***P<0.001 and ****P<0.0001 vs. NLS. NLS, non-lesional skin;
ns, not significant; TGM1, transglutaminase 1; NK, natural killer;
abs, absolute; cor, correlation coefficient. |
TGM1 aggravates the inflammatory
response and keratinocyte differentiation
To validate the prediction results of machine
learning techniques, transfection of HaCaT cells was performed to
support the effect of TGM1. The present study first performed an
overexpression experiment. The mRNA and protein expression levels
of TGM1 were significantly increased after transfection with the
overexpression plasmid compared with those in the negative control
group (Fig. 10A and B).
Additionally, mRNA expression levels of some inflammatory
cytokines, such as IL-1α, IL-1β, IL-6 and IL-23, were significantly
increased after TGM1 overexpression (Fig. 10C). Finally, the expression levels
of several markers of keratinocyte differentiation were also
increased following TGM1 overexpression, of which S100 calcium
binding protein (S100)A8, keratin (K)6, K10 and K17 showed
significant increases in expression compared with the control group
(Fig. 10D and E).
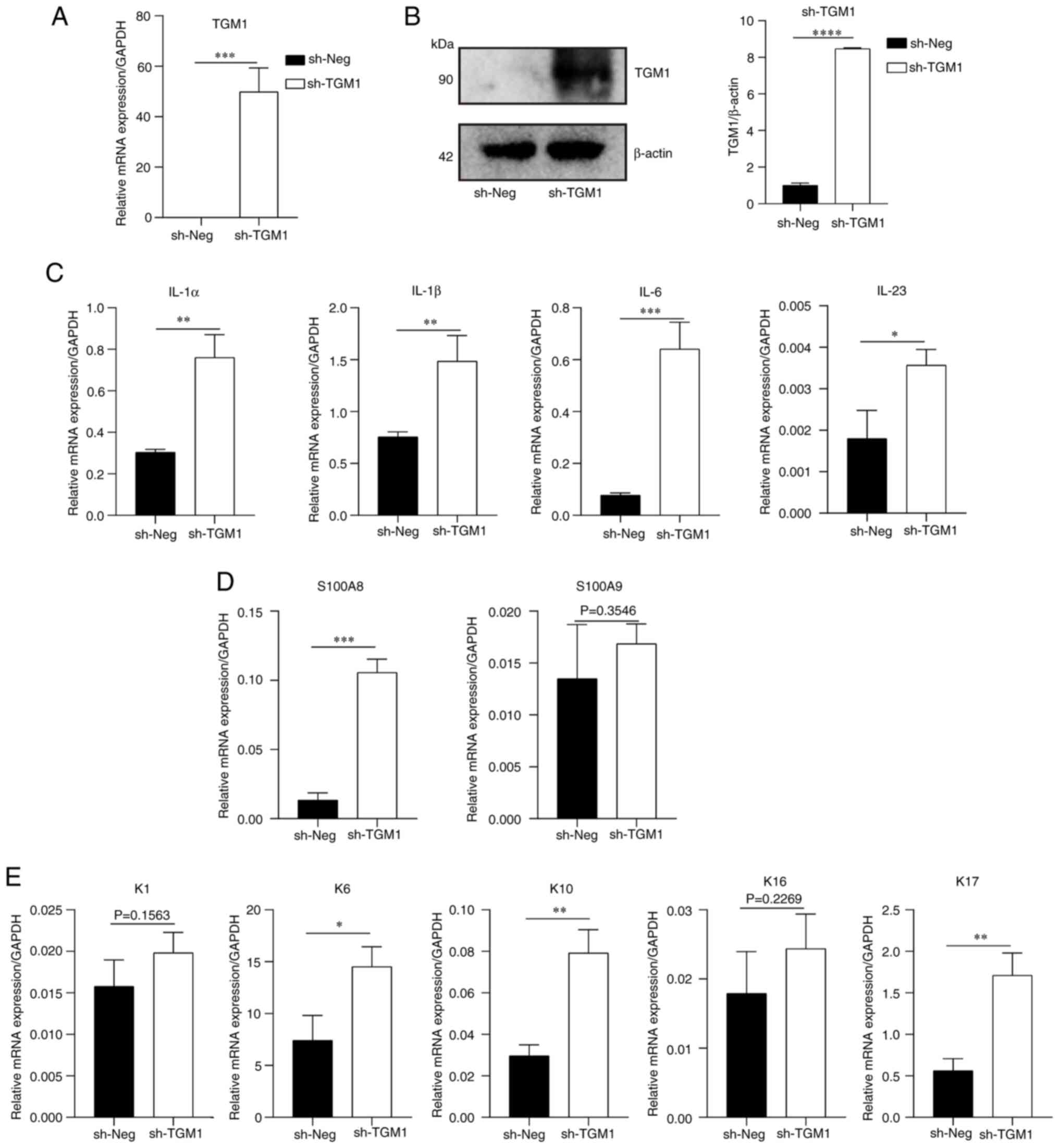 | Figure 10.mRNA expression levels of
inflammatory cytokines and differentiation markers of keratinocytes
after TGM1 overexpression. TGM1 (A) mRNA and (B) protein expression
following sh-TGM1 treatment. (C) mRNA expression levels of
inflammatory factors IL-1α, IL-1β, IL-6 and IL-23. mRNA expression
levels of (D) S100A8 and S100A9, as well as (E) K1, K6, K10, K16
and K17 after sh-TGM1 treatment. *P<0.05, **P<0.01,
***P<0.001 and ****P<0.0001. Neg, negative control; S100,
S100 calcium binding protein; K, keratin; TGM1, transglutaminase
1. |
To further verify the effect of TGM1, knockdown
experiments were subsequently performed using siRNA. As shown in
Fig. 11A and B, TGM1 mRNA and
protein expression levels were significantly reduced by si-TGM1
transfection compared with those in the negative control group.
mRNA expression levels of inflammatory cytokines, such as IL-1β,
was downregulated following TGM1 downregulation at 12 h (Fig. 11C). Similarly, the expression
levels of the keratinocyte differentiation markers S100A8, S100A9,
K1 were also decreased at 12 h (Fig.
11D and E).
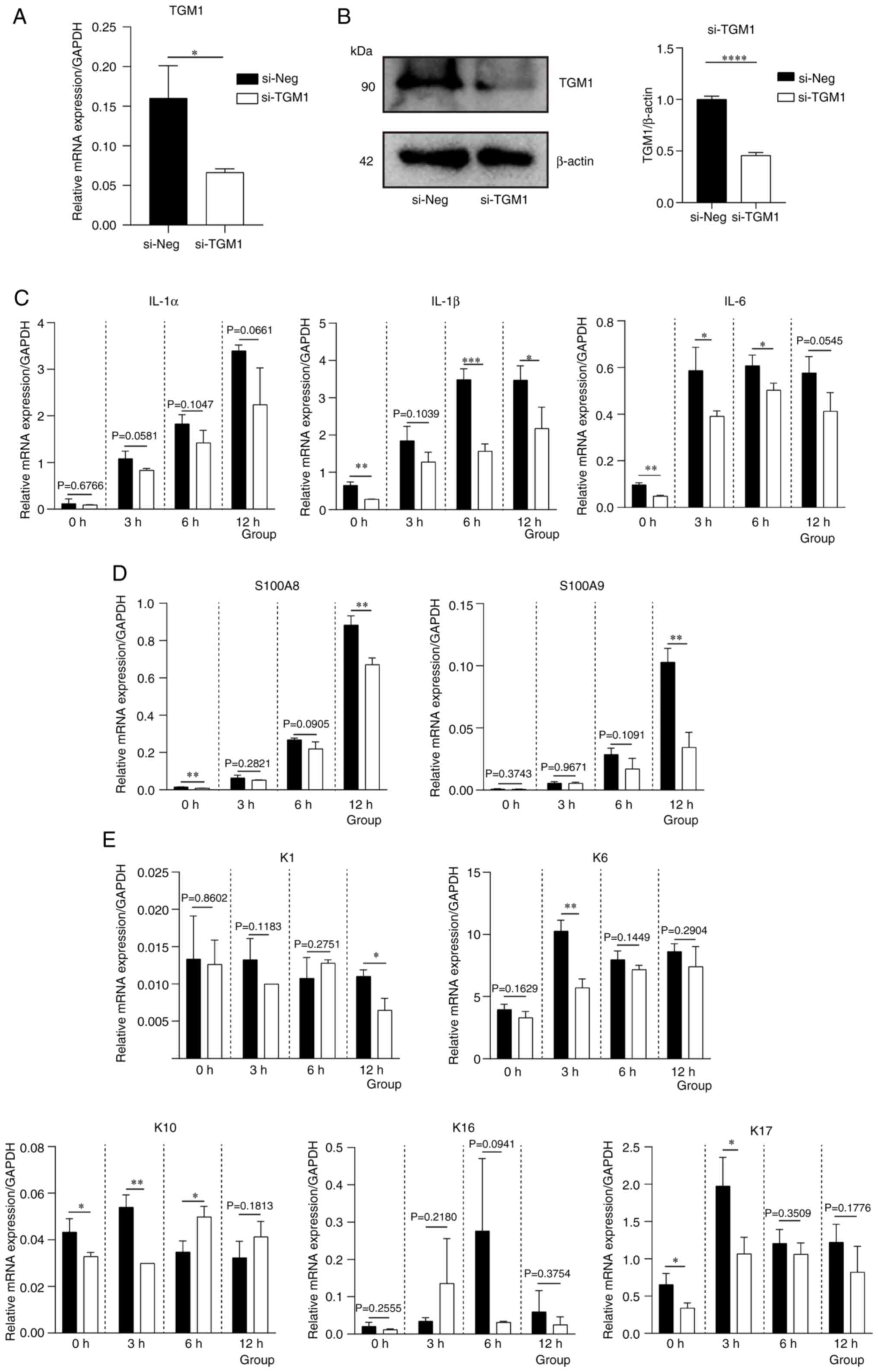 | Figure 11.mRNA expression levels of
inflammatory cytokines and differentiation markers of keratinocytes
after TGM1 knockdown. TGM1 (A) mRNA and (B) protein expression
after si-TGM1 transfection. (C) mRNA expression levels of
inflammatory factors IL-1α, IL-1β and IL-6. mRNA expression levels
of (D) S100A8 and S100A9, as well as (E) K1, K6, K10, K16 and K17
following M5 stimulation after si-TGM1 transfection. *P<0.05,
**P<0.01, ***P<0.001 and ****P<0.0001 vs. si-Neg. si,
small interfering RNA; Neg, negative control; TGM1,
transglutaminase 1; S100, S100 calcium binding protein; K,
keratin. |
Discussion
Psoriasis is a notable public health issue with
complex pathophysiology, with clinically relevant biomarkers
(4). Despite advancements, the
etiological mechanisms of psoriasis remain yet to be fully
elucidated, necessitating systematic exploration of disease
pathways to establish novel theoretical frameworks for early
diagnosis and targeted interventions.
The present study examined psoriasis-related
microarray data from the GEO database. The present study initially
performed GSEA on the GSE30999 dataset, uncovering significant
enrichment of the ‘Cytokine-JAK-STAT signaling pathway’ and
‘Keap1-Nrf2 signaling pathway’. The JAK/STAT pathway, a recognized
regulator of inflammatory and autoimmune disorders (29), facilitates the transcriptional
activation of psoriasis-associated cytokines such as IL-17 and
IL-23, driving keratinocyte hyperproliferation and disease symptoms
(25). Clinical evidence
additionally indicates that pharmacological inhibition of this
pathway reduces cutaneous cytokine production (30,31).
The Keap1/Nrf2 axis, an important regulator of redox homeostasis
and cutaneous barrier integrity (32), exhibits therapeutic significance in
psoriasis, with Nrf2 activators proving effective in
moderate-to-severe cases (33,34).
The enhancement of these pathways supports the biological validity
of the results of the present study.
Subsequent differential expression analysis
identified 163 significant DEGs with |log2FC|>3 from
1,845 candidates in the GSE30999 dataset. GO and KEGG functional
enrichment analyses identified two important pathways (‘Viral
protein interaction with cytokine and cytokine receptor’ and ‘IL-17
signaling pathway’). The etiology of psoriasis entails T
cell-mediated cytokine cascades, including cytokines such as TNF-α,
IL-17A, IL-23 and IL-36, which dysregulate keratinocyte dynamics
via receptor-mediated signaling (35). IL-17 receptor activation markedly
stimulates STAT3-dependent keratinocyte proliferation, intensifying
inflammatory responses (36). The
IL-17 family, mostly produced by T helper 17 cells, displays
isoform-specific functions in chronic inflammation and psoriasis
progression, with IL-17A exhibiting the most notable
pathophysiological impact (37–40).
Targeted biologics, such as secukinumab and ixekizumab, effectively
neutralize IL-17A, thus modulating cytokine networks and enhancing
clinical outcomes in moderate-to-severe psoriasis (41).
The present study utilized three machine learning
methods, LASSO, RF and SVM-RFE, to identify diagnostic biomarkers,
ultimately identifying TGM1 as a possible biomarker for psoriasis.
TGM1, a calcium-dependent enzyme in the epidermal differentiation
complex, catalyzes ε-(g-glutamyl) lysine crosslinks during
cornified envelope formation, a key process for epidermal barrier
function (42). Barrier
dysfunction, a characteristic of psoriasis and related dermatoses
(43,44), results from cytokine-driven
keratinocyte hyperproliferation (45) and differentiation imbalances
associated with disease severity (46). TGM1 mutations are associated with
laminar ichthyosis (47). However,
the upregulation of TGM1 in psoriatic lesions indicates a poorly
understood pathogenic function (48). TGM1 exhibits diagnostic utility in
multiple malignancies, such as adrenocortical carcinoma and bladder
cancer (49,50), underscoring its potential as a
dual-purpose biomarker in psoriasis.
Single-gene GSEA and immune infiltration analyses
further clarified the molecular roles of TGM1. Upregulated
pathways, including the ‘cytokine-JAK-STAT signaling pathway’,
‘TLR2/4-MAPK signaling pathway’ and ‘TNF-NFκB signaling pathway’,
and downregulated pathways, such as ‘GF-RTK-PI3K signaling pathway’
and ‘Wnt signaling modulation, LGR/RSPO’, correspond with
established psoriatic pathobiology (5). Immune profiling revealed positive
correlations between TGM1 expression and eosinophils, activated
dendritic cells and follicular T-helper cells, and a negative
correlation with resting mast cells, suggesting that TGM1 serves a
role in innate immune dysregulation. Cross-dataset validation using
both the GSE53552 (AUC, 1.000) and GSE13355 (AUC, 0.893) datasets
supported TGM1 upregulation, further validated by IHC revealing
increased protein levels in psoriatic lesions compared with
controls. Finally, cell experiments were conducted to verify the
potential effects of TGM1 on psoriasis and revealed that TGM1 may
aggravate the inflammation response and keratinocyte
differentiation. Collectively, these findings established TGM1 as
an important mediator and therapeutic target in psoriasis.
The present multimodal investigation integrating
bioinformatics, machine learning and experimental validation
identified TGM1 as a functionally significant biomarker in the
pathogenesis of psoriasis. The observed upregulation in affected
skin, pathway linkage and immunological correlations offer
mechanistic insights into disease progression. The findings of the
present study enhance the understanding of psoriatic
pathophysiology and suggest TGM1-targeted strategies for diagnostic
and therapeutic innovation.
Acknowledgements
The authors would like to thank Professor Litao
Zhang and Dr Lin Li (Tianjin Academy of Traditional Chinese
Medicine Affiliated Hospital, Tianjin, China) for providing the wax
blocks of the patients with psoriasis.
Funding
The present work was supported by The Science and Technology
Development Fund of Tianjin Education Commission for Higher
Education (grant no. 2022ZD047).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
PG and MS contributed to the literature search and
study design. JZ, QY, HC and JH analyzed and interpreted data. JL
and QZ assisted with the experiments and wrote the manuscript. BZ
was responsible for project administration and secured funding for
the study. BZ and LH conceived and designed the experiments,
revised the manuscript and confirmed the authenticity of all the
raw data. All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
For patient samples, written informed consent was
obtained from each patient and the study was approved by the Ethics
Committee of Tianjin Academy of Traditional Chinese Medicine
Affiliated Hospital (approval no. LLKY2022-39; Tianjin, China). The
study was performed in accordance with The Declaration of
Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Griffiths CE and Barker JN: Pathogenesis
and clinical features of psoriasis. Lancet. 370:263–271. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Stern RS, Nijsten T, Feldman SR, Margolis
DJ and Rolstad T: Psoriasis is common, carries a substantial burden
even when not extensive, and is associated with widespread
treatment dissatisfaction. J Investig Dermatol Symp Proc.
9:136–139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kurd SK and Gelfand JM: The prevalence of
previously diagnosed and undiagnosed psoriasis in US adults:
Results from NHANES 2003–2004. J Am Acad Dermatol. 60:218–224.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lee EB, Wu KK, Lee MP, Bhutani T and Wu
JJ: Psoriasis risk factors and triggers. Cutis. 102:18–20.
2018.PubMed/NCBI
|
|
5
|
Guo J, Zhang H, Lin W, Lu L, Su J and Chen
X: Signaling pathways and targeted therapies for psoriasis. Signal
Transduct Target Ther. 8:4372023. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Baliwag J, Barnes DH and Johnston A:
Cytokines in psoriasis. Cytokine. 73:342–350. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Michalak-Stoma A, Pietrzak A, Szepietowski
JC, Zalewska-Janowska A, Paszkowski T and Chodorowska G: Cytokine
network in psoriasis revisited. Eur Cytokine Netw. 22:160–168.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Singh R, Koppu S, Perche PO and Feldman
SR: The cytokine mediated molecular pathophysiology of psoriasis
and its clinical implications. Int J Mol Sci. 22:127932021.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kofoed K, Skov L and Zachariae C: New
drugs and treatment targets in psoriasis. Acta Derm Venereol.
95:133–139. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Furtunescu AR, Georgescu SR, Tampa M and
Matei C: Inhibition of the JAK-STAT Pathway in the treatment of
psoriasis: A review of the literature. Int J Mol Sci. 25:46812024.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mavropoulos A, Rigopoulou EI, Liaskos C,
Bogdanos DP and Sakkas LI: The role of p38 MAPK in the
aetiopathogenesis of psoriasis and psoriatic arthritis. Clin Dev
Immunol. 2013:5697512013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang M and Zhang X: The role of
PI3K/AKT/FOXO signaling in psoriasis. Arch Dermatol Res. 311:83–91.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Correa da Rosa J, Kim J, Tian S, Tomalin
LE, Krueger JG and Suárez-Fariñas M: Shrinking the psoriasis
assessment gap: Early Gene-expression profiling accurately predicts
response to Long-Term treatment. J Invest Dermatol. 137:305–312.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ding J, Gudjonsson JE, Liang L, Stuart PE,
Li Y, Chen W, Weichenthal M, Ellinghaus E, Franke A, Cookson W, et
al: Gene expression in skin and lymphoblastoid cells: Refined
statistical method reveals extensive overlap in cis-eQTL signals.
Am J Hum Genet. 87:779–789. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Russell CB, Rand H, Bigler J, Kerkof K,
Timour M, Bautista E, Krueger JG, Salinger DH, Welcher AA and
Martin DA: Gene expression profiles normalized in psoriatic skin by
treatment with brodalumab, a human anti-IL-17 receptor monoclonal
antibody. J Immunol. 192:3828–3836. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Guan S, Xu Z, Yang T, Zhang Y, Zheng Y,
Chen T, Liu H and Zhou J: Identifying potential targets for
preventing cancer progression through the PLA2G1B recombinant
protein using bioinformatics and machine learning methods. Int J
Biol Macromol. 276:1339182024. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wei C, Wei Y, Cheng J, Tan X, Zhou Z, Lin
S and Pang L: Identification and verification of diagnostic
biomarkers in recurrent pregnancy loss via machine learning
algorithm and WGCNA. Front Immunol. 14:12418162023. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z,
Feng T, Zhou L, Tang W, Zhan L, et al: clusterProfiler 4.0: A
universal enrichment tool for interpreting omics data. Innovation
(Camb. 2:1001412021.PubMed/NCBI
|
|
21
|
Ding R, Qu Y, Wu CH and Vijay-Shanker K:
Automatic gene annotation using GO terms from cellular component
domain. BMC Med Inform Decis Mak. 18 (Suppl 5):S1192018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
von Mering C, Huynen M, Jaeggi D, Schmidt
S, Bork P and Snel B: STRING: A database of predicted functional
associations between proteins. Nucleic Acids Res. 31:258–261. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Newman AM, Liu CL, Green MR, Gentles AJ,
Feng W, Xu Y, Hoang CD, Diehn M and Alizadeh AA: Robust enumeration
of cell subsets from tissue expression profiles. Nat Methods.
12:453–457. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Boehncke WH and Schön MP: Psoriasis.
Lancet. 386:983–994. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Langley RG and Ellis CN: Evaluating
psoriasis with psoriasis area and severity index, psoriasis global
assessment, and lattice system Physician's global assessment. J Am
Acad Dermatol. 51:563–569. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang J, Feng X, Zeng J, Zhang S, Zhang J,
Guo P, Yu H, Sun M, Wu J, Li M, et al: Aberrant HO-1/NQO1-Reactive
oxygen Species-ERK signaling pathway contributes to aggravation of
TPA-induced irritant contact dermatitis in Nrf2-deficient mice. J
Immunol. 208:1424–1433. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Banerjee S, Biehl A, Gadina M, Hasni S and
Schwartz DM: JAK-STAT signaling as a target for inflammatory and
autoimmune diseases: Current and future prospects. Drugs.
77:521–546. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kim BH, Na KM, Oh I, Song IH, Lee YS, Shin
J and Kim TY: Kurarinone regulates immune responses through
regulation of the JAK/STAT and TCR-mediated signaling pathways.
Biochem Pharmacol. 85:1134–1144. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Grabarek B, Krzaczyński J, Strzałka-Mrozik
B, Wcisło-Dziadecka D and Gola J: The influence of ustekinumab on
expression of STAT1, STAT3, STAT4, SOCS2, and IL17 in patients with
psoriasis and in a control. Dermatol Ther. 32:e130292019.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Baird L and Yamamoto M: The Molecular
mechanisms regulating the KEAP1-NRF2 pathway. Mol Cell Biol.
40:e00099–20. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Helwa I, Patel R, Karempelis P,
Kaddour-Djebbar I, Choudhary V and Bollag WB: The antipsoriatic
agent monomethylfumarate has antiproliferative, prodifferentiative,
and anti-inflammatory effects on keratinocytes. J Pharmacol Exp
Ther. 352:90–97. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bojanowski K, Ibeji CU, Singh P, Swindell
WR and Chaudhuri RK: A Sensitization-free Dimethyl fumarate
prodrug, isosorbide Di-(Methyl Fumarate), provides a topical
treatment candidate for psoriasis. JID Innov. 1:1000402021.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Afonina IS, Van Nuffel E and Beyaert R:
Immune responses and therapeutic options in psoriasis. Cell Mol
Life Sci. 78:2709–2727. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xu M, Lu H, Lee YH, Wu Y, Liu K, Shi Y, An
H, Zhang J, Wang X, Lai Y and Dong C: An Interleukin-25-Mediated
autoregulatory circuit in keratinocytes plays a pivotal role in
psoriatic skin inflammation. Immunity. 48:787–798.e784. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
McGeachy MJ, Cua DJ and Gaffen SL: The
IL-17 family of cytokines in health and disease. Immunity.
50:892–906. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Baker KJ, Brint E and Houston A:
Transcriptomic and functional analyses reveal a tumour-promoting
role for the IL-36 receptor in colon cancer and crosstalk between
IL-36 signalling and the IL-17/ IL-23 axis. Br J Cancer.
128:735–747. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kirkham BW, Kavanaugh A and Reich K:
Interleukin-17A: A unique pathway in immune-mediated diseases:
Psoriasis, psoriatic arthritis and rheumatoid arthritis.
Immunology. 141:133–142. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Martin DA, Towne JE, Kricorian G, Klekotka
P, Gudjonsson JE, Krueger JG and Russell CB: The emerging role of
IL-17 in the pathogenesis of psoriasis: Preclinical and clinical
findings. J Invest Dermatol. 133:17–26. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ly K, Smith MP, Thibodeaux Q, Reddy V,
Liao W and Bhutani T: Anti IL-17 in psoriasis. Expert Rev Clin
Immunol. 15:1185–1194. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Nemes Z, Marekov LN, Fésüs L and Steinert
PM: A novel function for transglutaminase 1: Attachment of
long-chain omega-hydroxyceramides to involucrin by ester bond
formation. Proc Natl Acad Sci USA. 96:8402–8407. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Baker P, Huang C, Radi R, Moll SB, Jules E
and Arbiser JL: Skin barrier function: The interplay of physical,
chemical, and immunologic properties. Cells. 12:27452023.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Montero-Vilchez T,
Segura-Fernández-Nogueras MV, Pérez-Rodríguez I, Soler-Gongora M,
Martinez-Lopez A, Fernández-González A, Molina-Leyva A and
Arias-Santiago S: Skin barrier function in psoriasis and atopic
dermatitis: Transepidermal water loss and temperature as useful
tools to assess disease severity. J Clin Med. 10:3592021.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Armstrong AW and Read C: Pathophysiology,
clinical presentation, and treatment of psoriasis: A review. JAMA.
323:1945–1960. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Maroto-Morales D, Montero-Vilchez T and
Arias-Santiago S: Study of skin barrier function in psoriasis: The
impact of emollients. Life (Basel). 11:6512021.PubMed/NCBI
|
|
47
|
Farasat S, Wei MH, Herman M, Liewehr DJ,
Steinberg SM, Bale SJ, Fleckman P and Toro JR: Novel
transglutaminase-1 mutations and genotype-phenotype investigations
of 104 patients with autosomal recessive congenital ichthyosis in
the USA. J Med Genet. 46:103–111. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kulski JK, Kenworthy W, Bellgard M, Taplin
R, Okamoto K, Oka A, Mabuchi T, Ozawa A, Tamiya G and Inoko H: Gene
expression profiling of Japanese psoriatic skin reveals an
increased activity in molecular stress and immune response signals.
J Mol Med (Berl). 83:964–975. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wu R, Li D, Zhang S, Wang J, Chen K, Tuo
Z, Miyamoto A, Yoo KH, Wei W, Zhang C, et al: A pan-cancer analysis
of the oncogenic and immunological roles of transglutaminase 1
(TGM1) in human cancer. J Cancer Res Clin Oncol. 150:1232024.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang J, Xiao Y, Wu R and Zhang C: TGM1
could predict overall survival for patients with urinary bladder
cancer. Asian J Surg. 46:5373–5375. 2023. View Article : Google Scholar : PubMed/NCBI
|