|
1
|
Liu C, Zhao ZL, Ji ZD, Jiang Y and Zheng
J: MiR-187-3p enhances propranolol sensitivity of hemangioma stem
cells. Cell Struct Funct. 44:41–50. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Acevedo LM and Cheresh DA: Suppressing
NFAT increases VEGF signaling in hemangiomas. Cancer Cell.
14:429–430. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang Q, Zhao C, Du Q, Cao Z and Pan J:
Non-coding RNA in infantile hemangioma. Pediatr Res. 96:1594–1602.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li Z, Cao Z, Li N, Wang L, Fu C, Huo R, Xu
G, Tian C and Bi J: M2 macrophage-derived exosomal lncRNA
MIR4435-2HG promotes progression of infantile hemangiomas by
targeting HNRNPA1. Int J Nanomedicine. 18:5943–5960. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li R, Liu Y, Liu J, Chen B, Ji Z, Xu A and
Zhang T: CCL2 regulated by the CTBP1-AS2/miR-335-5p axis promotes
hemangioma progression and angiogenesis. Immunopharmacol
Immunotoxicol. 46:385–394. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yu L, Shu H, Xing L, Lv MX, Li L, Xie YC,
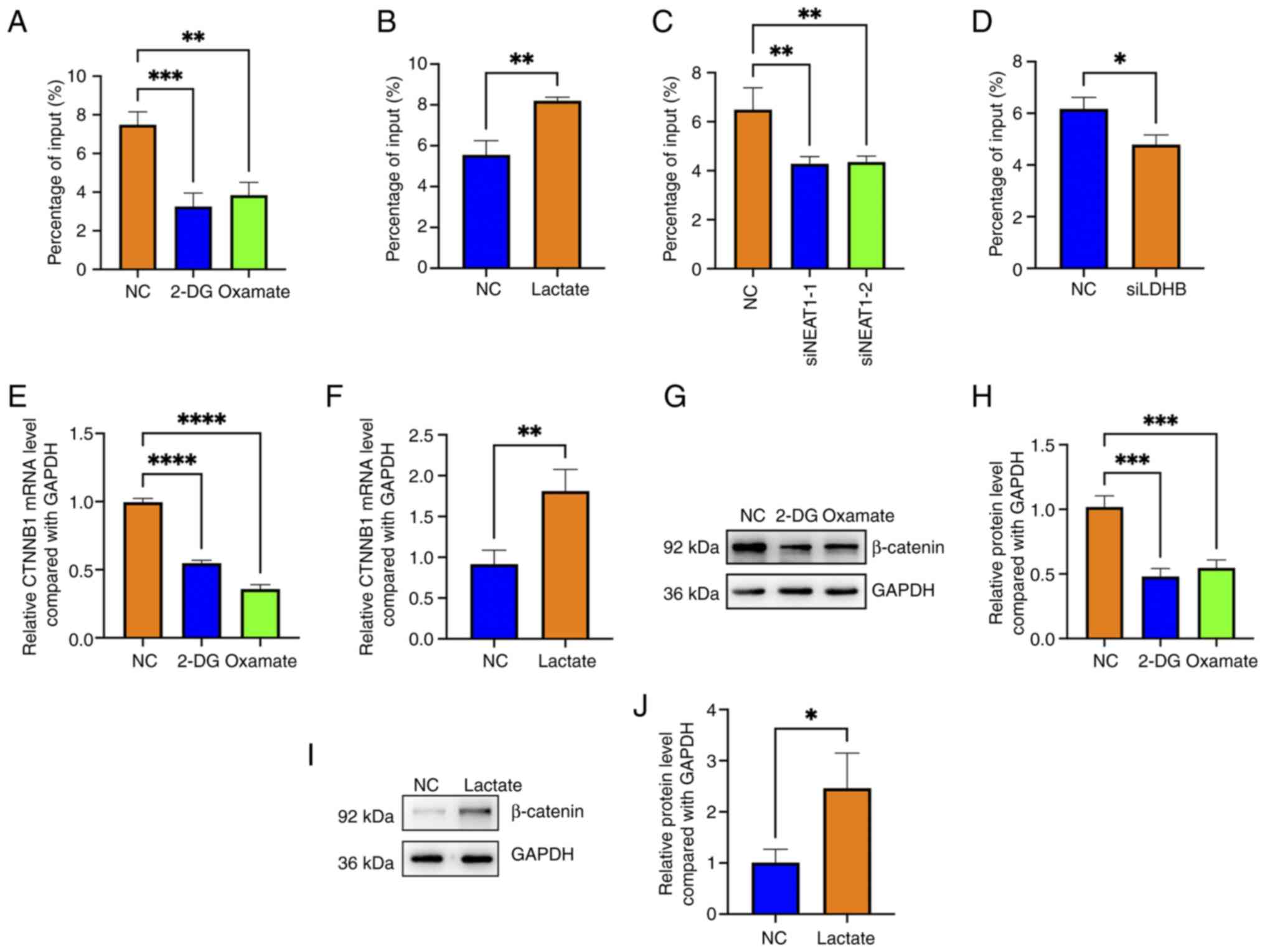
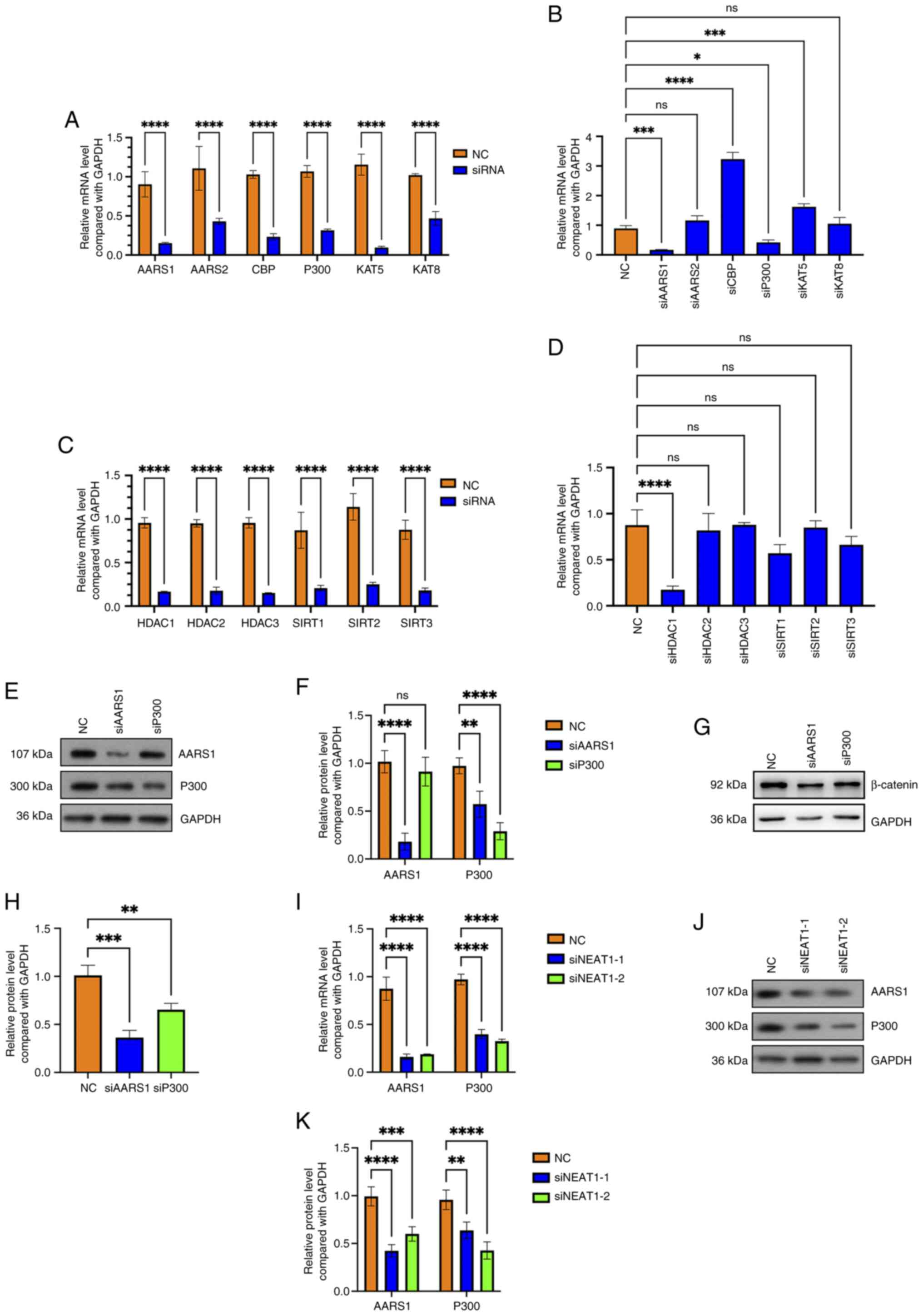
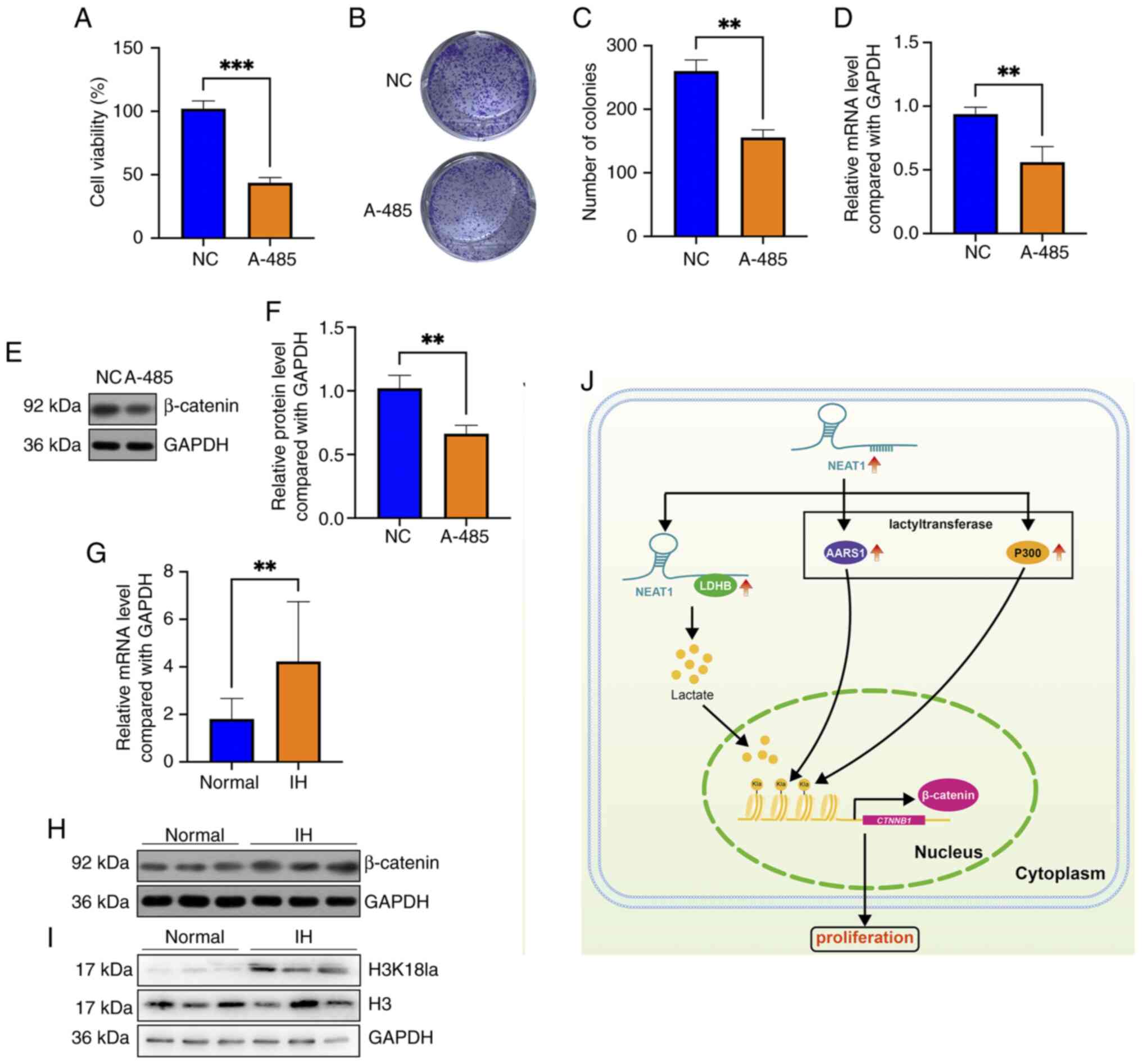
Zhang Z, Zhang L and Xie YY: Silencing long non-coding RNA NEAT1
suppresses the tumorigenesis of infantile hemangioma by
competitively binding miR-33a-5p to stimulate HIF1α/NF-κB pathway.
Mol Med Rep. 22:3358–3366. 2020.PubMed/NCBI
|
|
7
|
Yu X, Liu X, Wang R and Wang L: Long
non-coding RNA NEAT1 promotes the progression of hemangioma via the
miR-361-5p/VEGFA pathway. Biochem Biophys Res Commun. 512:825–831.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Peng K, Xia RP, Zhao F, Xiao Y, Ma TD, Li
M, Feng Y and Zhou CG: ALKBH5 promotes the progression of infantile
hemangioma through regulating the NEAT1/miR-378b/FOSL1 axis. Mol
Cell Biochem. 477:1527–1540. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang D, Tang Z, Huang H, Zhou G, Cui C,
Weng Y, Liu W, Kim S, Lee S, Perez-Neut M, et al: Metabolic
regulation of gene expression by histone lactylation. Nature.
574:575–580. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xiao SY, Zhang SH, Sun K, Huang Q and Hu
C: Lactate and lactylation: Molecular insights into histone and
non-histone lactylation in tumor progression, tumor immune
microenvironment, and therapeutic strategies. Biomark Res.
13:1342025. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fang N, Zhang N, Jiang X, Yan S, Wang Z,
Gao Q, Xu M, Mu L, Li X, Chen J, et al: PFKM-Driven lactate
overproduction promotes atrial fibrillation via triggering cardiac
fibroblasts histone lactylation. Adv Sci (Weinh). 12:e009632025.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gu JJ, Deng CC, An Q, Zhu DH, Xu XY, Fu
ZZ, Zhou ZY, Rong Z and Yang B: Lactate promotes collagen
expression, proliferation and migration through H3K18
lactylation-dependent stimulation of LTBP3/TGF-beta1 axis in keloid
fibroblasts. J Invest Dermatol. July 7–2025.(Epub ahead of print).
View Article : Google Scholar
|
|
13
|
Galle E, Wong CW, Ghosh A, Desgeorges T,
Melrose K, Hinte LC, Castellano-Castillo D, Engl M, de Sousa JA,
Ruiz-Ojeda FJ, et al: H3K18 lactylation marks tissue-specific
active enhancers. Genome Biol. 23:2072022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang H, Xu M, Zhang T, Pan J, Li C, Pan B,
Zhou L, Huang Y, Gao C, He M, et al: PYCR1 promotes liver cancer
cell growth and metastasis by regulating IRS1 expression through
lactylation modification. Clin Transl Med. 14:e700452024.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Khan ZA, Melero-Martin JM, Wu X, Paruchuri
S, Boscolo E, Mulliken JB and Bischoff J: Endothelial progenitor
cells from infantile hemangioma and umbilical cord blood display
unique cellular responses to endostatin. Blood. 108:915–921. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kong C, Li R, Wang X, Li L, Kang N, Zhen
X, Dong Y and Yan G: Environmental aminomethylphosphonic acid
(AMPA) exposure increased the risk of spontaneous abortion through
lactate-induced JunB lactylation in trophoblast. Ecotoxicol Environ
Saf. 302:1187432025. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lu Y, Zhu J, Zhang Y, Li W, Xiong Y, Fan
Y, Wu Y, Zhao J, Shang C, Liang H and Zhang W: Lactylation-Driven
IGF2BP3-mediated serine metabolism reprogramming and RNA
m6A-modification promotes lenvatinib resistance in HCC. Adv Sci
(Weinh). 11:e24013992024. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhao SS, Liu J, Wu QC and Zhou XL: Lactate
regulates pathological cardiac hypertrophy via histone lactylation
modification. J Cell Mol Med. 28:e700222024. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wei S, Zhang J, Zhao R, Shi R, An L, Yu Z,
Zhang Q, Zhang J, Yao Y, Li H and Wang H: Histone lactylation
promotes malignant progression by facilitating USP39 expression to
target PI3K/AKT/HIF-1α signal pathway in endometrial carcinoma.
Cell Death Discov. 10:1212024. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lu G, Yi J, Gubas A, Wang YT, Wu Y, Ren Y,
Wu M, Shi Y, Ouyang C, Tan HWS, et al: Suppression of autophagy
during mitosis via CUL4-RING ubiquitin ligases-mediated WIPI2
polyubiquitination and proteasomal degradation. Autophagy.
15:1917–1934. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Whitworth CP, Aw WY, Doherty EL, Handler
C, Ambekar Y, Sawhney A, Scarcelli G and Polacheck WJ: P300
modulates endothelial mechanotransduction of fluid shear stress.
Cell Mol Bioeng. 17:507–523. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen B, Deng S, Ge T, Ye M, Yu J, Lin S,
Ma W and Songyang Z: Live cell imaging and proteomic profiling of
endogenous NEAT1 lncRNA by CRISPR/Cas9-mediated knock-in. Protein
Cell. 11:641–660. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cai S, Xia Q, Duan D, Fu J, Wu Z, Yang Z
and Yu C: Creatine kinase mitochondrial 2 promotes the growth and
progression of colorectal cancer via enhancing Warburg effect
through lactate dehydrogenase B. PeerJ. 12:e176722024. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang C, Zhou L, Zhang M, Du Y, Li C, Ren
H and Zheng L: H3K18 Lactylation potentiates immune escape of
Non-small cell lung cancer. Cancer Res. 84:3589–3601. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu Z, Ma T, Li J, Ren W and Zhang Z:
IL13RA2 promotes progression of infantile haemangioma by activating
glycolysis and the Wnt/β-catenin signaling pathway. Oncol Res.
32:1453–1465. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhu Z, Luo J, Li L, Wang D, Xu Q, Teng J,
Zhou J, Sun L, Yu N and Zuo D: Fucoidan suppresses proliferation
and epithelial-mesenchymal transition process via Wnt/β-catenin
signalling in hemangioma. Exp Dermatol. 33:e150272024. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen J, Zhao D, Wang Y, Liu M, Zhang Y,
Feng T, Xiao C, Song H, Miao R, Xu L, et al: Lactylated
apolipoprotein C-II induces immunotherapy resistance by promoting
extracellular lipolysis. Adv Sci (Weinh). 11:e24063332024.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
van Schaijik B, Tan ST, Marsh RW and
Itinteang T: Expression of (pro)renin receptor and its effect on
endothelial cell proliferation in infantile hemangioma. Pediatr
Res. 86:202–207. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dai Y, Zheng H, Liu Z, Wang Y and Hu W:
The flavonoid luteolin suppresses infantile hemangioma by targeting
FZD6 in the Wnt pathway. Invest New Drugs. 39:775–784. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li X, Yang Y, Zhang B, Lin X, Fu X, An Y,
Zou Y, Wang JX, Wang Z and Yu T: Lactate metabolism in human health
and disease. Signal Transduct Target Ther. 7:3052022. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yu H, Zhu T, Ma D, Cheng X, Wang S and Yao
Y: The role of nonhistone lactylation in disease. Heliyon.
10:e362962024. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Brooks GA: Lactate as a fulcrum of
metabolism. Redox Biol. 35:1014542020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Raychaudhuri D, Singh P, Chakraborty B,
Hennessey M, Tannir AJ, Byregowda S, Natarajan SM, Trujillo-Ocampo
A, Im JS and Goswami S: Histone lactylation drives CD8+
T cell metabolism and function. Nat Immunol. 25:2140–2151. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chao J, Chen GD, Huang ST, Gu H, Liu YY,
Luo Y, Lin Z, Chen ZZ, Li X, Zhang B, et al: High histone H3K18
lactylation level is correlated with poor prognosis in epithelial
ovarian cancer. Neoplasma. 71:319–332. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sun J, Feng Q, He Y, Wang M and Wu Y:
Lactate activates CCL18 expression via H3K18 lactylation in
macrophages to promote tumorigenesis of ovarian cancer. Acta
Biochim Biophys Sin (Shanghai). 56:1373–1386. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li F, Si W, Xia L, Yin D, Wei T, Tao M,
Cui X, Yang J, Hong T and Wei R: Positive feedback regulation
between glycolysis and histone lactylation drives oncogenesis in
pancreatic ductal adenocarcinoma. Mol Cancer. 23:902024. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wu S, Li J and Zhan Y: H3K18 lactylation
accelerates liver fibrosis progression through facilitating SOX9
transcription. Exp Cell Res. 440:1141352024. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hou X, Ouyang J, Tang L, Wu P, Deng X, Yan
Q, Shi L, Fan S, Fan C, Guo C, et al: KCNK1 promotes proliferation
and metastasis of breast cancer cells by activating lactate
dehydrogenase A (LDHA) and up-regulating H3K18 lactylation. PLoS
Biol. 22:e30026662024. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen J, Wu D, Dong Z, Chen A and Liu S:
The expression and role of glycolysis-associated molecules in
infantile hemangioma. Life Sci. 259:1182152020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yang K, Zhang X, Chen L, Chen S and Ji Y:
Microarray expression profile of mRNAs and long noncoding RNAs and
the potential role of PFK-1 in infantile hemangioma. Cell Div.
16:12021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Rodriguez Bandera AI, Sebaratnam DF,
Wargon O and Wong LF: Infantile hemangioma. Part 1: Epidemiology,
pathogenesis, clinical presentation and assessment. J Am Acad
Dermatol. 85:1379–1392. 2021. View Article : Google Scholar : PubMed/NCBI
|