Introduction
Osteonecrosis of the femoral head (ONFH) is a
degenerative hip joint disease that worsens over time (1). ONFH is characterized by bone cell
death due to a lack of blood supply, which can lead to joint
collapse and hip dysfunction (1).
The causes of ONFH are complex and involve mechanical injury,
genetic predisposition and biochemical imbalances (2,3).
Among these factors, steroid use is reported in ~51% of ONFH cases
(4). Prolonged or high-dose
steroid therapy markedly increases the risk of ONFH, leading to
steroid-induced ONFH (SONFH) (1).
For example, in patients with systemic lupus erythematosus, the
corticosteroid dosage is positively associated with the occurrence
of ONFH. Daily doses as low as 10 mg have been associated with a
3.6% increase in risk, while doses >40 mg greatly increase the
chance of developing ONFH (5).
Steroids damage bone and blood vessel health through various
mechanisms, including endothelial cell dysfunction and reduced
angiogenesis, ultimately impairing blood flow supply (3,6).
Endothelial cells serve an important role in angiogenesis, as their
growth, migration and tube formation are important for repairing
bone tissue (6,7). When angiogenesis is impaired, bone
regeneration often fails (8).
VEGF, a key factor that promotes angiogenesis, has been shown to
enhance bone repair by stimulating angiogenesis, making it a
potential treatment for ONFH (9–11).
Angiogenesis and osteogenesis are closely linked processes that
occur throughout bone development, maturation, aging and disease
progression (12). Notably,
activation of the VEGF-Notch signaling pathway has been reported to
restore the proliferation of mesenchymal stem cells in aplastic
anemia (13), while the mTOR/AP-1
complex subunit µ-1/VEGF axis regulates endothelial cell growth
(14). Additionally, increasing
hypoxia-inducible factor (HIF)-1α/VEGF levels can help to protect
against bone loss in models of nasal obstruction (15). Therefore, understanding the
molecular mechanisms of angiogenesis and endothelial cell function
is important for advancing treatments for ONFH.
Ubiquitination has become an important
post-translational modification for regulating protein stability,
signal transduction and cellular homeostasis (16–19).
Neural precursor cell expressed developmentally downregulated
protein 4 (NEDD4) is an E3 ubiquitin ligase that facilitates
substrate ubiquitination and degradation, thereby influencing
multiple biological processes such as cell proliferation,
migration, angiogenesis and endothelial cell protection (20–22).
NEDD4 has been demonstrated to promote endothelial cell
proliferation, migration and angiogenesis (23). In addition to its role in
endothelial regulation, NEDD4 also affects macrophage autophagy and
tumor cell proliferation, highlighting its versatile function in
maintaining the cellular balance (21,24).
NEDD4 subfamily influences osteogenesis and bone
cell biology (25). By mediating
PTEN degradation, NEDD4 activates the PI3K/AKT pathway, which
promotes cell proliferation and survival. This pathway also
supports angiogenesis through PTEN inactivation and VEGF
upregulation (26–29). Additionally, NEDD4 boosts osteocyte
proliferation and is associated with improved outcomes in
postmenopausal osteoporosis (30,31).
Overall, these findings emphasize the potential of NEDD4 as a
regulator of both angiogenesis and osteogenesis, making it a
promising target in ONFH therapy. The present study was based on
the concept that NEDD4 may serve a role in SONFH. To explore this,
the present study observed how overexpression of NEDD4 affected the
viability, migration and tube formation of bone microvascular
endothelial cells (BMECs). The present study also investigated how
glucocorticoids influenced NEDD4 expression and promoter
methylation, shedding light on how NEDD4 contributes to
angiogenesis and repair in SONFH.
Materials and methods
Construction of the NEDD4
overexpression (OE) vector
Plasmids pcDNA3.1-NEDD4-3×HA (cat. no. HG-HO284338;
HonorGene Biotechnology Co., Ltd.) and the negative control (NC)
were transformed into competent E. coli cells (AoLu
Biotechnology Co., Ltd.). A total of 2 µl plasmid was combined with
100 µl competent cells, on ice for 30 min. The mixture was
heat-shocked at 42°C for 90 sec and cooled on ice for 3 min.
Subsequently, 700 µl LB (Luria-Bertani) medium (cat. no. L3022;
Thermo Fisher Scientific, Inc.) was added, and the mixture was
incubated at 37°C with shaking at 200 × g for 1 h. This was
followed by plating 60 µl cells onto LB agar containing ampicillin
(Amp) and incubation at 37°C for 12–14 h. A single colony was
selected and inoculated into 20 ml LB medium with 50 µg/ml Amp,
followed by incubation overnight at 37°C. Subsequently, the culture
was scaled up in 50 ml LB/Amp medium and incubated for 12–16 h.
Plasmids were extracted using the HiBind DNA plasmid extraction kit
(Omega Bio-Tek, Inc.) according to the manufacturer's protocol. DNA
concentration was determined using an Implen NanoPhotometer (Implen
GmbH) and the sequence was verified by Sanger sequencing (performed
by AoNuo Gene Technology Co., Ltd.). The purified plasmids were
used for cell transfection at a final amount of 2 µg DNA per well
in a 6-well plate, mixed with 6–8 µl PEI-40K transfection reagent
(Wuhan Servicebio Technology Co., Ltd.) and incubated at room
temperature for 15 min before being added to BMECs. Cells were
cultured at 37°C, 5% CO2 for 5 h, then refreshed with
complete medium and harvested 48 h post-transfection for subsequent
experiments. OE-NC consisted of the corresponding empty vector
backbone (pcDNA3.1 empty vector; cat. no. V79020; Thermo Fisher
Scientific, Inc.), ensuring that any observed effects were
specifically attributable to NEDD4 OE. siRNA sequences, including
the NC, are listed in Table SI.
The purified plasmids were used for transfection at a final amount
of 2 µg DNA per well in a 6-well plate.
Transient transfection of BMECs
For the present study, BMECs were obtained from AoLu
Biotechnology Co., Ltd. (cat. no. ORC0562). BMECs were seeded at a
density of 6×105 cells per well in 6-well plates. BMECs
were cultured in complete Endothelial Cell Medium (ScienCell
Research Laboratories) supplemented with 10% fetal bovine serum
(FBS) and 1% penicillin-streptomycin (100 U/ml and 100 µg/ml,
respectively; all Gibco; Thermo Fisher Scientific, Inc.) at 37°C.
After washing with PBS twice, cells were cultured in 1.8 ml
serum-free basal medium (Endothelial Cell Medium). To prepare the
transfection complexes, solution A, consisting of 100 µl serum-free
medium with 20 nM siRNA, was mixed with solution B comprising 100
µl serum-free medium with 6–8 µl PEI 40K (Wuhan Servicebio
Technology Co., Ltd.). Subsequently, this mixture was added
dropwise to each well. The cells were incubated at 37°C for 5 h.
The cells were harvested 48 h after transfection for subsequent
experiments. Cells were centrifuged at 200 × g for 5 min at 4°C,
resuspended in Cell Freezing Medium; Gibco; cat. no. 12648010),
transferred to cryovials and stored at −80°C overnight before being
transferred to liquid nitrogen for long-term storage.
Reverse transcription-quantitative PCR
(RT-qPCR)
BMECs were divided into eight groups: OE-NC,
OE-NEDD4, control, si-NEDD4, si-mTOR-1, si-mTOR-2, si-mTOR-3 and
si-NEDD4 + si-mTOR. Total RNA was isolated using TRIzol reagent
(Thermo Fisher Scientific, Inc.). according to the manufacturer's
protocol. The quality and concentration of the RNA were then
determined spectrophotometrically. First-strand cDNA synthesis was
performed using the SureScript First-Strand cDNA Synthesis Kit
(Wuhan Servicebio Technology Co., Ltd.). qPCR was performed with
SYBR Green Master Mix (Takara Bio, Inc.) on a StepOnePlus™
Real-Time PCR System (Bio-Rad Laboratories, Inc.). RT was performed
at 42°C for 15 min, followed by 85°C for 5 sec. qPCR thermocycling
conditions were as follows: initial denaturation at 95°C for 30
sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 30 sec.
Relative mRNA expression was calculated using the
2^-ΔΔCq method (32).
The primers used are listed in Table
SII. GAPDH was used as the reference gene.
Western blot analysis of NEDD4
expression
Cells were harvested 48 h after transfection with
si-NEDD4, pcDNA3.1-NEDD4-3×HA or the respective controls. Total
proteins were extracted using RIPA lysis buffer (Beyotime
Biotechnology) supplemented with protease inhibitors (cat. no.
78430; Thermo Fisher Scientific, Inc.). Protein concentrations were
quantified using the BCA Protein Assay Kit (cat. no. 23225; Thermo
Fisher Scientific, Inc.). Equal quantities of protein (20–30
µg/lane) were separated via 8–10% SDS-PAGE and subsequently
transferred onto PVDF membranes (MilliporeSigma). Membranes were
blocked with 5% BSA (Beijing Solarbio Science & Technology Co.,
Ltd.) for 1 h at room temperature, followed by overnight incubation
at 4°C with the following primary antibodies: Anti-NEDD4 (1:2,000;
cat. no. AF4636; Affinity Biosciences) and anti-β-actin (1:25,000;
cat. no. 66009-1-Ig; Proteintech Group, Inc.) as the loading
control. After washing with TBST (0.1% Tween-20), membranes were
incubated with HRP-conjugated secondary antibodies: Goat
anti-rabbit IgG (H+L) (1:3,000; cat. no. GB23303; Wuhan Servicebio
Technology Co., Ltd.) and goat anti-mouse IgG (H+L) (1:5,000; cat.
no. GB23301; Wuhan Servicebio Technology Co., Ltd.) for 1 h at room
temperature. Protein bands were visualized using the Pierce ECL
Western Blotting Substrate (Thermo Fisher Scientific, Inc.) and
band intensities were semi-quantified using ImageJ software
(version 1.53; National Institutes of Health).
MTT assay
Cells were incubated in 90 µl DMEM/F12 (Meilunbio)
supplemented with 10% FBS and 1% penicillin-streptomycin with 10 µl
5 mg/ml MTT solution (Beijing Solarbio Science & Technology
Co., Ltd.) for 4 h. The supernatants were then removed, and 110 µl
DMSO formazan solubilization solution was added. The absorbance was
measured at 560 nm using a Bio-Rad microplate reader.
5-ethynyl-2′-deoxyuridine (EdU)
proliferation assay
Cell proliferation was evaluated using the EdU Cell
Proliferation Kit (Wuhan Servicebio Technology Co., Ltd.) according
to the manufacturer's instructions. In brief, cells from various
experimental groups were seeded into 24-well plates at a density of
1×105 cells/well and incubated with EdU working solution
for 2 h at 37°C. After fixation with 4% paraformaldehyde (Beijing
Solarbio Science & Technology Co., Ltd.) at room temperature
for 15 min and permeabilization with 0.5% Triton X-100, cells were
stained using the Apollo® 567 fluorescent dye at room
temperature for 30 min, according to the manufacturer's
instructions.. Images were captured using a Nikon fluorescence
microscope (Nikon Corporation) Cells that incorporated nuclear EdU
exhibited green fluorescence. The percentage of EdU-positive cells
was calculated from five randomly chosen fields per group and the
data were statistically analyzed using ImageJ software (version
1.53; National Institutes of Health).
Tube formation assay
Matrigel® (150 µl/well; Corning, Inc.)
was added to 48-well plates and allowed to solidify at 37°C for 30
min. Cells (1×105/well) were then seeded and incubated
at 37°C for 4 h. Cells were stained with Calcein-AM (2 µM; Beyotime
Biotechnology) at 37°C for 30 min to visualize the tubular network.
The formation of tubes was observed and images were captured under
an inverted fluorescence microscope (Motic Biological Technology
Co., Ltd.).
Wound healing assay
BMECs were seeded into 6-well plates in triplicate
and cultured until reaching 100% confluence. Cells were
serum-starved overnight prior to the assay. Cells were cultured in
DMEM/F12 with 10% fetal bovine serum (Gibco, USA) and 1%
penicillin-streptomycin. A linear scratch was created across the
cell monolayer using a sterile 200-µl pipette tip. Detached cells
were removed by rinsing the wells three times with PBS, after which
fresh culture medium containing NEDD4 overexpression plasmid,
si-NEDD4 and their respective negative controls. Images were
captured at 0, 24 and 48 h using an inverted light microscope
(Motic Biological Technology Co., Ltd.). The scratch width was
measured in pixels using ImageJ software (version 1.53; National
Institutes of Health), with the width at 0 h defined as 100%
(baseline); the widths at subsequent time points were expressed as
a percentage relative to this baseline. The mean value from three
independent experiments was used for analysis.
Methylation-specific PCR
Genomic DNA was extracted from BMECs using a DNA
extraction kit (Omega Bio-Tek, Inc.) and then converted to
bisulfite using the EZ DNA Methylation-Gold Kit (Zymo Research
Corp.). The methylation status was detected through PCR using Taq
DNA Polymerase (Takara Bio, Inc.) with the following primers:
NEDD4-M (methylated): Forward, 5′-ATTTTTTGTAGAAAGATTTGAAGGC-3′ and
reverse, 5′-CGCAACTCTATAATTAAATTTAACGAT-3′; NEDD4-U (unmethylated):
Forward, 5′-TTTTTGTAGAAAGATTTGAAGGTGT-3′ and Reverse,
5′-CACAACTCTATAATTAAATTTAACAAT-3′. The thermocycling conditions
were as follows: Initial denaturation at 95°C for 5 min, followed
by 40 cycles of 95°C for 30 sec, 58°C for 30 sec and 72°C for 30
sec and final extension at 72°C for 10 min. PCR products were
separated on 2% agarose gels and visualized using an E-Gel Imager
system (Thermo Fisher Scientific, Inc.) with ethidium bromide
staining The experiment included five groups as follows: i)
H2O (blank negative control); ii) BMECs treated with PBS
(vehicle control); iii) BMECs treated with 1 µM methylprednisolone
(MePr; MedChemExpress); iv) BMECs treated with 5 µM 5-azacytidine
(5-AZA; Sigma-Aldrich; Merck KGaA); and v) BMECs co-treated with
MePr (1 µM) and 5-AZA (5 µM). All treatments were performed at 37°C
for 48 h under standard cell-culture conditions.
Immunoprecipitation (IP) assay
Proteins were extracted from the cells using RIPA
buffer (Beyotime Biotechnology; cat. no. P0013B) that contained a
protease inhibitor cocktail (cat. no. 04693132001; Roche
Diagnostics). The cell lysates were then cleared by centrifugation
at 12,000 × g for 15 min at 4°C. The protein concentrations were
measured using a BCA Protein Assay Kit (cat. no. 23225; Thermo
Fisher Scientific, Inc.). A total of 500 µg lysate (300 µl) was
incubated with Protein A/G magnetic beads (50 µl; cat. no. 88802;
Thermo Fisher Scientific, Inc.) prebound to anti-ubiquitin antibody
(2 µg; 1:2,000; cat. no. ER65617; Hangzhou HuaAn Biotechnology Co.,
Ltd.) overnight at 4°C. Beads were washed three times with cold PBS
containing 0.1% Tween-20, followed by centrifugation at 2,000 × g
for 3 min at 4°C. The immunocomplexes were eluted by boiling for 5
min in SDS loading buffer (Beyotime Institute of Biotechnology).
Subsequently, 30 µg of total protein per lane was separated using
8–10% SDS-PAGE, transferred onto PVDF membranes, and blocked with
5% BSA (Servicebio) for 1 h at room temperature. Membranes were
incubated with primary antibodies overnight at 4°C, followed by 1 h
incubation with secondary antibodies at room temperature, and then
incubated with the following primary antibodies:, anti-mTOR (1:20;
cat. no. AF6308; Affinity Biosciences); anti-ubiquitin (1:2,000;
cat. no. ER65617; Hangzhou HuaAn Biotechnology Co., Ltd.). This was
followed by incubation with HRP-conjugated goat anti-rabbit IgG
(H+L) (1:3,000; cat. no. GB23303; Servicebio);Goat anti-mouse IgG
(H+L) (1:5,000; cat. no. GB23301; Servicebio), and visualization
using ECL reagents (Thermo Fisher Scientific, Inc.). Images of
bands were captured the ChemiDoc™ Touch Imaging System (cat. no.
1708370; Bio-Rad Laboratories, Inc.) and band intensities were
semi-quantified using ImageJ software version 1.54 (National
Institutes of Health). Statistical analysis was carried out using
GraphPad Prism 8 (Dotmatics). Western blotting was performed as
aforementioned.
Bioinformatics analysis
The GSE123568 dataset (33) from the Gene Expression Omnibus
(GEO) database (ncbi.nlm.nih.gov/geo/), which includes gene
expression profiles from 30 patients with SONFH and 10 controls,
was analyzed. Differential expression analysis was performed using
the ‘limma’ package (v3.58.1; bioconductor.org/packages/limma/) in
R software (version 4.3.2; R Foundation for Statistical Computing,
Vienna, Austria), with the cut-off criteria of |log2FC| ≥1 and
adjusted P<0.05. Additional analyses included weighted gene
co-expression network analysis (WGCNA; version 1.72–5;
horvath.genetics.ucla.edu/html/CoexpressionNetwork/), single-sample
Gene Set Enrichment Analysis (ssGSEA; GSVA version 1.46.0;
http://bioconductor.org/packages/GSVA/), and random
forest and support vector machine (SVM) modeling (MetaboAnalyst
version 4.0.0; http://www.metaboanalyst.ca/). Principal component
analysis (PCA) was performed using the ‘FactoMineR’ package
(version 2.8; http://cran.r-project.org/package=FactoMineR) to
visualize sample clustering. Enrichment analysis was conducted
using the Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthology
Based Annotation System (http://bioinfo.org/kobas/) Gene Ontology (GO)
annotation. Protein–protein interaction networks were constructed
using STRING (version 12.0; string-db.org/). Pathway enrichment
analysis was performed using the REACTOME database (reactome.org/).
All data visualization was performed in R using the ‘ggplot2’
package (version 3.5.1; cran.r-project.org/package=ggplot2).
Receiver operating characteristic (ROC) curve
analysis was performed using the pROC package in R to evaluate the
diagnostic performance of key ubiquitination-related genes and
multivariate predictive models.
Gene selection frequencies were calculated as the
number of times each gene was identified as an important variable
across repeated resampling steps during model training.
The present study retrieved the gene expression
dataset GSE123568 from the GEO database, a publicly available
functional genomics repository. This dataset was created using the
GPL15207 Affymetrix Human Gene Expression Array platform (Thermo
Fisher Scientific, Inc.) and contained peripheral blood mononuclear
cell samples from 40 individuals, including 30 patients with SONFH
and 10 controls without SONFH. For more details on clinical
information and experimental protocols, please refer to the
original submission. In the present study, the dataset was used to
analyze differential gene expression and then subjected to
bioinformatics techniques such as functional enrichment and network
analysis to pinpoint candidate genes and signaling pathways
associated with SONFH.
Statistical analysis
Data are presented as the mean ± standard error of
the mean. The data were analyzed using GraphPad Prism 8
(Dotmatics). For comparisons between two groups, unpaired Student's
t-tests were used. For comparisons among multiple groups, one-way
ANOVA was performed followed by Tukey's multiple comparisons test
as the post hoc test. Pearson's correlation test was performed to
evaluate association between gene expression levels. All
experiments were independently repeated ≥3 times to ensure
reproducibility. All statistical analyses included the necessary
corrections for multiple testing using the Benjamini-Hochberg false
discovery rate method. P<0.05 was considered to indicate a
statistically significant difference.
Results
Changes in gene expression after SONFH
are closely associated with ubiquitination in terms of functional
enrichment
The principal component (PC) analysis performed on
the GSE123568 dataset revealed partial separation between the
control group and the SONFH group, with notable separation in PC1
and PC2, highlighting notable differences in gene expression
patterns (Fig. 1A). Analysis of
differential gene expression revealed 229 upregulated genes and 349
downregulated genes, with significant differences in TSTA3, RIOK3,
STOM, HEPACAM2, RUNX2, ARAP1, TYK2 and SIGLEC7 (Fig. 1B). KEGG enrichment analysis
revealed pathways that were significantly enriched, including,
‘protein processing in endoplasmic reticulum’, ‘lysosome’, ‘cell
cycle’ and ‘autophagy-animal’. The enriched REACTOME pathways
included ‘Deubiquitination’ and ‘Antigen processing: Ubiquitination
& Proteasome degradation’. Gene Ontology (GO) enrichment
analysis showed significant enrichment in processes related to
ubiquitination, such as ‘proteasome mediated ubiquitin dependent
protein catabolic process’, ‘ubiquitin protein ligase activity’,
‘ubiquitin-dependent protein catabolic process’, ‘protein
polyubiquitination’, ‘ubiquitin ligase complex’, ‘ubiquitin
conjugating enzyme binding’, ‘positive regulation of
ubiquitin-protein transferase activity’ and ‘protein
deubiquitination’. These findings indicated significant differences
in gene expression and biological functions between the SONFH and
control groups, particularly in pathways related to ubiquitination
(Fig. 1C).
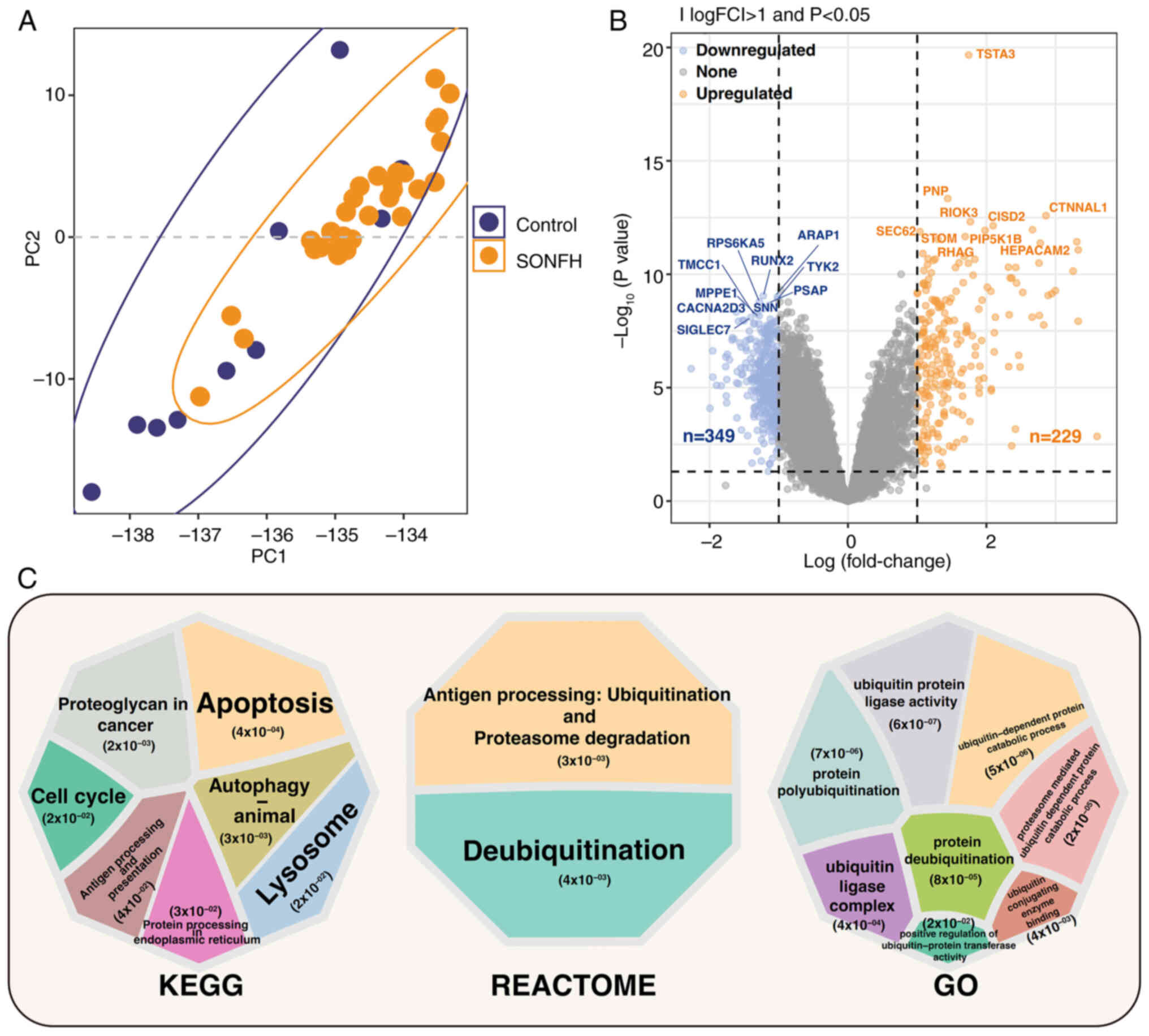 | Figure 1.Differential gene and pathway
enrichment analysis between the control and SONFH groups. (A) PC
analysis showing the separation between control and SONFH groups.
(B) Results of differential analysis. The x-axis represents
log(fold change) and the y-axis represents-log10
(P-value). Differentially expressed genes are categorized as
upregulated, downregulated or not significantly changed. Genes with
|log(FC)|>1 and P<0.05 were considered significantly
differentially expressed (orange for upregulated, blue for
downregulated and gray for not significantly changed). (C) KEGG,
REACTOME and GO enrichment results, with significant pathways
highlighted. The pathways were selected based on P<0.05 and
|log(FC)| >1). Each circle represents one pathway; the size of
the circle corresponds to the number of genes, while the color
indicates-log10 (P-value), with darker colors
representing higher significance. SONFH, steroid-induced
osteonecrosis of the femoral head; KEGG, Kyoto Encyclopedia of
Genes and Genomes; GO, Gene Ontology; FC, fold change; PC,
principal component. |
Association of ubiquitination with
SONFH
To investigate the role of ubiquitination in SONFH,
ssGSEA was first performed on pathways related to ubiquitination,
cell proliferation and the cell cycle using the KEGG, GO and
REACTOME databases. This revealed marked differences in scores
between the control and SONFH groups (Fig. S1). Subsequently, key
ubiquitination-related pathway ssGSEA scores were chosen for
co-expression network analysis, using control and SONFH as
phenotypes in WGCNA. Numerous module pathways, including those
associated with ubiquitination, angiogenesis and cell
proliferation, exhibited significant differences between the SONFH
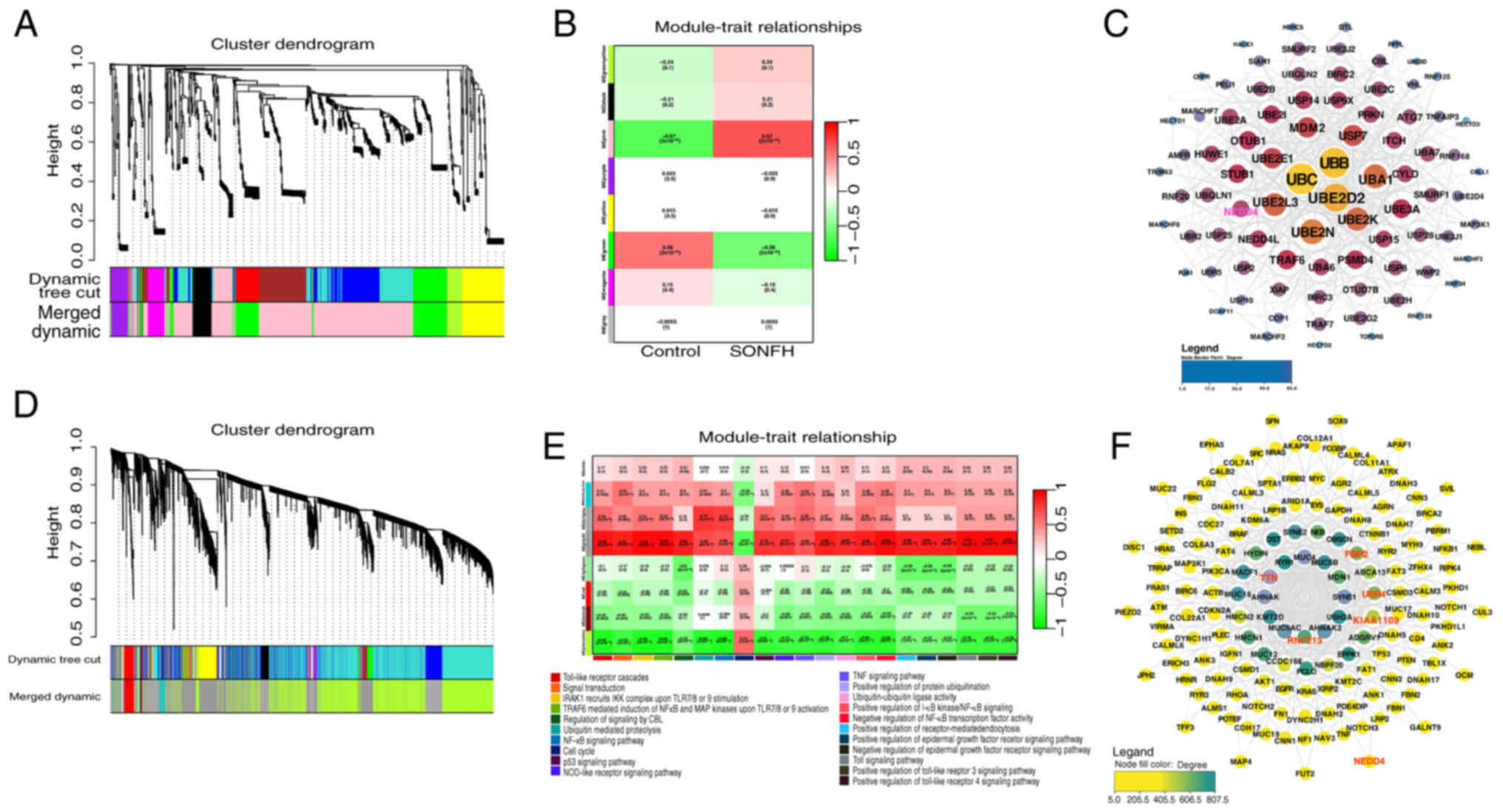
and control phenotypes (Fig. 2A and
B). Genes from 489 pathways in the ME (module eigengene (ME)
pink module were selected for network interaction analysis,
identifying UBB and UBC at the core of the network (Fig. 2C). Further analysis showed the
effect of ubiquitination on SONFH. Using ssGSEA scores from
ubiquitination-related pathways as phenotypes, WGCNA was performed
again, revealing significant roles for these pathways in different
gene modules. In the MEdarkgrey module, pathways such as ‘positive
regulation of protein ubiquitination’ (R=0.47), ‘ubiquitin mediated
proteolysis’ (R=0.77) and ‘ubiquitin-ubiquitin ligase activity’
(R=0.41) exhibited significant correlations with module genes
(Fig. 2D and E). Network
interaction analysis revealed the interactions between these genes
in the context of ubiquitination, with RNF213, TTN, FSIP2, KIAA1109
and UBR4 at the network center (Fig.
2F), and demonstrated that the E3 ubiquitin ligase NEDD4 was
connected to two core network genes, RNF213 and FSIP2, within the
ubiquitination-associated network.
Role of NEDD4 in ubiquitination
related to SONFH
The next step was to investigate genes closely
associated with ubiquitination pathways in SONFH. The present study
used multivariate exploratory receiver operating characteristic
(ROC) analysis on specific pathway genes to achieve this. In the
ubiquitin-ubiquitin ligase activity pathway, the two-factor model
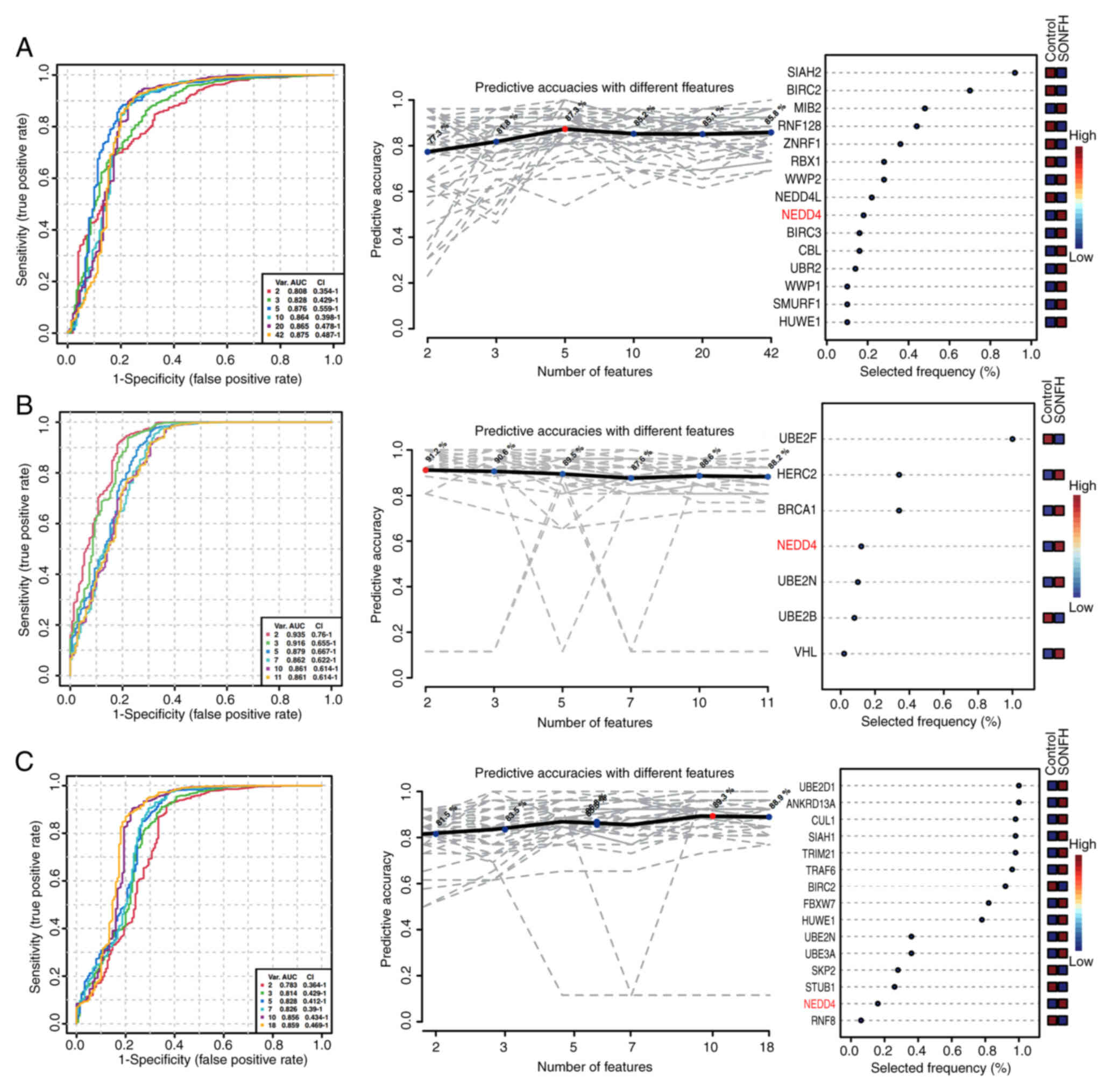
achieved an AUC of 0.876 and a predictive accuracy of 87.3%
(Fig. 3A, right), indicating a
strong ability to distinguish between the control and SONFH groups.
The five-factor model was demonstrated to be the most accurate
predictor, with an AUC value of 0.876. Here, gene selection
frequency reflects the relative stability and importance of each
gene during model construction. Gene selection frequencies were
ranked from highest to lowest as follows: SIAH2, BIRC2, MIB2,
RNF128, ZNRF1, RBX1, WWP2, NEDD4L, NEDD4, BIRC3, CBL, UBR2, WWP1,
SMURF1 and HUWE1 (Fig. 3A). Within
the ubiquitin-mediated proteolysis pathway, all AUC values were
>0.8, with the two-factor model yielding the highest AUC at
0.935. Gene selection frequencies, ranked from highest to lowest,
were: UBE2F, HERC2, BRCA1, NEDD4, UBE2N, UBE2B and VHL (Fig. 3B). In the positive regulation of
protein ubiquitination pathway, ROC curves exhibited AUC values
>0.75, indicating high sensitivity. The 10-factor model
demonstrated high specificity and accuracy, with an AUC of 0.893.
Gene selection frequencies, ranked from highest to lowest, were:
UBE2D1, ANKRD13A, CUL1, SIAH1, TRIM21, TRAF6, BIRC2, FBXW7, HUWE1,
UBE2N, UBE3A, SKP2, STUB1, NEDD4 and RNF8 (Fig. 3C). ROC analysis revealed that NEDD4
had high AUC values and high selection frequencies across multiple
pathways, especially in the ubiquitin-ubiquitin ligase activity and
ubiquitin-mediated proteolysis pathways. This suggested a
significant role for NEDD4 in distinguishing between control and
SONFH groups. Gene contribution analysis, random forest analysis
and SVM analyses also supported this finding (Fig. S2). NEDD4 consistently ranked among
the top genes by feature importance, accuracy contribution and
selection frequency, underscoring its robust predictive value.
NEDD4 potentially regulates the cell
cycle, angiogenesis and cell proliferation in SONFH
Based on the analyses showing the potential
regulatory role of NEDD4 in SONFH, correlation analyses were
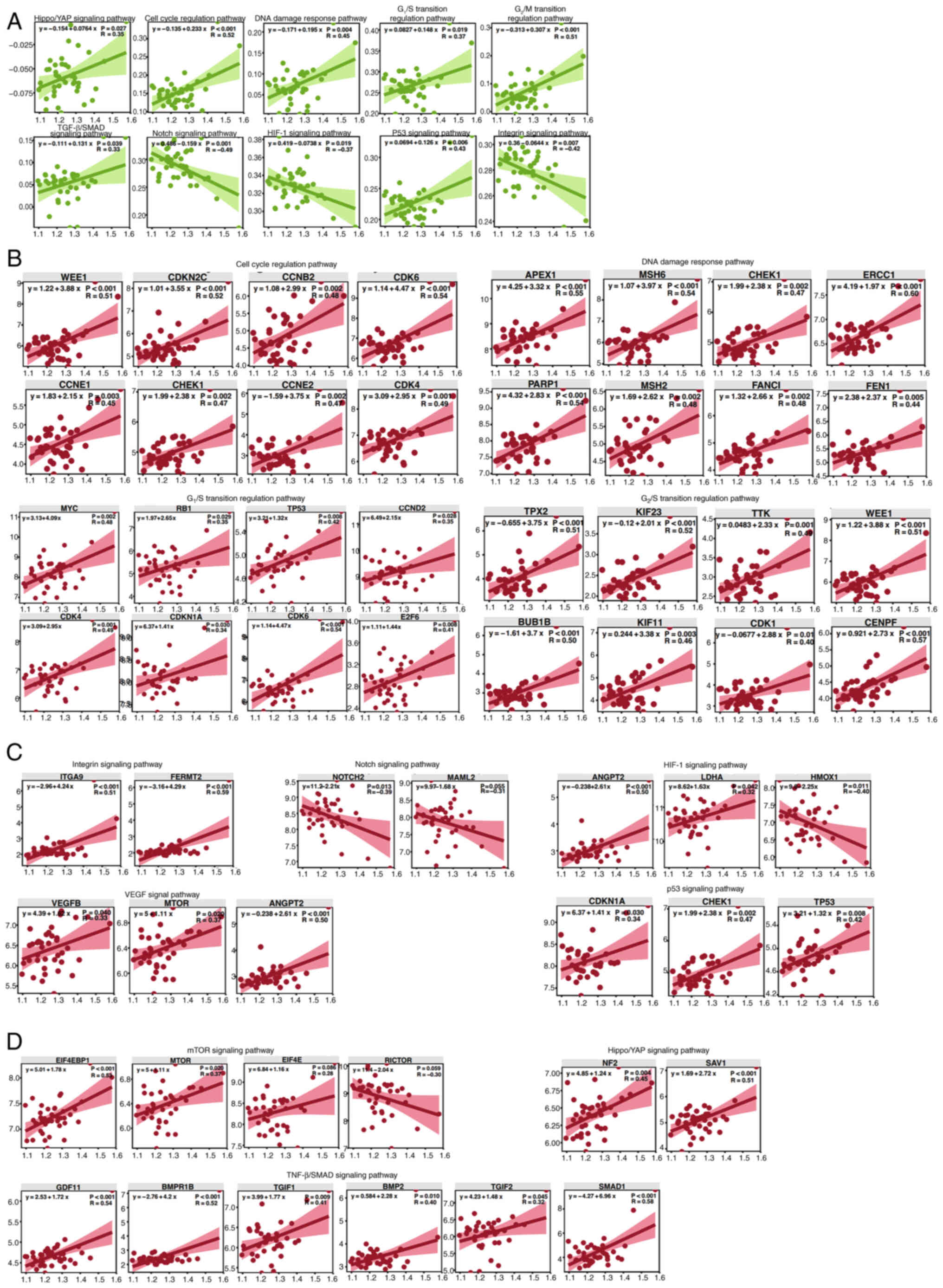
performed. The results of the present study revealed significant
associations between NEDD4 and pathways related to the cell cycle,
angiogenesis and cell growth. Specifically, NEDD4 was negatively
correlated with key pathways, including the ‘HIF-1 signaling
pathway’ (R=−0.37), ‘integrin signaling pathway’ (R=−0.42) and
‘Notch signaling pathway’ (R=−0.49). Additionally, NEDD4 was
positively correlated with multiple pathways, including the ‘p53
signaling pathway’ (R=0.43), ‘DNA damage response pathway’ (R=0.45)
and the ‘G2/M transition regulation pathway’ (R=0.51;
Fig. 4A). Among cell cycle-related
genes, NEDD4 showed significant positive correlations with key
genes such as WEE1 (R=0.51), CCNE1 (R=0.45), MSH6 (R=0.54), ERCC1
(R=0.60), CDK6 (R=0.54) and CENPF (R=0.57), further supporting the
role of NEDD4 in regulating the cell cycle (Figs. 4B and S3). NEDD4 also showed positive
correlations with multiple angiogenesis-associated genes, including
ANGPT2 (R=0.50), VEGFB (R=0.33) and FERMT2 (R=0.59), suggesting a
potential role for NEDD4 in regulating angiogenesis (Figs. 4C and S3). Within cell proliferation-related
pathways, NEDD4 displayed significant positive correlations with
multiple genes, including EIF4EBP1 (R=0.53), SAV1 (R=0.51), GDF11
(R=0.54) and MTOR (R=0.37), further highlighting the key role of
NEDD4 in cell proliferation and metabolic regulation (Figs. 4D and S3). These findings indicated that NEDD4
serves an important role in regulating the cell cycle, angiogenesis
and cell proliferation in SONFH.
OE of NEDD4 promotes proliferation,
angiogenesis and wound healing in BMECs
To determine the role of NEDD4 in SONFH, a NEDD4 OE
vector was created and tested in BMECs. BMECs displayed clear
boundaries, no trailing and appeared healthy (Fig. 5A and B), following transfection
with si-NEDD4 or si-NC. BMECs also remained in healthy condition,
characterized by intact cell morphology, clear cell-cell boundaries
and absence of cytoplasmic blebbing, following transient
overexpression of NEDD4 (pcDNA3.1-NEDD4-3×HA) compared with the
OE-NC group (Fig. 5C). RT-qPCR
demonstrated significantly increased NEDD4 mRNA levels in the
OE-NEDD4 group compared with the OE-NC group, indicating successful
transfection (Fig. 5D). After 48
h, the optical density (OD)560 values of BMECs in the OE-NEDD4
group were significantly increased compared with those in the
control group (Fig. 5E). Western
blot analysis supported notable OE of NEDD4 in
pcDNA3.1-NEDD4-3×HA-transfected BMECs compared with controls and
effective knockdown of NEDD4 and mTOR in si-NEDD4- and
si-mTOR-transfected cells, respectively. (Fig. 5F). The tube formation assay
revealed a significant increase in the total tube length and number
of junctions and meshes in BMECs in the OE-NEDD4 group compared
with the control group (Fig. 5G and
I). The wound healing assay indicated a significant decrease in
wound area in the OE-NEDD4 compared with the control group at 24
and 48 h (Fig. 5H). Subsequently,
the present study evaluated how NEDD4 affected the viability of
BMECs using an MTT assay; OE-NEDD4 group exhibited significantly
higher OD (560 nm) values compared with the control group,
indicating enhanced cell viability (Fig. 5J).
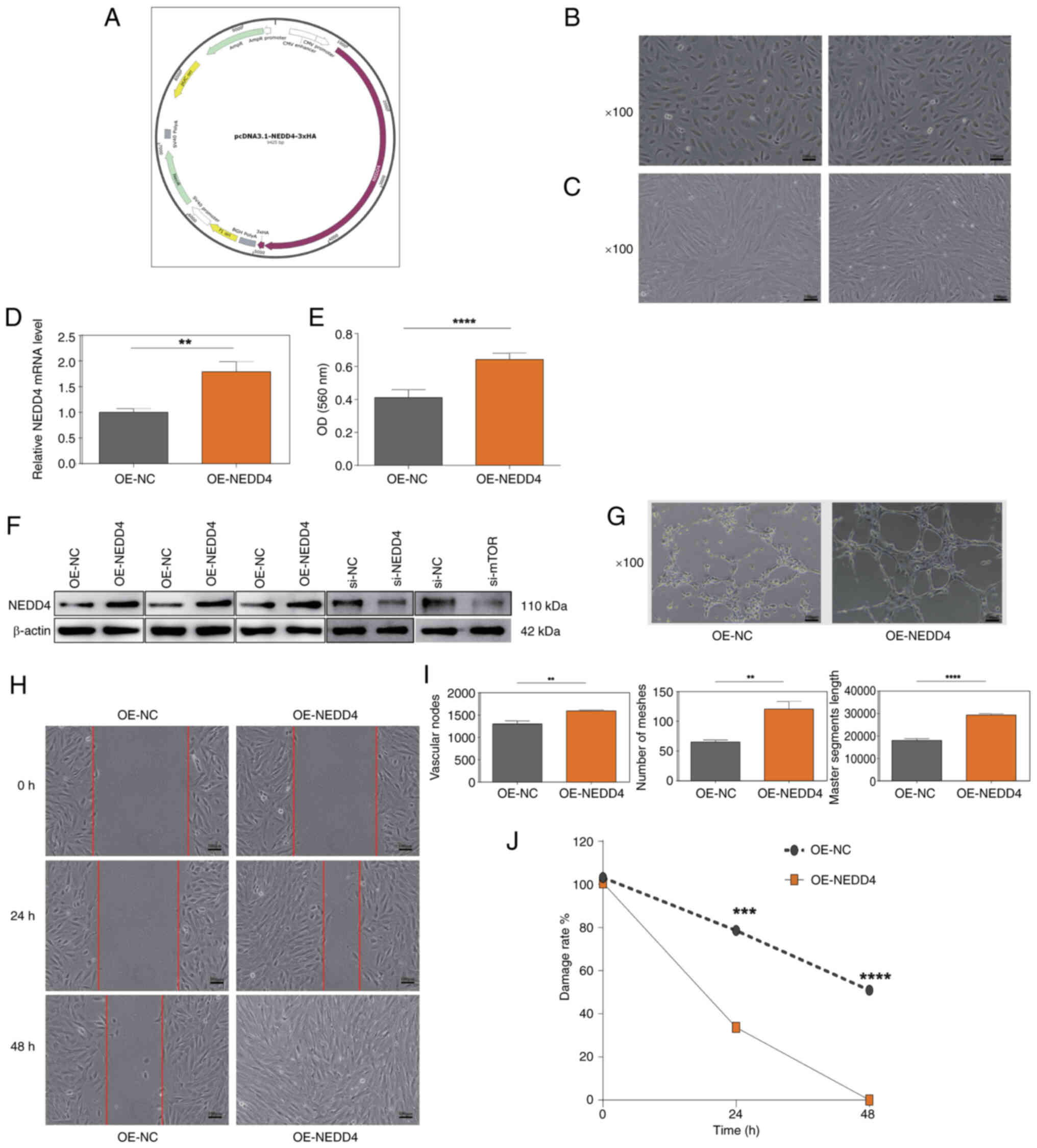 | Figure 5.Effects of NEDD4 overexpression on
BMECs. (A) Morphology of BMECs in the control group showing normal
cell boundaries and morphology. (B) Morphology of BMECs in the
OE-NEDD4 group showing no visible damage. (C) BMECs maintained good
condition after transient NEDD4 transfection. Magnification, ×100.
(D) Reverse transcription-quantitative PCR confirmed significantly
elevated NEDD4 mRNA expression in the OE-NEDD4 group. (E) An MTT
assay showed increased viability of BMECs after NEDD4
overexpression. (F) Western blot analysis demonstrating efficient
NEDD4 overexpression and knockdown in OE-NEDD4- and
siRNA-2-transfected cells, respectively. (G) Quantitative analysis
of tube formation indicating increased number of meshes, greater
master segment length, and more vascular nodes in the OE-NEDD4
group. (H) Representative wound-healing assay showing smaller
scratch areas in the OE-NEDD4 group. Magnification, ×100; scale
bar, 100 µm. (I) Quantitative analysis of wound-healing rates,
indicating significantly improved migration capacity in the
OE-NEDD4 group. (J) Decreased scratch area in the OE-NEDD4 group at
0, 24 and 48 h, consistent with enhanced migration ability.
Magnification, ×100. OE, overexpression; NC, negative control; si,
small interfering RNA; NEDD4, neural precursor cell expressed
developmentally downregulated protein 4; BMEC, bone microvascular
endothelial cell; OD, optical density. *P<0.05; **P<0.01;
***P<0.001; ****P<0.0001 vs. OE-NC or 0 h. |
NEDD4 regulates vascular endothelial
cell function by activating the VEGF signaling pathway while
functionally interacting with mTOR signaling in BMECs
To verify the associations between NEDD4, mTOR and
the VEGF signaling pathway, the present study used RT-qPCR to
measure the mRNA levels of mTOR, VEGF and VEGFR2. NEDD4 OE
significantly increased the mRNA levels of mTOR, VEGF and VEGFR2
(Fig. 6A). To explore the role of
NEDD4 in BMEC function via mTOR regulation, siRNA transfection was
performed to generate si-NEDD4- and si-mTOR-transfected BMECs.
RT-qPCR identified si-NEDD4-1 and si-mTOR-3 as the most effective
interference sequences (Fig. 6B).
IP was performed to assess the ubiquitination level of mTOR,
revealing a notable decrease in mTOR ubiquitination in
NEDD4-overexpressing cells compared with the control group
(Fig. 6C).. The morphology and
fluorescence of BMECs following siRNA transfection confirmed
successful transfection efficiency (Fig. 6D). Wound healing assay results
showed a significant reduction in wound healing rates in the
si-NEDD4 and si-mTOR groups compared with the control group, with a
greater decrease observed in the si-NEDD4 + si-mTOR group at 48 h,
aligning with the results from the MTT and EdU assays, where
si-NEDD4 and si-NEDD4 + si-mTOR groups showed significantly reduced
proliferation while the si-mTOR group exhibited a non-significant
downward trend (Fig. 6E-G). In the
tube formation assay, total tube length and number of junctions and
meshes were significantly decreased in the si-NEDD4 and si-mTOR
groups compared with the control group, with further significant
reductions observed in the si-NEDD4 + si-mTOR group compared with
the si-NEDD4 group, consistent with the findings from the MTT and
scratch assays (Fig. 6H). In the
MTT assay, OD560 values were significantly decreased in the
si-NEDD4- and si-mTOR-transfected groups compared with the control
group, and further decreased in the si-NEDD4 + si-mTOR compared
with the si-NEDD4 group (Fig.
6I).
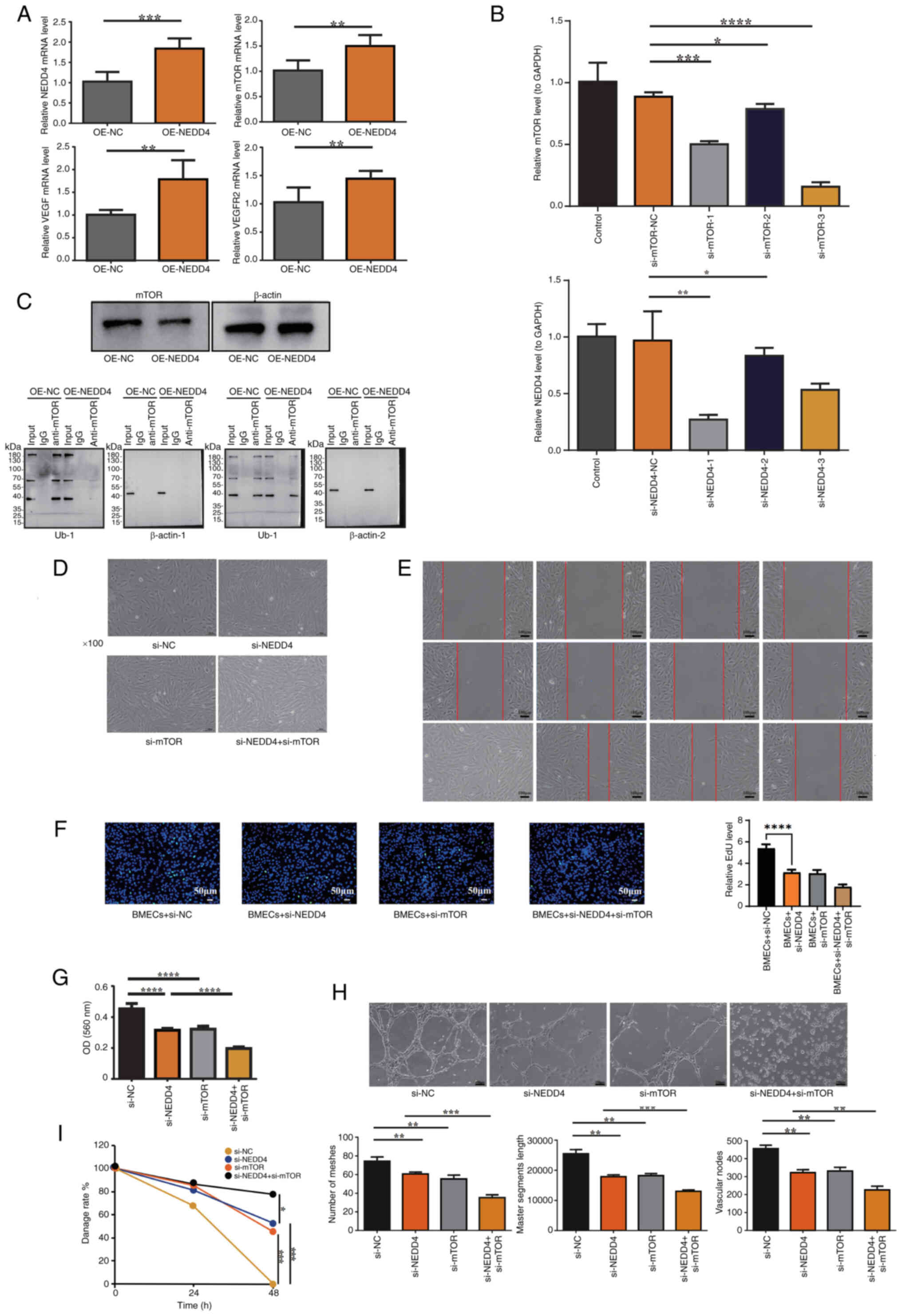 | Figure 6.Functional validation of NEDD4 in the
VEGF signaling pathway. (A) RT-qPCR showed increased mTOR, VEGF and
VEGFR2 mRNA levels after NEDD4 overexpression. (B) RT-qPCR
identified si-NEDD4-1 and si-mTOR-3 as the most effective
interference sequences. (C) Immunoprecipitation analysis showing
that NEDD4 overexpression decreased the ubiquitination level of
mTOR. (D) Validation of transfection efficiency in BMECs.
Representative microscopic images show cells transfected with
si-NEDD4, si-mTOR and their respective controls (si-NC). BMECs
maintained a normal morphology and healthy growth state after
transfection, without observable cytotoxicity or detachment,
confirming that the transfection was effective and did not
adversely affect cell viability. Magnification, ×100. (E) Scratch
assay showing decreased wound closure rates in the si-NEDD4 and
si-mTOR groups compared with the control group, with the lowest
migration observed in the si-NEDD4 + si-mTOR group. Scale bar, 100
µm. (F) Representative images from the EdU proliferation assay
demonstrating reduced EdU-positive nuclei in the si-NEDD4 and
si-mTOR groups, with further reductions in the si-NEDD4 + si-mTOR
group. Scale bar, 50 µm. (G) MTT assay showed reduced viability in
si-NEDD4 and si-mTOR groups, with the greatest reduction in the
si-NEDD4 + si-mTOR group. (H) Tube formation assay demonstrating
reduced angiogenesis parameters in the si-NEDD4 and si-mTOR groups,
with further reductions in the si-NEDD4 + si-mTOR group.
Magnification, ×100. (I) Quantitative analysis of wound-healing
rates, indicating significantly reduced migration ability in the
si-NEDD4 and si-mTOR groups, with further reduction in the si-NEDD4
+ si-mTOR group. NC, negative control; si, small interfering RNA;
NEDD4, neural precursor cell expressed developmentally
downregulated protein 4; OE, overexpression; RT-qPCR, reverse
transcription-quantitative PCR; BMEC, bone microvascular
endothelial cell; OD, optical density; Ub-1, first ubiquitination
site assay. *P<0.05, **P<0.01, ***P<0.001 and
****P<0.0001. |
Glucocorticoids inhibit NEDD4
expression by upregulating methylation levels in its promoter
region
Correlation analysis of NEDD4 and
methylation-related genes showed significant positive and negative
correlations with genes such as KDM6B, PCNA, DNMT3A, SUV39H2,
UHRF1, EZH2, TET2, DMAP1 and PRMT1, further highlighting the
complex role of NEDD4 in methylation regulation (Fig. 7A). BMECs exhibited healthy
proliferation after treatment with PBS, MePr, 5-AZA, or MePr +
5-AZA (Fig. 7B).
Methylation-specific PCR results revealed increased methylation
levels following MePr addition and decreased levels following 5-AZA
treatment. Compared with the 5-AZA group, combined treatment with
MePr and 5-AZA increased methylation levels in the NEDD4 promoter
region (Fig. 7C). RT-qPCR results
indicated that combined treatment with MePr + 5-AZA significantly
suppressed NEDD4 mRNA expression compared with 5-AZA treatment
alone (Fig. 7D).
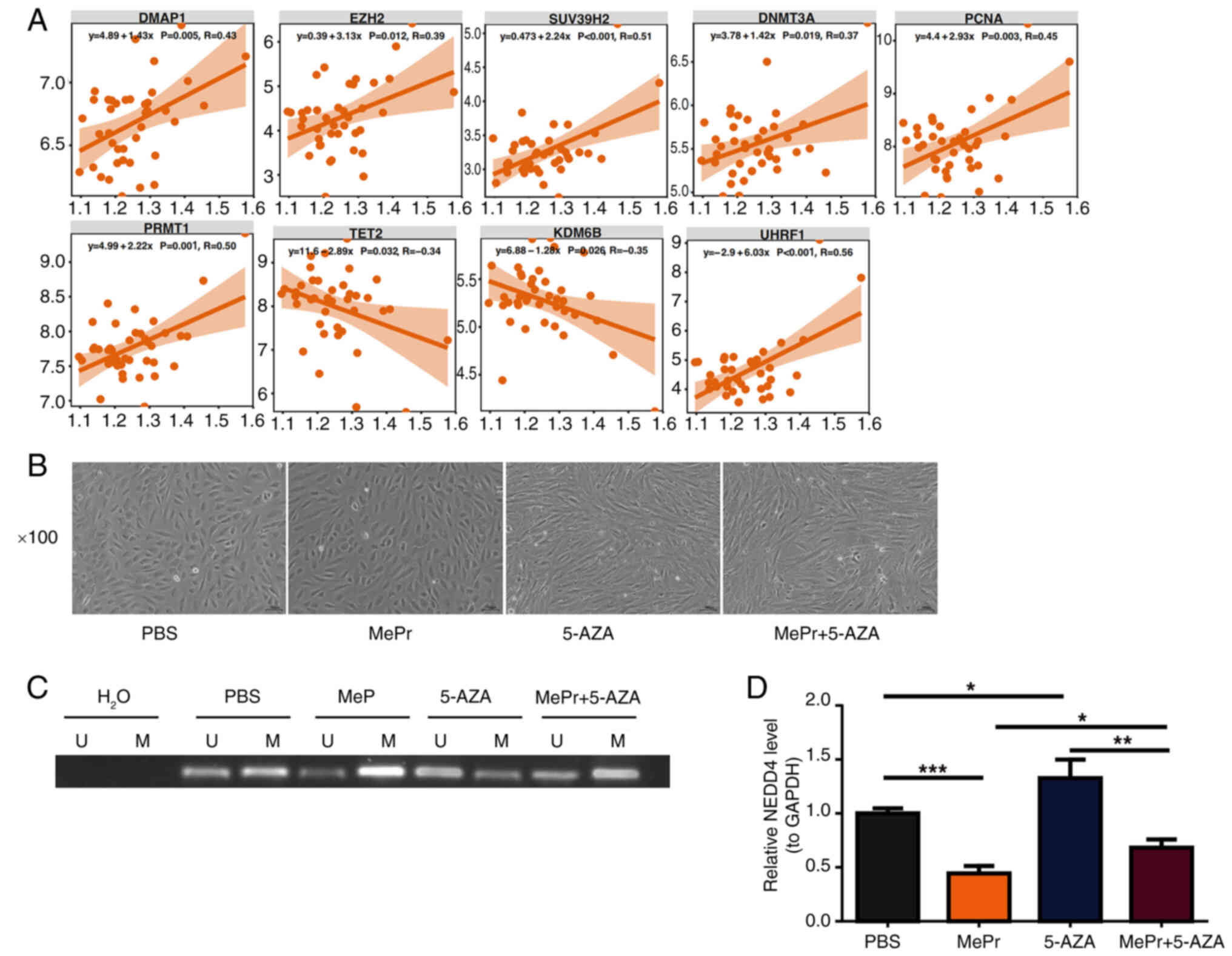 | Figure 7.Glucocorticoids inhibit NEDD4
expression by increasing promoter methylation levels in BMECs. (A)
Correlation between NEDD4 and methylation-related genes, with
significance and correlation marked. (B) BMEC culture, including
PBS, MePr, 5-AZA and MePr + 5-AZA groups. Magnification, ×100. (C)
Methylation PCR, with H2O as the system negative control
to exclude contamination. (D) NEDD4 mRNA expression level, with
significant differences marked. U, unmethylated control; M,
methylated sample; NEDD4, neural precursor cell expressed
developmentally downregulated protein 4; BMEC, bone microvascular
endothelial cell; MePr, methylprednisolone; 5-AZA, 5-azacytidine.
*P<0.05, **P<0.01 and ***P<0.001. |
Discussion
Research highlights the important role of NEDD4 in
ONFH. The p53 signaling pathway is important for regulating the
cell cycle and apoptosis by slowing down cell proliferation and
killing abnormal cells, while protecting healthy cells (34). P53 is controlled by
post-translational modifications such as ubiquitination,
phosphorylation, acetylation and methylation (34). The positive link between NEDD4 and
the p53 signaling pathway, as well as tumor protein p53, suggests
that NEDD4 may impact the cell cycle and apoptosis by affecting p53
activity (34). The DNA damage
response (DDR) pathway is important for fixing DNA damage and
preserving genome stability. When cellular DNA is harmed, the DDR
pathway is activated, triggering cellular responses such as DNA
repair (35,36). Important genes such as APEX1, MSH6,
CHEK1, ERCC1, PARP1, MSH2, FANC and FEN1 are involved in DNA
repair, influencing the cell cycle and apoptosis (37–42).
The positive link between NEDD4 and key DDR genes suggests that
NEDD4 may be involved in DNA repair and cell cycle control through
its association with these genes. Additionally, CCNE1 and CCNE2 are
regulatory subunits of CDK2 that control promote the re-entry of
quiescent (G0-phase) cells into the cell cycle and regulate the
G1/S phase transition (43). WEE1
encodes a kinase that negatively regulates the G2/M
transition of the cell cycle (44). Upregulation of this protein helps
to strengthen the G2/M DNA damage checkpoint (45). CHEK1 serves an important role in
maintaining and restoring spermatogonial cells during mitosis
(44). In meiosis, CHK1
selectively weakens the DNA damage signal on autosomes at specific
stages, ensuring the normal progression of meiosis (46). The positive correlation between
NEDD4 and cell cycle regulatory genes such as CCNE1, CCNE2, WEE1
and CHEK1 highlights its important role in the G1/S and
G2/M transitions, as well as in mitosis and meiosis. In
the present study, the significance of NEDD4 in cell cycle
regulation and apoptosis was emphasized through its positive
association with multiple key genes and signaling pathways. These
results suggested that NEDD4 may serve an important role in the
cell cycle and DNA damage repair in ONFH.
Integrins, as transmembrane receptors, can detect
the physical properties of the extracellular matrix and organize
the cytoskeleton while transmitting bidirectional cellular signals.
They mediate signaling pathways such as the TGF-β, Hippo, Wnt and
Notch pathways, thereby serving important roles in biological
processes such as cell adhesion, spreading, migration and
mechanotransduction (47,48). The Notch signaling pathway could
have therapeutic benefits for bone regeneration and osteoporosis
treatment (49). The analysis in
the present study showed that NEDD4 expression was positively
correlated with ssGSEA scores of the TGF-β/SMAD and
Hippo/yes-associated protein (YAP) pathways but negatively
correlated with the Notch signaling pathway. Although NEDD4
expression was positively correlated with ITGA9 and FERMT2
expression in the integrin pathway, the ssGSEA score for this
pathway was negative, suggesting that the regulatory role of NEDD4
is complex and this may influence pathways through different
mechanisms. As an E3 ubiquitin ligase, NEDD4 may serve a key role
in the progression and repair of ONFH. NEDD4 may contribute to bone
tissue repair and remodeling by boosting the activity of the
TGF-β/SMAD and Hippo/YAP signaling pathways (25,31).
For example, NEDD4 may promote the growth and differentiation of
bone cells by stabilizing TGF-β receptors or SMAL proteins
(31). At the same time, by
enhancing the Hippo/YAP pathway, it may help regulate the
mechanical adaptability and repair of bone tissue (25). Conversely, the negative correlation
between NEDD4 and the Notch signaling pathway genes such as NOTCH2
and MAML2 indicated that NEDD4 may affect the pathological process
of SONFH by inhibiting the Notch pathway and influencing osteoblast
differentiation (49). The
interaction of the integrin signaling pathway with NEDD4 could
impact the regulation of these pathways, further affecting the
progression and repair of SONFH (47). In summary, the regulatory roles of
NEDD4 in the integrin, TGF-β/SMAD, Hippo/YAP and Notch signaling
pathways highlighted its complex function in cell signal
transduction. This diverse regulatory mechanism offers important
insights for further exploring the functions and mechanisms of
NEDD4 in SONFH.
Research has shown that ANGPT2 serves an important
role in the vascular niche within the bone marrow, helping to
maintain the balance and regeneration of hematopoietic stem cells
(50). Although VEGFB does not
directly contribute to angiogenesis, it indirectly supports
vascular growth and cell survival by affecting the actions of VEGFA
(51). Bone morphogenetic protein
receptor type-1B (BMPR1B) also serves a role, as it can prevent
chondrocyte hypertrophy and act as a stabilizer for cartilage
during joint development (52).
mTOR is involved in tissue regeneration across neurons, muscles,
liver and intestines, helping restore tissue homeostasis (53). In the present study, NEDD4
expression was positively associated with the expression of genes
involved in angiogenesis, such as ANGPT2, VEGFB, MTOR and BMPR1B.
This finding further emphasizes the notable role of NEDD4 in
regulating both angiogenesis and bone tissue homeostasis. Notably,
the positive correlation between NEDD4 and ANGPT2 supported the
role of ANGPT2 in coordinating hematopoietic stem cell homeostasis
within the vascular niche (50).
Additionally, this correlation suggested that NEDD4 may indirectly
affect the function of VEGFA through interactions with VEGFB and
mTOR, thereby encouraging vascular growth, and cell proliferation
and survival (51,53). The positive correlation between
NEDD4 and BMPR1B suggested that NEDD4 may prevent chondrocyte
hypertrophy and support cartilage health and stability (52).
The present study demonstrated that NEDD4 OE
promoted the viability, migration and angiogenesis of BMECs,
indicating its key role in endothelial cell function, and
maintaining angiogenesis and homeostasis. NEDD4 regulates processes
such as proliferation and angiogenesis via ubiquitination (23). Glucocorticoid-induced apoptosis of
BMECs promotes the onset and progression of ONFH (54). The present research sheds light on
the role and mechanism of NEDD4 in ONFH. The present study found
that NEDD4 enhanced the cell function of BMECs by activating the
VEGF signaling pathway and working with mTOR. When NEDD4 was
overexpressed, the mRNA levels of mTOR, VEGF and VEGFR2 were
significantly increased, demonstrating that NEDD4 promoted
angiogenesis by triggering the VEGF signaling pathway. Notably,
overexpression of NEDD4 did not markedly affect the ubiquitination
level of mTOR, suggesting that NEDD4 did not directly mediate the
ubiquitination of mTOR but instead affected it by activating the
mTOR signaling pathway. In the siRNA transfection experiments, a
synergistic effect of NEDD4 and mTOR on BMEC proliferation and
migration was observed. Additionally, glucocorticoids inhibited the
expression of NEDD4 by increasing the methylation level of the
NEDD4 promoter region. After adding the methylation inhibitor
5-AZA, the methylation levels of the NEDD4 promoter notably
decreased, while glucocorticoid MePr treatment markedly increased
the methylation level of the NEDD4 promoter and significantly
suppressed NEDD4 mRNA expression. This further supported the
important role of NEDD4 in methylation regulation and showed the
substantial influence of glucocorticoids on NEDD4 expression.
In conclusion, the present study revealed the
diverse roles of NEDD4 in SONFH, particularly in angiogenesis,
cellular homeostasis, cell cycle regulation and DNA damage repair.
NEDD4 enhanced BMEC function by activating the VEGF signaling
pathway and working with mTOR, with its expression regulated by
methylation. This offers novel insights into NEDD4 as a potential
therapeutic target for SONFH. Further research into the functions
of NEDD4 in bone pathology and angiogenesis will provide valuable
insights for developing novel treatment strategies. These findings
not only add to the existing literature but also provide novel
perspectives on the multiple roles of NEDD4 in SONFH and
angiogenesis, indicating that NEDD4 may affect angiogenesis, bone
tissue repair and bone regeneration through various mechanisms.
NEDD4 may represent a promising therapeutic target
for SONFH. By mediating PTEN degradation and activating the
PI3K/AKT/mTOR pathway, NEDD4 promoted cell proliferation and
survival, while also enhancing VEGF signaling to support
angiogenesis and bone repair (26,31).
These dual effects on osteogenesis and angiogenesis highlight its
translational potential, and future studies on pharmacological or
genetic modulation of NEDD4 could lead to novel treatment
strategies.
The present study had several limitations.
Primarily, the bioinformatics analysis was based on a single
publicly available dataset without external validation, which may
limit the generalizability of the present findings. Secondly,
although the present study validated key molecular alterations
in vitro, in vivo animal experiments were not performed,
which reduced the translational value of the results. Furthermore,
the relatively small sample size and limited types of control may
weaken the conclusions of the present study. Additionally, the
present study did not explore how NEDD4 regulates the mTOR and VEGF
pathways at a mechanistic level. Finally, the present study did not
include clinical relevance data, and thus, future studies should
incorporate patient samples and correlation analyses to increase
the translational significance of the present work. Overall, future
research with larger cohorts, more comprehensive controls, in
vivo validation, more in-depth mechanistic studies and clinical
correlation analyses is needed to confirm and extend the findings
of the present study.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was funded by The Guizhou Provincial Natural
Science Foundation [grant no. Qiankehebasis-ZK (2024) general 241],
the Start-up Fund for Doctoral Research at the Affiliated Hospital
of Guizhou Medical University (grant no. gyfybsky-2022-38), the
National Natural Science Foundation of China Cultivation Program of
Affiliated Hospital of Guizhou Medical University [grant no.
gyfynsfc(2023)-62], the Key Medical Discipline Construction Project
of Guizhou Provincial Health Commission during 2025–2026, and the
2025 Annual Hospital-Level Scientific Research Fund of Guizhou
Hospital of Beijing Jishuitan Hospital [grant no.
JGYYK(2025)-24].
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
JL conceived and designed the experiments, performed
the experiments, analyzed the data, authored and reviewed drafts of
the article, and approved the final draft. DZ performed the
experiments. YQ analyzed and interpreted data CG supervised the
study and interpreted data. All authors read and approved the final
version of the manuscript, JL and CG confirm the authenticity of
all the raw data.
Ethics approval and consent to
participate
The use of primary human cells in the present study
was conducted in accordance with The Declaration of Helsinki. The
protocol was approved by the Ethics Committee of Beijing Jishuitan
Hospital Guizhou Hospital (approval no. KT2025022804; Guiyang,
China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhang W, Du H, Liu Z, Zhou D, Li Q and Liu
W: Worldwide research trends on femur head necrosis (2000–2021): A
bibliometrics analysis and suggestions for researchers. Ann Transl
Med. 11:1552023. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang T, Azeddine B, Mah W, Harvey EJ,
Rosenblatt D and Séguin C: Osteonecrosis of the femoral head:
Genetic basis. Int Orthop. 43:519–530. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shao W, Wang P, Lv X, Wang B, Gong S and
Feng Y: Unraveling the role of endothelial dysfunction in
osteonecrosis of the femoral head: A pathway to new therapies.
Biomedicines. 12:6642024. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fukushima W, Fujioka M, Kubo T, Tamakoshi
A, Nagai M and Hirota Y: Nationwide epidemiologic survey of
idiopathic osteonecrosis of the femoral head. Clin Orthop Relat
Res. 468:2715–2724. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mont MA, Pivec R, Banerjee S, Issa K,
Elmallah RK and Jones LC: High-dose corticosteroid use and risk of
hip osteonecrosis: Meta-analysis and systematic literature review.
J Arthroplasty. 30:1506–1512.e5. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Radke S, Battmann A, Jatzke S, Eulert J,
Jakob F and Schütze N: Expression of the angiomatrix and angiogenic
proteins CYR61, CTGF, and VEGF in osteonecrosis of the femoral
head. J Orthop Res. 24:945–952. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shao W, Li Z, Wang B, Gong S, Wang P, Song
B, Chen Z and Feng Y: Dimethyloxalylglycine attenuates
steroid-associated endothelial progenitor cell impairment and
osteonecrosis of the femoral head by regulating the HIF-1α
signaling pathway. Biomedicines. 11:9922023. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Huard J: Stem cells, blood vessels, and
angiogenesis as major determinants for musculoskeletal tissue
repair. J Orthop Res. 37:1212–1220. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang Y, Xia CJ, Wang BJ, Ma XW and Zhao
DW: The association between VEGF-634C/G polymorphisms and
osteonecrosis of femoral head: A meta-analysis. Int J Clin Exp Med.
8:9313–9319. 2015.PubMed/NCBI
|
|
10
|
Hang D, Wang Q, Guo C, Chen Z and Yan Z:
Treatment of osteonecrosis of the femoral head with VEGF165
transgenic bone marrow mesenchymal stem cells in mongrel dogs.
Cells Tissues Organs. 195:495–506. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Grosso A, Burger MG, Lunger A, Schaefer
DJ, Banfi A and Di Maggio N: It takes two to tango: Coupling of
angiogenesis and osteogenesis for bone regeneration. Front Bioeng
Biotechnol. 5:682017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Portal-Núñez S, Lozano D and Esbrit P:
Role of angiogenesis on bone formation. Histol Histopathol.
27:559–566. 2012.PubMed/NCBI
|
|
13
|
Deng S, Xiang JJ, Shen YY, Lin SY, Zeng YQ
and Shen JP: Effects of VEGF-notch signaling pathway on
proliferation and apoptosis of bone marrow MSC in patients with
aplastic anemia. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 27:1925–1932.
2019.(In Chinese). PubMed/NCBI
|
|
14
|
Wang S, Lu J, You Q, Huang H, Chen Y and
Liu K: The mTOR/AP-1/VEGF signaling pathway regulates vascular
endothelial cell growth. Oncotarget. 7:53269–53276. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu Z and Li Y: Expression of the
HIF-1α/VEGF pathway is upregulated to protect alveolar bone density
reduction in nasal-obstructed rats. Histol Histopathol.
39:1053–1063. 2024.PubMed/NCBI
|
|
16
|
Gao L, Zhang W, Shi XH, Chang X, Han Y,
Liu C, Jiang Z and Yang X: The mechanism of linear ubiquitination
in regulating cell death and correlative diseases. Cell Death Dis.
14:6592023. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Popovic D, Vucic D and Dikic I:
Ubiquitination in disease pathogenesis and treatment. Nat Med.
20:1242–1253. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang Y, Huang S, Xu P and Li Y: Progress
in atypical ubiquitination via K6-linkages. Sheng Wu Gong Cheng Xue
Bao. 38:3215–3227. 2022.(In Chinese). PubMed/NCBI
|
|
19
|
Yan J, Qiao G, Yin Y, Wang E, Xiao J, Peng
Y, Yu J, Du Y, Li Z, Wu H, et al: Black carp RNF5 inhibits
STING/IFN signaling through promoting K48-linked ubiquitination and
degradation of STING. Dev Comp Immunol. 145:1047122023. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu J, Sheng Z, Li F, Wang S, Yuan Y, Wang
M and Yu Z: NEDD4 protects vascular endothelial cells against
Angiotensin II-induced cell death via enhancement of XPO1-mediated
nuclear export. Exp Cell Res. 383:1115052019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Eide PW, Cekaite L, Danielsen SA,
Eilertsen IA, Kjenseth A, Fykerud TA, Ågesen TH, Bruun J, Rivedal
E, Lothe RA and Leithe E: NEDD4 is overexpressed in colorectal
cancer and promotes colonic cell growth independently of the
PI3K/PTEN/AKT pathway. Cell Signal. 25:12–18. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang ZJ, Zhu JJ, Yang XY and Biskup E:
NEDD4 promotes cell growth and migration via PTEN/PI3K/AKT
signaling in hepatocellular carcinoma. Oncol Lett. 14:2649–2656.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Guo Y, Wang Y, Liu H, Jiang X and Lei S:
High glucose environment induces NEDD4 deficiency that impairs
angiogenesis and diabetic wound healing. J Dermatol Sci.
112:148–157. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun W, Lu H, Cui S, Zhao S, Yu H, Song H,
Ruan Q, Zhang Y, Chu Y and Dong S: NEDD4 ameliorates myocardial
reperfusion injury by preventing macrophages pyroptosis. Cell
Commun Signal. 21:292023. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xu K, Chu Y, Liu Q, Fan W, He H and Huang
F: NEDD4 E3 Ligases: Functions and mechanisms in bone and tooth.
Int J Mol Sci. 23:99372022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Drinjakovic J, Jung H, Campbell DS,
Strochlic L, Dwivedy A and Holt CE: E3 ligase Nedd4 promotes axon
branching by downregulating PTEN. Neuron. 65:341–357. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Han X, Zhang G, Chen G, Wu Y, Xu T, Xu H,
Liu B and Zhou Y: Buyang huanwu decoction promotes angiogenesis in
myocardial infarction through suppression of PTEN and activation of
the PI3K/Akt signalling pathway. J Ethnopharmacol. 287:1149292022.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou YJ, Xiong YX, Wu XT, Shi D, Fan W,
Zhou T, Li YC and Huang X: Inactivation of PTEN is associated with
increased angiogenesis and VEGF overexpression in gastric cancer.
World J Gastroenterol. 10:3225–3229. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lindberg ME, Stodden GR, King ML, MacLean
JA II, Mann JL, DeMayo FJ, Lydon JP and Hayashi K: Loss of CDH1 and
Pten accelerates cellular invasiveness and angiogenesis in the
mouse uterus. Biol Reprod. 89:82013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jeon SA, Lee JH, Kim DW and Cho JY:
E3-ubiquitin ligase NEDD4 enhances bone formation by removing
TGFβ1-induced pSMAD1 in immature osteoblast. Bone. 116:248–258.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zheng HL, Xu WN, Zhou WS, Yang RZ, Chen
PB, Liu T, Jiang LS and Jiang SD: Beraprost ameliorates
postmenopausal osteoporosis by regulating Nedd4-induced Runx2
ubiquitination. Cell Death Dis. 12:4972021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liang XZ, Luo D, Chen YR, Li JC, Yan BZ,
Guo YB, Wen MT, Xu B and Li G: Identification of potential
autophagy-related genes in steroid-induced osteonecrosis of the
femoral head via bioinformatics analysis and experimental
verification. J Orthop Surg Res. 17:862022. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu Y, Su Z, Tavana O and Gu W:
Understanding the complexity of p53 in a new era of tumor
suppression. Cancer Cell. 42:946–967. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chatterjee N and Walker GC: Mechanisms of
DNA damage, repair, and mutagenesis. Environ Mol Mutagen.
58:235–263. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ciccia A and Elledge SJ: The DNA damage
response: Making it safe to play with knives. Mol Cell. 40:179–204.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li J, Zhao H, McMahon A and Yan S: APE1
assembles biomolecular condensates to promote the ATR-Chk1 DNA
damage response in nucleolus. Nucleic Acids Res. 50:10503–10525.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Provasek V, Kodavati M, Kim B, Mitra J and
Hegde ML: TDP43 Interacts with MLH1 and MSH6 Proteins in A DNA
Damage-Inducible Manner. Mol Brain. 17:322024. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pfeiffer C, Grandits AM, Asnagli H,
Schneller A, Huber J, Zojer N, Schreder M, Parker AE, Bolomsky A,
Beer PA and Ludwig H: CTPS1 is a novel therapeutic target in
multiple myeloma which synergizes with inhibition of CHEK1, ATR or
WEE1. Leukemia. 38:181–192. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li WH, Wang F, Song GY, Yu QH, Du RP and
Xu P: PARP-1: A critical regulator in radioprotection and
radiotherapy-mechanisms, challenges, and therapeutic opportunities.
Front Pharmacol. 14:11989482023. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Olazabal-Herrero A, He B, Kwon Y, Gupta
AK, Dutta A, Huang Y, Boddu P, Liang Z, Liang F, Teng Y, et al: The
FANCI/FANCD2 complex links DNA damage response to R-loop regulation
through SRSF1-mediated mRNA export. Cell Rep. 43:1136102024.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zheng L, Jia J, Finger LD, Guo Z, Zer C
and Shen B: Functional regulation of FEN1 nuclease and its link to
cancer. Nucleic Acids Res. 39:781–794. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sonntag R, Giebeler N, Nevzorova YA,
Bangen JM, Fahrenkamp D, Lambertz D, Haas U, Hu W, Gassler N,
Cubero FJ, et al: Cyclin E1 and cyclin-dependent kinase 2 are
critical for initiation, but not for progression of hepatocellular
carcinoma. Proc Natl Acad Sci USA. 115:9282–9287. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ghelli Luserna di Rorà A, Cerchione C,
Martinelli G and Simonetti G: A WEE1 family business: Regulation of
mitosis, cancer progression, and therapeutic target. J Hematol
Oncol. 13:1262020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sokhi S, Lewis CW, Bukhari AB, Hadfield J,
Xiao EJ, Fung J, Yoon YJ, Hsu WH, Gamper AM and Chan GK: Myt1
overexpression mediates resistance to cell cycle and DNA damage
checkpoint kinase inhibitors. Front Cell Dev Biol. 11:12705422023.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Abe H, Alavattam KG, Kato Y, Castrillon
DH, Pang Q, Andreassen PR and Namekawa SH: CHEK1 coordinates DNA
damage signaling and meiotic progression in the male germline of
mice. Hum Mol Genet. 27:1136–1149. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Li S, Sampson C, Liu C, Piao HL and Liu
HX: Integrin signaling in cancer: Bidirectional mechanisms and
therapeutic opportunities. Cell Commun Signal. 21:2662023.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Huveneers S and Danen EH: Adhesion
signaling-crosstalk between integrins, Src and Rho. J Cell Sci.
122:1059–1069. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Nobta M, Tsukazaki T, Shibata Y, Xin C,
Moriishi T, Sakano S, Shindo H and Yamaguchi A: Critical regulation
of bone morphogenetic protein-induced osteoblastic differentiation
by Delta1/Jagged1-activated Notch1 signaling. J Biol Chem.
280:15842–15848. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Rafii S and Lis R: Angiocrine ANGPTL2
executes HSC functions in endothelial niche. Blood. 139:1433–1434.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Lal N, Puri K and Rodrigues B: Vascular
endothelial growth factor B and its signaling. Front Cardiovasc
Med. 5:392018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Mang T, Kleinschmidt-Doerr K, Ploeger F,
Schoenemann A, Lindemann S and Gigout A: BMPR1A is necessary for
chondrogenesis and osteogenesis, whereas BMPR1B prevents
hypertrophic differentiation. J Cell Sci. 133:jcs2469342020.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wei X, Luo L and Chen J: Roles of mTOR
signaling in tissue regeneration. Cells. 8:10752019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhang F, Wei L, Wang L, Wang T, Xie Z, Luo
H, Li F, Zhang J, Dong W, Liu G, et al: FAR591 promotes the
pathogenesis and progression of SONFH by regulating Fos expression
to mediate the apoptosis of bone microvascular endothelial cells.
Bone Res. 11:272023. View Article : Google Scholar : PubMed/NCBI
|