Introduction
Infantile hemangioma (IH) is the most common
vascular tumor in children, with an incidence of 3–10% (1). Due to its rapid growth, some IHs lead
to long-term residual permanent skin damage and might affect the
physical and mental health of children (2). Although a number of studies have
focused on the mechanisms underlying IH progression, the detailed
regulatory mechanisms are still not fully understood.
Long non-coding RNAs (lncRNAs), with a length of
>200 nucleotides, have crucial roles in hemangioma cell
proliferation and invasion (3,4). A
recent study reported that CTBP1-AS2 sponges microRNA (miR)-335-5p,
upregulates C-C motif chemokine ligand 2 and subsequently enhances
hemangioma angiogenesis and progression (5). MIR4435-2HG, which is secreted by M2
macrophages, targets Heterogeneous nuclear ribonucleoprotein A1 and
promotes IH progression (4). Most
notably, our previous study revealed that knocking down lncRNA
NEAT1 repressed IH progression by sponging miR-33a-5p and
regulating the downstream hypoxia inducible factor 1 subunit α
(HIF1α)/NF-κB pathway (6).
Additional studies have further reported that in hemangioma, lncRNA
NEAT1 is positively regulated by alkB homolog 5, RNA demethylase in
an m6A modification-dependent manner and promotes hemangioma
development by regulating the miR-361-5p/VEGFA pathway (7,8).
However, the exact regulatory roles of lncRNA NEAT1 in hemangioma
progression are still largely unknown.
Histone lactylation (addition of a lactyl group to a
lysine residue) by P300 and KAT8 is a protein modification newly
discovered in 2019 that affects transcription (9,10),
and has been found to widely participate in disease progression
including cancer, atrial fibrillation and keloids (11,12).
Recent studies have shown that H3 lysine 18 lactylation (H3K18la)
can activate gene transcription (13,14).
However, the roles of protein lactylation in hemangioma development
are still unclear.
In the present study, the effects of the lncRNA
NEAT1/H3K18la/β-catenin signaling pathway on the colony formation
ability and viability of hemangioma cells were investigated.
Additionally, the detailed mechanism by which NEAT1 regulates
β-catenin in hemangioma cells was further investigated.
Materials and methods
Tissue sample information
IH tissues (proliferating stage; median age, 7
months; n=10, Table I) and normal
adjacent subcutaneous tissues (n=10) were collected from the
Kunming Children's Hospital (Kunming, China) between February 2022
and September 2024. Samples were kept at −80°C until analysis.
Informed consent was obtained from the parents/legal guardians of
each patient. The patients received no treatment before surgery.
The Ethics Committee of Kunming Children's Hospital approved the
present study (approval no. 2021-03-181-K01). The inclusion
criteria were as follows: i) The patients were diagnosed with IH
(proliferating stage); ii) age of <1 years old; and iii) The
hemangioma lesion was located in concealed areas such as the trunk
and limbs, and the parents voluntarily opted for surgical removal.
The exclusion criteria were as follows: i) Multiple hemangiomas;
ii) vascular malformation; and iii) the patients had received any
other treatment.
 | Table I.Clinical features of the patients
with infantile hemangioma included in the present study. |
Table I.
Clinical features of the patients
with infantile hemangioma included in the present study.
| Patient no. | Sex | Age (months) | Location of
tumor | Growth phase |
|---|
| 1 | Male | 9 | Abdominal wall | Proliferating |
| 2 | Female | 4 | Waist | Proliferating |
| 3 | Female | 3 | Neck | Proliferating |
| 4 | Male | 6 | Abdominal wall | Proliferating |
| 5 | Female | 7 | Abdominal wall | Proliferating |
| 6 | Male | 7 | Neck | Proliferating |
| 7 | Female | 9 | Waist | Proliferating |
| 8 | Female | 4 | Neck | Proliferating |
| 9 | Male | 8 | Abdominal wall | Proliferating |
| 10 | Female | 8 | Waist | Proliferating |
Cell culture and treatments
Hemangioma endothelial cells (HemECs) were prepared
following the protocol described in a previous study (15) using the hemangioma sample from
patient no. 4 in Table I. The
HemECs were cultured in human endothelial serum-free medium (Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco;
Thermo Fisher Scientific, Inc.) at 5% CO2 and 37°C.
Lipofectamine 3000 (Thermo Fisher Scientific, Inc.)
was used to transfect small interfering RNAs (siRNA; 50 nM;
Shanghai GenePharma Co., Ltd.) into the cells at 37°C for 48 h.
Then, the cells were collected for reverse
transcription-quantitative PCR (RT-qPCR), western blotting,
chromatin immunoprecipitation (ChIP)-PCR and cell viability assays.
Table SI lists the sequences of
the siRNAs.
2-Deoxy-D-glucose (2-DG), oxamate, lactate, MG132,
bafilomycin A1 (BafA1), cycloheximide (CHX) and A-485 were
purchased from Selleck Chemicals. In the present study, 10 mM 2-DG,
20 mM oxamate and 10 mM lactate were used to treat the HemECs
(16–19) (RT-qPCR, western blotting, ChIP-PCR
and cell viability assays: 37°C for 48 h; colony formation assay:
37°C for 7–9 days), and the protein lactylation was inactivated
(2-DG and oxamate) and activated (lactate). At 46.5 and 46 h after
transfection of NEAT1 siRNA, 20 µM MG132 (treatment: 1.5 h at 37°C)
and 100 nM BafA1 (treatment: 2 h at 37°C) were used to suppress the
proteosome and lysosome, respectively (20), and the cells were harvested for
western blotting after a total time of 48 h of transfection with
NEAT1 siRNA. At 33, 36, 39, 42 and 45 h after transfection of NEAT1
siRNA (corresponding to the 15, 12, 9, 6 and 3 h treatment group),
100 µg/ml CHX was used to treat the cells (20), and the cells were harvested for
western blotting after a total time of 48 h of transfection with
NEAT1 siRNA. Additionally, 5 µM A-485 (RT-qPCR, western blotting,
cell viability assays: 37°C for 48 h; colony formation assay: 37°C
for 7–9 days) were applied to block p300 (21).
RT-qPCR assay
Total RNA from tissues and cells was extracted using
RNAiso Esay Plus (Wuhan Servicebio Technology Co., Ltd.) according
to the manufacturer's instructions. First-strand cDNA synthesis and
qPCR were performed using the HifiScript cDNA Synthesis Kit
(Cwbiotech) and PowerUp™ SYBR™ Green Master Mix (Thermo Fisher
Scientific, Inc.), respectively, according to the manufacturer's
protocol. The qPCR program was as follows: 95°C for 2 min, followed
by 40 cycles of 95°C for 15 sec and 60°C for 1 min. The
2−ΔΔcq method was used to calculate the relative
expression of the genes (22).
GAPDH was used for normalization. Table SII lists the primer sequences.
Colony formation and cell viability
assays
A total of 500 cells per well were plated in 12-well
plate and then incubated for 7–9 days [lactate dehydrogenase B
(LDHB)-knock down cells: cells were seeded after transfection for
24 h; 2-DG/oxamate/lactate/A-485 treated cells: drugs were used to
treat after seeding for 24 h]. At the end of the experiment, the
cells were fixed with 4% paraformaldehyde for 15 min at room
temperature and then stained with 0.1% crystal violet (Beyotime
Biotechnology) for 30 min at room temperature. Colonies (>50
cells) were visualized and quantified by light microscopy. The cell
viability was detected using Cell Counting Kit-8 (Dojindo
Laboratories, Inc.) assays as previously described (6).
RNA immunoprecipitation (RIP)
assay
According to the protocol previously described
(23), the hemangioma cells were
harvested and washed twice with ice-cold PBS. The hemangioma cells
were suspended in RIP buffer (1 ml) containing 50 mmol/l Tris pH
7.4, 0.5% NP-40, 1X RNasin plus (Promega Corporation), 150 mmol/l
NaCl, 2 mmol/l ribonucleoside vanadyl complex (New England BioLabs,
Inc.), 1X protease inhibitor cocktail (MilliporeSigma) and 1 mmol/l
PMSF. After brief sonication, cell lysates were centrifuged at
10,000 × g for 10 min at 4°C and the supernatants were precleared
using 10 µl Dynabeads Protein G (Thermo Fisher Scientific, Inc.).
The precleared supernatants were divided into two equal parts, and
one part was incubated with 1 µg LDHB antibody (cat. no.
14824-1-AP; Proteintech Group, Inc.) and 20 µl Protein A/G Magnetic
Beads (cat. No. HY-K0202; MedChemExpress) overnight at 4°C. The
other part was incubated with 1 µg IgG control antibody (cat. no.
30000-0-AP; Proteintech Group, Inc.) and 20 µl Protein A/G Magnetic
Beads overnight at 4°C. After washing the beads three times with
RIP buffer, the samples were decrosslinked by proteinase K buffer
(50 mmol/l Tris pH 7.4, 150 mmol/l NaCl, 1X RNasin plus, 0.5% SDS
and 200 µg/ml proteinase K) at room temperature for 30 min. Then,
the RNA was extracted and RT-qPCR was performed.
ChIP-PCR
The SimpleChIP Enzymatic Chromatin IP Kit (Magnetic
Beads) (cat. no. CST-9003; Cell Signaling Technology, Inc.) were
used to analyze the H3K18 lactylation level of the promoter of the
β-catenin gene (CTNNB1) following the manufacturer's
protocol. The 2-DG/oxamate-treated cells, lactate-treated cells,
NEAT1-knockdown cells and LDHB-knockdown cells were crosslinked
with 1% formaldehyde for 10 min at room temperature, then collected
after washing twice using PBS (containing 0.5 mM EDTA). The cell
pellet was lysed with 0.3 ml of cell lysis buffer (50 mM Tris-HCl
pH 8.1, 10 mM EDTA, 1% SDS and protease inhibitor) and incubated
for 10 min on ice. The cell lysates were sonicated (sonication for
15 sec, with a 10-sec interval, totaling 3 min of sonication) to
obtain DNA fragments (150–900 base pair in length) and ~50 µg of
cross-linked sheared chromatin solution was then used for
immunoprecipitation. The L-lactyl-histone H3 (Lys18) antibody (cat.
no. PTM-1427RM; PTM BIO LLC; 6 µg/5×106 cells) was
incubated with the sample overnight at 4°C on a rotating shaker.
Magnetic beads were added to the solution, incubated at 4°C for 1 h
and then washed with washing buffer. The cross-linking was reversed
by adding NaCl at a final concentration of 200 mM and heating at
65°C for 30 min. The DNA fragments were purified using a spin
column and qPCR was performed. The sequences of the primers used
were as follows: CTNNB1 promoter forward primer,
5′-CCTAGTGACAAGTGGAACCAGA-3′; and reverse primer,
5′-GAACTCTCCGTAGAACGGGC-3′.
Western blot assay
The western blot procedure was described in our
previous study (6). The primary
antibodies used were LDHB (cat. no. 14824-1-AP; 1:5,000), β-catenin
(cat. no. 51067-2-AP; 1:5,000), alanyl-tRNA synthetase 1 (AARS1;
cat. no. 17394-1-AP; 1:2,000), P300 (cat. no. 20695-1-AP; 1:1,000)
and GAPDH (cat. no. 60004-1-Ig; 1:50,000) from Proteintech Group,
Inc., and L-lactyl-Histone H3 (Lys18) (cat. no. PTM-1406RM;
1:1,000), L-lactyl lysine (pan-lac; cat. no. PTM-1401RM; 1:1,000)
and Histone H3 (cat. no. PTM-1002RM; 1:1,000) from PTM BIO LLC. All
primary antibodies were incubated overnight at 4°C. The secondary
antibodies used were HRP-conjugated Goat Anti-Mouse IgG (H+L) (cat.
no. SA00001-1; 1:10,000; Proteintech Group, Inc.) and
HRP-conjugated Goat Anti-Rabbit IgG (H+L) (cat, no. SA00001-2;
1:10,000; Proteintech Group, Inc.), which were incubated for 1 h at
room temperature.
Cellular lactate detection
The cellular lactate concentrations were detected
using a Lactic Acid assay kit (cat. no. BC2230; Beijing Solarbio
Science & Technology Co., Ltd.) according to the manufacturer's
protocol.
Bioinformatics analysis
The NEAT1-interacting proteins were predicted using
catRAPID software (http://s.tartaglialab.com/page/catrapid_group;
accessed on September 5, 2024). Based on the NEAT1-interacting
proteins (predicted by catRAPID; Interaction Propensity >50),
Gene Oncology (GO) and Kyoto Encyclopedia of Genes and Genomes
(KEGG) analyses were performed using the DAVID platform (24,25).
Statistical analysis
GraphPad Prism 10 software (Dotmatics) was used for
data analysis. The quantitative data are shown as the mean ± SD and
were analyzed via one-way with Tukey's multiple comparisons test
(multiple groups) or unpaired Student's t-test (two groups).
P<0.05 was considered to indicate a statistically significant
difference.
Results
Identification of lncRNA
NEAT1-interacting proteins
Our previous study reported that NEAT1 was highly
expressed in IH tissues (6). In
the present study, to investigate the regulatory mechanisms
underlying NEAT1-affected hemangioma progression, the proteins that
interact with NEAT1 were analyzed using catRAPID software. In
total, 2,064 proteins (such as HSP90B1 and CDK11B) were predicted
to interact with NEAT1. GO and pathway enrichment analyses of these
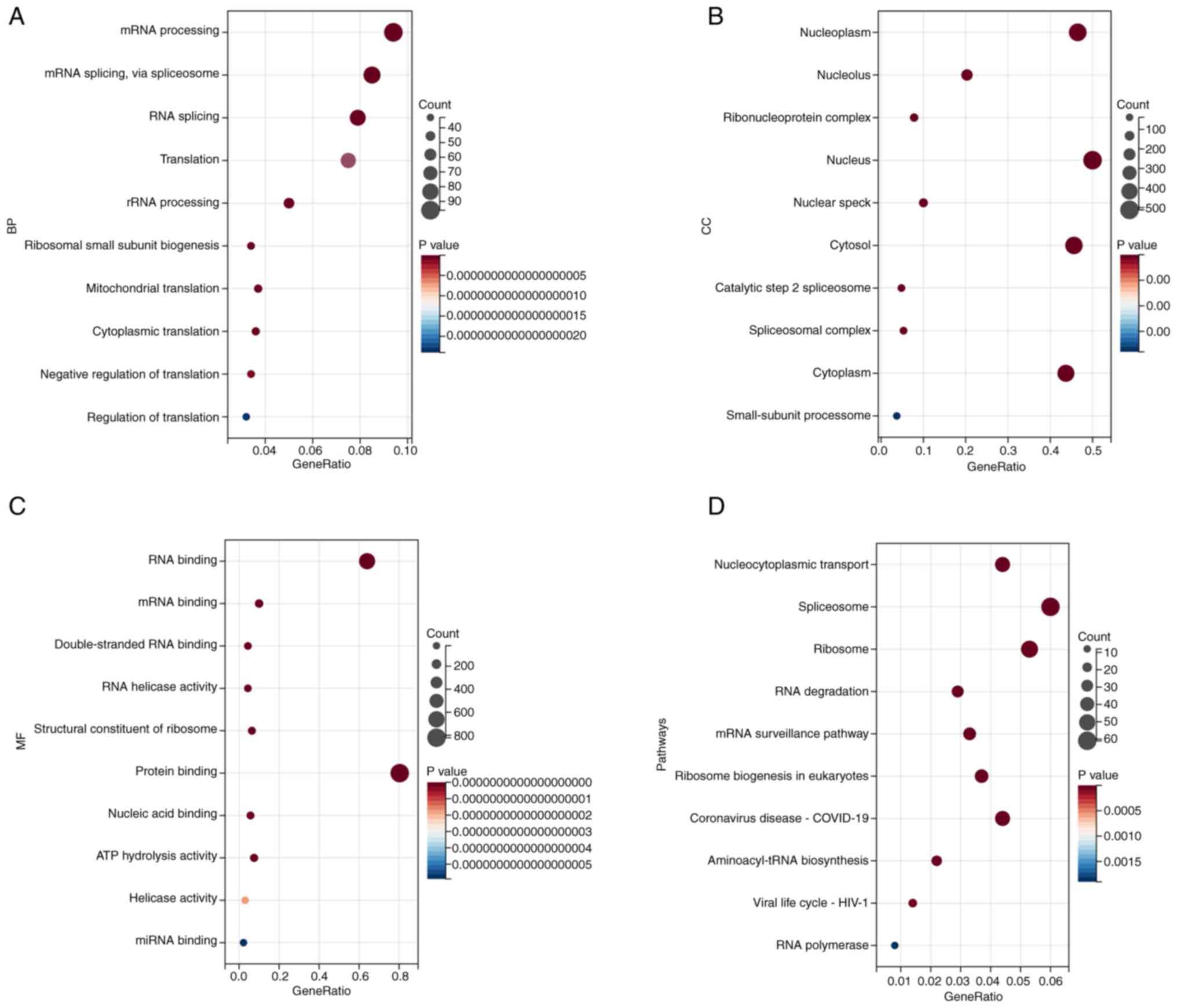
genes were subsequently performed using DAVID. The biological
process enrichment analysis revealed that NEAT1-interacting
proteins were associated with processes such as ‘mRNA processing’,
‘mRNA splicing, via spliceosome’, ‘RNA splicing’, ‘Translation’ and
‘rRNA processing’ (Fig. 1A).
Cellular component enrichment analysis revealed that
NEAT1-interacting proteins were associated with components such as
‘Nucleoplasm’, ‘Nucleolus’, ‘Ribonucleoprotein complex’, ‘Nucleus’
and ‘Nuclear speck’ (Fig. 1B).
Molecular function enrichment analysis revealed that
NEAT1-interacting proteins were associated with functions such as
‘RNA binding’, ‘mRNA binding’, ‘Double-stranded RNA binding’, ‘RNA
helicase activity’ and ‘Structural constituent of ribosome’
(Fig. 1C). KEGG enrichment
analysis revealed that NEAT1-interacting proteins were associated
with ‘Nucleocytoplasmic transport’, ‘Spliceosome,’ ‘Ribosome’, ‘RNA
degradation’ and ‘mRNA surveillance pathway’ (Fig. 1D).
lncRNA NEAT1 interacts with and
positively regulates LDHB in hemangioma cells
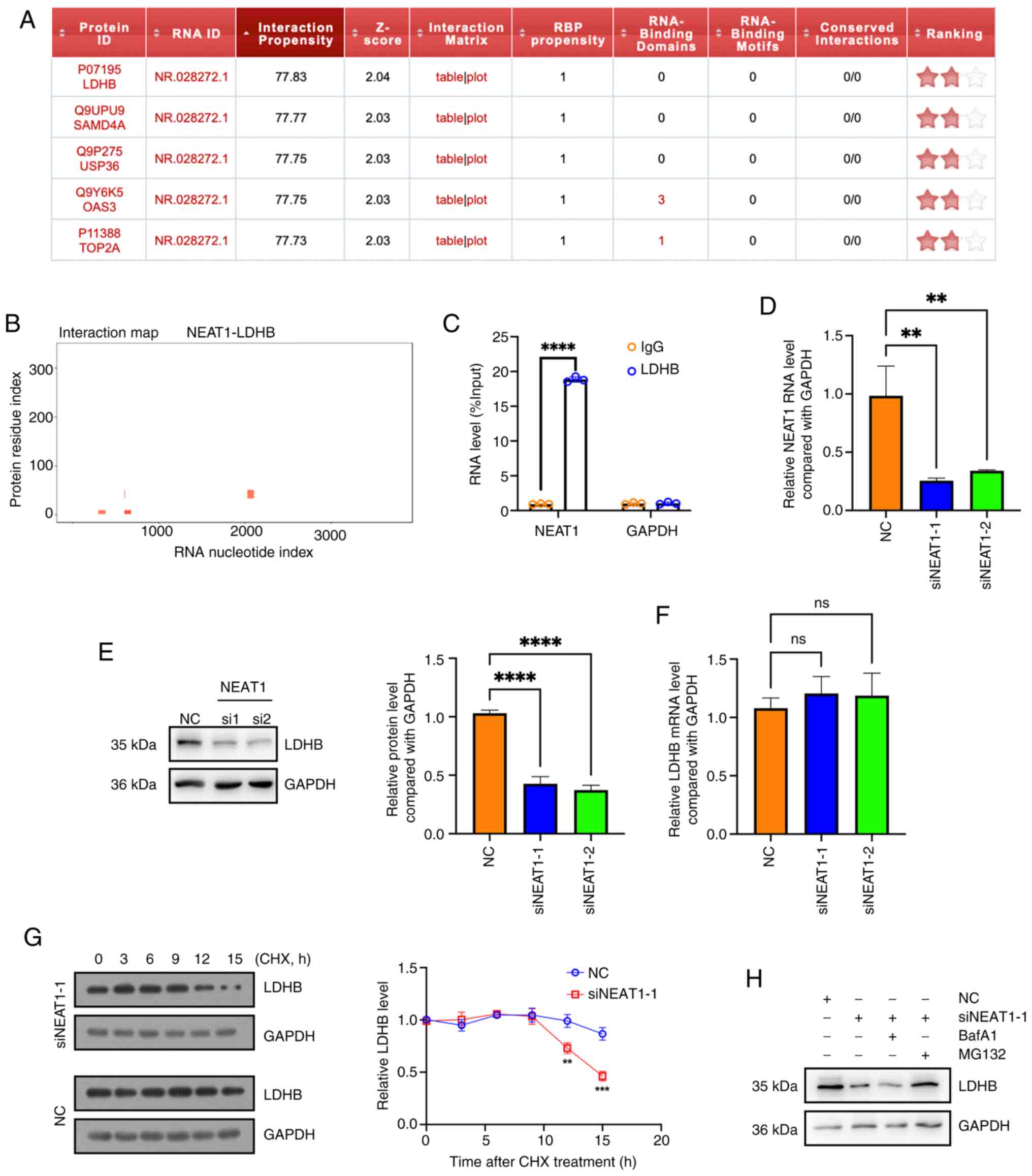
Notably, it was found that LDHB was predicted to
bind with NEAT1 using catRAPID software (Fig. 2A and B). The interaction between
NEAT1 and LDHB in hemangioma cells was validated using a RIP assay
(Fig. 2C). Additionally, after
knocking down NEAT1 expression (Fig.
2D), the protein level (Fig.
2E) but not the mRNA level of LDHB (Fig. 2F) was reduced, indicating that
NEAT1 may regulate LDHB at the protein level. After protein
translation was inhibited via CHX, LDHB degradation occurred faster
in NEAT1-knock down cells than in the negative control cells
(Fig. 2G). The proteasome
inhibitor MG132, but not the lysosome inhibitor BafA1, reversed the
effect of NEAT1 knockdown on the protein level of LDHB, indicating
that NEAT1 knockdown reduced the LDHB protein levels via activation
of the proteasome activity (Fig.
2H). These results suggested that the lncRNA NEAT1 interacted
with and positively regulated LDHB in hemangioma cells.
Knockdown of lncRNA NEAT1 inhibits
cellular protein lactylation and downregulates β-catenin in
hemangioma cells
LDHB and LDHA have critical roles in converting
pyruvate to lactate during glycolysis and are involved in
regulating protein lactylation (26,27).
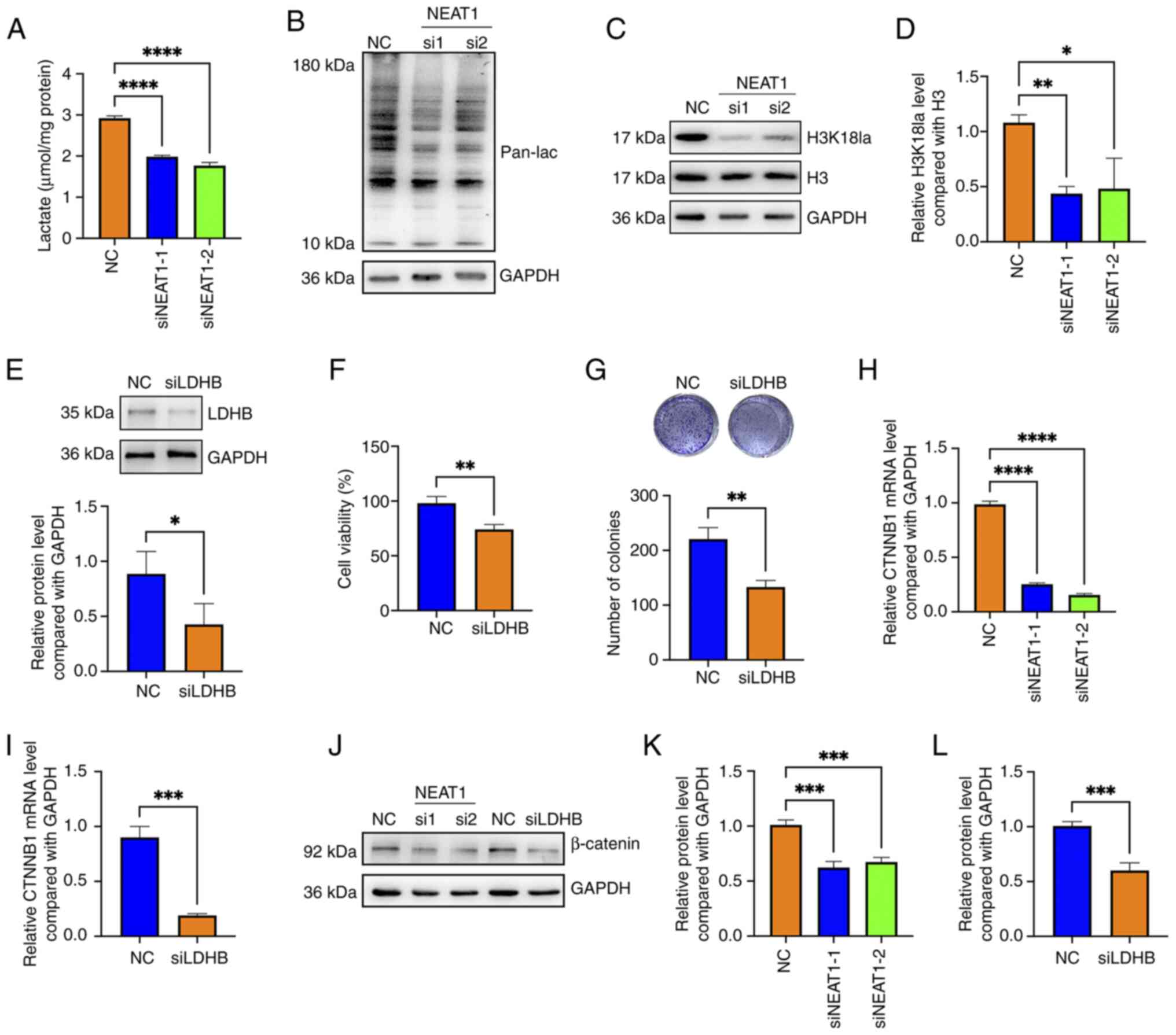
In the present study, the knockdown of NEAT1 reduced the cellular
lactate concentration and attenuated the levels of both pan
lactylation (pan-lac) and H3K18 lactylation in HemECs (Fig. 3A-D). After LDHB expression was
knocked down (Fig. 3E), the
viability and colony formation ability of HemECs was decreased
(Fig. 3F and G).
Activation of the Wnt/β-catenin pathway potentiates
the invasion, migration, proliferation and epithelial-mesenchymal
transition (EMT) of HemECs (28,29).
Therefore, it was next evaluated whether lncRNA NEAT1 promotes the
colony formation and viability of HemECs by regulating the
Wnt/β-catenin signaling pathway. The results revealed that knocking
down NEAT1 downregulated β-catenin at both the mRNA and protein
levels, and that knocking down LDHB also reduced the mRNA and
protein levels of β-catenin in HemECs (Fig. 3H-L). These data revealed that
knocking down of lncRNA NEAT1 inhibited cellular protein
lactylation and downregulated β-catenin in hemangioma cells.
Blockade of lactylation using 2-DG and
oxamate attenuates the viability and colony formation of hemangioma
cells and lactate treatment has the opposite effects
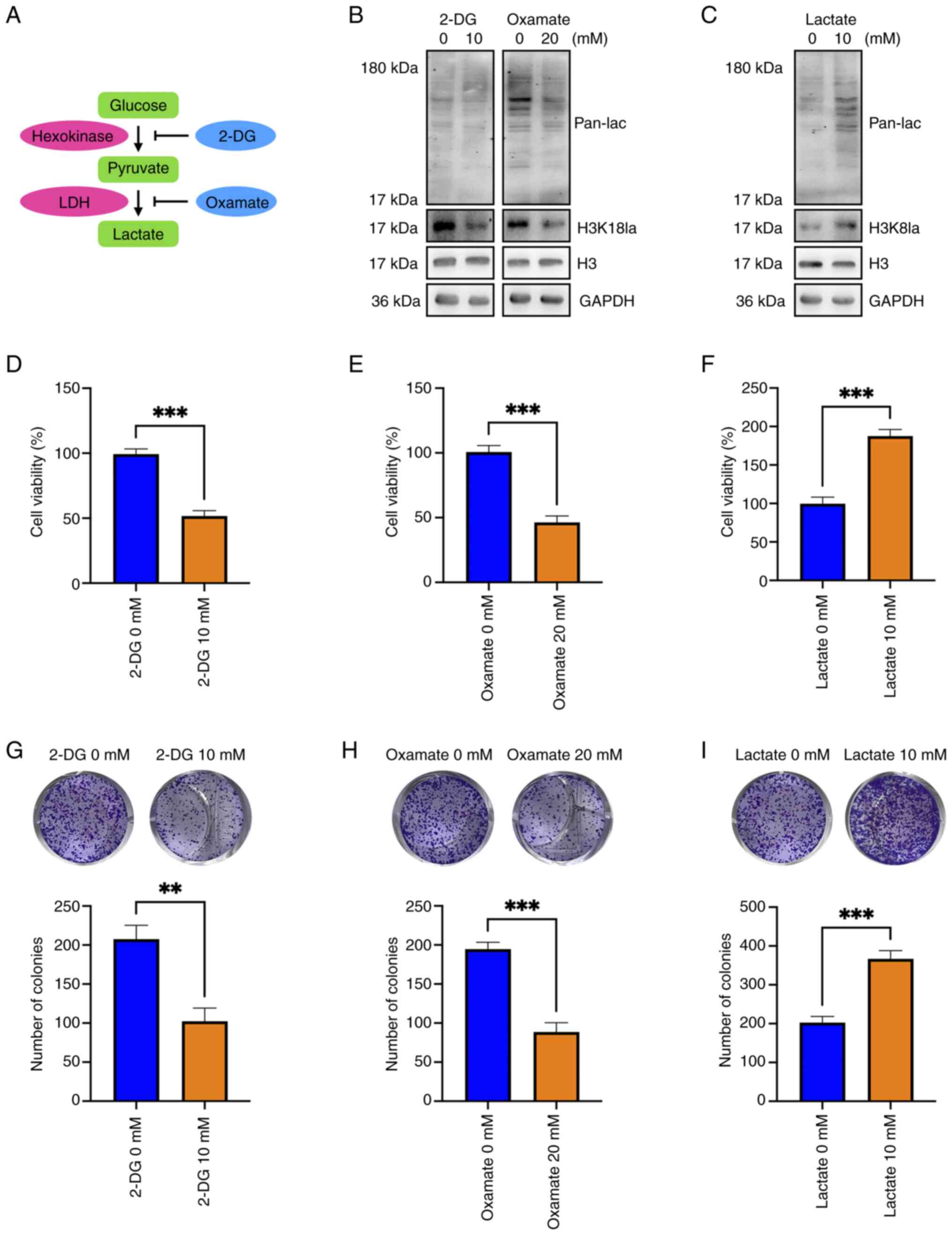
Oxamate suppressed cellular protein and H3K18
lactylation in hemangioma cells and 2-DG slightly reduced the H3K18
lactylation level and inhibited cellular protein lactylation in
hemangioma cells (Fig. 4A and B).
Conversely, lactate addition markedly increased cellular protein
lactylation and H3K18 lactylation in hemangioma cells (Fig. 4C). Both 2-DG and oxamate
significantly impeded the viability and colony formation of
hemangioma cells (Fig. 4D, E, G and
H), and lactate significantly enhanced the viability and colony
formation of hemangioma cells (Fig. 4F
and I). These data indicated that inhibition of lactylation
suppressed the viability and colony formation of hemangioma cells
and that promotion of lactylation exhibited promotive effects.
lncRNA NEAT1 promotes H3K18
lactylation of the CTNNB1 promoter and the blockade of lactylation
downregulates β-catenin in hemangioma cells
Lactylation of promoters or enhancers is an
indication of transcription activation (13); hence, whether NEAT1 regulates
β-catenin by affecting lactylation of the CTNNB1 promoter
was investigated. The results showed that both 2-DG and oxamate
treatment reduced the H3K18 lactylation level of the CTNNB1
promoter; conversely, lactate addition increased the H3K18
lactylation level (Fig. 5A and B).
Notably, knocking down NEAT1 or LDHB inhibited the H3K18
lactylation of the CTNNB1 promoter (Fig. 5C and D).
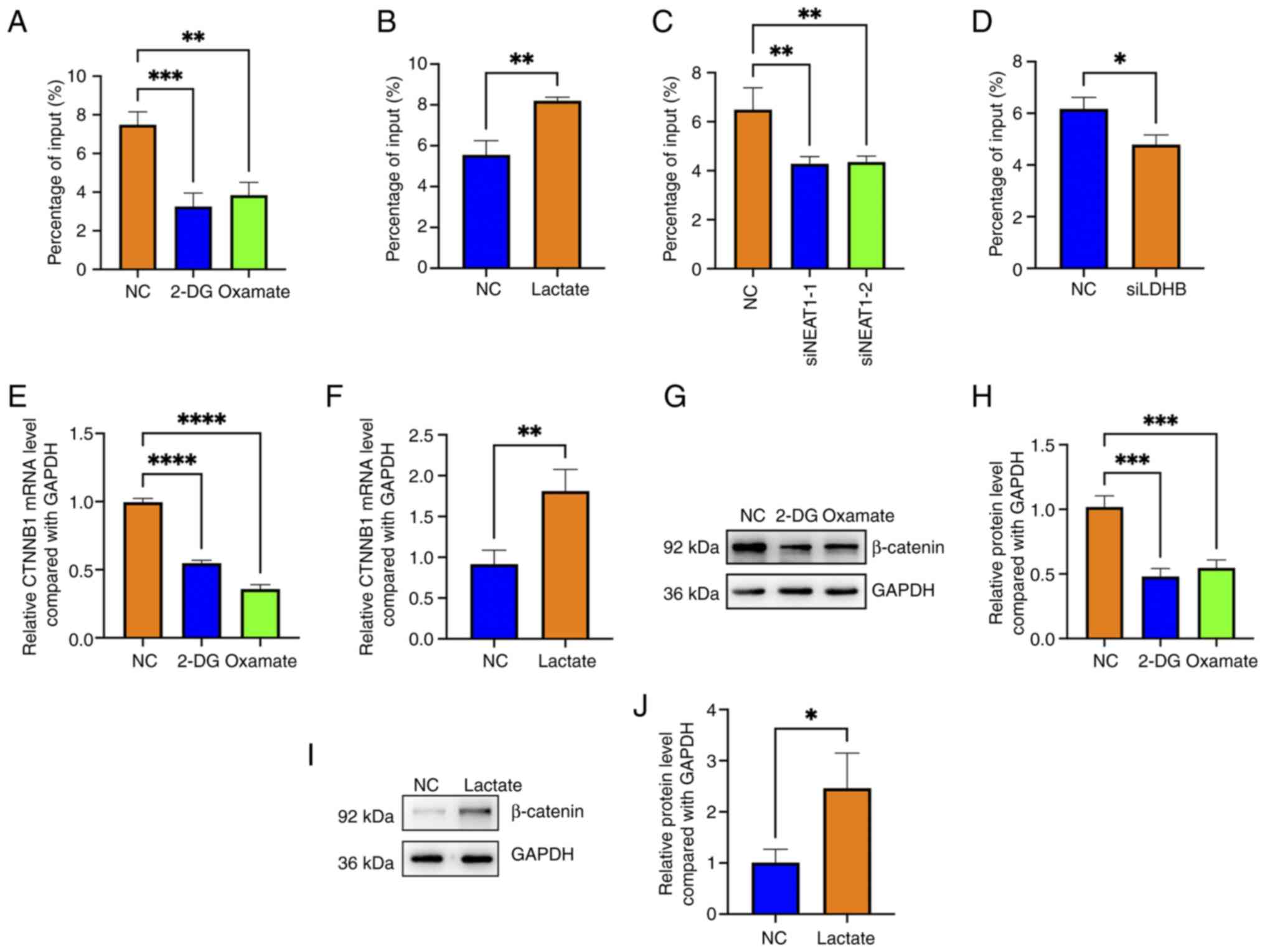 | Figure 5.Long non-coding RNA NEAT1 promotes
the H3K18 lactylation of the CTNNB1 promoter and the
blockade of lactylation downregulates β-catenin in hemangioma
cells. H3K18 lactylation levels of the CTNNB1 promoter in
(A) 2-DG- or oxamate-treated cells and (B) lactate-treated, (C)
NEAT1-knock down and (D) LDHB-knock down hemangioma cells were
evaluated using ChIP-PCR; *P<0.05, **P<0.01, ***P<0.001
vs. NC (n=3). The CTNNB1 mRNA levels in (E) 2-DG- and
oxamate-treated cells as well as (F) lactate-treated cells were
detected by RT-qPCR; **P<0.01, ****P<0.0001 vs. NC (n=3). The
protein levels of β-catenin in (G) 2-DG- and oxamate-treated cells
were detected by western blotting (n=3). (H) Semi-quantitative
analysis of the levels of β-catenin in 2-DG and oxamate-treated
cells detected by western blotting; ***P<0.001 vs. NC (n=3). (I)
The protein levels of β-catenin in lactate-treated cells were
detected by western blotting (n=3). (J) Semi-quantitative analysis
of the levels of β-catenin in lactate-treated cells detected by
western blotting; *P<0.05 vs. NC (n=3). 2-DG, 2-Deoxy-D-glucose;
NC, negative control; CTNNB1, β-catenin gene; LDHB, lactate
dehydrogenase B; ChIP, chromatin immunoprecipitation; RT-qPCR,
reverse transcription-quantitative polymerase chain reaction; si,
small interfering (RNA). |
Both 2-DG and oxamate treatment reduced the
CTNNB1 mRNA level; by contrast, lactate increased
CTNNB1 mRNA expression (Fig. 5E
and F). Additionally, western blotting revealed that 2-DG and
oxamate treatment downregulated β-catenin at the protein level
(Fig. 5G and H) and that lactate
upregulated β-catenin at the protein level (Fig. 5I and J). These findings suggested
that NEAT1 promoted H3K18 lactylation of the CTNNB1 promoter
and that blocking lactylation downregulated β-catenin expression in
hemangioma cells.
Lactyltransferases AARS1 and P300 are
indirectly regulated by lncRNA NEAT1 and further regulate β-catenin
in hemangioma cells
AARS1, AARS2, CREB binding lysine acetyltransferase
(CBP), lysine acetyltransferase (KAT) 5, KAT8 and P300 are reported
to positively regulate protein lactylation as lactyltransferases,
and Sirtuin (SIRT) 1, SIRT2, SIRT3, Histone deacetylase (HDAC) 1,
HDAC2 and HDAC3 have been found to negatively affect protein
lactylation as delactyltransferases (30). Hence, which molecules regulate
β-catenin in hemangioma cells were screened next. Notably, after
separately knocking down every lactyltransferase (Fig. 6A) and delactyltransferase (Fig. 6C), it was found that the knockdown
of AARS1 and P300 reduced the mRNA level of CTNNB1 (Fig. 6B). Additionally, knocking down
HDAC1 also downregulated CTNNB1 at the mRNA level in
hemangioma cells (Fig. 6D). The
knockdown efficiency of AARS1 and P300 was further confirmed using
western blotting (Fig. 6E and F).
It was subsequently found that the knocking down of AARS1 and P300
in hemangioma cells also downregulated β-catenin at the protein
level (Fig. 6G and H).
Furthermore, knockdown of NEAT1 significantly decreased the mRNA
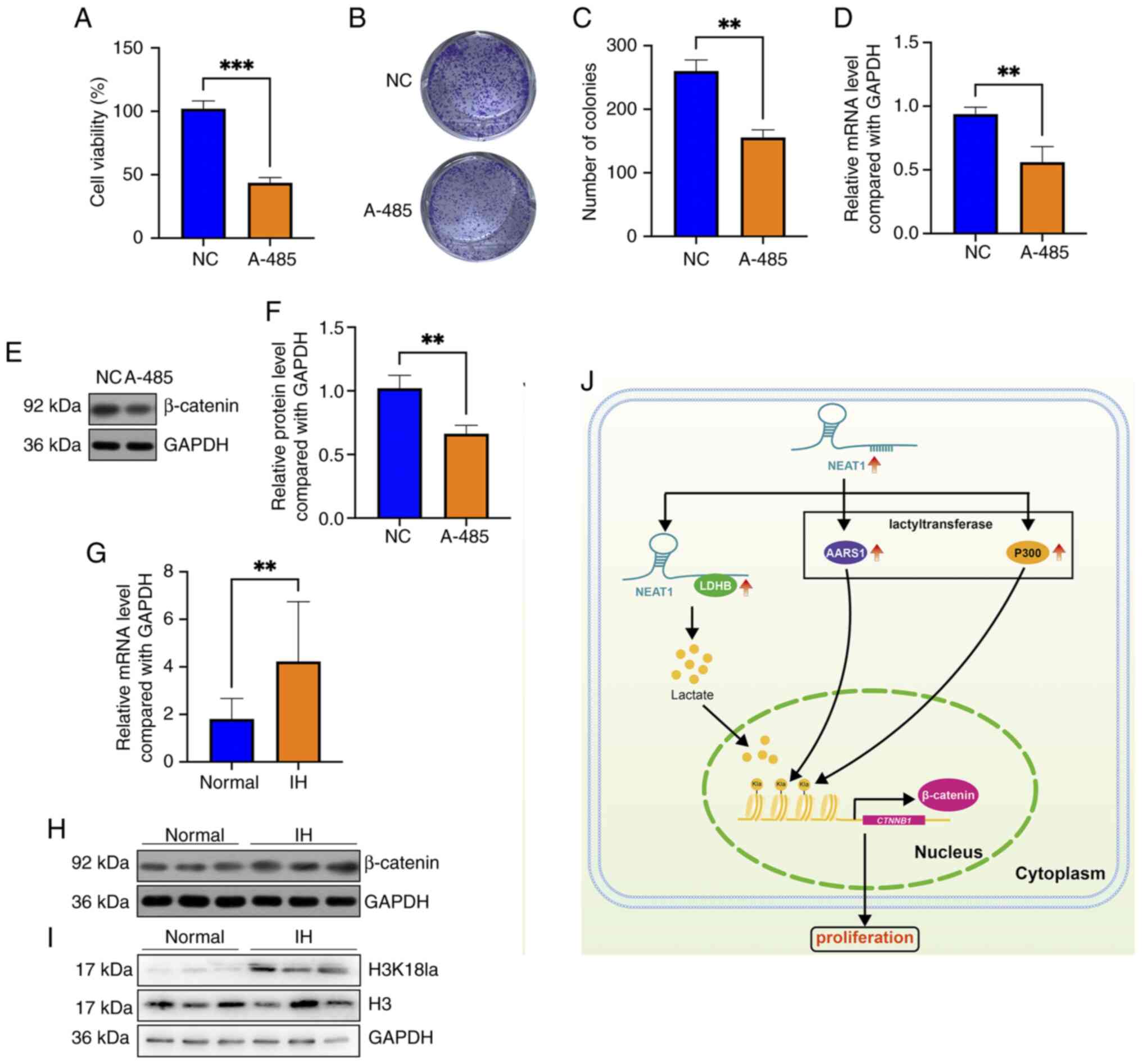
and protein levels of AARS1 and P300 (Fig. 6I-K). Notably, blocking P300 using
A-485 suppressed the viability and colony formation of HemECs
(Fig. 7A-C), and A-485 treatment
also decreased the mRNA and protein levels of β-catenin (Fig. 7D-F). These results indicated that
NEAT1 regulates β-catenin in hemangioma cells possibly by
positively affecting the lactyltransferases AARS1 and P300.
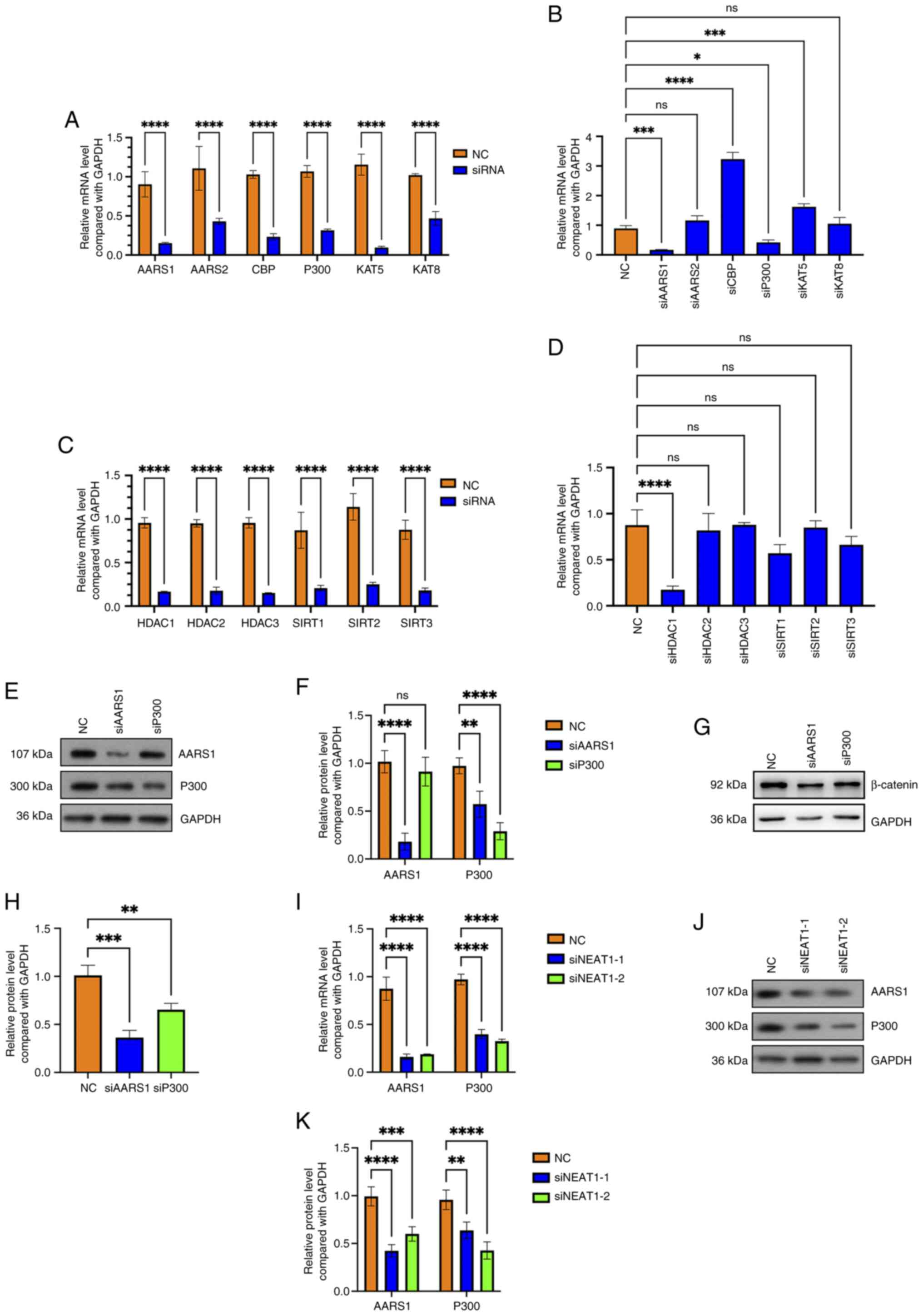 | Figure 6.Long non-coding RNA NEAT1 regulates
β-catenin by positively affecting the lactyltransferases AARS1 and
P300 in hemangioma cells. (A) The knockdown efficiencies of AARS1,
AARS2, CBP, KAT5, KAT8 and P300 were confirmed by RT-qPCR;
****P<0.0001 vs. NC (n=3). (B) After AARS1, AARS2, CBP, KAT5,
KAT8 and P300 were knocked down, the CTNNB1 mRNA level was
detected using RT-qPCR; *P<0.05, ***P<0.001, ****P<0.0001
vs. NC (n=3). (C) The knockdown efficiencies of SIRT1, SIRT2,
SIRT3, HDAC1, HDAC2 and HDAC3 were confirmed by RT-qPCR;
****P<0.0001 vs. NC (n=3). (D) After SIRT1, SIRT2, SIRT3, HDAC1,
HDAC2 and HDAC3 were knocked down, the CTNNB1 mRNA level was
detected using RT-qPCR; ****P<0.0001 vs. NC (n=3). (E) The
knockdown efficiency of AARS1 and P300 was confirmed by western
blotting (n=3). (F) Semi-quantitative analysis of the levels of
AARS1 and P300 in AARS1-knock down and P300-knock down cells
detected by western blotting; **P<0.01, ****P<0.0001 vs. NC
(n=3). (G) The protein level of β-catenin after AARS1 and P300
knockdown was detected using western blotting (n=3). (H)
Semi-quantitative analysis of the levels of β-catenin in
AARS1-knock down and P300-knock down cells detected by western
blotting; **P<0.01, ***P<0.001 vs. NC (n=3). (I) The mRNA
levels of AARS1 and P300 after NEAT1 knockdown were detected using
RT-qPCR; ****P<0.0001 vs. NC (n=3). (J) The protein levels of
AARS1 and P300 after NEAT1 knockdown were detected using western
blotting (n=3). (K) Semi-quantitative analysis of the levels of
AARS1 and P300 in NEAT1-knock down cells detected by western
blotting; **P<0.01, ***P<0.001, ****P<0.0001 vs. NC (n=3).
ns, not significant; NC, negative control; AARS, alany-tRNA
synthetase; CBP, CREB binding lysine acetyltransferase; KAT, lysine
acetyltransferase; RT-qPCR, reverse transcription-quantitative
polymerase chain reaction; CTNNB1, β-catenin gene; SIRT, sirtuin;
HDAC, histone deacetylase; siRNA, small interfering RNA. |
β-catenin and H3K18 lactylation levels
are elevated in IH tissues
The status of β-catenin expression and H3K18
lactylation levels in IH tissues were further evaluated. The
RT-qPCR and western blotting results revealed that both the mRNA
and protein levels of β-catenin were greater in the IH tissues than
in normal tissues (Fig. 7G and H).
Additionally, western blotting indicated that the H3K18 lactylation
level was greater in IH tissues than in normal tissues (Fig. 7I). In summary, the results revealed
that lncRNA NEAT1, which is upregulated in hemangioma, binds with
and upregulates LDHB, subsequently elevates the levels of cellular
lactate and H3K18 lactylation, potentiates β-catenin transcription
and ultimately enhances the proliferation of hemangioma cells
(Fig. 7G).
Discussion
Our previous study revealed that lncRNA NEAT1 is
highly expressed in IH tissues and promotes the proliferation,
migration and invasion of hemangioma cells by sponging miR-33a-5p
and regulating the downstream HIF1α/NF-κB pathway (6). Nevertheless, the functional roles and
detailed mechanisms of NEAT1 in hemangioma are still not fully
understood.
The Wnt/β-catenin pathway plays critical promotional
roles in hemangioma progression, and inactivation of the
Wnt/β-catenin pathway has therapeutic effects on hemangioma
(28,29). IL13RA2 was overexpressed in IH
tissues, and exogenous expression of IL13RA2 potentiated hemangioma
progression by interacting with and activating the β-catenin
pathway (28). Renin has been
shown to promote the proliferation of IH cells by activating the
Wnt pathway (31). Additionally,
blockade of the Wnt/β-catenin pathway by fucoidan has been shown to
inhibit the proliferation and EMT of hemangioma cells (29). Dai et al (32) reported that luteolin suppresses IH
progression by blocking the Wnt signaling pathway. Notably, the
results of the present study indicated that lncRNA NEAT1, which is
upregulated in IH tissues, upregulated β-catenin expression in an
H3K18 lactylation-dependent manner.
Lactic acid was first discovered in 1780 and was
formerly considered a byproduct of metabolism (33). However, at present, lactate is
considered to act as both a metabolic substance and a signaling
molecule (34,35). In 2019, Zhang et al
(9) discovered protein lysine
lactylation, a new posttranslational modification, and provided a
new insight into the novel function of lactate. H3K18 lactylation
is a critical type of histone lactylation that typically activates
gene transcription (36) and is
recognized as a biomarker for poor prognosis in epithelial ovarian
cancer (37). In non-small cell
lung carcinoma, H3K18 lactylation directly transcriptionally
activates POM121 transmembrane nucleoporin and ultimately induces
programmed cell death protein 1 expression and enhances immune
escape (27). In ovarian cancer,
lactate-induced H3K18 lactylation increases CCL18 expression
(38). In pancreatic ductal
adenocarcinoma (PDAC), H3K18 lactylation, which is elevated in
PDAC, promotes the transcription of BUB1 mitotic checkpoint
serine/threonine kinase B and TTK protein kinase and eventually
causes tumorigenesis (39). H3K18
lactylation transcriptionally activates SOX9 and facilitates liver
fibrosis development (40).
Notably, the present study revealed that H3K18 lactylation
transcriptionally activates β-catenin in hemangioma.
H3K18 lactylation is typically regulated by
glycolysis, lactyltransferases and delactyltransferases. In breast
cancer, potassium two pore domain channel subfamily K member 1
binds with LDHA and affects histone lysine lactylation (41). In PDAC, HDAC2 and P300 regulate
histone lactylation as the eraser and writer of protein lactylation
(39). The present study revealed
that NEAT1 binds to LDHB and positively regulates LDHB expression
in hemangioma cells. A previous study reported that LDHA was highly
expressed in HemECs compared with human umbilical vein endothelial
cells (42). Another study
revealed that the protein expression of phosphofructokinase-1
(FPK-1) is greater in proliferating IHs than in involuting IHs, and
that suppression of FPK-1 impedes the proliferation and migration
of HemECs and reduces lactate production (43). However, the role of LDHB in
hemangioma progression is still unclear.
The proliferative (first 6–12 months after birth),
involuting (starting at ~13 months) and involuted (4–7 years of
age) phases are the three stages of the self-limiting disease
course in IH (44). Unraveling the
mechanisms underlying IH development from proliferation to
involution is crucial for developing a new therapeutic strategy for
IH. The findings of the present study suggest that the levels of
H3K18la and β-catenin are increased in proliferating IH tissues.
Molecular analysis further revealed that H3K18la-driven
transcriptional activation of CTNNB1 enhances the
proliferation of hemangioma cells. Therefore, blocking this
mechanism might be a benefit for IH therapy.
Taken together, the results of the present study
revealed that lncRNA NEAT1, which is upregulated in hemangioma,
binds with and stabilizes LDHB, subsequently elevates the levels of
cellular lactate and H3K18 lactylation, potentiates CTNNB1
transcription and finally enhances the proliferation of hemangioma
cells. Nevertheless, several limitations of the present study
remain. First, when the candidate lactyltransferases or
delactyltransferases that may regulate β-catenin expression were
screened, the changes in expression were only examined at the mRNA
level and not the protein level. Second, in addition to H3K18
lactylation, NEAT1 also regulates the lactylation of non-histone
proteins; therefore, whether the lactylation of non-histone
proteins is also involved in the NEAT1-affected hemangioma cell
proliferation still needs to be investigated. Third, how NEAT1
regulates AARS1 and P300 requires further exploration.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was funded by the Joint Special Funds for the
Department of Science and Technology of Yunnan Province-Kunming
Medical University (grant no. 202201AY070001-202).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
HS conceived and designed the research. LY performed
the experiments and wrote the manuscript. NZ, XLZ, XJP, LX, YJP, LZ
and JNW analyzed the data. All authors read and approved the final
version of the manuscript. HS and LY confirm the authenticity of
all the raw data.
Ethics approval and consent to
participate
The Ethics Committee of Kunming Children's Hospital
(Kunming, China) approved this study (approval no.
2021-03-181-K01). Informed consent was obtained from the
parents/legal guardians of each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Liu C, Zhao ZL, Ji ZD, Jiang Y and Zheng
J: MiR-187-3p enhances propranolol sensitivity of hemangioma stem
cells. Cell Struct Funct. 44:41–50. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Acevedo LM and Cheresh DA: Suppressing
NFAT increases VEGF signaling in hemangiomas. Cancer Cell.
14:429–430. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang Q, Zhao C, Du Q, Cao Z and Pan J:
Non-coding RNA in infantile hemangioma. Pediatr Res. 96:1594–1602.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li Z, Cao Z, Li N, Wang L, Fu C, Huo R, Xu
G, Tian C and Bi J: M2 macrophage-derived exosomal lncRNA
MIR4435-2HG promotes progression of infantile hemangiomas by
targeting HNRNPA1. Int J Nanomedicine. 18:5943–5960. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li R, Liu Y, Liu J, Chen B, Ji Z, Xu A and
Zhang T: CCL2 regulated by the CTBP1-AS2/miR-335-5p axis promotes
hemangioma progression and angiogenesis. Immunopharmacol
Immunotoxicol. 46:385–394. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yu L, Shu H, Xing L, Lv MX, Li L, Xie YC,
Zhang Z, Zhang L and Xie YY: Silencing long non-coding RNA NEAT1
suppresses the tumorigenesis of infantile hemangioma by
competitively binding miR-33a-5p to stimulate HIF1α/NF-κB pathway.
Mol Med Rep. 22:3358–3366. 2020.PubMed/NCBI
|
|
7
|
Yu X, Liu X, Wang R and Wang L: Long
non-coding RNA NEAT1 promotes the progression of hemangioma via the
miR-361-5p/VEGFA pathway. Biochem Biophys Res Commun. 512:825–831.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Peng K, Xia RP, Zhao F, Xiao Y, Ma TD, Li
M, Feng Y and Zhou CG: ALKBH5 promotes the progression of infantile
hemangioma through regulating the NEAT1/miR-378b/FOSL1 axis. Mol
Cell Biochem. 477:1527–1540. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang D, Tang Z, Huang H, Zhou G, Cui C,
Weng Y, Liu W, Kim S, Lee S, Perez-Neut M, et al: Metabolic
regulation of gene expression by histone lactylation. Nature.
574:575–580. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xiao SY, Zhang SH, Sun K, Huang Q and Hu
C: Lactate and lactylation: Molecular insights into histone and
non-histone lactylation in tumor progression, tumor immune
microenvironment, and therapeutic strategies. Biomark Res.
13:1342025. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fang N, Zhang N, Jiang X, Yan S, Wang Z,
Gao Q, Xu M, Mu L, Li X, Chen J, et al: PFKM-Driven lactate
overproduction promotes atrial fibrillation via triggering cardiac
fibroblasts histone lactylation. Adv Sci (Weinh). 12:e009632025.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gu JJ, Deng CC, An Q, Zhu DH, Xu XY, Fu
ZZ, Zhou ZY, Rong Z and Yang B: Lactate promotes collagen
expression, proliferation and migration through H3K18
lactylation-dependent stimulation of LTBP3/TGF-beta1 axis in keloid
fibroblasts. J Invest Dermatol. July 7–2025.(Epub ahead of print).
View Article : Google Scholar
|
|
13
|
Galle E, Wong CW, Ghosh A, Desgeorges T,
Melrose K, Hinte LC, Castellano-Castillo D, Engl M, de Sousa JA,
Ruiz-Ojeda FJ, et al: H3K18 lactylation marks tissue-specific
active enhancers. Genome Biol. 23:2072022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang H, Xu M, Zhang T, Pan J, Li C, Pan B,
Zhou L, Huang Y, Gao C, He M, et al: PYCR1 promotes liver cancer
cell growth and metastasis by regulating IRS1 expression through
lactylation modification. Clin Transl Med. 14:e700452024.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Khan ZA, Melero-Martin JM, Wu X, Paruchuri
S, Boscolo E, Mulliken JB and Bischoff J: Endothelial progenitor
cells from infantile hemangioma and umbilical cord blood display
unique cellular responses to endostatin. Blood. 108:915–921. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kong C, Li R, Wang X, Li L, Kang N, Zhen
X, Dong Y and Yan G: Environmental aminomethylphosphonic acid
(AMPA) exposure increased the risk of spontaneous abortion through
lactate-induced JunB lactylation in trophoblast. Ecotoxicol Environ
Saf. 302:1187432025. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lu Y, Zhu J, Zhang Y, Li W, Xiong Y, Fan
Y, Wu Y, Zhao J, Shang C, Liang H and Zhang W: Lactylation-Driven
IGF2BP3-mediated serine metabolism reprogramming and RNA
m6A-modification promotes lenvatinib resistance in HCC. Adv Sci
(Weinh). 11:e24013992024. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhao SS, Liu J, Wu QC and Zhou XL: Lactate
regulates pathological cardiac hypertrophy via histone lactylation
modification. J Cell Mol Med. 28:e700222024. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wei S, Zhang J, Zhao R, Shi R, An L, Yu Z,
Zhang Q, Zhang J, Yao Y, Li H and Wang H: Histone lactylation
promotes malignant progression by facilitating USP39 expression to
target PI3K/AKT/HIF-1α signal pathway in endometrial carcinoma.
Cell Death Discov. 10:1212024. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lu G, Yi J, Gubas A, Wang YT, Wu Y, Ren Y,
Wu M, Shi Y, Ouyang C, Tan HWS, et al: Suppression of autophagy
during mitosis via CUL4-RING ubiquitin ligases-mediated WIPI2
polyubiquitination and proteasomal degradation. Autophagy.
15:1917–1934. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Whitworth CP, Aw WY, Doherty EL, Handler
C, Ambekar Y, Sawhney A, Scarcelli G and Polacheck WJ: P300
modulates endothelial mechanotransduction of fluid shear stress.
Cell Mol Bioeng. 17:507–523. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen B, Deng S, Ge T, Ye M, Yu J, Lin S,
Ma W and Songyang Z: Live cell imaging and proteomic profiling of
endogenous NEAT1 lncRNA by CRISPR/Cas9-mediated knock-in. Protein
Cell. 11:641–660. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cai S, Xia Q, Duan D, Fu J, Wu Z, Yang Z
and Yu C: Creatine kinase mitochondrial 2 promotes the growth and
progression of colorectal cancer via enhancing Warburg effect
through lactate dehydrogenase B. PeerJ. 12:e176722024. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang C, Zhou L, Zhang M, Du Y, Li C, Ren
H and Zheng L: H3K18 Lactylation potentiates immune escape of
Non-small cell lung cancer. Cancer Res. 84:3589–3601. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu Z, Ma T, Li J, Ren W and Zhang Z:
IL13RA2 promotes progression of infantile haemangioma by activating
glycolysis and the Wnt/β-catenin signaling pathway. Oncol Res.
32:1453–1465. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhu Z, Luo J, Li L, Wang D, Xu Q, Teng J,
Zhou J, Sun L, Yu N and Zuo D: Fucoidan suppresses proliferation
and epithelial-mesenchymal transition process via Wnt/β-catenin
signalling in hemangioma. Exp Dermatol. 33:e150272024. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen J, Zhao D, Wang Y, Liu M, Zhang Y,
Feng T, Xiao C, Song H, Miao R, Xu L, et al: Lactylated
apolipoprotein C-II induces immunotherapy resistance by promoting
extracellular lipolysis. Adv Sci (Weinh). 11:e24063332024.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
van Schaijik B, Tan ST, Marsh RW and
Itinteang T: Expression of (pro)renin receptor and its effect on
endothelial cell proliferation in infantile hemangioma. Pediatr
Res. 86:202–207. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dai Y, Zheng H, Liu Z, Wang Y and Hu W:
The flavonoid luteolin suppresses infantile hemangioma by targeting
FZD6 in the Wnt pathway. Invest New Drugs. 39:775–784. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li X, Yang Y, Zhang B, Lin X, Fu X, An Y,
Zou Y, Wang JX, Wang Z and Yu T: Lactate metabolism in human health
and disease. Signal Transduct Target Ther. 7:3052022. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yu H, Zhu T, Ma D, Cheng X, Wang S and Yao
Y: The role of nonhistone lactylation in disease. Heliyon.
10:e362962024. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Brooks GA: Lactate as a fulcrum of
metabolism. Redox Biol. 35:1014542020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Raychaudhuri D, Singh P, Chakraborty B,
Hennessey M, Tannir AJ, Byregowda S, Natarajan SM, Trujillo-Ocampo
A, Im JS and Goswami S: Histone lactylation drives CD8+
T cell metabolism and function. Nat Immunol. 25:2140–2151. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chao J, Chen GD, Huang ST, Gu H, Liu YY,
Luo Y, Lin Z, Chen ZZ, Li X, Zhang B, et al: High histone H3K18
lactylation level is correlated with poor prognosis in epithelial
ovarian cancer. Neoplasma. 71:319–332. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sun J, Feng Q, He Y, Wang M and Wu Y:
Lactate activates CCL18 expression via H3K18 lactylation in
macrophages to promote tumorigenesis of ovarian cancer. Acta
Biochim Biophys Sin (Shanghai). 56:1373–1386. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li F, Si W, Xia L, Yin D, Wei T, Tao M,
Cui X, Yang J, Hong T and Wei R: Positive feedback regulation
between glycolysis and histone lactylation drives oncogenesis in
pancreatic ductal adenocarcinoma. Mol Cancer. 23:902024. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wu S, Li J and Zhan Y: H3K18 lactylation
accelerates liver fibrosis progression through facilitating SOX9
transcription. Exp Cell Res. 440:1141352024. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hou X, Ouyang J, Tang L, Wu P, Deng X, Yan
Q, Shi L, Fan S, Fan C, Guo C, et al: KCNK1 promotes proliferation
and metastasis of breast cancer cells by activating lactate
dehydrogenase A (LDHA) and up-regulating H3K18 lactylation. PLoS
Biol. 22:e30026662024. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen J, Wu D, Dong Z, Chen A and Liu S:
The expression and role of glycolysis-associated molecules in
infantile hemangioma. Life Sci. 259:1182152020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yang K, Zhang X, Chen L, Chen S and Ji Y:
Microarray expression profile of mRNAs and long noncoding RNAs and
the potential role of PFK-1 in infantile hemangioma. Cell Div.
16:12021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Rodriguez Bandera AI, Sebaratnam DF,
Wargon O and Wong LF: Infantile hemangioma. Part 1: Epidemiology,
pathogenesis, clinical presentation and assessment. J Am Acad
Dermatol. 85:1379–1392. 2021. View Article : Google Scholar : PubMed/NCBI
|