Introduction
Renal cell carcinoma (RCC) is the 14th most
frequently diagnosed cancer during 2022, Globally in 2022, RCC
accounted for an estimated 434,000 newly diagnosed cases,
corresponding to age-standardized incidence rates of 5.9 per
100,000 males and 3.0 per 100,000 females (1). A major clinical challenge in RCC is
its high metastatic potential. It has been reported that ~30% of
patients have distant metastasis at the time of the first diagnosis
and the most common sites of metastasis are the lungs, bones, liver
and brain (2). The prognosis for
patients with metastatic RCC is generally unfavorable, therefore
suggesting the key clinical importance of understanding the
metastatic mechanisms in the development of effective therapeutic
strategies for RCC in the future. RCC originates from the
epithelium of the proximal convoluted tubule and accounts for
80–90% of kidney cancer cases (3).
The etiology of RCC is not fully understood and the risk factors
that can be identified include smoking, obesity, hypertension,
chronic kidney disease and certain hereditary diseases, such as von
Hippel-Lindau (VHL) disease and hereditary papillary renal
carcinoma (4). Understanding these
mechanisms is key for developing targeted therapies to improve
patient outcomes.
The epithelial-mesenchymal transition (EMT) is a
fundamental biological process, by which epithelial cells organized
in tight junctions can transform into mesenchymal cells with
increased motility and invasiveness. Through this process, RCC
cells acquire mesenchymal stem cell-like features, which enhance
their metastatic potential and invasive capacity, thereby
facilitating entry into the bloodstream and colonization of distant
organs to form secondary tumors. Furthermore, EMT has been
implicated in drug resistance (5,6) and
serves a key role in renal fibrosis (7), which is a common pathological
manifestation of chronic kidney disease and is an independent risk
factor for RCC. In summary, EMT is one of the primary mechanisms of
distant metastasis in RCC and its activation is frequently
associated with poor prognosis.
Non-coding RNAs (ncRNAs) are functional RNA
molecules that do not encode proteins. Although a limited number of
ncRNAs have recently been reported to encode peptides or proteins,
the majority of these RNAs perform their biological functions
through other functions (8,9).
Major types of ncRNAs include microRNAs (miRNAs/miR), long ncRNAs
(lncRNAs), circular RNAs (circRNAs), small interfering RNAs
(siRNAs) and small nucleolar RNAs (snoRNAs). ncRNAs serve key roles
in regulating gene expression and are involved in diverse
biological processes, including growth, development, organ function
and EMT. ncRNAs are reported to be associated with various human
diseases such as cardiac hypertrophy and spinal motor neuron
disease (10), particularly
cancer, including RCC (11).
Decades of research have reported that ncRNAs can regulate EMT and
affect the occurrence, progression and metastasis of RCC through
multiple pathways (12–15). For example, downregulation of the
miR-200 family promotes EMT and enhances the invasive and
metastatic potential of cancer cells (16).
Unlike previous reports that have only partially
addressed the roles of individual ncRNA species in EMT (17) of miscellaneous cancer types like
breast cancer (18) and colorectal
carcinoma (19), to the best of
our knowledge, the present review introduces the first integrated
analysis of miRNAs, lncRNAs, circRNAs, transcribed-ultra conserved
regions and pseudogenes specifically within RCC context By
synthesizing evidence published until March 2025, the present
review reports an updated regulatory landscape that highlights
ncRNA crosstalk, most notably miRNA-lncRNA-circRNA axes, and
clarifies how these interactions drive both EMT and emerging
therapy resistance (20,21). Furthermore, the present review
translates these mechanistic insights into clinically actionable
prospects, notably evaluating the diagnostic accuracy of
circulating ncRNA signatures and the therapeutic potential of
circRNA-based vaccines together with exosome-directed
interventions, aspects that previous reviews have, to the best of
our knowledge, rarely contemplated (22,23).
Therefore, the present review comprehensively
summarizes current advances in understanding how ncRNAs regulate
EMT in RCC and explores their clinical potential as diagnostic
biomarkers and therapeutic targets to improve the prognosis of
patients with RCC in the future.
Literature search strategy
A systematic literature search was conducted using
the PubMed database (https://pubmed.ncbi.nlm.nih.gov) to identify all
relevant studies published up to March 2025. The search strategy
was designed to encompass three key concepts: ncRNAs, RCC and EMT.
Medical Subject Headings (MeSH; http://www.ncbi.nlm.nih.gov/mesh) terms were primarily
used to ensure retrieval of high-quality, relevant studies.
The core search strategy that yielded the most
comprehensive results was: [‘RNA, Untranslated’ (Mesh)] AND
‘Carcinoma, Renal Cell’(Mesh) AND ‘Epithelial-Mesenchymal
Transition’ (Mesh). This search returned 88 results. These results
were sorted by publication date to prioritize the most recent
evidence. To ensure comprehensiveness, a broader search without the
EMT term [‘RNA, Untranslated’ (Mesh)] AND ‘Carcinoma, Renal Cell’
(Mesh) was also executed, which returned 1,501 results.
Furthermore, a filter for systematic reviews was applied to the
core search string, which returned 0 results, confirming the
novelty of the synthesis approach in the present review. The
reference lists of all retrieved articles and relevant reviews were
manually screened to identify any additional publications that the
electronic search might have missed.
Inclusion criteria for the studies were as follows:
i) Original research articles or reviews focusing on ncRNAs
regulating EMT in RCC; and ii) studies published in English.
The exclusion criteria were as follows: i) Studies
not associated with RCC; ii) studies not investigating EMT or
ncRNAs; and iii) conference abstracts, editorials and letters
without original data.
Due to the predominantly mechanistic nature of the
studies reviewed, which address molecular rather than clinical
endpoints, an objective evidence hierarchy analogous to that
employed in large-scale clinical research could not be rigorously
applied; therefore, formal evidence grading akin to meta-analytical
methodology was not feasible. Although the present review is
structured as a narrative review rather than a systematic review or
meta-analysis, the present review adopted a comprehensive search
strategy with clearly defined exclusion criteria to minimize
selection bias. The present analysis incorporated all relevant
studies identified by the search, with the exception of retracted
publications. Therefore, it can be considered that the potential
for selection bias in the present review is minimal. However, the
present review acknowledges that the scope and emphasis of the
synthesis may be influenced by the current research focus within
the field, which inherently shapes the available evidence.
Overview of EMT
EMT is a biological process in which epithelial
cells undergo morphological and functional transformation into
mesenchymal cells. EMT underlies key biological processes such as
embryonic development and wound healing, and also serves a key role
in the metastasis of malignant tumors (24).
Epithelial cells are usually present in the
superficial layers of the skin or lumen, such as the small
intestinal epithelium, lung epithelium and renal tubular
epithelium. Common features of epithelial cells include abundant
intercellular connections, intercellular communication through
tight junctions and desmosomes and adhesion to each other and
tightly latch onto the basement membrane (25). In cancer, epithelial cells lose
their tissue structure and cell polarity, which suppress cancer
malignancy by controlling asymmetric cell division and the
integrity of the apical junctional complex (26). The main biomarkers of epithelial
cells are E-cadherin, β-catenin and other key proteins for
inter-epithelial cell junctions and epithelial cell adhesion
molecule (27). By contrast,
mesenchymal stem/stromal cells are extensively distributed in
various types of tissues, including bone marrow, adipose and blood.
These cells are non-polarized, often exhibit an irregular
morphology and are loosely arranged with connections mediated by
cytoplasmic protrusions. They also possess certain stem cell
properties such as self-renewal (28), CSC-marker expression (29), multi-lineage potential (28) and enhanced tumorigenicity (30). The main markers of mesenchymal
stromal cells include N-cadherin and vimentin (VIM) (31).
The EMT process in tumors typically does not occur
spontaneously but is triggered by various internal and external
factors, including growth factors such as fibroblast growth factor
(FGF), epidermal growth factor and hepatocyte growth factor.
Furthermore, several signaling pathways, including the TGF-β,
Wnt/β-catenin, NF-κB, Notch and PI3K-AKT pathways, can regulate EMT
(32). The regulatory network of
EMT is highly complex, involving multiple transcription factors
[for example, Twist, Snail and zinc finger E-box binding homeobox
(ZEB)], ncRNAs and epigenetic factors such as EZH2 (33) and HOTAIR. Numerous studies have
demonstrated that ncRNAs serve a notable role in regulating the EMT
process in cancer (8,10,11).
During EMT, epithelial cells detach from the
basement membrane and acquire a partial mesenchymal phenotype to
varying degrees, entering an intermediate state between epithelium
and mesenchyme. This transition endows cells with enhanced invasive
and migratory capabilities. Throughout this process, cells undergo
multiple changes, including cytoskeletal remodeling and
downregulation of adhesion molecules. Morphologically, epithelial
cells transform from a homogeneous, tightly packed structure to
various irregular shapes, such as spindle-like and elongated forms.
The apical-basal polarity of cells is lost and the cytoskeleton
shifts to be dominated by VIM, a wave-like protein. These changes
confer a stronger capacity for locomotion and migration,
facilitating entry into the bloodstream and subsequent colonization
of distant organs (34). However,
there is ongoing debate regarding the necessity of the EMT process
for tumor metastasis. Some studies suggest that EMT may not be key
to metastasis but can markedly enhance chemoresistance. For
instance, research using spontaneous breast-to-lung metastasis
models and genetically engineered mouse models of pancreatic ductal
adenocarcinoma has indicated that EMT is not a prerequisite for
metastasis but can contribute to drug resistance in tumor cells
(5,35). These findings highlight the complex
role of EMT in tumor biology, where its primary function may extend
beyond facilitating metastasis to enhancing therapeutic resistance.
The resistance mechanisms associated with EMT are considered to
involve increased drug efflux, reduced cell proliferation and the
evasion of apoptosis signaling pathways (36). Furthermore, numerous clinical
studies have reported that EMT is associated with immunosuppression
within the tumor microenvironment (TME), with immune cells in the
TME facilitating the progression of EMT in tumor cells (37–39).
EMT transcription factors (EMT-TF) such as Twist, Snail and ZEB are
activated, leading to the accumulation of immunosuppressive cells
within the TME (40). Furthermore,
EMT-TF inducers, including TGF-β, also contribute to the
immunosuppressive milieu of the TME (41). Therefore, it is widely recognized
that the EMT process enables cancer cells to acquire stem cell-like
properties, which not only promote the proliferation of metastatic
foci but also increase resistance to targeted therapies and
immunotherapy (42). The major
regulatory mechanisms governing EMT and the functional roles of the
ncRNAs highlighted in this review are schematically depicted in
Fig. 1.
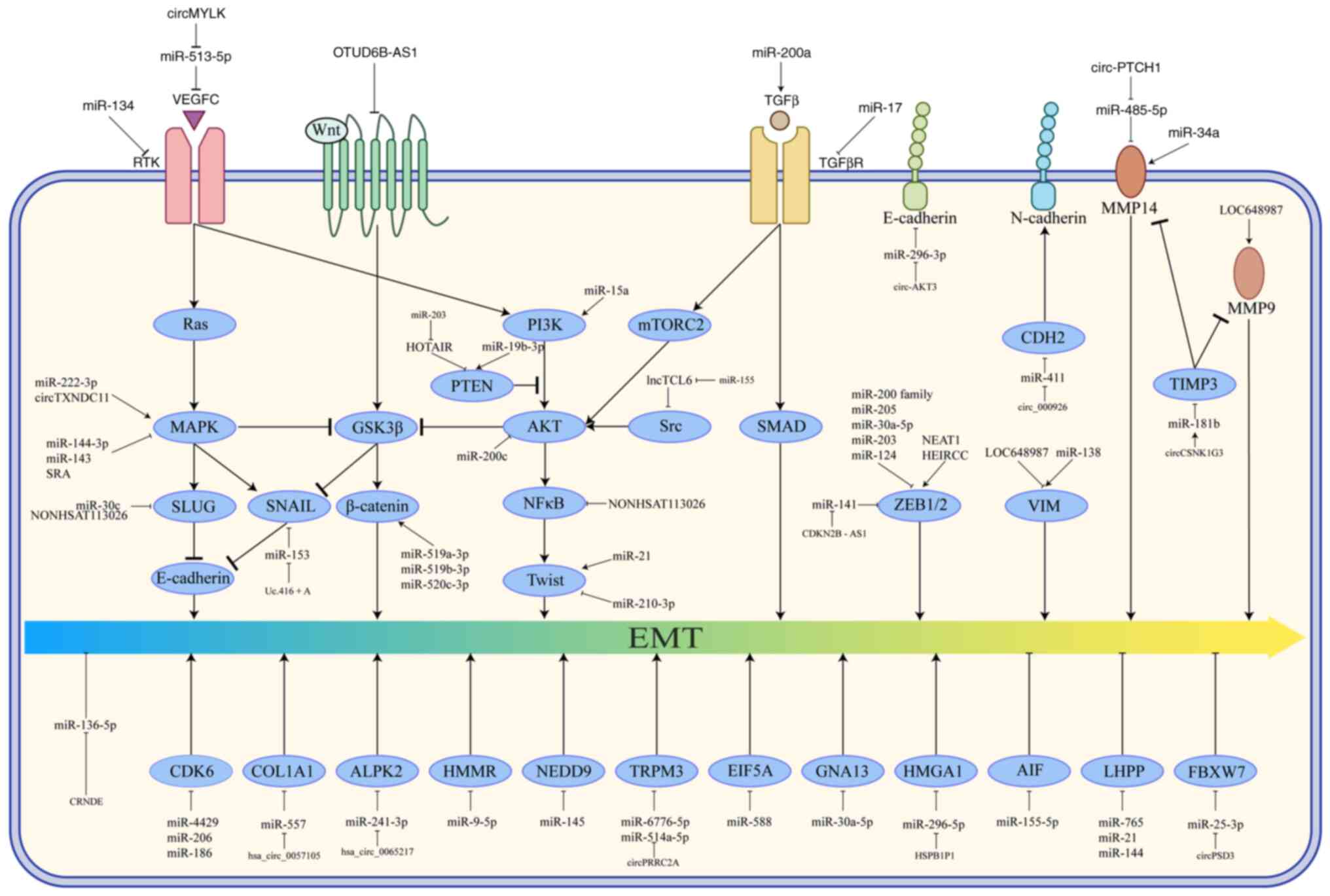 | Figure 1.Regulatory mechanisms of EMT
progression associated with ncRNA in RCC. A complex regulatory
network of ncRNAs modulates EMT in cancer cells through selective
activation or inhibition of key signaling pathway components. →,
promotion; ⊥, inhibition; EMT, epithelial-mesenchymal transition;
ncRNA, non-coding RNA; RCC, renal cell carcinoma; miRNA, microRNA;
circRNA, circular RNA; HOTAIR, HOX transcript antisense intergenic
RNA; NEAT1, nuclear enriched abundant transcript 1; SNHG, small
nuclear RNA host gene; CDKN2B-AS1, CDK inhibitor 2B-antisense RNA
1; OTUD6B, ovarian tumor domain deubiquitinase 6B. |
Therefore, the EMT process represents a key
mechanism by which cancer cells gain increased invasiveness and
metastatic potential. Numerous studies have demonstrated that the
cell populations in advanced epithelial tumors and metastatic
cancer exhibit varying degrees of mesenchymal characteristics when
compared with early-stage cancer (24,43),
suggesting that epithelial-to-mesenchymal transition is a key step
and hallmark event in cancer metastasis. Furthermore, EMT is a
reversible process and cells can undergo reversion to an epithelial
phenotype through the mesenchymal-to-epithelial transition (MET).
This MET process after metastasis may facilitate the colonization
of tumors in distant organs (7).
Regulatory mechanisms of ncRNAs
There are various types of ncRNAs, such as miRNAs,
lncRNAs, circRNAs, siRNAs, transfer RNAs, ribosomal RNAs, snoRNAs
and small nuclear RNAs. However, due to the extensive research on
the regulatory roles of miRNAs, lncRNAs and circRNAs in RCC, the
present review primarily focused on their roles. As shown in
Fig. 2, several ncRNAs influence
the progression of renal cancer by regulating the EMT process.
miRNAs constitute the hub of this regulatory network; all other
ncRNAs exert their effects principally by modulating these miRNAs,
whereas select circRNAs and lncRNAs can additionally regulate the
EMT programme directly in the absence of a downstream miRNA
intermediate.
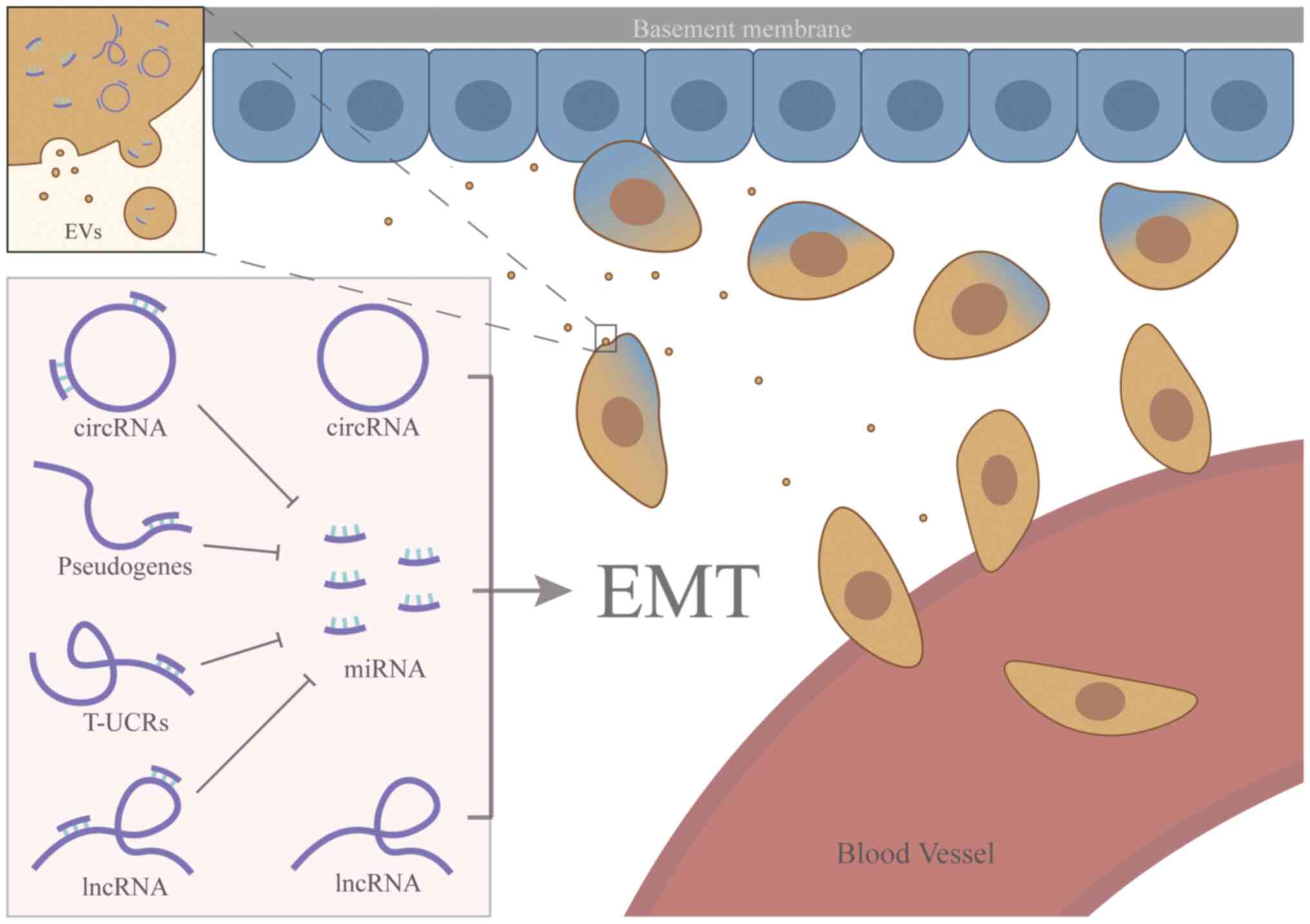 | Figure 2.ncRNAs regulate EMT in RCC. ncRNAs
regulating the EMT progression and miRNAs are the center of this
system. Pseudogenes, circRNAs, lncRNAs and T-UCRs can exert their
downstream effects by sequestering miRNAs, which establishes miRNAs
as the central hub of this regulatory network. However, certain
circRNAs and lncRNAs may also regulate EMT directly, independent of
the miRNA pathway. RNAs can be located in the cytoplasm and
extracellular vesicles of RCC. Certain tumor cells release various
EVs, including exosomes and microparticles containing ncRNAs, which
further promote the EMT process and facilitate tumor metastasis.
After EMT, the carcinoma cells become more invasive and spread to
distant organs through the circulation. EMT, epithelial-mesenchymal
transition; ncRNA, non-coding RNA; RCC, renal cell carcinoma; EVs,
extracellular vesicles; miRNA, microRNA; circRNA, circular RNA;
T-UCRs, transcribed-ultra conserved regions; lncRNA, long
non-coding RNA. |
miRNAs
miRNAs are a family of single-stranded ncRNA
molecules, ~22 nucleotides in length. These small RNAs are
ubiquitously present in eukaryotes and primarily function in
regulating gene expression in plants, animals and humans.
Dysregulation of miRNAs has been observed in nearly all types of
cancer cells such as A549 (44)
and 786-O (45), highlighting
their key role in cancer pathogenesis (46).
The primary biological function of miRNAs is to
regulate post-transcriptional gene expression. A single miRNA can
regulate multiple mRNAs, while several miRNAs can target the
expression of a single mRNA, creating a ‘many-to-many’
relationship. miRNAs interact with argonaute and other functional
proteins to form the miRNA-induced silencing complex, which is key
to their function (47). miRNAs
can influence mRNA translation through several mechanisms. If a
miRNA is complementary to its target mRNA, it will induce direct
cleavage of the transcript. This mechanism is common in plants but
rarely occurs in animals and humans, as the complementarity between
miRNAs and target mRNAs is often insufficient. In animals and
humans, miRNAs typically exert their effects by binding to
sequences in the 3′-untranslated region of mRNAs, thereby
preventing ribosome-mediated translation or promoting mRNA
destabilization (48).
Occasionally, miRNAs directly enter the nucleus and enhance the
expression of target genes. In addition, miRNAs can interact with
other ncRNAs, such as circRNAs, lncRNAs and other miRNAs (49).
lncRNAs
lncRNAs are a class of ncRNAs typically longer than
200 nucleotides. Most of the lncRNAs possess an mRNA-like
structure, including a 5′-end cap, a poly(A) tail and exon-intron
organization. However, they generally lack open reading frames of
sufficient length to encode proteins, representing a fundamental
distinction between lncRNAs and mRNAs. Despite their inability to
be translated into proteins, several lncRNAs still exert notable
biological functions. lncRNAs are involved in nearly all stages of
gene expression, including transcription, post-transcriptional
regulation and epigenetic modifications. In cancer cells, lncRNAs
regulate genes with specific functions, thereby influencing
biological processes such as proliferation, invasion and
metastasis. The mechanisms underlying these actions are complex and
diverse, including miRNA sponging, RNA-protein scaffolding,
recruitment of specific proteins, chromatin remodeling and the
production of micropeptides (50,51).
lncRNAs can function as competing endogenous RNAs (ceRNAs) when
they contain specific miRNA binding sites. Therefore, lncRNAs
inhibit the activity of their target mRNAs by sequestering miRNAs
(52). Furthermore, lncRNAs
modulate transcriptional activation or repression by influencing
chromatin topology. Certain lncRNAs can recruit various
chromatin-modifying enzymes such as PRC2 and G9a to specific loci
or directly bind to chromatin structural proteins (53), thereby altering chromatin structure
and influencing gene expression at those sites. A subset of lncRNAs
is transcribed from cis-regulatory elements, such as promoters and
enhancers, and regulate the expression levels of downstream genes
(50).
circRNAs
circRNAs are a class of ncRNAs that form covalently
closed continuous loops, with covalent bonds at both ends, thus
lacking 3′- and 5′-ends (54).
This unique structure grants circRNAs higher stability and makes
them more resistant to degradation by ribonuclease R (55,56).
Initially, circRNAs were considered byproducts of transcriptional
errors, but recent studies have revealed their notable regulatory
functions as ncRNAs.
The main mechanisms of circRNA action are as
follows: i) circRNAs can inhibit miRNA activity by binding large
numbers of miRNAs. A single circRNA may contain multiple miRNA
binding sites, allowing it to sequester numerous miRNAs. Once
bound, these miRNAs are unable to exert their regulatory effects,
thereby influencing their downstream targets. CircRNAs are often
referred to as ‘molecular sponges’ due to this function (57). Since miRNAs typically exert their
functions by binding to mRNAs, circRNAs act as ceRNAs, competing
with mRNAs for miRNA binding and thereby indirectly regulating gene
expression (58). This is the
predominant mechanism by which circRNAs function (22).
ii) In addition to miRNA binding, circRNAs can also
function as ‘protein sponges’. By binding to RNA-binding proteins,
circRNAs prevent the proper functioning of these proteins (59). Furthermore, recent studies
indicated that certain circRNAs bind to proteins and form
circRNA-protein complexes (60,61).
These complexes can serve as scaffolds facilitating interactions
between proteins and mRNAs or other proteins or participate in
subcellular protein trafficking. The various ways in which circRNAs
interact with proteins warrant further research (19,62).
iii) Although most circRNAs do not encode proteins,
a small subset of circRNAs has been reported to serve a role in
biological functions through translation into proteins (63).
miRNAs in EMT of RCC
Among all ncRNAs, miRNAs are the most extensively
studied class, with several regulatory processes of EMT involving
the participation of miRNAs. Based on their roles in the EMT
process of RCC, miRNAs can be broadly classified into several
categories: One category includes miRNAs that are downregulated
during EMT, typically functioning as tumor suppressors (6,12,13,64–88).
Another category comprises miRNAs that are upregulated during EMT,
generally promoting cancer progression. There are also miRNAs with
more complex mechanisms of action, which can simultaneously exert
both pro-tumor and antitumor effects; these will be discussed
separately. Based on the extant literature accessible to date, the
preponderance of reported miRNAs fall into the first category.
Tumor suppressor miRNAs
Numerous miRNAs are closely associated with EMT,
with the earliest identified miRNA being the miRNA-200 family
(miR-200a, miR-200b, miR-200c, miR-141, miR-429 and miR-205)
(16,64–66).
The roles of the miR-200 family in various cancer types such as
lung adenocarcinoma (LUAD) (89),
endometrial cancer (EC) (90) and
RCC (16) have been extensively
documented and they are among the most well-established miRNAs
associated with EMT. This family can be categorized into two groups
based on the chromosomal regions where it is located:
miR-200b/c/429 and miR-200a/141 (91).
It has been observed that all six of the
aforementioned miRNAs are markedly downregulated in RCC cells.
Reintroducing miRNA clusters consisting of these miRNAs inhibits
EMT development. These findings suggest a strong association
between these miRNAs and the EMT process (16,64–66).
Mechanistically, these miRNAs inhibit the expression of ZEB1/ZEB2
genes, which are direct targets of miR-200c and miR-200b,
respectively. ZEB1/2 functions to downregulate E-cadherin and
β-catenin expression by reducing inter-epithelial cell adhesion and
the interaction between epithelial cells and the basement membrane,
thereby promoting EMT (64,66).
Under normal conditions, miR-200a, miR-200b and miR-205 work
synergistically to suppress ZEB1/2 expression (64,67).
The underlying mechanism may be associated with chromosome
instability triggered by telomere shortening (92).
It is worth noting that RNAs from this family may
have distinct roles in different cancer types. For instance,
miR-200c expression is upregulated in pancreatic cancer and
non-small cell lung cancer, although the exact mechanism remains to
be elucidated. High expression of miR-144 in prostate cancer may
also indicate a relatively poor prognosis (93). Numerous other miRNAs exhibit
similar behaviors, performing different functions across various
tumors, for example, miR-106b-5p is oncogenic in gastric cancer
(GC), hepatocellular carcinoma (HCC), and RCC, yet
tumor-suppressive in non-small cell lung cancer (NSCLC) and
colorectal cancer (CRC) (94).
However, in RCC, this family is still considered oncogenic.
In addition to the classical ZEB pathway, other
mechanisms of action for the miR-200 family have been identified
through studies in RCC. In RCC, the miR-200 family can also
modulate the PI3K/AKT pathway by regulating AKT proteins, which in
turn influences the production of E-cadherin, thereby affecting the
EMT process (66). miR-429 can
directly target B-cell-specific Moloney murine leukemia virus
integration region 1 (BMI1) and E2F transcription factor 3, leading
to their downregulation. BMI1 inhibits E-cadherin expression and
the reduced levels of miR-429 in RCC result in the loss of its
inhibitory effect, thus promoting EMT (68). A previous study on disease-free
survival and overall survival in patients with clear cell RCC
(ccRCC) suggested that low miR-429 levels are associated with a
relatively poor prognosis (69).
In the context of RCC, miR-200a directly targets TGF-β2 to inhibit
RCC progression (65). miR-141 has
been reported to reverse the EMT process and enhance the
sensitivity of tumor cells to hypoxia. Furthermore, miR-141
promotes resistance to sunitinib in tumor cells (6).
Another miRNA family markedly associated with EMT is
the miR-30 family (miR-30a, miR-30b, miR-30c, miR-30a-5p,
miR-30a-3p, miR-30e and miR-30e-3p), which is also notably
downregulated in renal tumors (95). Low expression level of miR-30a-5p,
which targets ZEB2, is associated with poor patient survival and,
under normal conditions, this miRNA markedly reduces ZEB2
expression. Furthermore, this miRNA may be regulated by
lncRNA-deleted in lymphocytic leukemia 2, although this requires
further verification (70).
Compared with normal renal tissues, miR-30b-5p is downregulated in
both RCC cell lines and tissue samples. Upregulation of miR-30b-5p
in vitro markedly inhibits the proliferation and metastasis
of RCC cells. Mechanistically, miR-30b-5p directly targets and
inhibits the expression of guanine nucleotide-binding protein α 13,
which is upregulated in RCC and promotes tumor cell proliferation
and metastasis; its tumorigenic role has been validated in a
variety of other cancer types (71). miR-30c is typically an oncogenic
miRNA, serving a key role in various cancer types, including
endometrial, ovarian and breast cancer (96). In RCC, the downregulation of
miR-30c expression reduces its inhibitory effect on Snail family
transcriptional repressor 2 (SLUG), leading to increased SLUG
expression and excessive production of E-cadherin, which triggers
the EMT process. The decrease in miR-30c expression is induced by
hypoxia, a process dependent on hypoxia inducible factor (HIF) and
VHL deficiency in RCC hampers HIF degradation, resulting in
excessive HIF accumulation (97).
Similar to the aforementioned RNAs, miRNAs with
inhibitory effects on cancer are typically downregulated in RCC and
a notable proportion of them decrease with the progression of tumor
stage. Specifically, their expression is lower in advanced and
metastatic cancer types compared with early-stage and primary
cancer types (6,12,13,64–88).
The specific RNA names and targets are presented in Table I.
 | Table I.EMT-associated miRNAs in RCC. |
Table I.
EMT-associated miRNAs in RCC.
|
|
|
| Function |
|
|
|
|---|
|
|
|
|
|
|
|
|
|---|
| First author,
year | miRNA | Expression | Proliferation | Metastasis | Target | Verified in
vivo | (Refs.) |
|---|
| Gregory et
al, 2008 | miR-200 family | Down | ↓ | ↓ | ZEB1/2 | No | (64) |
| Lu et al,
2015 | miR-200a | Down | ↓ | ↓ | TGFB2 | No | (65) |
| Wang et al,
2013 | miR-200c | Down | NA | ↓ | ZEB1 and AKT | No | (66) |
| Machackova et
al, 2016; | miR-429 | Down | ↓ | ↓ | BMI1/E2F3 | No | (69,68) |
| Qiu et al,
2015 |
|
|
|
|
|
|
|
| Gregory et
al, 2008; | miR-205 | Down | ↓ | ↓ | ZEB1/2 | No | (64,67) |
| Xu et al,
2018 |
|
|
|
|
|
|
|
| Yamasaki et
al, 2012 | miR-138 | Down | ↓ | ↓ | VIM | No | (73) |
| Berkers et
al, 2013; | miR-141 | Down | ↓ | ↓ | ZEB, CDKN2B- | No | (6,74) |
| Dasgupta et
al, 2020 |
|
|
|
| AS1, cyclin-D1 and
cyclin-D2 |
|
|
| Huang et al,
2013 | miR-30c | Down | NA | ↓ | SLUG | No | (97) |
| Chen et al,
2017 | miR-30a-5p | Down | ↓ | ↓ | ZEB2 | Yes | (70) |
| Liu et al,
2017 | miR-30b-5p | Down | ↓ | ↓ | GNA13 | No | (71) |
| Lu et al,
2014 | miR-145 | Down | ↓ | ↓ | ANGPT2 and
NEDD9 | No | (75) |
| Lichner et
al, 2015 | miR-17 | Down | ↓ | ↓ | TGFBR2 | Yes | (76) |
| Liu et al,
2015 | miR-134 | Down | ↓ | ↓ | KRAS | No | (77) |
| Liu et al,
2016 | miR-144-3p | Down | ↓ | ↓ | MAP3K8 | No | (78) |
| Chen et al,
2020; | miR-203 | Down | ↓ | ↓ | HOTAIR and | Yes | (13,79,72) |
| Dasgupta et
al, 2018; |
|
|
|
| CAV1 |
|
|
| Han et al,
2020 |
|
|
|
|
|
|
|
| Dong et al,
2019 | miR-588 | Down | ↓ | ↓ | EIF5A2 | No | (80) |
| Chen et al,
2020 | miR-124 and
miR-203 | Down | ↓ | ↓ | ZEB2 | No | (13) |
| Pan et al,
2019 | miR-4429 | Down | ↓ | ↓ | CDK6 | No | (81) |
| Guo et al,
2020 | miR-206 | Down | ↓ | ↓ | CDK6 | Yes | (82) |
| Guo et al,
2020; | miR-186 | Down | ↓ | ↓ | CDK6 | Yes | (83,88) |
| Wang et al,
2021 |
|
|
|
|
|
|
|
| Xu et al,
2020 | miR-143 | Down | ↓ | ↓ | ABL2 | No | (84) |
| Sekino et
al, 2018 | miR-153 | Down | ↓ | ↓ | Snail | No | (85) |
| Chen et al,
2021 | miR-296-5p | Down | ↓ | ↓ | HMGA1 | No | (87) |
| Niu et al,
2025 | miR-9-5p | Down | ↓ | ↓ | HMMR | Yes | (12) |
| Xue et al,
2019 | miR-296-3p | Up | → | ↑ | E-cadherin | Yes | (86) |
| Cao et al,
2016; | miR-21 | Up | ↑ | ↑ | HIF-1α | No | (102,103) |
| Wu et al,
2019 |
|
|
|
|
|
|
|
| Wang et al,
2019 | miR-19b-3p | Up | ↑ | ↑ | PTEN | Yes | (14) |
| Lyu et al,
2020 | miR-222-3p | Up | ↑ | ↑ | TIMP2 and
ERK1/2 | No | (104) |
| Gorka et al,
2021 | miR-519a-3p,
miR-519b-3p and miR-520c-3p | Up | ↑ | ↑ | Wnt pathway
inhibitors (SFRP4, KREMEN1, CXXC4, CSNK1A1 and ZNFR3) | No | (105) |
| Lei et al,
2021 | miR-155-5p | Up | ↑ | ↑ | AIF | Yes | (106) |
| Kulkarni et
al, 2021 | miR-155 | Up | ↑ | ↑ | lncTCL6 | Yes | (15) |
| Li et al,
2021 | miR-15a | Up | ↑ | ↑ | BTG2, PI3K and
AKT | No | (101) |
| Meng et al,
2021 | miR-765, miR-21 and
miR-144 | Up | ↑ | ↑ | LHPP | No | (107) |
| Yoshino et
al, 2017 | miR-210-3p | Up | ↓ | ↓ | TWIST1 | Yes | (109) |
| Landolt et
al, 2017 | miR-34a | Up | NA | NA | MMP14 and AXL | No | (108) |
Certain miRNAs can influence EMT through multiple
pathways. An example of this is a study by Han et al
(72), in which miR-203,
previously described as targeting FGF2, was reported to inhibit the
EMT process by another mechanism: Inactivation of the PI3K/AKT/mTOR
signaling pathway via the inhibition of caveolin-1 (CAV1).
In addition to acting individually, miRNAs can
cooperate with each other. Multiple miRNAs can work together to
regulate a common target, as seen in the case of miR-124 and
miR-203. miR-124 is a key miRNA that targets CAV1 and flotillin 1
to promote ccRCC invasiveness and its downregulation can be a
predictor of relatively poor prognosis (98). miR-203 targets FGF2 and reduces
tumor invasiveness (99); it is
regulated by lncRNA snRNA host gene (SNHG) 14 and HOX transcript
antisense intergenic RNA (HOTAIR) and is also considered an
oncogene and indicator of poor prognosis in certain studies
(13). Both miR-124 and miR-203
have been identified to be downregulated in RCC cell lines, where
they have a synergistic effect in regulating EMT, with ZEB2 being
their direct target. When miR-124 and miR-203 were overexpressed in
RCC cell lines, ZEB2 expression was reduced by 11 and 27%,
respectively. Co-overexpression of both miRNAs resulted in a 45%
reduction in ZEB2 expression. Co-transfection of miR-124 and
miR-203 inhibited the invasion and metastasis of RCC cells more
effectively compared with the transfection of either miRNA alone
(13). This study suggested that a
single RNA may lack efficacy, but a synergistic combination of
multiple RNAs can form a regulatory network with notable
effects.
miRNAs can also serve a role in intercellular
communication. Grange et al (100) reported that microvesicles (MVs)
derived from CD105+ RCC cells stimulate angiogenesis and
the formation of a lung premetastatic niche in vitro and
in vivo. These MVs contain miRNAs involved in tumor
progression and metastasis, including miR-19b, miR-29c and miR-151.
These miRNAs were reported to be upregulated in renal carcinomas
compared with normal renal tissue, although the specific roles of
these miRNAs were not further investigated (100). Nevertheless, miRNAs can regulate
the metastasis of RCC via extracellular vesicles, as these MVs are
released from mesenchymal stem cells, where EMT may serve a role.
This was exemplified in the work of Wang et al (14), which demonstrated that exosomes
derived from ccRCC cancer stem cells (CSCs) had higher expression
levels of miR-19b-3p compared with that in normal tissues. This
miRNA can promote the EMT process by targeting PTEN, thus
facilitating distant tumor metastasis. Experiments in vivo
and in vitro have demonstrated that this miRNA markedly
enhances the malignancy of ccRCC (14). Furthermore, exosomal miR-15a from
adenocarcinoma human nasal (ACHN) cells has been reported to
enhance EMT activity and aggravate ccRCC via the B-cell
translocation gene 2/PI3K/AKT axis (101).
Several miRNAs have been identified as promoters of
EMT in kidney cancer. For instance, miR-138 is downregulated in
renal cancer cells and under normal physiological conditions, this
miRNA inhibits the synthesis of VIM, a key component of the
mesenchymal cytoskeleton. The downregulation of miR-138 facilitates
the EMT process in cancer cells (73). Similarly, miR-134 directly targets
KRAS, thereby suppressing the ERK signaling pathway, which enhances
cell proliferation and inhibits EMT in RCC (77). Furthermore, miR-4429 exerts its
effects by inhibiting CDK6 and its downregulation in RCC leads to
relative upregulation of CDK6, which promotes tumor cell
proliferation, invasion and metastasis, thereby predicting a worse
prognosis in patients (81). The
mechanisms underlying the EMT-inhibitory roles of certain miRNAs,
such as miR-145 and miR-17, remain to be elucidated. The latter may
regulate the EMT process through modulation of TGF-β signaling
(75,76).
Oncogenic miRNAs
In comparison with cancer-inhibiting miRNAs, the
number of identified oncogenic miRNAs is relatively limited in
current reports (14,15,86,101–107). miR-21 is known to exhibit
oncogenic properties and can promote the formation of ccRCC tumor
spheres (102). Other oncogenic
miRNAs include miR-34a, although its exact mechanism is not fully
understood. miR-34a may exert its effects by regulating MMP14
(108).
However, oncogenic miRNAs that are highly expressed
in RCC tissues do not always promote the EMT process. For example,
miR-210-3p is highly expressed in RCC cell lines and clinical ccRCC
tissues; however, its expression is lower in high-grade and
advanced ccRCC with metastasis compared with low-grade and
early-stage ccRCC. Knockdown or overexpression of miR-210-3p
promoted or suppressed tumor invasiveness, respectively.
Furthermore, it was reported that miR-210-3p inhibited the EMT
process, as evidenced by changes in cell morphology. Knockdown of
miR-210-3p enhanced Twist-related protein 1 expression, promoting
tumor metastasis. This study suggested that miR-210-3p is necessary
for tumorigenesis and its inhibitory effect on EMT should be
relieved in advanced metastatic stages (109).
lncRNAs in EMT of RCC
lncRNAs are predominantly oncogenic in RCC (74,79,110–118). For example, lncRNA HEIRCC acts as
a positive regulator of EMT by suppressing the production of
E-cadherin through the inhibition of ZEB1. This lncRNA is
upregulated in RCC and its silencing leads to decreased tumor cell
proliferation and increased apoptosis (113).
Several lncRNAs interact with miRNAs. A well-known
example is HOTAIR, which has been implicated in various types of
cancer like breast carcinoma (119) and prostate cancer (120). In RCC, miR-203 exerts an
oncostatic effect by interacting with HOTAIR, inhibiting EMT as
well as tumor cell proliferation, invasion and metastasis, as
demonstrated in both in vivo and in vitro experiments
(79). Similarly, CDK inhibitor 2B
antisense RNA 1 (AS1) is markedly overexpressed in RCC, with its
expression increasing from early to advanced stages of the disease.
This lncRNA can bind to miR-141 and influence its regulation of
cyclin D, subsequently regulating the EMT process (74).
By contrast, certain lncRNAs function through
independent mechanisms. For instance, SNHG15 is highly expressed in
RCC tissues and cell lines compared with that in adjacent non-tumor
tissues and a proximal tubule epithelial cell line. Knockdown of
SNHG15 markedly inhibits tumor cell proliferation. SNHG15 promotes
the expression of Snail family transcriptional repressor 1, SLUG
and ZEB1 through the NF-κB pathway, thereby facilitating the EMT
process and promoting invasion and metastasis in RCC (114). Similarly, nuclear enriched
abundant transcript 1 (NEAT1) is highly expressed in ccRCC samples
and cell lines and is associated with poor patient prognosis
(112). Knockdown or
overexpression of NEAT1 in ccRCC cell lines markedly inhibits or
promotes tumor cell proliferation, invasion and metastasis.
Mechanistically, NEAT1 may act by promoting ZEB1, NEAT1 knockdown
repressed ZEB1 expression in 786-O and Caki-1 cells, thereby
impairing the subsequent epithelial-mesenchymal transition
(112). lncRNA Z38 is highly
expressed in patients with RCC, with its transcriptional level
being higher in late-stage RCC. Z38 promotes tumor invasion and
metastasis through the EMT process in vitro; however, the
upstream and downstream mechanisms remain to be elucidated in
further research (111).
However, certain lncRNAs exert antitumor effects
(15,121–123). lncRNA NONHSAT113026 is markedly
downregulated in RCC and its low expression is associated with poor
prognosis. Upregulation of this RNA inhibits tumor cell
proliferation in vivo and in vitro and suppresses the
EMT process, reducing invasiveness and metastatic potential.
Mechanistically, NONHSAT113026 functions by binding to the 3′-UTR
of NF-κB/p50 and SLUG in RCC cells, inhibiting their expression
(121). Similarly, lncRNA-ovarian
tumor domain deubiquitinase 6B-AS1 exerts its effects by inhibiting
the aberrantly activated Wnt/β-catenin pathway in tumors (122).
Due to the complex biological mechanisms of lncRNAs,
the targets of certain lncRNAs remain to be elucidated. Certain
lncRNAs exert their effects by targeting downstream miRNAs, such as
SNHG12 and colorectal neoplasia differentially expressed. Others
directly target factors associated with EMT, including
high-expressed in RCC and LOC648987. Further research is warranted
to elucidate the mechanisms through which these RNAs function.
Detailed information on these lncRNAs is provided in Table II.
 | Table II.EMT-associated lncRNAs in RCC. |
Table II.
EMT-associated lncRNAs in RCC.
|
|
|
| Function |
|
|
|
|
|---|
|
|
|
|
|
|
|
|
|
|---|
| First author,
year | lncRNA | Expression | Proliferation | Metastasis | Target | miRNA target
genes/proteins | Verified in
vivo | (Refs.) |
|---|
| Xiong et al,
2016 | lncRNA-ATB | Up | ↑ | ↑ | NA | NA | No | (110) |
| Dasgupta et
al, 2018 | HOTAIR | Up | ↑ | ↑ | PTEN | - | Yes | (79) |
| He et al,
2017 | Z38 | Up | ↑ | ↑ | NA | NA | No | (111) |
| Ning et al,
2017 | NEAT1 | Up | ↑ | ↑ | ZEB1 | - | No | (112) |
| Xiong et al,
2017 | HEIRCC | Up | ↑ | ↑ | ZEB1 | - | No | (113) |
| Du et al,
2018 | SNHG15 | Up | ↑ | ↑ | NF-κB | - | No | (114) |
| Dasgupta et
al, 2020 | CDKN2B-AS1 | Up | ↑ | ↑ | miR-141 | cyclin-D1 and
cyclin-D2 | Yes | (74) |
| Su et al,
2021 | LOC648987 | Up | ↑ | ↑ | Vimentin and
MMP-9 | - | Yes | (115) |
| Yu et al,
2021 | SNHG12 | Up | ↑ | ↑ | miR-30a-3p | RUNX2, IGF-1R and
Wnt2 | Yes | (116) |
| Zhang et al,
2021 | CRNDE | Up | ↑ | ↑ | miR-136-5p | NA | No | (117) |
| Shao et al,
2023 |
RP11-367G18.1V2 | Up | ↑ | ↑ | H4K16Ac | - | Yes | (118) |
| Pu et al,
2019 | NONHSAT-113026 | Down | ↓ | ↓ | NF-κB/p50 and
SLUG | - | Yes | (121) |
| Wang et al,
2019 | OTUD6B-AS1 | Down | ↓ | ↓ | Wnt/β-catenin | - | Yes | (122) |
| Zhang et al,
2020 | SRA | Down | ↓ | ↓ | ERK | - | No | (123) |
| Kulkarni et
al, 2021 | lncTCL6 | Down | ↓ | ↓ | Src | Src/AKT | Yes | (15) |
circRNAs in EMT of RCC
As presented in Table
III, circRNAs primarily function as miRNA sponges during EMT in
RCC. The circRNA-miRNA-gene regulatory axis is commonly observed in
circRNA studies (86,124–133). When circRNAs target oncogenic
miRNAs, they exert pro-oncogenic effects and are often upregulated
in tumor cell lines and RCC tissues. For example, circPRRC2A, by
competitively binding to miR-514a-5p and miR-6776-5p, disrupts
their inhibitory effect on the target gene transient receptor
potential melastatin-3, resulting in increased expression of this
oncogene and promotion of EMT, tumor cell proliferation and
metastasis (128). Similarly,
circRNAs that act as sponges for pro-oncogenic miRNAs can exert
oncogenic effects. For instance, circ-AKT3 upregulates the
expression level of its downstream target, E-cadherin, by adsorbing
miR-296-3p, thereby inhibiting the EMT process and suppressing RCC
cell proliferation, invasion and migration both in vitro and
in vivo (86).
 | Table III.EMT-associated circRNAs in RCC. |
Table III.
EMT-associated circRNAs in RCC.
|
|
|
| Function |
|
|
|
|
|---|
|
|
|
|
|
|
|
|
|
|---|
| First author,
year | circRNA | Expression | Proliferation | Metastasis | Target | miRNA target
genes/proteins | Verified in
vivo | (Refs.) |
|---|
| Zhang et al,
2019 | circ_000926 | Up | ↑ | ↑ | miR-411 | CDH2 | Yes | (126) |
| Li et al,
2020 | circMYLK | Up | ↑ | ↑ | miR-513a-5p | VEGFC | Yes | (127) |
| Li et al,
2020 | circPRRC2A | Up | ↑ | ↑ | miR-514a-5p and
miR-6776-5p | TRPM3 | Yes | (128) |
| Liu et al,
2020 | circPTCH1 | Up | NA | ↑ | miR-485-5p | MMP14 | Yes | (129) |
| Yan et al,
2021 |
hsa_circ_0065217 | Up | ↑ | ↑ | miR-214-3p | ALPK2 | No | (130) |
| Li et al,
2022 | circCSNK1G3 | Up | ↑ | ↑ | miR-181b | TIMP3 | Yes | (131) |
| Cen et al,
2023 |
has_circ_0057105 | Up | ↑ | ↑ | miR-557 | COL1A1/VDAC2 | Yes | (132) |
| Yang et al,
2021 |
hsa_circ_101705/circTXNDC11 | Up | ↑ | ↑ | p-ERK/p-MEK | - | Yes | (134) |
| Xue et al,
2019 |
hsa_circ_0017252/circ-AKT3 | Down | → | ↓ | miR-296-3p | E-cadherin | Yes | (86) |
| Xie et al,
2022 | circPSD3 | Down | ↓ | ↓ | miR-25-3p | FBXW7 | Yes | (133) |
However, circTXNDC11 regulates EMT through direct
modulation of the ERK/MEK pathway. In this study, the researchers
observed changes in the expression levels of phosphorylated (p)-ERK
and p-MEK proteins without involving any miRNA molecules (134). Further studies on how circRNAs
exert effects on EMT through other mechanisms are warranted.
Although circRNAs have only recently gained notable
research attention, they have been reported to serve key roles in
regulating gene expression. Therefore, future research in this area
has a lot of potential.
Other ncRNAs in EMT of RCC
A number of ncRNAs also contribute to the EMT
process in RCC, such as transcribed-ultra conserved regions
(T-UCRs) and pseudogenes, which require further investigation.
T-UCRs represent a novel class of ncRNAs, typically
located at fragile sites and cancer-associated genomic regions. One
such T-UCR, Uc.416+A, which has been reported to be upregulated in
gastric cancer and downregulated in prostate cancer, exhibits high
expression levels in RCC, particularly in metastatic RCC tissues.
Knockdown of Uc.416+A in RCC cells resulted in reduced cell
proliferation and migration activity, along with notable changes in
EMT biomarker levels. Luciferase reporter assays confirmed that
Uc.416+A is directly regulated by miR-153, a miRNA known to promote
EMT through Snail. Uc.416+A promotes RCC EMT by binding to and
sequestering miR-153, thus preventing its regulatory functions. To
the best of our knowledge, this is the first T-UCR confirmed to
modulate EMT (85).
Pseudogenes, traditionally regarded as ‘junk DNA’,
share sequence similarity with ≥1 paralogous genes but lack
functional protein-coding ability. However, a subset of pseudogenes
can regulate gene expression by acting as decoys or ceRNAs
(135). For instance, Chen et
al (87) identified that the
pseudogene heat shock protein family B (small) member 1 pseudogene
1 promotes RCC proliferation and metastasis by targeting the
miR-296-5p/high mobility group AT-hook 1 axis. This finding has
expanded the understanding of the regulatory functions of ncRNAs in
RCC.
In certain cases, pseudogenes and T-UCRs have been
classified as lncRNAs due to their unique characteristics. However,
due to their distinct features, pseudogenes and T-UCRs were chosen
to be discussed separately in the present review.
Potential of EMT-associated ncRNAs in the
diagnosis and treatment of RCC
RCC is an aggressive carcinoma with a rising
incidence, the age-standardized incidence rate (ASIR) rose from
2.88 to 4.37 per 100,000, an increase of ~52% (136), and 25–30% of patients with RCC
present with metastasis at the time of diagnosis (137,138). Therefore, identifying appropriate
biomarkers and therapeutic targets is of notable importance for the
clinical treatment of RCC.
Currently, it is commonly considered that the EMT
process may be a key step in tumorigenesis and progression
(139,140). After the EMT process in RCC,
cells enhance their invasiveness and metastatic potential and
increase their drug resistance (141). Therefore, the early detection of
ongoing EMT via ncRNA signatures presents a key target for
therapeutic intervention. Halting or reversing the EMT process at
this stage could prevent the acquisition of invasive and metastatic
capabilities, potentially improving patient outcomes in the future
(142).
The EMT process in RCC is heavily regulated by
ncRNAs. Exploring the mechanisms through which ncRNAs modulate EMT
is key to identify novel therapeutic strategies and targets in RCC.
As previously discussed, several EMT-associated ncRNAs are
differentially expressed in tumor tissues compared with normal
tissues, and the expression levels of certain ncRNAs are associated
with tumor stage, malignancy and metastasis. These observations
highlight the potential of using these ncRNAs as diagnostic and
prognostic markers for RCC in the future (112,143). For example, Silva-Santos et
al (144) demonstrated that
miR-141 and miR-200b could effectively distinguish renal tumors
from normal renal tissues and miR-21, miR-141 and miR-155 could
serve as prognostic markers for patients with RCC. Furthermore,
Zhang et al (145)
constructed a risk-scoring system using EMT-associated lncRNAs,
which can predict overall survival and immune microenvironment
status in patients with ccRCC.
Since the EMT process is reversible to a certain
extent, EMT-associated ncRNAs also offer potential as therapeutic
targets for RCC in the future. Approaches such as the synthesis of
mimics of oncogenic ncRNAs or antisense oligonucleotides targeting
oncogenic ncRNAs may prove effective. For example, cerebellar
degeneration-related protein 1 antisense (ciRS-7), an oncogenic RNA
in RCC, is highly expressed in tumors and its expression is
associated with poor prognosis. Researchers have developed
poly(β-amino ester)/si-ci-ciRS-7 nanocomplexes, which can markedly
inhibit its oncogenic effects both in vitro and in
vivo (143). This approach
also illustrates the potential to combine RNA-based therapies with
nanomaterials in RCC treatment. Furthermore, CSCs may induce EMT by
secreting exosomes containing ncRNAs, which contribute to TME
formation and distant metastasis. Removing CD103+ CSC
exosomes may represent a novel strategy for the treatment of
patients with metastatic RCC in the future (14).
A notable portion of advanced RCC treatment relies
on targeted therapies, such as sunitinib. However, several patients
are either insensitive to these treatments or develop resistance
over time, leading to poor prognosis (146). A previous study reported that the
level of EMT-associated miR-141 can influence the therapeutic
response to sunitinib in patients with RCC (6). Therefore, altering drug resistance by
modulating the EMT process through ncRNAs may offer a promising
adjuvant therapeutic strategy in the future.
Furthermore, as miRNAs can be regulated by other
ncRNAs, such as circRNAs and lncRNAs, these molecules can serve as
natural inhibitors of oncogenic miRNAs. Compared with linear RNAs,
circRNAs have a more stable molecular structure, making them less
susceptible to degradation by RNases and prolonging their half-life
(147). Furthermore, circRNAs
typically contain numerous RNA-binding sites, which allow them to
efficiently sponge miRNAs. Unmodified circRNAs are less immunogenic
compared with linear mRNAs and N6-methyladenosine modification of
exogenous circRNAs almost entirely abrogates their immunogenicity
(148). Furthermore, lipid
nanoparticle-delivered (LNP) circRNAs provoke minimal immune
responses following injection, highlighting their safety profile
(149). These properties make
circRNAs promising candidates for reliable cancer biomarkers in the
future and anticancer drugs with fewer side effects, however, their
clinical translation necessitates further investigation. A
circRNA-based vaccine platform has demonstrated notable efficacy in
B16 orthotopic melanoma and B16 lung metastasis models (150). Thus, it is conceivable that
circRNA-based vaccines targeting EMT could be a viable therapeutic
approach for RCC.
Study limitations and future
perspectives
Despite comprehensively summarizing the regulatory
role of ncRNAs in RCC EMT, the present review also highlighted
several inherent limitations in the current body of literature that
should be addressed in future research.
Firstly, the mechanistic insights discussed in the
present review are predominantly derived from in vitro cell
line experiments and murine xenograft models. Although these
studies were valuable, these models cannot fully recapitulate the
intricate TME, immune cell interactions and physiological pressures
present in human patients. Potential confounding factors, such as
the selective pressure of cell culture and the lack of a fully
functional immune system in immunodeficient mice, may oversimplify
the complex role of ncRNAs in vivo (151). Future studies should prioritize
more physiologically relevant models, such as patient-derived
organoids, orthotopic implantation models and genetically
engineered mouse models of RCC, to validate these findings
(152).
Secondly, there is a notable heterogeneity across
published studies regarding RCC subtypes (for example, ccRCC vs.
papillary), sampling techniques and methodological approaches (for
example, RNA extraction, normalization methods in reverse
transcription-quantitative PCR). This heterogeneity poses a
challenge for direct comparison and meta-analysis. The clinical
translation of ncRNA biomarkers will require rigorous
standardization of protocols and validation in large-scale,
multi-center, prospective patient cohorts to ensure reliability and
reproducibility (153).
Thirdly, to the best of our knowledge, due to the
paucity of data on EMT-associated ncRNAs in RCC, molecules other
than the well-characterized miR-200 family are typically described
in single reports, precluding longitudinal or cross-study
comparisons. As the majority of these publications remain
methodologically constrained, most notably by the absence of in
vivo validation, the present review contended that systematic
re-evaluation of previously identified ncRNAs constitutes a
clinically relevant and underexplored avenue of investigation.
Lastly, while the present review outlined numerous
ncRNAs with therapeutic potential, their delivery to tumor sites
in vivo remains a notable hurdle. Future research should
focus on developing efficient and targeted delivery systems, such
as LNPs or exosome-based vectors, conjugated with ligands specific
to RCC cells to potentially minimize off-target effects and
maximize therapeutic efficacy in the future.
Addressing these limitations will be key to moving
the field forward from descriptive mechanistic studies to the
clinical realization of ncRNA-based diagnostics and therapies for
patients with RCC.
Conclusions
The study of the metastatic mechanisms of RCC is of
notable clinical importance, as a large proportion of patients with
RCC present with metastasis at the time of their initial diagnosis.
EMT serves as a pivotal driver of RCC metastasis and the present
review has systematically elucidated the regulatory roles of
ncRNAs, particularly miRNAs, lncRNAs and circRNAs, in this process.
miRNAs are central to these ncRNA-mediated regulatory networks in
RCC, with the most extensive research conducted in this area. Most
circRNAs, T-UCRs, pseudogenes and certain lncRNAs regulate EMT by
targeting miRNAs, primarily via the ceRNA mechanism.
Beyond summarizing current knowledge, the present
review highlighted the notable clinical potential of EMT-associated
ncRNAs. Their differential expression across tumor stages and
metastatic states positions them as promising diagnostic and
prognostic biomarkers. Furthermore, their involvement in
therapeutic resistance and metastasis underscores their potential
value as novel therapeutic targets. For instance, strategies such
as synthetic miRNA mimics, antisense oligonucleotides or
circRNA-based vaccines offer potential avenues for intervention in
the future. The stability, low immunogenicity and efficient
regulatory capacity of circRNAs, make them promising candidates for
next-generation RNA therapeutics.
In conclusion, RCC can enhance its malignancy
through the EMT process, which is tightly regulated by various
ncRNAs. This highlights the notable potential of these molecules in
RCC diagnosis, prognosis and treatment in the future. Through
further research on these ncRNAs, novel biomarkers for RCC may be
identified and novel therapeutic targets may be revealed that
potentially improve patient outcomes in the future.
Acknowledgements
The authors would like to thank Dr Mariya
Rangarajan (International School of Medicine, Zhejiang University,
Yiwu, China) for their review of the language in the
manuscript.
Funding
Funding: No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
ZL and HZ conceptualized the present review. ZL
reviewed the data and wrote the manuscript. CZ provided
supervision. CZ, ZL, JS, YQ and XG reviewed and edited the
manuscript. All authors read and approved the final manuscript.
Data authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263.
2024.PubMed/NCBI
|
|
2
|
Rouprêt M, Seisen T, Birtle AJ, Capoun O,
Compérat EM, Dominguez-Escrig JL, Gürses Andersson I, Liedberg F,
Mariappan P, Hugh Mostafid A, et al: European association of
urology guidelines on upper urinary tract urothelial carcinoma:
2023 Update. Eur Urol. 84:49–64. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Motzer RJ, Jonasch E, Agarwal N, Alva A,
Baine M, Beckermann K, Carlo MI, Choueiri TK, Costello BA, Derweesh
IH, et al: Kidney cancer, version 3.2022, NCCN clinical practice
guidelines in oncology. J Natl Compr Cancer Netw. 20:71–90. 2022.
View Article : Google Scholar
|
|
4
|
Young M, Jackson-Spence F, Beltran L, Day
E, Suarez C, Bex A, Powles T and Szabados B: Renal cell carcinoma.
Lancet. 404:476–491. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zheng X, Carstens JL, Kim J, Scheible M,
Kaye J, Sugimoto H, Wu CC, LeBleu VS and Kalluri R:
Epithelial-to-mesenchymal transition is dispensable for metastasis
but induces chemoresistance in pancreatic cancer. Nature.
527:525–530. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Berkers J, Govaere O, Wolter P, Beuselinck
B, Schöffski P, van Kempen LC, Albersen M, Van den Oord J, Roskams
T, Swinnen J, et al: A possible role for microRNA-141
down-regulation in sunitinib resistant metastatic clear cell renal
cell carcinoma through induction of epithelial-to-mesenchymal
transition and hypoxia resistance. J Urol. 189:1930–1938. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Galichon P, Finianos S and Hertig A:
EMT-MET in renal disease: Should we curb our enthusiasm? Cancer
Lett. 341:24–29. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fu XD: Non-coding RNA: A new frontier in
regulatory biology. Natl Sci Rev. 1:190–204. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xiao Y, Ren Y, Hu W, Paliouras AR, Zhang
W, Zhong L, Yang K, Su L, Wang P, Li Y, et al: Long non-coding
RNA-encoded micropeptides: Functions, mechanisms and implications.
Cell Death Discov. 10:4502024. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Slack FJ and Chinnaiyan AM: The role of
non-coding RNAs in oncology. Cell. 179:1033–1055. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Niu X, Lu D, Zhan W, Sun J, Li Y, Shi Y,
Yu K, Huang S, Ma X, Liu X and Liu B: miR-9-5p/HMMR regulates the
tumorigenesis and progression of clear cell renal cell carcinoma
through EMT and JAK1/STAT1 signaling pathway. J Transl Med.
23:362025. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen J, Zhong Y and Li L: miR-124 and
miR-203 synergistically inactivate EMT pathway via coregulation of
ZEB2 in clear cell renal cell carcinoma (ccRCC). J Transl Med.
18:692020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang L, Yang G, Zhao D, Wang J, Bai Y,
Peng Q, Wang H, Fang R, Chen G, Wang Z, et al: CD103-positive CSC
exosome promotes EMT of clear cell renal cell carcinoma: Role of
remote MiR-19b-3p. Mol Cancer. 18:862019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kulkarni P, Dasgupta P, Hashimoto Y,
Shiina M, Shahryari V, Tabatabai ZL, Yamamura S, Tanaka Y, Saini S,
Dahiya R and Majid S: A lncRNA TCL6-miR-155 interaction regulates
the Src-Akt-EMT network to mediate kidney cancer progression and
metastasis. Cancer Res. 81:1500–1512. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Saleeb R, Kim SS, Ding Q, Scorilas A, Lin
S, Khella HW, Boulos C, Ibrahim G and Yousef GM: The miR-200 family
as prognostic markers in clear cell renal cell carcinoma. Urol
Oncol. 37:955–963. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang ZH, Wang Y, Zhang Y, Zheng SF, Feng
T, Tian X, Abudurexiti M, Wang ZD, Zhu WK, Su JQ, et al: The
function and mechanisms of action of circular RNAs in urologic
cancer. Mol Cancer. 22:612023. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Javdani H, Mollaei H, Karimi F, Mahmoudi
S, Farahi A, Mirzaei-Parsa MJ and Shahabi A: Review article
epithelial to mesenchymal transition-associated microRNAs in breast
cancer. Mol Biol Rep. 49:9963–9973. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ,
Ma XD, Han K, Chen JW, Judde JG, Deas O, et al:
N6-methyladenosine modification of circNSUN2 facilitates
cytoplasmic export and stabilizes HMGA2 to promote colorectal liver
metastasis. Nat Commun. 10:46952019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Entezari M, Taheriazam A, Orouei S, Fallah
S, Sanaei A, Hejazi ES, Kakavand A, Rezaei S, Heidari H,
Behroozaghdam M, et al: LncRNA-miRNA axis in tumor progression and
therapy response: An emphasis on molecular interactions and
therapeutic interventions. Biomed Pharmacother. 154:1136092022.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen B, Dragomir MP, Yang C, Li Q, Horst D
and Calin GA: Targeting non-coding RNAs to overcome cancer therapy
resistance. Signal Transduct Target Ther. 7:1212022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen L and Shan G: CircRNA in cancer:
Fundamental mechanism and clinical potential. Cancer Lett.
505:49–57. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu Y, Liu H, Zhang Y, Luo J, Li H, Lai C,
Shi L and Heng B: piRNAs and circRNAs acting as diagnostic
biomarkers in clear cell renal cell carcinoma. Sci Rep.
15:77742025. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nieto MA: Epithelial plasticity: A common
theme in embryonic and cancer cells. Science. 342:12348502013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Glauert AM, Daniel MR, Lucy JA and Dingle
JT: Studies on the mode of action of excess of vitamin A. VII.
Changes in the fine structure of erythrocytes during haemolysis by
vitamin A. J Cell Biol. 17:111–121. 1963. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Royer C and Lu X: Epithelial cell
polarity: A major gatekeeper against cancer? Cell Death Differ.
18:1470–1477. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Han L, Luo H, Huang W, Zhang J, Wu D, Wang
J, Pi J, Liu C, Qu X, Liu H, et al: Modulation of the EMT/MET
process by E-cadherin in airway epithelia stress injury.
Biomolecules. 11:6692021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang SS, Jiang J, Liang XH and Tang YL:
Links between cancer stem cells and epithelial-mesenchymal
transition. OncoTargets Ther. 8:2973–2980. 2015.PubMed/NCBI
|
|
29
|
Banales JM, Marin JJG, Lamarca A,
Rodrigues PM, Khan SA, Roberts LR, Cardinale V, Carpino G, Andersen
JB, Braconi C, et al: Cholangiocarcinoma 2020: The next horizon in
mechanisms and management. Nat Rev Gastroenterol Hepatol.
17:557–588. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan
A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al: The
epithelial-mesenchymal transition generates cells with properties
of stem cells. Cell. 133:704–715. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lan T, Luo M and Wei X: Mesenchymal
stem/stromal cells in cancer therapy. J Hematol Oncol. 14:1952021.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Polyak K and Weinberg RA: Transitions
between epithelial and mesenchymal states: Acquisition of malignant
and stem cell traits. Nat Rev Cancer. 9:265–273. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Galassi C, Manic G, Esteller M, Galluzzi L
and Vitale I: Epigenetic regulation of cancer stemness. Signal
Transduct Target Ther. 10:2432025. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dongre A and Weinberg RA: New insights
into the mechanisms of epithelial-mesenchymal transition and
implications for cancer. Nat Rev Mol Cell Biol. 20:69–84. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fischer KR, Durrans A, Lee S, Sheng J, Li
F, Wong STC, Choi H, El Rayes T, Ryu S, Troeger J, et al:
Epithelial-to-mesenchymal transition is not required for lung
metastasis but contributes to chemoresistance. Nature. 527:472–476.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
De Las Rivas J, Brozovic A, Izraely S,
Casas-Pais A, Witz IP and Figueroa A: Cancer drug resistance
induced by EMT: Novel therapeutic strategies. Arch Toxicol.
95:2279–2297. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Spranger S, Koblish HK, Horton B, Scherle
PA, Newton R and Gajewski TF: Mechanism of tumor rejection with
doublets of CTLA-4, PD-1/PD-L1, or IDO blockade involves restored
IL-2 production and proliferation of CD8(+) T cells directly within
the tumor microenvironment. J Immunother Cancer. 2:32014.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fridlender ZG, Sun J, Kim S, Kapoor V,
Cheng G, Ling L, Worthen GS and Albelda SM: Polarization of
tumor-associated neutrophil phenotype by TGF-beta: ‘N1’ versus ‘N2’
TAN. Cancer Cell. 16:183–194. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhai L, Spranger S, Binder DC, Gritsina G,
Lauing KL, Giles FJ and Wainwright DA: Molecular pathways:
Targeting IDO1 and other tryptophan dioxygenases for cancer
immunotherapy. Clin Cancer Res. 21:5427–5433. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Taki M, Abiko K, Ukita M, Murakami R,
Yamanoi K, Yamaguchi K, Hamanishi J, Baba T, Matsumura N and Mandai
M: Tumor immune microenvironment during epithelial-mesenchymal
transition. Clin Cancer Res. 27:4669–4679. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang F, Wang H, Wang X, Jiang G, Liu H,
Zhang G, Wang H, Fang R, Bu X, Cai S and Du J: TGF-β induces
M2-like macrophage polarization via SNAIL-mediated suppression of a
pro-inflammatory phenotype. Oncotarget. 7:52294–52306. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Mittal V: Epithelial mesenchymal
transition in tumor metastasis. Annu Rev Pathol. 13:395–412. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Aparicio LA, Blanco M, Castosa R, Concha
Á, Valladares M, Calvo L and Figueroa A: Clinical implications of
epithelial cell plasticity in cancer progression. Cancer Lett.
366:1–10. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lin S, Sun JG, Wu JB, Long HX, Zhu CH,
Xiang T, Ma H, Zhao ZQ, Yao Q, Zhang AM, et al: Aberrant microRNAs
expression in CD133+/CD326+ human lung
adenocarcinoma initiating cells from A549. Mol Cells. 33:277–283.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yuan J, Dong R, Liu F, Zhan L, Liu Y, Wei
J and Wang N: The miR-183/182/96 cluster functions as a potential
carcinogenic factor and prognostic factor in kidney renal clear
cell carcinoma. Exp Ther Med. 17:2457–2464. 2019.PubMed/NCBI
|
|
46
|
Bartel DP: Metazoan MicroRNAs. Cell.
173:20–51. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Seyhan AA: Trials and tribulations of
MicroRNA therapeutics. Int J Mol Sci. 25:14692024. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hill M and Tran N: miRNA interplay:
Mechanisms and consequences in cancer. Dis Model Mech.
14:dmm0476622021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ransohoff JD, Wei Y and Khavari PA: The
functions and unique features of long intergenic non-coding RNA.
Nat Rev Mol Cell Biol. 19:143–157. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
McCabe EM and Rasmussen TP: lncRNA
involvement in cancer stem cell function and epithelial-mesenchymal
transitions. Semin Cancer Biol. 75:38–48. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yoon JH, Abdelmohsen K and Gorospe M:
Post-transcriptional gene regulation by long noncoding RNA. J Mol
Biol. 425:3723–3730. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kristensen LS, Andersen MS, Stagsted LVW,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nat Rev Genet. 20:675–691. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Mauer C, Paz S and Caputi M: Backsplicing
of the HIV-1 transcript generates multiple circRNAs to promote
viral replication. Npj Viruses. 3:212025. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ma Y, Wang T, Zhang X, Wang P and Long F:
The role of circular RNAs in regulating resistance to cancer
immunotherapy: Mechanisms and implications. Cell Death Dis.
15:3122024. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Thamjamrassri P and Ariyachet C: Circular
RNAs in cell cycle regulation of cancers. Int J Mol Sci.
25:60942024. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Hu X, Wu D, He X, Zhao H, He Z, Lin J,
Wang K, Wang W, Pan Z, Lin H and Wang M: circGSK3β promotes
metastasis in esophageal squamous cell carcinoma by augmenting
β-catenin signaling. Mol Cancer. 18:1602019. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Fang L, Du WW, Awan FM, Dong J and Yang
BB: The circular RNA circ-Ccnb1 dissociates Ccnb1/Cdk1 complex
suppressing cell invasion and tumorigenesis. Cancer Lett.
459:216–226. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Fei T, Chen Y, Xiao T, Li W, Cato L, Zhang
P, Cotter MB, Bowden M, Lis RT, Zhao SG, et al: Genome-wide CRISPR
screen identifies HNRNPL as a prostate cancer dependency regulating
RNA splicing. Proc Natl Acad Sci USA. 114:E5207–E5215. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang
Y, Jin Y, Yang Y, Chen LL, Wang Y, et al: Extensive translation of
circular RNAs driven by N6-methyladenosine. Cell Res.
27:626–641. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Gregory PA, Bert AG, Paterson EL, Barry
SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y and Goodall GJ:
The miR-200 family and miR-205 regulate epithelial to mesenchymal
transition by targeting ZEB1 and SIP1. Nat Cell Biol. 10:593–601.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Lu R, Ji Z, Li X, Qin J, Cui G, Chen J,
Zhai Q, Zhao C, Zhang W and Yu Z: Tumor suppressive microRNA-200a
inhibits renal cell carcinoma development by directly targeting
TGFB2. Tumour Biol. 36:6691–6700. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Wang X, Chen X, Wang R, Xiao P, Xu Z, Chen
L, Hang W, Ruan A, Yang H and Zhang X: microRNA-200c modulates the
epithelial-to-mesenchymal transition in human renal cell carcinoma
metastasis. Oncol Rep. 30:643–650. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Xu XW, Li S, Yin F and Qin LL: Expression
of miR-205 in renal cell carcinoma and its association with
clinicopathological features and prognosis. Eur Rev Med Pharmacol
Sci. 22:662–670. 2018.PubMed/NCBI
|
|
68
|
Qiu M, Liang Z, Chen L, Tan G, Wang K, Liu
L, Liu J and Chen H: MicroRNA-429 suppresses cell proliferation,
epithelial-mesenchymal transition, and metastasis by direct
targeting of BMI1 and E2F3 in renal cell carcinoma. Urol Oncol.
33:332.e9–e18. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Machackova T, Mlcochova H, Stanik M,
Dolezel J, Fedorko M, Pacik D, Poprach A, Svoboda M and Slaby O:
MiR-429 is linked to metastasis and poor prognosis in renal cell
carcinoma by affecting epithelial-mesenchymal transition. Tumour
Biol. 37:14653–14658. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Chen Z, Zhang J, Zhang Z, Feng Z, Wei J,
Lu J, Fang Y, Liang Y, Cen J, Pan Y, et al: The putative tumor
suppressor microRNA-30a-5p modulates clear cell renal cell
carcinoma aggressiveness through repression of ZEB2. Cell Death
Dis. 8:e28592017. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Liu W, Li H, Wang Y, Zhao X, Guo Y, Jin J
and Chi R: MiR-30b-5p functions as a tumor suppressor in cell
proliferation, metastasis and epithelial-to-mesenchymal transition
by targeting G-protein subunit α-13 in renal cell carcinoma. Gene.
626:275–281. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Han N, Li H and Wang H: MicroRNA-203
inhibits epithelial-mesenchymal transition, migration, and invasion
of renal cell carcinoma cells via the inactivation of the PI3K/AKT
signaling pathway by inhibiting CAV1. Cell Adhes Migr. 14:227–241.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Yamasaki T, Seki N, Yamada Y, Yoshino H,
Hidaka H, Chiyomaru T, Nohata N, Kinoshita T, Nakagawa M and
Enokida H: Tumor suppressive microRNA-138 contributes to cell
migration and invasion through its targeting of vimentin in renal
cell carcinoma. Int J Oncol. 41:805–817. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Dasgupta P, Kulkarni P, Majid S, Hashimoto
Y, Shiina M, Shahryari V, Bhat NS, Tabatabai L, Yamamura S, Saini
S, et al: LncRNA CDKN2B-AS1/miR-141/cyclin D network regulates
tumor progression and metastasis of renal cell carcinoma. Cell
Death Dis. 11:6602020. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Lu R, Ji Z, Li X, Zhai Q, Zhao C, Jiang Z,
Zhang S, Nie L and Yu Z: miR-145 functions as tumor suppressor and
targets two oncogenes, ANGPT2 and NEDD9, in renal cell carcinoma. J
Cancer Res Clin Oncol. 140:387–397. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Lichner Z, Saleh C, Subramaniam V,
Seivwright A, Prud'homme GJ and Yousef GM: miR-17 inhibition
enhances the formation of kidney cancer spheres with stem
cell/tumor initiating cell properties. Oncotarget. 6:5567–5581.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Liu Y, Zhang M, Qian J, Bao M, Meng X,
Zhang S, Zhang L, Zhao R, Li S, Cao Q, et al: miR-134 functions as
a tumor suppressor in cell proliferation and
epithelial-to-mesenchymal transition by targeting KRAS in renal
cell carcinoma cells. DNA Cell Biol. 34:429–436. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Liu F, Chen N, Xiao R, Wang W and Pan Z:
miR-144-3p serves as a tumor suppressor for renal cell carcinoma
and inhibits its invasion and metastasis by targeting MAP3K8.
Biochem Biophys Res Commun. 480:87–93. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Dasgupta P, Kulkarni P, Majid S, Shahryari
V, Hashimoto Y, Bhat NS, Shiina M, Deng G, Saini S, Tabatabai ZL,
et al: MicroRNA-203 inhibits long noncoding RNA HOTAIR and
regulates tumorigenesis through epithelial-to-mesenchymal
transition pathway in renal cell carcinoma. Mol Cancer Ther.
17:1061–1069. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Dong JS, Wu B and Zha ZL: MicroRNA-588
regulates migration capacity and invasiveness of renal cancer cells
by targeting EIF5A2. Eur Rev Med Pharmacol Sci. 23:10248–10256.
2019.PubMed/NCBI
|
|
81
|
Pan H, Hong Y, Yu B, Li L and Zhang X:
miR-4429 inhibits tumor progression and epithelial-mesenchymal
transition via targeting CDK6 in clear cell renal cell carcinoma.
Cancer Biother Radiopharm. 34:334–341. 2019.PubMed/NCBI
|
|
82
|
Guo Z, Jia H and Ge J: MiR-206 suppresses
proliferation and epithelial-mesenchymal transition of renal cell
carcinoma by inhibiting CDK6 expression. Hum Cell. 33:750–758.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Guo Z, Lv X and Jia H: MiR-186 represses
progression of renal cell cancer by directly targeting CDK6. Hum
Cell. 33:759–767. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Xu B, Wang C, Wang YL, Chen SQ, Wu JP, Zhu
WD, Wang CY, Guan H, Guan C, You ZH and Chen M: miR-143 inhibits
renal cell carcinoma cells metastatic potential by suppressing
ABL2. Kaohsiung J Med Sci. 36:592–598. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Sekino Y, Sakamoto N, Goto K, Honma R,
Shigematsu Y, Quoc TP, Sentani K, Oue N, Teishima J, Kawakami F, et
al: Uc.416 + A promotes epithelial-to-mesenchymal transition
through miR-153 in renal cell carcinoma. BMC Cancer. 18:9522018.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Xue D, Wang H, Chen Y, Shen D, Lu J, Wang
M, Zebibula A, Xu L, Wu H, Li G and Xia L: Circ-AKT3 inhibits clear
cell renal cell carcinoma metastasis via altering
miR-296-3p/E-cadherin signals. Mol Cancer. 18:1512019. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Chen Z, Wang Z, Chen Z, Fu F, Huang X and
Huang Z: Pseudogene HSPB1P1 contributes to renal cell carcinoma
proliferation and metastasis by targeting miR-296-5p to regulate
HMGA1 expression. Cell Biol Int. 45:2479–2489. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Wang YB, Zhang ZL, Shao JK and Li RS:
Effect of miR-186 targeting E-cadherin on proliferation and
metastasis of renal cell carcinoma. Zhonghua Yi Xue Za Zhi.
101:1020–1025. 2021.(In Chinese). PubMed/NCBI
|
|
89
|
Sharma A, Singh P, Jha R, Almatroodi SA,
Alrumaihi F, Rahmani AH, Alharbi HO, Dohare R and Syed MA:
Exploring the role of miR-200 family in regulating CX3CR1 and CXCR1
in lung adenocarcinoma tumor microenvironment: Implications for
therapeutic intervention. Sci Rep. 13:163332023. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
He L, Jiang Z, Wang J and Han Z: Mechanism
of miR-200b-3p-induced FOSL2 inhibition of endometrial cancer cell
proliferation and metastasis. Sci Rep. 15:157422025. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Jo H, Shim K and Jeoung D: Potential of
the miR-200 family as a target for developing anti-cancer
therapeutics. Int J Mol Sci. 23:58812022. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Castro-Vega LJ, Jouravleva K, Liu WY,
Martinez C, Gestraud P, Hupé P, Servant N, Albaud B, Gentien D, Gad
S, et al: Telomere crisis in kidney epithelial cells promotes the
acquisition of a microRNA signature retrieved in aggressive renal
cell carcinomas. Carcinogenesis. 34:1173–1180. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Richardsen E, Andersen S, Melbø-Jørgensen
C, Rakaee M, Ness N, Al-Saad S, Nordby Y, Pedersen MI, Dønnem T,
Bremnes RM and Busund LT: MicroRNA 141 is associated to outcome and
aggressive tumor characteristics in prostate cancer. Sci Rep.
9:3862019. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Yang C, Dou R, Yin T and Ding J:
MiRNA-106b-5p in human cancers: Diverse functions and promising
biomarker. Biomed Pharmacother. 127:1102112020. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Mlcochova H, Machackova T, Rabien A,
Radova L, Fabian P, Iliev R, Slaba K, Poprach A, Kilic E, Stanik M,
et al: Epithelial-mesenchymal transition-associated microRNA/mRNA
signature is linked to metastasis and prognosis in clear-cell renal
cell carcinoma. Sci Rep. 6:318522016. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Han W, Cui H, Liang J and Su X: Role of
MicroRNA-30c in cancer progression. J Cancer. 11:2593–2601. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Huang J, Yao X, Zhang J, Dong B, Chen Q,
Xue W, Xue W, Liu D and Huang Y: Hypoxia-induced downregulation of
miR-30c promotes epithelial-mesenchymal transition in human renal
cell carcinoma. Cancer Sci. 104:1609–1617. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Butz H, Szabó PM, Khella HWZ, Nofech-Mozes
R, Patocs A and Yousef GM: miRNA-target network reveals miR-124as a
key miRNA contributing to clear cell renal cell carcinoma
aggressive behaviour by targeting CAV1 and FLOT1. Oncotarget.
6:12543–12557. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
He S, Zhang G, Dong H, Ma M and Sun Q:
miR-203 facilitates tumor growth and metastasis by targeting
fibroblast growth factor 2 in breast cancer. OncoTargets Ther.
9:6203–6210. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Grange C, Tapparo M, Collino F, Vitillo L,
Damasco C, Deregibus MC, Tetta C, Bussolati B and Camussi G:
Microvesicles released from human renal cancer stem cells stimulate
angiogenesis and formation of lung premetastatic niche. Cancer Res.
71:5346–5356. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Li DY, Lin FF, Li GP and Zeng FC: Exosomal
microRNA-15a from ACHN cells aggravates clear cell renal cell
carcinoma via the BTG2/PI3K/AKT axis. Kaohsiung J Med Sci.
37:973–982. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Cao J, Liu J, Xu R, Zhu X, Liu L and Zhao
X: MicroRNA-21 stimulates epithelial-to-mesenchymal transition and
tumorigenesis in clear cell renal cells. Mol Med Rep. 13:75–82.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Wu TK, Wei CW, Pan YR, Hsu RJ, Wu CY and
Yu YL: The uremic toxin p-cresyl sulfate induces proliferation and
migration of clear cell renal cell carcinoma via microRNA-21/
HIF-1α axis signals. Sci Rep. 9:32072019. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Lyu J, Zhu Y and Zhang Q: An increased
level of MiR-222-3p is associated with TMP2 suppression, ERK
activation and is associated with metastasis and a poor prognosis
in renal clear cell carcinoma. Cancer Biomark. 28:141–149. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Gorka J, Marona P, Kwapisz O, Waligórska
A, Pospiech E, Dobrucki JW, Rys J, Jura J and Miekus K: MCPIP1
inhibits Wnt/β-catenin signaling pathway activity and modulates
epithelial-mesenchymal transition during clear cell renal cell
carcinoma progression by targeting miRNAs. Oncogene. 40:6720–6735.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Lei QQ, Huang Y, Li B, Han L and Lv C:
MiR-155-5p promotes metastasis and epithelial-mesenchymal
transition of renal cell carcinoma by targeting apoptosis-inducing
factor. Int J Biol Markers. 36:20–27. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Meng K, Li Z and Cui X: Three LHPP
gene-targeting co-expressed microRNAs (microRNA-765, microRNA-21,
and microRNA-144) promote proliferation, epithelial-mesenchymal
transition, invasion, and are independent prognostic biomarkers in
renal cell carcinomas patients. J Clin Lab Anal. 35:e240772021.
View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Landolt L, Eikrem Ø, Strauss P, Scherer A,
Lovett DH, Beisland C, Finne K, Osman T, Ibrahim MM, Gausdal G, et
al: Clear cell renal cell carcinoma is linked to
epithelial-to-mesenchymal transition and to fibrosis. Physiol Rep.
5:e133052017. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Yoshino H, Yonemori M, Miyamoto K,
Tatarano S, Kofuji S, Nohata N, Nakagawa M and Enokida H:
microRNA-210-3p depletion by CRISPR/Cas9 promoted tumorigenesis
through revival of TWIST1 in renal cell carcinoma. Oncotarget.
8:20881–20894. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Xiong J, Liu Y, Jiang L, Zeng Y and Tang
W: High expression of long non-coding RNA lncRNA-ATB is correlated
with metastases and promotes cell migration and invasion in renal
cell carcinoma. Jpn J Clin Oncol. 46:378–384. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
He X, Liu H, Guo F, Feng Y, Gao Y, Sun F,
Song B, Lu H and Li Y: Long non-coding RNA Z38 promotes cell
proliferation and metastasis in human renal cell carcinoma. Mol Med
Rep. 16:5489–5494. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Ning L, Li Z, Wei D, Chen H and Yang C:
LncRNA, NEAT1 is a prognosis biomarker and regulates cancer
progression via epithelial-mesenchymal transition in clear cell
renal cell carcinoma. Cancer Biomark. 19:75–83. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Xiong J, Liu Y, Luo S, Jiang L, Zeng Y,
Chen Z, Shi X, Lv B and Tang W: High expression of the long
non-coding RNA HEIRCC promotes renal cell carcinoma metastasis by
inducing epithelial-mesenchymal transition. Oncotarget.
8:6555–6563. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Du Y, Kong C, Zhu Y, Yu M, Li Z, Bi J, Li
Z, Liu X, Zhang Z and Yu X: Knockdown of SNHG15 suppresses renal
cell carcinoma proliferation and EMT by regulating the NF-κB
signaling pathway. Int J Oncol. 53:384–394. 2018.PubMed/NCBI
|
|
115
|
Su Y, Zhou L, Yu Q, Lu J and Liu W: Long
non-coding RNA LOC648987 promotes proliferation and metastasis of
renal cell carcinoma by regulating epithelial-mesenchymal
transition. Technol Cancer Res Treat. 20:15330338219978342021.
View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Yu H, Liu J, Zhang Z, Zhu Y, Bi J and Kong
C: SNHG12 promotes carcinogenesis of human renal cell cancer via
functioning as a competing endogenous RNA and sponging miR-30a-3p.
J Cell Mol Med. 25:4696–4708. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Zhang Y, Lan X, Wang Y, Liu C and Cui T:
CRNDE mediates the viability and epithelial-mesenchymal transition
of renal cell carcinoma via miR-136-5p. J Recept Signal Transduct
Res. 41:234–244. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Shao IH, Peng PH, Wu HH, Chen JL, Lai JCY,
Chang JS, Wu HT, Wu KJ, Pang ST and Hsu KW: RP11-367G18.1 V2
enhances clear cell renal cell carcinoma progression via induction
of epithelial-mesenchymal transition. Cancer Med. 12:9788–9801.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Zhang A, Zhao JC, Kim J, Fong KW, Yang YA,
Chakravarti D, Mo YY and Yu J: LncRNA HOTAIR enhances the
androgen-receptor-mediated transcriptional program and drives
castration-resistant prostate cancer. Cell Rep. 13:209–221. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Pu Y, Dong Z, Xia Y, Zhang M, Song J, Han
J and Liu H: LncRNA NONHSAT113026 represses renal cell carcinoma
tumorigenesis through interacting with NF-κB/p50 and SLUG. Biomed
Pharmacother. 118:1093822019. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Wang G, Zhang ZJ, Jian WG, Liu PH, Xue W,
Wang TD, Meng YY, Yuan C, Li HM, Yu YP, et al: Novel long noncoding
RNA OTUD6B-AS1 indicates poor prognosis and inhibits clear cell
renal cell carcinoma proliferation via the Wnt/β-catenin signaling
pathway. Mol Cancer. 18:152019. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Zhang C, Zhu N, Liu C, Wu H, Yin Y, Shi Y,
Liao D and Qin L: Steroid receptor RNA activator inhibits the
migration, invasion and stemness characteristics of renal cell
carcinoma cells. Int J Mol Med. 46:1765–1776. 2020.PubMed/NCBI
|
|
124
|
Singh D, Kesharwani P, Alhakamy NA and
Siddique HR: Accentuating CircRNA-miRNA-transcription factors axis:
A conundrum in cancer research. Front Pharmacol. 12:7848012022.
View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Rong D, Sun H, Li Z, Liu S, Dong C, Fu K,
Tang W and Cao H: An emerging function of circRNA-miRNAs-mRNA axis
in human diseases. Oncotarget. 8:73271–73281. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Zhang D, Yang XJ, Luo QD, Fu DL, Li ZL,
Zhang P and Chong T: Down-regulation of circular RNA_000926
attenuates renal cell carcinoma progression through
miRNA-411-dependent CDH2 inhibition. Am J Pathol. 189:2469–8246.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Li J, Huang C, Zou Y, Yu J and Gui Y:
Circular RNA MYLK promotes tumour growth and metastasis via
modulating miR-513a-5p/VEGFC signalling in renal cell carcinoma. J
Cell Mol Med. 24:6609–6621. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Li W, Yang FQ, Sun CM, Huang JH, Zhang HM,
Li X, Wang GC, Zhang N, Che JP, Zhang WT, et al: circPRRC2A
promotes angiogenesis and metastasis through epithelial-mesenchymal
transition and upregulates TRPM3 in renal cell carcinoma.
Theranostics. 10:4395–4409. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Liu H, Hu G, Wang Z, Liu Q, Zhang J, Chen
Y, Huang Y, Xue W, Xu Y and Zhai W: circPTCH1 promotes invasion and
metastasis in renal cell carcinoma via regulating miR-485-5p/MMP14
axis. Theranostics. 10:10791–10807. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Yan JS, Chen Q, Li YL and Gao YQ:
Hsa_circ_0065217 promotes growth and metastasis of renal cancer
through regulating the miR-214-3p-ALPK2 axis. Cell Cycle.
20:2519–2530. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Li W, Song YYY, Rao T, Yu WM, Ruan Y, Ning
JZ, Yao XB, Yang SY and Cheng F: CircCSNK1G3 up-regulates miR-181b
to promote growth and metastasis via TIMP3-mediated epithelial to
mesenchymal transitions in renal cell carcinoma. J Cell Mol Med.
26:1729–1741. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Cen J, Liang Y, Feng Z, Chen X, Chen J,
Wang Y, Zhu J, Xu Q, Shu G, Zheng W, et al: Hsa_circ_0057105
modulates a balance of epithelial-mesenchymal transition and
ferroptosis vulnerability in renal cell carcinoma. Clin Transl Med.
13:e13392023. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Xie X, Li H, Gao C, Lai Y, Liang J, Chen
Z, Chen Z, Heng B, Yao N and Lai C: Downregulation of circular RNA
circPSD3 promotes metastasis by modulating FBXW7 expression in
clear cell renal cell carcinoma. J Oncol. 2022:50846312022.
View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Yang CY, Wang J, Zhang JQ and Cai HM:
Human circular RNA hsa_circRNA_101705 (circTXNDC11) regulates renal
cancer progression by regulating MAPK/ERK pathway. Bioengineered.
12:4432–4441. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Mighell AJ, Smith NR, Robinson PA and
Markham AF: Vertebrate pseudogenes. FEBS Lett. 468:109–914. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Ayubi E, Shahbazi F and Khazaei S:
Decomposing difference in the kidney cancer burden measures between
1990 and 2019 based on the global burden of disease study. Sci Rep.
14:103902024. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Choi J, Bang S, Suh J, Choi CI, Song W,
Yuk HD, Lee CH, Kang M, Choo SH, Kim JK, et al: Survival pattern of
metastatic renal cell carcinoma patients according to WHO/ISUP
grade: A long-term multi-institutional study. Sci Rep. 14:47402024.
View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Logan JE, Rampersaud EN, Sonn GA, Chamie
K, Belldegrun AS, Pantuck AJ, Slamon DJ and Kabbinavar FF: Systemic
therapy for metastatic renal cell carcinoma: A review and update.
Rev Urol. 14:65–78. 2012.PubMed/NCBI
|
|
139
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Yang J, Antin P, Berx G, Blanpain C,
Brabletz T, Bronner M, Campbell K, Cano A, Casanova J, Christofori
G, et al: Guidelines and definitions for research on
epithelial-mesenchymal transition. Nat Rev Mol Cell Biol.
21:341–352. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Zhang L, Lin H, Liang J, Liu X, Zhang C,
Man Q, Li R, Zhao Y and Liu B: Programmed death-ligand 1 regulates
ameloblastoma growth and recurrence. Int J Oral Sci. 17:292025.
View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Zhong W and Sun T: Editorial:
Epithelial-mesenchymal transition (EMT) as a therapeutic target in
cancer, volume II. Front Oncol. 13:12188552023. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Mao W, Wang K, Xu B, Zhang H, Sun S, Hu Q,
Zhang L, Liu C, Chen S, Wu J, et al: ciRS-7 is a prognostic
biomarker and potential gene therapy target for renal cell
carcinoma. Mol Cancer. 20:1422021. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Silva-Santos RM, Costa-Pinheiro P, Luis A,
Antunes L, Lobo F, Oliveira J, Henrique R and Jerónimo C: MicroRNA
profile: A promising ancillary tool for accurate renal cell tumour
diagnosis. Br J Cancer. 109:2646–2653. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Zhang YP, Cheng YB, Li S, Zhao N and Zhu
ZH: An epithelial-mesenchymal transition-related long non-coding
RNA signature to predict overall survival and immune
microenvironment in kidney renal clear cell carcinoma.
Bioengineered. 12:555–564. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Jin J, Xie Y, Zhang JS, Wang JQ, Dai SJ,
He WF, Li SY, Ashby CR Jr, Chen ZS and He Q: Sunitinib resistance
in renal cell carcinoma: From molecular mechanisms to predictive
biomarkers. Drug Resist Updat. 67:1009292023. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Enuka Y, Lauriola M, Feldman ME, Sas-Chen
A, Ulitsky I and Yarden Y: Circular RNAs are long-lived and display
only minimal early alterations in response to a growth factor.
Nucleic Acids Res. 44:1370–1383. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Chen YG, Chen R, Ahmad S, Verma R, Kasturi
SP, Amaya L, Broughton JP, Kim J, Cadena C, Pulendran B, et al:
N6-methyladenosine modification controls circular RNA immunity. Mol
Cell. 76:96–109.e9. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Wesselhoeft RA, Kowalski PS, Parker-Hale
FC, Huang Y, Bisaria N and Anderson DG: RNA circularization
diminishes immunogenicity and can extend translation duration in
vivo. Mol Cell. 74:508–520.e4. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Li H, Peng K, Yang K, Ma W, Qi S, Yu X, He
J, Lin X and Yu G: Circular RNA cancer vaccines drive immunity in
hard-to-treat malignancies. Theranostics. 12:6422–6436. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Liu Y, Wu W, Cai C, Zhang H, Shen H and
Han Y: Patient-derived xenograft models in cancer therapy:
Technologies and applications. Signal Transduct Target Ther.
8:1602023. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Zhu J, Ding Y and Xu Q: Exosomal noncoding
RNAs in renal cell carcinoma: Mechanisms, roles, and therapeutic
potential. Crit Rev Oncol Hematol. 213:1048292025. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Yu X, Du Z, Zhu P and Liao B: Diagnostic,
prognostic, and therapeutic potential of exosomal microRNAs in
renal cancer. Pharmacol Rep. 76:273–286. 2024. View Article : Google Scholar : PubMed/NCBI
|
















