Introduction
Myocardial infarction (MI), a critical condition
among cardiovascular diseases, is primarily caused by inadequate
blood supply to the myocardium, resulting in ischemia and
subsequent necrosis (1). This
pathology is markedly influenced by numerous risk factors,
including obesity, smoking, hypertension, hypercholesterolemia and
poor dietary habits (2).
Clinically, MI is typically manifested through chest pain, along
with symptoms such as chest tightness, dyspnoea, nausea and
vomiting (3). The incidence of MI
varies regionally and by population, typically ranging from a few
hundred to several thousand cases per 100,000 individuals per year
(4,5). The mortality associated with MI is
influenced by several factors, including age, sex, preexisting
cardiac health and the quality of care received (6). Timely and effective interventions,
including emergency resuscitation, anticoagulant and antiplatelet
therapies, vasodilators or coronary artery bypass grafting (CABG),
are key and have been revealed to reduce notably mortality
(7). In addition, a previous study
suggested that the use of volatile anaesthetics during CABG may
reduce postoperative MI rates and long-term cardiac mortality
(8).
Mepivacaine, a local anesthetic, is commonly used to
reduce pain during various medical procedures through the
inhibition of sodium channels in nerve cells (9). This function hinders the transmission
of nerve impulses, thereby facilitating local anaesthesia. Although
considered relatively safe, mepivacaine can cause side effects such
as overdose, allergic reactions and nerve damage (10). A protocol study by Dell'Olio et
al (11) suggested that
light-conscious sedation with mepivacaine may effectively manage
postoperative pain in patients with acute MI (AMI). Further
experimental evidence revealed that mepivacaine markedly reduces
calcium transients in adult mouse cardiomyocytes (12). This is achieved through sodium
channel blockade and enhancement of the reverse mode activity of
the sodium-calcium exchanger, suggesting a novel mechanism by which
mepivacaine may reduce myocardial contractility. In addition, a
clinical case report by de Groot-van der Mooren et al
(13) described an infant with
unexplained cardiorespiratory distress and neurological symptoms
shortly after birth, possibly related to maternal exposure to
mepivacaine. This case study raised concerns regarding the possible
function of mepivacaine in the emergence of cardiovascular and
neurological adverse effects.
The calcium voltage-gated channel auxiliary subunit
β1 (CACNB1) gene, located on chromosome 12 of the human
genome, encodes the β1 subunit of the voltage-gated calcium channel
(14). In cardiomyocytes,
voltage-gated calcium channels are essential for controlling
cardiac muscle contraction (15).
The β subunit encoded by the CACNB1 gene forms a complex
with the α1 subunit in cardiomyocytes, contributing to the
functionality of the calcium channel complex (16). Abnormalities in this function of
the complex may affect calcium channel activity, thereby
influencing cardiomyocyte excitability and contractility. Previous
studies have reported that abnormal expression of CACNB1 in
cardiac muscle cells is associated with such as arrhythmias,
myocardial hypertrophy and heart failure (17,18).
A study revealed that the CACNB1 gene encodes the CaVβ1
protein, which regulates T-cell function (17). After infection with the lymphocytic
choriomeningitis virus, deletion of CACNB1 increased T-cell
death and inhibited clonal proliferation. However, CACNB1 is
not necessary for T-cell proliferation, cytokine production or
Ca2+ signal transduction. Xuan et al (18) demonstrated that the levels of
potassium voltage-gated channel subfamily D member 3 (KCND3),
CACNB1 and potassium inwardly rectifying channel subfamily J member
2 (KCNJ2) proteins can be regulated by microRNA 195 through
direct interactions with KCND3, CACNB1 and KCNJ2
genes. Therefore, further exploration of the potential role of
CACNB1 in MI is warranted.
In the context of MI, understanding the molecular
mechanisms of cardiomyocyte injury is key for developing effective
treatment plans. The present study aimed to elucidate the impacts
of mepivacaine on apoptosis, cell viability, cell cycle regulation,
inflammatory responses and oxidative stress in H9c2 cells.
Furthermore, through CACNB1 knockdown experiments, the
protective effect of CACNB1 against mepivacaine- and
hypoxia/reoxygenation (H/R)-induced injury was investigated,
highlighting its potential as a therapeutic target to attenuate
myocardial injury. The present study reveals an innovative view
into the pathogenic processes and therapeutic interventions of
mepivacaine in the context of MI.
Materials and methods
Differential expression analysis of
the GSE19339 dataset
The GSE19339 dataset, available in the Gene
Expression Omnibus database (https://www.ncbi.nlm.nih.gov/gds/), was used to
identify genes associated with MI. This data set comprises
transcriptomic data from four patients with MI and four normal
individuals. The ‘limma’ package in the R programming language
(version 3.5.3; http://www.r-project.org/) was used to carry out the
differential expression analysis. The fold change (FC) criterion
was employed to assess differentially expressed genes (DEGs), with
FC ≤0.77 designating downregulated DEGs and FC ≥1.3 designating
upregulated DEGs. P<0.05 was considered to indicate a
statistically significant difference.
Functional enrichment analysis of DEGs
and expression analysis of CACNB1 in MI
The Database for Annotation, Visualization, and
Integrated Discovery (DAVID) (https://david.ncifcrf.gov/tools.jsp) was employed to
elucidate the functional characteristics of MI-related DEGs.
Analysis also included the Kyoto Encyclopedia of Genes and Genomes
(KEGG; http://www.kegg.jp/) pathway and Gene
Ontology (GO; http://geneontology.org/) term. GO classification
facilitates the categorization of gene and protein functions into
three hierarchical classifications: Biological Process (BP),
Molecular Function (MF) and Cell Component (CC). Finally, the
CACNB1 gene expression in the MI and normal groups was
assessed using the GSE19339 dataset, which was analyzed using the R
software (version 3.5.3).
Culturing and maintenance of H9c2
cells
H9c2 cells (cat. no. GNR 5, National Collection of
Authenticated Cell Cultures), a rat ventricular cardiomyocyte cell
line, were purchased from the Shanghai Institute of Biochemistry
and Cell Biology. H9c2 cells were cultured under conventional cell
culture conditions, which include a humidified environment at 37°C
with 5% CO2, using DMEM (cat. no. 10566016; Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% foetal bovine
serum (Gibco; Thermo Fisher Scientific, Inc.). The cell culture
solution was refreshed regularly to maintain cell viability and
growth. All experiments were carried out using cells within
passages 3–5 after cell recovery. The H9c2 cell line was
authenticated through short tandem repeat analysis and evaluated
for mycoplasma contamination, with negative results.
Cell transfection and treatment
Transfection was carried out on H9c2 cells utilizing
the following specific small interfering RNA (siRNA) targeting
CACNB1: si-CACNB1-1, sense 5′-CAAGACCAAGCCAGUGGCAUUUGCU-3′ and
antisense, 5′-AGCAAAUGCCACUGGCUUGGUCUUG-3′; si-CACNB1-2 sense,
5′-CAAUGUCCAAAUAGCGGCCUCGGAA-3′ and antisense,
5′-UUCCGAGGCCGCUAUUUGGACAUUG-3′; and negative control siRNA
(si-NC), sense 5′-CAAACCUAUAAGGCGCUCCGGUGAA-3′ and antisense,
5′-UUCACCGGAGCGCCUUAUAGGUUUG-3′. All siRNAs, including CACNB1-siRNA
and negative control siRNA, were purchased from Shanghai GenePharma
Co., Ltd. H9c2 cells were transfected with siRNAs (at a final
concentration of 50 nM) using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) after being cultured
in 24-well plates. Cells were incubated with the transfection
mixture at 37°C in a humidified atmosphere with 5% CO2
for 6 h, after which the medium was replaced with fresh DMEM
supplemented with 10% FBS. All transfections were carried out using
the same procedure. At 48 h post-transfection, to explore the
impact of mepivacaine on H9c2 cells, the cells were subjected to
different mepivacaine concentrations (0.5, 1.0, 2.0, 5.0, 7.0 and
10.0 mM; cat. no. HY-B0517; MedChemExpress; CAS No. 96-88-8) at
37°C in a humidified atmosphere with 5% CO2 for 24 h.
After mepivacaine treatment, cells were harvested for subsequent
analyses.
Cell Counting Kit-8 (CCK-8) assay
A CCK-8 assay (cat. no. CK04; Dojindo Laboratories,
Inc.) was employed to measure cell viability. H9c2 cells were
seeded in a 96-well plate at a density of 5×103
cells/well. Subsequently, each well was treated with 10 µl CCK-8
reagent and the wells were then incubated at 37°C with 5%
CO2 for 4 h. Subsequently, the absorbance at 450 nm was
measured utilizing a microplate reader (Thermo Fisher Scientific,
Inc.).
Reverse transcription-quantitative PCR
(RT-qPCR)
H9c2 cells were treated with the TRIzol®
Reagent (Invitrogen; Thermo Fisher Scientific, Inc.) to extract
total RNA. The purity and concentration of the RNA were evaluated
through spectrophotometric analysis. The SuperScript III
First-Strand Synthesis System (Invitrogen; Thermo Fisher
Scientific, Inc.) was used for the synthesis of complementary DNA,
with 1 µg of the extracted RNA being reverse-transcribed.
Subsequently, the Applied Biosystems 7500 Fast Real-Time PCR System
(Applied Biosystems; Thermo Fisher Scientific, Inc.) was employed
to carry out qPCR analysis using the following primer pairs:
CACNB1, forward, 5′-AGGGCTATGAGGTAACTGAC-3′ and reverse,
5′-GGAAATCCTGCCATCAAACC-3′); Bax forward,
5′-TGAAGACAGGGGCCTTTTTG-3′ and reverse, 5′-AATTCGCCTGAGACACTCG-3′);
Bcl-2 forward, 5′-ATGCCTTTGTGGAACTATATGGC-3′ and reverse,
5′-GGTATGCACCCAGAGTGATGC-3′; Caspase-3 forward,
5′-AGCATTTCCCATAAGCCTCCT-3′ and reverse,
5′-AGCACCAAAAGTATCATTGGC-3′; P21 forward,
5′-ACCCCAGATAACCAAGGATGC-3′ and reverse,
5′-AGAACACCATCTTGCCCTTGT-3′; Cyclin E1 forward,
5′-GACACAGCTTCGGGTACGG-3′ and reverse, 5′-TTGCTGTGGTCCTTCGAGTC-3′;
CDK4 forward, 5′-CTTCCCGTCAGCACAGTTC-3′ and reverse,
5′-GGTCAGCATTTCCAGCAGC-3′; and GAPDH forward,
5′-ACGGGAAACCCATCACCATC-3′ and reverse, 5′-CTCGTGGTTCACACCCATCA-3′.
GAPDH was used as an internal reference. The thermocycling
conditions were as follows: Initial denaturation at 95°C for 10
min, followed by 40 cycles of denaturation at 95°C for 15 sec and
annealing/extension at 60°C for 1 min. A melt curve analysis
(60–95°C) was performed to verify amplification specificity. The
expression was examined utilizing the 2−ΔΔCq method
(19).
Western blotting (WB)
Total protein lysates from H9c2 cells were prepared
utilizing RIPA lysis buffer (Thermo Fisher Scientific, Inc.)
containing protease and phosphatase inhibitors (Thermo Fisher
Scientific, Inc.). The BCA Protein Assay Kit (Beyotime Institute of
Biotechnology) was used to measure the protein content. Equal
amounts of protein (30 µg per lane) were separated on 10% gels
using SDS-PAGE and then transferred onto PVDF membranes
(MilliporeSigma). The membranes were blocked with 5% non-fat milk
in TBS-Tween (TBST; 0.1% (v/v) Tween-20) at room temperature for 1
h and then incubated overnight at 4°C with the following primary
antibodies: CACNB1 (1:1,000; cat. no. 13039-1-AP; Proteintech
Group, Inc.), Bax (1:20,000; cat. no. 50599-2-Ig; Proteintech
Group, Inc.), Caspase-3 (1:1,000; cat. no. 19677-1-AP; Proteintech
Group, Inc.), Bcl-2 (1:2,000; cat no. 26593-1-AP; Proteintech
Group, Inc.), P21 (1:1,000; cat no. 28248-1-AP; Proteintech Group,
Inc.), CDK4 (1:1,000; cat no. 11026-1-AP; Proteintech Group, Inc.),
Cyclin E1 (1:1,000; cat no. AF0144; Affinity Biosciences), NOD-like
receptor protein 3 (NLRP3; 1:2,000; cat no. 30109-1-AP; Proteintech
Group, Inc.), Caspase-1 (1:2,000; cat no. 31020-1-AP; Proteintech
Group, Inc.), C-Caspase-1 (1:1,000; cat no. AF4005; Affinity
Biosciences), GSDMD (1:2,000; cat no. 20770-1-AP; Proteintech
Group, Inc.), IL-18 (1:2,000; cat no. DF6252; Affinity
Biosciences), ASC (1:5,000; cat no. 30641-1-AP; Proteintech Group,
Inc.), IL-1β (1:2,000; cat no. AF5103; Affinity Biosciences),
nuclear factor erythroid 2-related factor 2 (Nrf2; 1:1,000; cat no.
AF0639; Affinity Biosciences), GAPDH (1:5,000; cat no. 10494-1-AP;
Proteintech Group, Inc.) and Lamin B (1:2,000, cat no. 12987-1-AP;
Proteintech Group, Inc.). After secondary antibody incubation with
HRP-conjugated goat anti-rabbit IgG (1:5,000; cat. no. SA00001-2;
Proteintech Group, Inc.) at room temperature for 1 h, bands were
developed using an ECL kit (Cytiva) and captured on ImageJ software
(version 1.8.0; National Institutes of Health).
Flow cytometry analysis
Flow cytometry analysis was carried out to evaluate
apoptosis in H9c2 cells. After being seeded in 24-well plates at a
density of 1×104 cells/well, H9c2 cells were grown for
24 h at 37°C in a humidified 5% CO2 incubator. Following
cell detachment with trypsin-EDTA (0.25% trypsin and 0.02% EDTA),
the cells were collected by centrifugation at 300 × g for 5 min at
4°C, washed twice with PBS, and resuspended. Subsequently, the
cells were cultured with 5 µl Annexin V-fluorescein isothiocyanate
and 5 µl of PI solution at ambient temperature for 15 min for
apoptosis analysis. The staining with PI and Annexin V was then
evaluated using a flow cytometer (CyFlow® Cube 6;
Sysmex-Partec; Guangzhou Jiyuan Biotechnology Co., Ltd.) and the
FlowJo program (version 7.4.1; BD Biosciences) was employed to
analyze the data.
Measurement of oxidative stress
biomarkers in H9c2 cells
H9c2 cells were plated in 96-well plates at a
density of 1×104 cells/well and allowed to adhere for 24
h at 37°C in a humidified incubator with 5% CO2. After
adhesion, the cells were rinsed twice with PBS and then lysed. The
supernatants were separated from the lysates through centrifugation
at 10,000 × g for 5 min at 4°C. To aid homogeneity, the
supernatants were sonicated on ice at 20 kHz for 3×10 sec bursts
with 10 sec intervals. The prepared samples were utilized to
measure the expression of lactate dehydrogenase (LDH) activity,
reactive oxygen species (ROS), superoxide dismutase (SOD) activity
and malondialdehyde (MDA) content. The assays for SOD and MDA were
conducted utilizing commercial kits (Nanjing Jiancheng
Bioengineering Institute). The ROS assay and LDH activity were
evaluated utilizing a kit from Beyotime Institute of Biotechnology.
All measurements were carried out in triplicate and the data were
standardized to protein content in the samples to account for
variations in cell density across wells.
ELISA
In the present study, the secretion of IL-1β, TNF-α
and IL-8 in the culture supernatants of H9c2 cells, either
untreated or treated with mepivacaine (0.5, 1 and 2 mM for 24 h),
was determined using ELISA kits for IL-1β (cat. no. CSB-E08055r;
Cusabio Technology, LLC), TNF-α (cat. no. CSB-E11987r; Cusabio
Technology, LLC), and IL-8 (cat. no. SEKR-0014; Beijing Solarbio
Science & Technology Co., Ltd.). The absorbance at 450 nm was
measured using an ELISA reader. The obtained values were compared
with standard curves established using standard titration. The BCA
test was utilized to ascertain the protein content in each culture
bottle. All concentrations were standardized to the respective
cellular protein content and expressed as pg/ml (cellular
protein).
H/R model and treatment in H9c2
cells
After being moved to the cell culture equipment,
H9c2 cells were treated with a hypoxic buffer solution (1%
O2 + 5% CO2 + 94% N2) to exhaust
any remaining oxygen for 2 h at 37°C. Cells were then exposed to 3
h of hypoxia in a constant temperature CO2 incubator at
37°C with 95% humidity. After hypoxic exposure, the cells were
reoxygenated under normal oxygen conditions (21% O2, 74%
N2 and 5% CO2) at 37°C for 2 h following the
replacement of the buffer solution with a standard culture medium.
Cell samples were randomly divided into three groups: i) Control,
with normal incubation (37°C, 5% CO2, 95% humidity) for
6 h; ii) HR, pre-treated with 2 mM mepivacaine for 24 h before H/R
induction, followed by 2 h of reoxygenation after 3 h of hypoxia;
iii) si-CACNB1−1 + HR, pre-treated with 2 mM mepivacaine for
24 h before H/R induction and then transfected with CACNB1-1
siRNA, as described in the ‘Cell transfection and treatment’
section.
Statistical analysis
R software was employed for statistical analysis of
the current dataset. The normality of data distribution was
assessed using the Shapiro-Wilk normality test. P>0.05 indicated
that the data were normally distributed. Comparisons between two
independent groups of normally distributed data were analyzed using
an unpaired, two-tailed Student's t-test. For comparisons involving
more than two groups, a one-way ANOVA followed by Tukey's post-hoc
test was applied to control the familywise error rate. All
quantitative data are presented as mean ± standard deviation.
P<0.05 was considered to indicate a statistically significant
difference. To ensure equal protein loading in western blot
analyses, GAPDH was used as the loading control for cytoplasmic
proteins, while Lamin B was used for nuclear proteins. In the
RT-qPCR analyses, GAPDH was used as the internal reference. Each
experiment was independently repeated at least three times with
separate biological samples to ensure reproducibility and
reliability of statistical analysis.
Results
Functional enrichment analysis of
GSE19339-DEGs and expression verification of CACNB1
DEG analysis was carried out on samples from the
GSE19339 dataset using the ‘limma’ package, resulting in the
identification of 2,396 upregulated DEGs and 1,230 downregulated
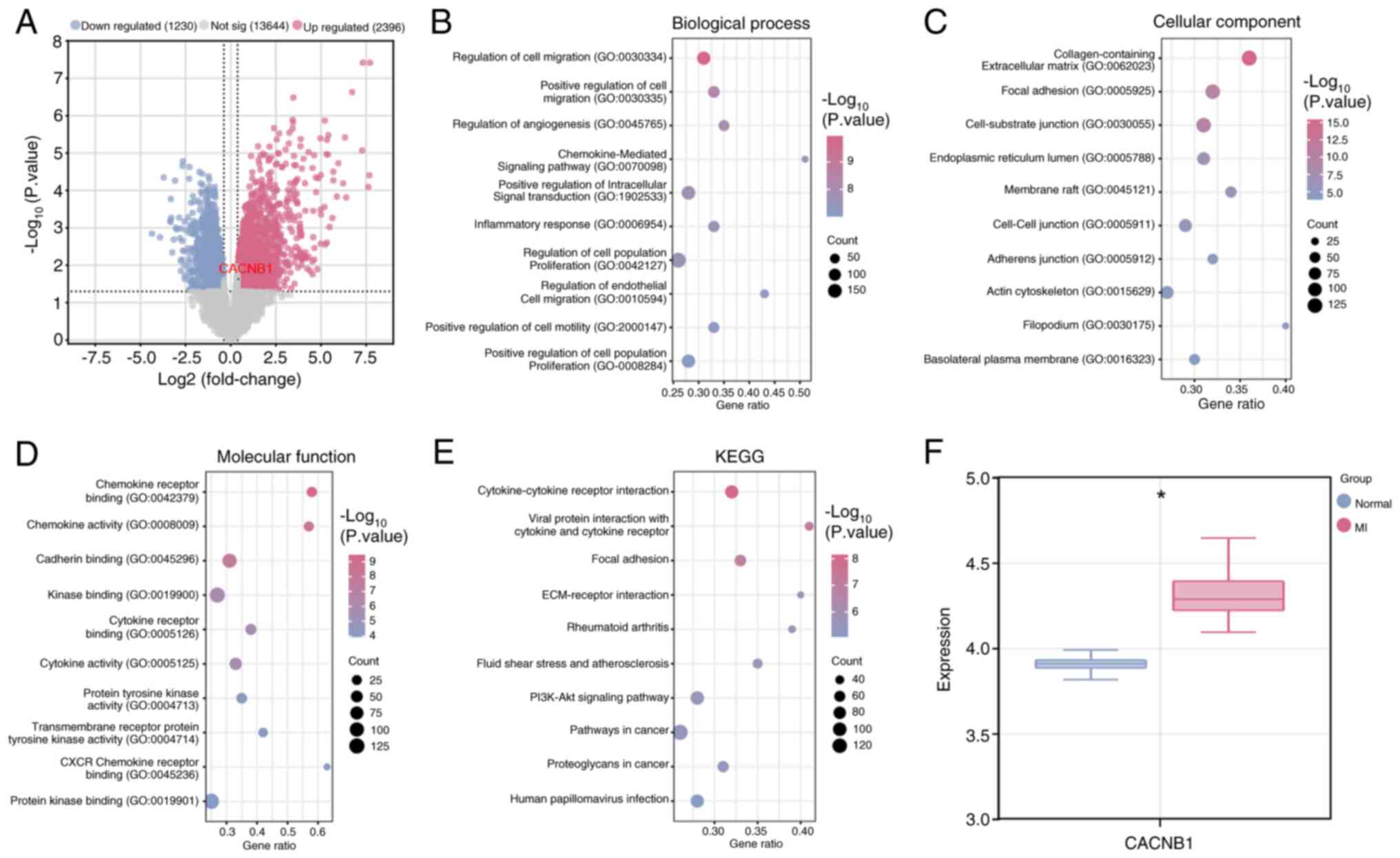
DEGs (Fig. 1A). Subsequently,
functional enrichment analysis was conducted on the DEGs. Analysis
of GO terms revealed significant enrichment of DEGs in BP such as
‘Chemokine-Mediated Signalling Pathway’, ‘Inflammatory Response’
and ‘Regulation Of Cell Migration’ (Fig. 1B). Moreover, in CC terms, DEGs
demonstrated notable enrichment in ‘Endoplasmic Reticulum Lumen’,
‘Adherens Junction’ and ‘Actin Cytoskeleton’ (Fig. 1C). For MF terms, DEGs exhibited
significant enrichment in ‘Protein Kinase Binding’, ‘Protein
Tyrosine Kinase Activity’ and ‘Cadherin Binding’ (Fig. 1D). As depicted in Fig. 1E, in the KEGG pathways, the
majority of DEGs were enriched in pathways such as ‘PI3K-Akt
signalling pathway’, ‘Fluid shear stress and atherosclerosis’ and
‘Proteoglycans in cancer’. Given that CACNB1 has been
previously associated with cardiovascular diseases (20,21)
and encodes the β1 subunit of the voltage-gated calcium channel,
which is essential for regulating calcium influx and modulating
cardiac muscle contraction, CACNB1 was selected as the
target gene. Further investigation revealed that the expression of
CACNB1 was upregulated in MI samples of the GSE19339
dataset, suggesting a potential role of this gene in enhancing
cardiac dysfunction or exacerbating disease pathology in MI
(Fig. 1F).
Mepivacaine induces apoptosis in H9c2
cells in a dose-dependent manner
Clinically relevant plasma concentrations of
mepivacaine may vary depending on the specific application and
patient population. For example, in a study on plasma mepivacaine
concentrations in patients undergoing anesthesia, the peak plasma
concentration ranged from 0.77–8.31 µg/ml (22). Based on preliminary experiments
(Fig. S1), the concentrations of
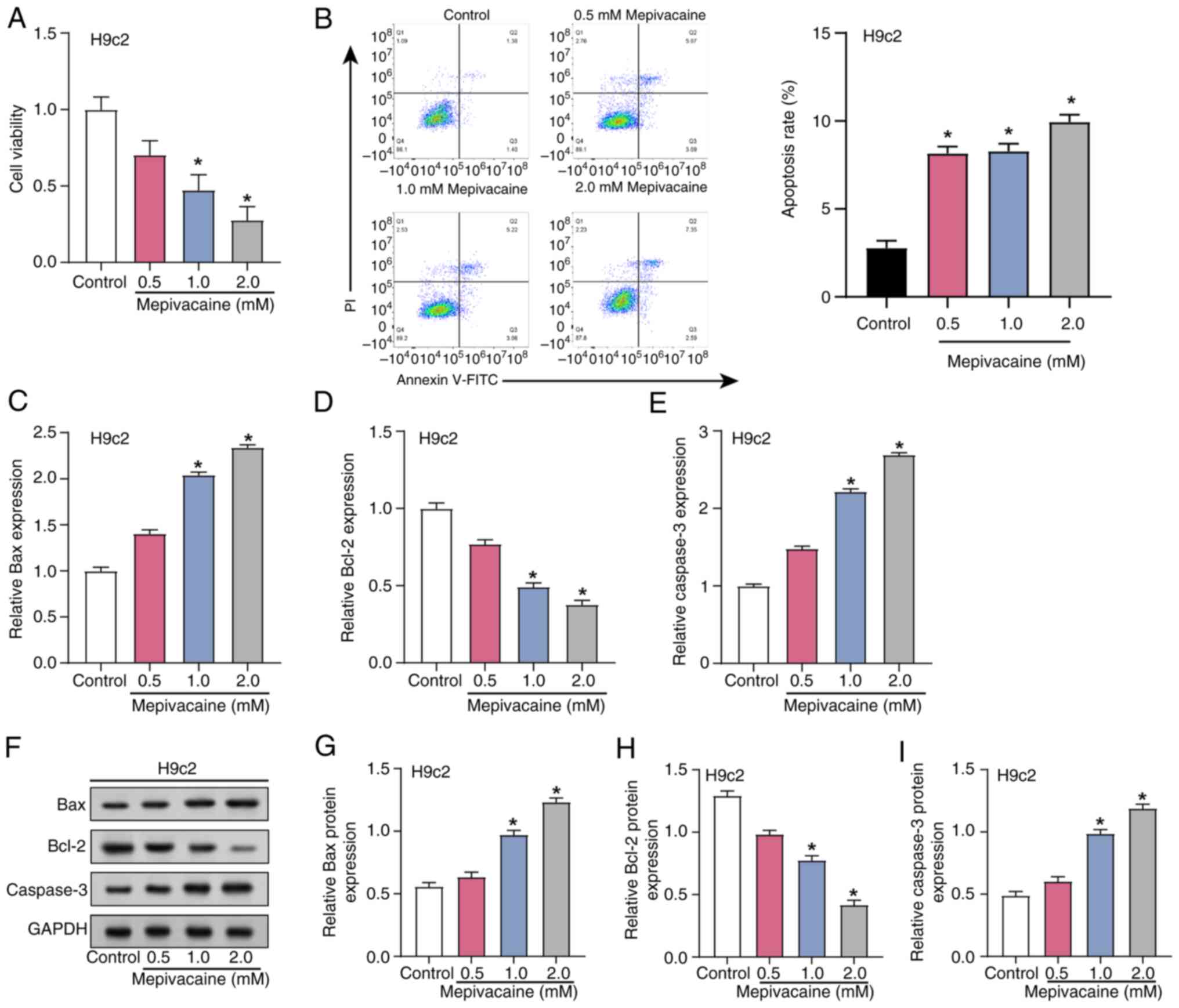
0.5, 1.0 and 2.0 mM for mepivacaine were used (23). Cell viability and apoptosis in H9c2
cells subjected to different concentrations (0.5, 1.0 and 2.0 mM)
of mepivacaine were assessed using flow cytometry, as well as CCK-8
assays (Fig. 2A and B). The
outcomes demonstrated that there was a dose-dependent reduction in
cell viability of H9c2 cells with increasing concentrations of
mepivacaine, while the apoptosis rate increased. Subsequent
analysis using RT-qPCR examined the effects of mepivacaine
treatment on apoptotic factors (caspase-3, Bax and Bcl-2) in H9c2
cells (Fig. 2C-E). Compared with
the control group, levels of Caspase-3 and Bax increased, whereas
the expression of Bcl-2 decreased with increasing concentrations of
mepivacaine. These changes were further confirmed through
semi-quantification of protein expression (Fig. 2F-I). Collectively, these findings
suggested that mepivacaine induces apoptosis in H9c2 cells in a
concentration-dependent manner.
Mepivacaine modulates cell cycle
regulatory factors to induce G1 arrest in H9c2
cells
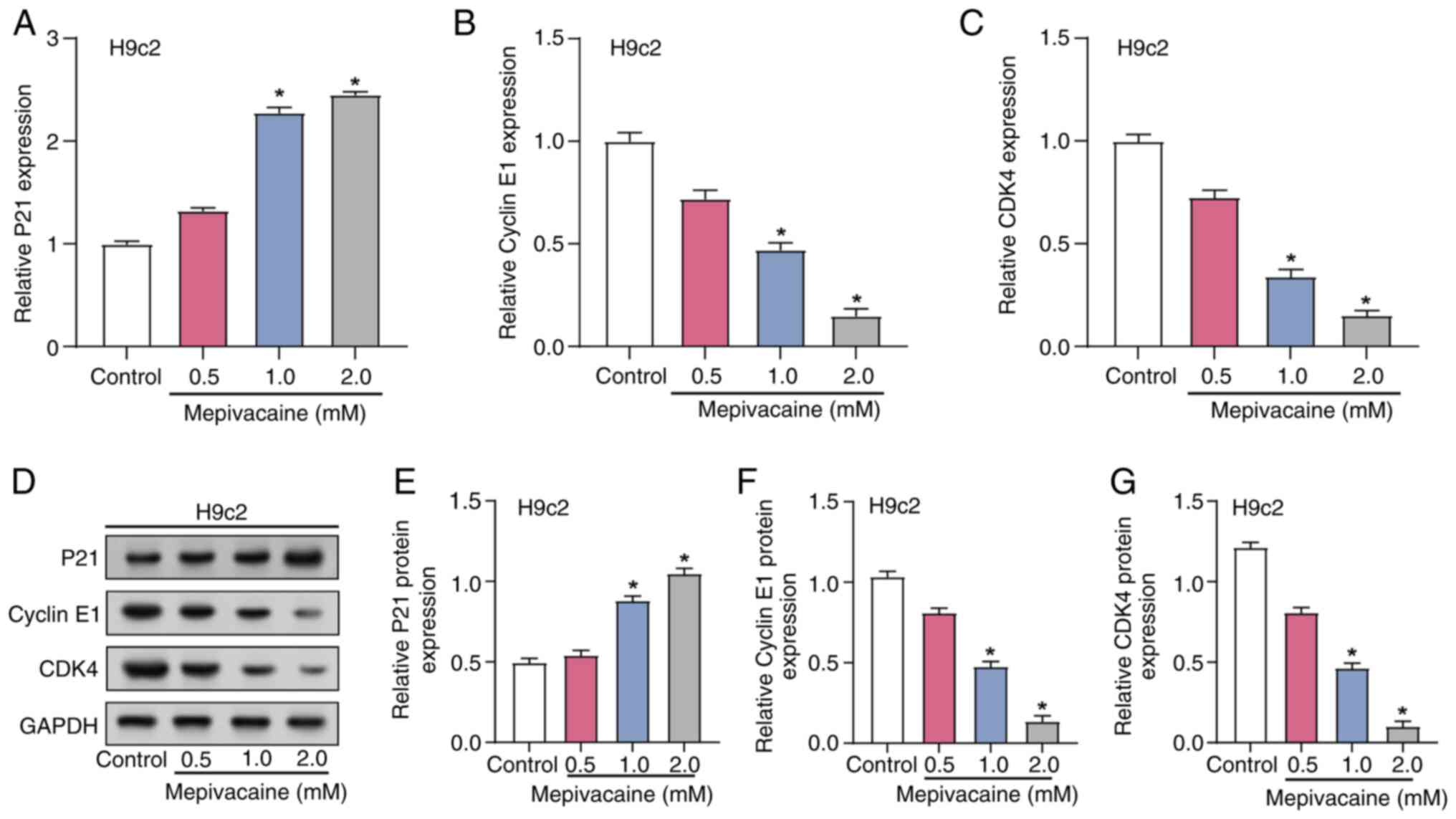
The influence of mepivacaine on cell cycle dynamics
in H9c2 cells was investigated by assessing the expression of key
cell cycle regulators, including Cyclin E1, P21 and CDK4. RT-qPCR
and WB analysis were utilized to measure changes in the levels of
cell cycle regulators after exposure to varying concentrations of
mepivacaine (0.5, 1.0 and 2.0 mM; Fig.
3A-G). The data revealed a dose-dependent increase in P21
expression, with significant elevations at 1 and 2 mM mepivacaine,
suggesting an enhanced inhibitory effect on the cell cycle
(Fig. 3A and E). Conversely, the
levels of CDK4 and Cyclin E1, key promoters of the G1 to
S phase transition, revealed a marked decrease across the same
mepivacaine concentrations (Fig. 3B,
C, F and G). This reciprocal regulation of cell cycle proteins
suggested that mepivacaine induces G1 phase arrest in
H9c2 cells by disrupting the balance of cell cycle regulators
required for progression into the S phase.
Mepivacaine induces oxidative stress
and inflammation in H9c2 cells
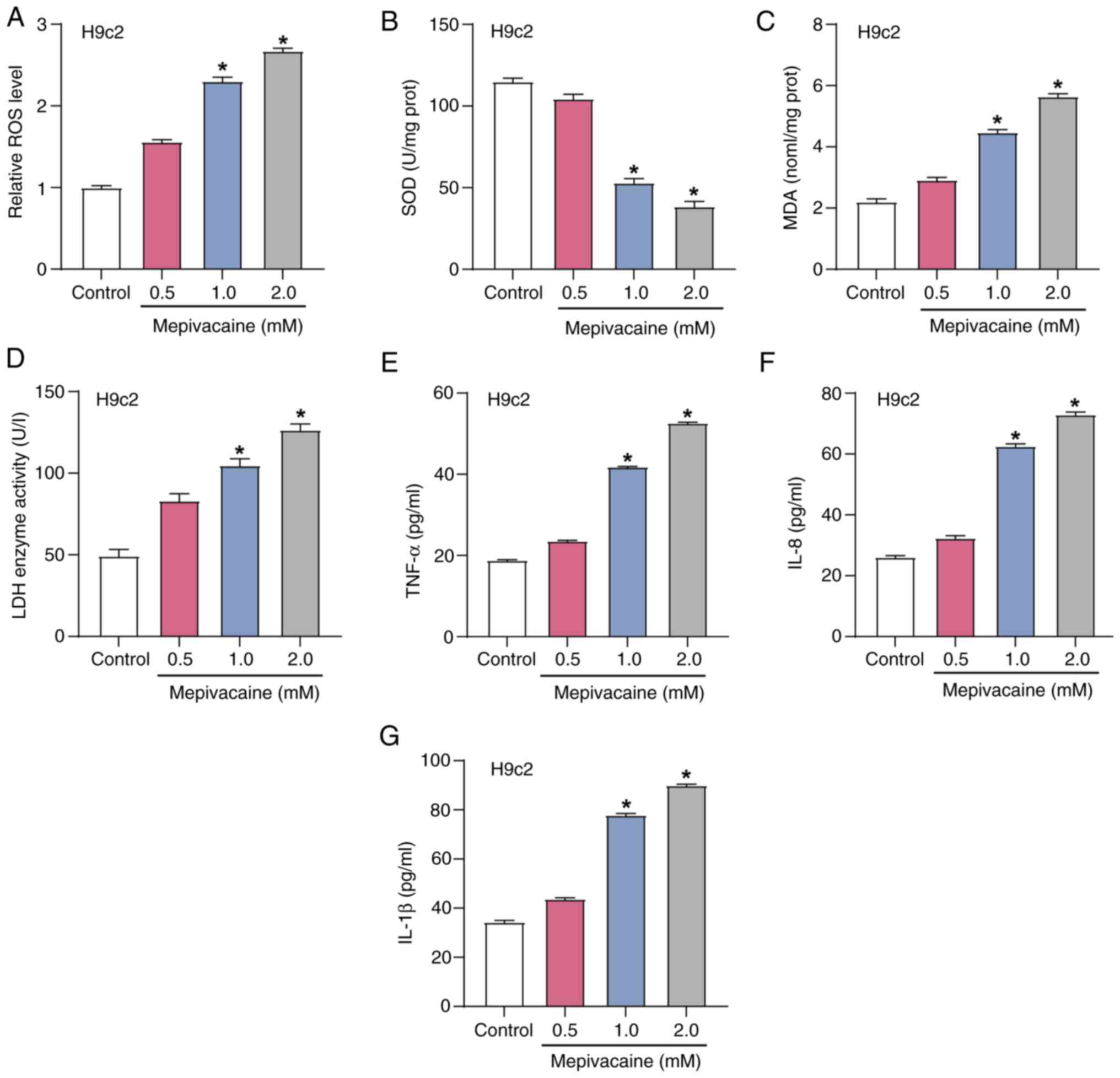
To explore the impact of mepivacaine on oxidative
stress in H9c2 cells, the expression levels of ROS, SOD, MDA and
LDH were assessed using the corresponding assay kits after exposure
to varying concentrations of mepivacaine (Fig. 4A-D). The outcomes indicated that
compared with the control group, the ROS, MDA and LDH levels
increased with increasing concentrations of mepivacaine, while SOD
levels decreased. These findings suggested that mepivacaine induces
oxidative stress and cell damage in H9c2 cells. Subsequently, the
secretion levels of inflammatory cytokines (IL-1β, TNF-α and IL-8)
in H9c2 cells after exposure to different concentrations of
mepivacaine were measured using ELISA experiments (Fig. 4E-G). The current findings
demonstrated that, compared with the control group, the expression
levels of IL-1β, TNF-α and IL-8 increased with increasing
concentrations of mepivacaine. This indicated a significant
pro-inflammatory effect of mepivacaine in H9c2 cells.
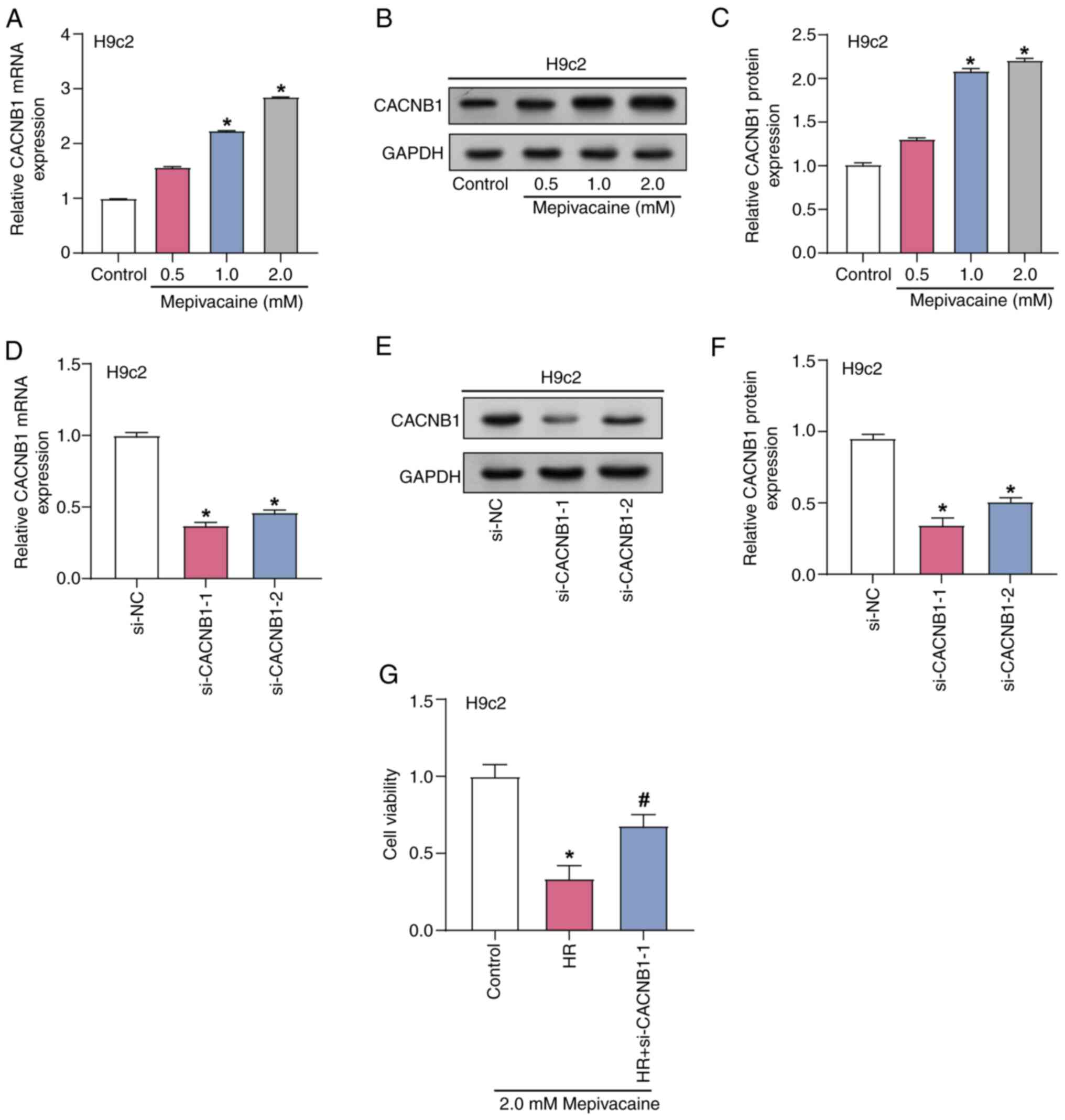
CACNB1 knockdown protects H9c2 cells
from mepivacaine and H/R-induced injury
The effect of mepivacaine treatment at several
concentrations on the expression of CACNB1 in H9c2 cells was
investigated using RT-qPCR and WB analyses (Fig. 5A-C). Analysis revealed that as the
mepivacaine concentration increases, the level of CACNB1 in
H9c2 cells also increases. Furthermore, CACNB1 expression
was highest under treatment with 2 mM mepivacaine; therefore, this
particular concentration was used for further experiments.
Subsequently, the transfection efficiency of CACNB1
knockdown plasmids was assessed with RT-qPCR and WB analyses, with
si-CACNB1−1 demonstrating the greatest efficacy. Therefore,
si-CACNB1−1 was used for knockdown experiments (Fig. 5D-F). The viability of H/R model
H9c2 cells subjected to 2 mM mepivacaine for 24 h, combined with
CACNB1 knockdown, was assessed with the CCK-8 assay
(Fig. 5G). The findings indicated
that the viability of H/R-treated cells was notably reduced
compared with the control group, whereas knockdown of CACNB1
significantly improved cell viability compared with the H/R group.
This suggested that CACNB1 knockdown protects cardiomyocytes
from injury induced by mepivacaine and H/R.
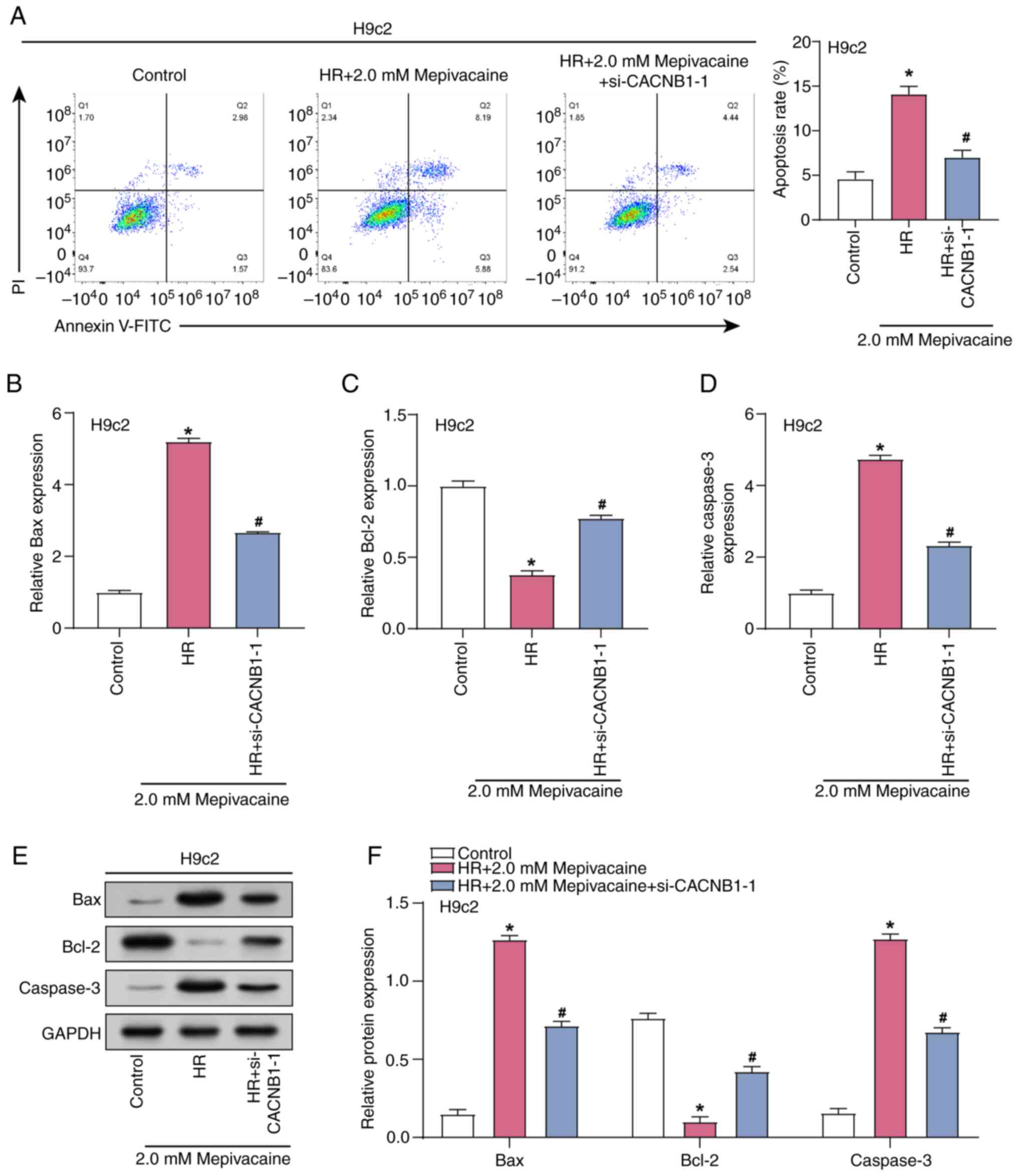
CACNB1 knockdown attenuates apoptosis
in H/R model H9c2 cells treated with mepivacaine
Flow cytometry was used to measure the level of
apoptosis in H/R model H9c2 cells subjected to 2 mM mepivacaine in
combination with CACNB1 knockdown (Fig. 6A). The results indicated a notable
increase in the apoptosis rate in the H/R group in comparison with
the control group, while the apoptosis rate was decreased in the
H/R group with CACNB1 knockdown compared with the H/R group.
Subsequently, RT-qPCR and WB analyses were conducted to examine the
impact of combined treatment with 2 mM mepivacaine and
CACNB1 knockdown on the levels of apoptotic proteins in H/R
model H9c2 cells (Fig. 6B-F).
Analysis revealed that, compared with the Control group, the
expression of Caspase-3 and Bax was upregulated, whereas the
expression of Bcl-2 was downregulated in the H/R group. However,
upon additional CACNB1 knockdown, the expression of
Caspase-3 and Bax decreased, while the expression of Bcl-2
increased compared with the H/R group. These experiments
demonstrated that CACNB1 knockdown exerts anti-apoptotic
effects, alleviating the damage induced by H/R and mepivacaine.
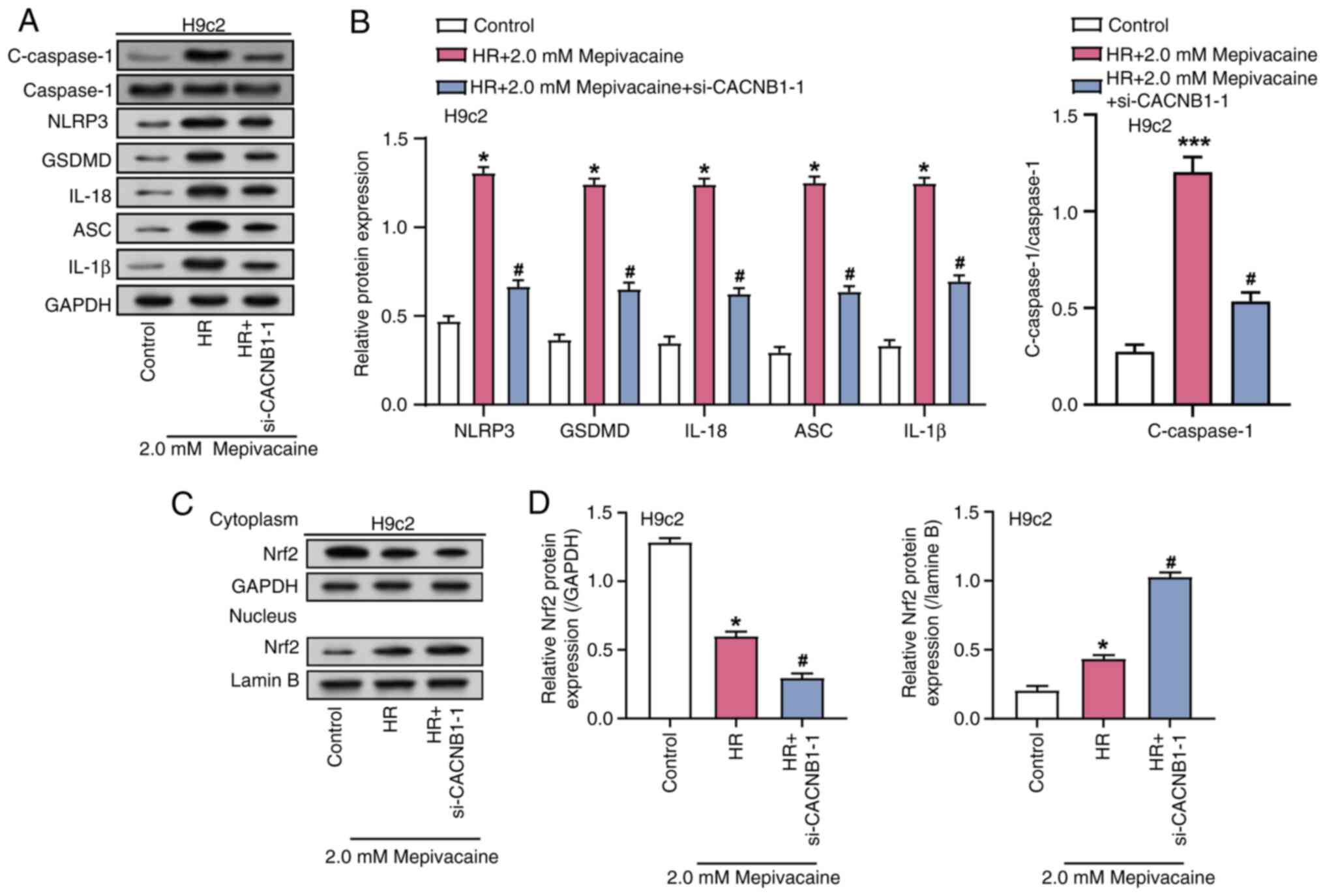
CACNB1 knockdown modulates the
expression of inflammatory mediators and Nrf2 nuclear translocation
in the H/R model treated with mepivacaine
WB analysis was employed to assess the impact of
combined treatment with 2 mM mepivacaine and CACNB1
knockdown on the levels of inflammatory-related factors (NLRP3,
IL-1β, GSDMD, ASC, IL-18, Caspase-1 and C-Caspase-1) in H/R model
H9c2 cells (Fig. 7A and B).
Analysis revealed a noteworthy rise in the level of these
inflammation-related factors in the H/R group, compared with the
control group, while their expression was notably reduced in the
H/R group with CACNB1 knockdown compared with the H/R group.
Previous research suggested that Nrf2 is involved in maintaining
redox homeostasis and exhibits protective effects against MIRI
(24). To investigate whether the
anti-apoptotic effect induced by CACNB1 knockdown is
associated with Nrf2 nuclear translocation, the levels of nuclear
Nrf2 and cytoplasmic Nrf2 proteins were assessed through WB
analysis (Fig. 7C and D). Analysis
revealed that compared with the Control group, the levels of
cytoplasmic Nrf2 decreased, while the levels of nuclear Nrf2
increased in the H/R group. Furthermore, in the H/R group with
CACNB1 knockdown, the level of nuclear Nrf2 increased and the level
of cytoplasmic Nrf2 further decreased. These findings indicated
that CACNB1 knockdown advances the translocation of Nrf2
into the nucleus, indicating that CACNB1 inhibits
inflammation and apoptosis through the CACNB1/NLRP3/Nrf2
axis, thereby alleviating the damage induced by H/R and
mepivacaine.
Discussion
MI, as a globally prevalent and highly lethal
disease, has attracted considerable attention from clinicians and
researchers. Zhao et al (25) identified genes such as IL1R2,
IRAK3 and THBD as diagnostic biomarkers for AMI, which
are associated with immune cell infiltration. Scholars have
summarized signaling pathways associated with MI, including
inflammatory pathways such as TLR4/MyD88/NF-κB and NLRP3/Caspase-1,
oxidative stress and apoptosis-related pathways such as Sonic
hedgehog and Nrf2/HO-1 and key regulators of angiogenesis such as
PI3K/Akt and MAPK pathways, all carrying out key roles in the
development of MI (26). The
bioinformatic analysis of the present study, leveraging the
GSE19339 dataset, supported these findings by identifying
significant enrichment of ‘Inflammatory Response’ and ‘PI3K-Akt
signaling pathway’ among the GSE19339-DEGs. Paolisso et al
(27) revealed that patients with
obstructive AMI with hyperglycaemia exhibited increased
inflammation markers and larger infarct areas, whereas patients
with non-obstructive AMI demonstrated less myocardial damage.
Additionally, the hypothesis by Zhao et al (28) that the long non-coding RNA MI
Associated Transcript regulates the PI3K/Akt pathway, which
potentially influences myocardial fibrosis and heart failure,
underscores the complexity of post-MI remodeling processes.
Notably, the findings of the present study also revealed an
upregulation of CACNB1 in MI samples, indicating its pivotal role
in cardiac dysfunction and the progression of disease pathology.
This positions CACNB1 as a potential target for therapeutic
intervention, aiming to modulate cardiac responses to ischemic
stress and improve outcomes in patients with MI.
Mepivacaine, an amide-type local anesthetic known
for its rapid diffusion and brief duration, is widely used in
surgery and dentistry (29,30).
While essential for effective pain management, its potential for
cardiac toxicity requires careful use, especially in patients with
cardiovascular risks (31). The
current research on H9c2 cardiomyocytes revealed that mepivacaine
may exacerbate myocardial injury, characterized by notable cell
cycle arrest and activation of apoptosis. This effect is
characterized by a reduction in the anti-apoptotic Bcl-2 and
increases in the pro-apoptotic markers Caspase-3 and Bax,
suggesting that mepivacaine could intensify cell death pathways
that are important under ischemic conditions. Furthermore,
mepivacaine treatment disrupted cell cycle progression by inducing
a G1 phase arrest, as demonstrated by increased P21
expression and decreased levels of Cyclin E1 and CDK4. Importantly,
mepivacaine elevates oxidative stress markers such as ROS and MDA,
indicators of increased oxidative damage (32). Oxidative stress is a major factor
in cellular damage during MI, where it contributes to cell membrane
damage through mechanisms such as lipid peroxidation, evidenced by
elevated MDA levels. This oxidative damage is particularly key
during IR events, characterized by a rapid increase in oxygen that
leads to excessive ROS production. This surge can overwhelm the
antioxidant defenses of a cell, such as SOD, which was observed to
be diminished in the present study. This imbalance between
pro-oxidants and antioxidants exacerbates cellular injury,
underscoring the deleterious impact of mepivacaine on myocardial
health, especially under ischemic conditions. Inflammation is
another key factor in the pathophysiology of MI, with cytokines
such as IL-1β, IL-8 and TNF-α carrying out key roles in mediating
inflammatory responses (33,34).
These cytokines are known to influence the infarct size and
intensity of myocardial injury by promoting further inflammatory
cell infiltration and activating additional cytotoxic pathways
(35). The present study revealed
that mepivacaine elevates these cytokines, potentially increasing
the inflammatory response in myocardial tissues, which could
exacerbate the damage in ischemic conditions. These findings
highlight the ability of mepivacaine to amplify cardiomyocyte
oxidative stress and inflammatory responses.
As an integral component of calcium ion channels,
CACNB1 carries out a key role in regulating calcium ion
permeability across cell membranes and maintaining intracellular
calcium levels (36). In the
present study, CACNB1 expression increased as mepivacaine
concentration increased, suggesting its involvement in the cellular
response to mepivacaine exposure. MIRI is a considerable
pathophysiological component of MI, where, despite the restoration
of blood supply alleviating ischemia, reperfusion itself can induce
further myocardial damage (37).
Reperfusion is known to trigger inflammatory responses, activate
inflammatory cells and release mediators that exacerbate myocardial
damage and remodeling (38). A
recent study by Algoet et al (39) emphasized the intertwined mechanisms
of IR injury and inflammatory changes and explored dual
intervention strategies as potential therapeutic approaches. It is
well-documented that MIRI can lead to calcium overload (40–42),
worsening MIRI. Specifically, myocardial cell-specific loss of the
NBCe1 Na+-HCO3− cotransporter was revealed to
confer cardioprotective effects during IR injury (43). Additionally, disruptions in
Ca2+ homeostasis, commonly observed in cardiac IR and
heart failure, were associated with mitochondrial-associated
membrane between the endoplasmic reticulum and mitochondria,
affecting Ca2+ transport and suggesting new therapeutic
targets for cardiac diseases (44,45).
In the present study, experiments using the H/R model demonstrated
that knocking down CACNB1 could alleviate cell damage
induced by H/R and mepivacaine, providing novel insights into
myocardial cell protection mechanisms. These results emphasize the
importance of understanding the role of CACNB1 in calcium
regulation and its potential as a focus to lessen myocardial injury
in the setting of IR.
NLRP3 and Nrf2 represent two key regulatory pathways
involved in inflammation and oxidative stress responses (46,47).
The multi-protein complex known as the NLRP3 inflammasome is
primarily produced by immunological cells, including dendritic and
macrophage cells (48).
Conversely, Nrf2 acts as a major regulator of cellular antioxidant
defense mechanisms. Emerging evidence suggests a complex interplay
between the NLRP3 inflammasome and the Nrf2 pathway (49). While triggering the NLRP3
inflammasome promotes inflammation and oxidative stress, activation
of Nrf2 exerts anti-inflammatory and antioxidative effects. Nie
et al (50) revealed that
the SIRT1/NRF2 signaling pathway is the true target of resveratrol
and carries out a key part in the cardioprotective effect against
inflammation induced by resveratrol. Mauro et al (51) highlighted that NLRP3 senses
intracellular danger signals during ischemia and extracellular or
intracellular alarm molecules during tissue injury and highlighted
its role in initiating inflammation and cell death through
activation of caspase-1. Shen et al (52) further emphasized the role of Nrf2
in maintaining redox balance and demonstrated that the
Nrf2/Keap1/ARE pathway is key in reducing the size of MI and
protecting the cardiac function after MIRI.
In line with these insights, the present study
investigated the effects of CACNB1 knockdown in an H/R model
treated with mepivacaine. WB analysis revealed that CACNB1
knockdown modulates inflammatory mediators, such as NLRP3 and
IL-1β, and notably promotes Nrf2 translocation into the nucleus.
This suggested a protective mechanism against H/R-induced
myocardial injury, where CACNB1 knockdown not only reduces
inflammatory and apoptotic signaling but also enhances the
antioxidant response. The decreased expression of cytoplasmic Nrf2
and increased Nrf2 translocation to the nucleus in the H/R model
with CACNB1 knockdown demonstrated a dynamic shift towards
enhancing cellular resilience against oxidative stress. These
findings highlight the CACNB1/NLRP3/Nrf2 axis as a strategic
approach to alleviate inflammation and enhance antioxidant
defenses, offering potential therapeutic pathways to reduce
myocardial injury during IR, particularly in treatments involving
mepivacaine and other stressors.
The use of the H9c2 cell line in the present study
provided a valuable model for investigating the effects of
mepivacaine on cardiomyocytes. However, it is essential to
acknowledge the potential limitations associated with using a
single cell line rather than primary cardiomyocytes. H9c2 cells,
derived from embryonic rat heart tissue, may not fully recapitulate
the mature phenotype and complex physiological responses of adult
cardiomyocytes. Additionally, the genetic homogeneity of the cell
line may limit the generalizability of the current findings to a
broader population. While H9c2 cells offer reproducibility and ease
of use, their response to experimental conditions may differ from
that of primary cardiomyocytes, which are embedded in a native
extracellular matrix and exhibit more diverse genetic backgrounds.
Future studies should aim to validate these findings in primary
cardiomyocytes or in vivo models to further elucidate the
physiological relevance of present observations.
In conclusion, the present study elucidated the
potential cardiotoxic effects of mepivacaine, particularly in the
context of MIRI. CACNB1 was identified as a key player in
mediating mepivacaine-induced cardiomyocyte injury and demonstrated
its involvement in apoptosis and inflammation pathways. Through
CACNB1 knockdown, a notable protection against mepivacaine
and H/R-induced damage was observed, evidenced by improved cell
viability, reduced apoptosis and modulation of inflammatory
mediators. Importantly, the current findings revealed a novel
mechanism involving the CACNB1/NLRP3/Nrf2 axis, wherein
CACNB1 knockdown facilitates Nrf2 nuclear translocation,
thereby mitigating inflammation and apoptosis in cardiomyocytes.
These insights shed light on potential therapeutic targets for
mitigating mepivacaine-induced cardiotoxicity and offer novel
perspectives for the development of cardioprotective strategies,
particularly within the framework of perioperative cardiac care.
Further investigation into the precise molecular mechanisms
underlying CACNB1-mediated cardioprotection is warranted to
fully harness its therapeutic potential in clinical settings.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
QS, JZ and HW contributed to the conception and
design of the present study. QS and JZ contributed to acquisition
of data. QS and JZ contributed to analysis and interpretation of
data. QS contributed to statistical analysis. QS and JZ contributed
to drafting the manuscript. HW contributed to revision of
manuscript for important intellectual content. QS and HW confirm
the authenticity of all the raw data. All the authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Katari V, Kanajam LSP, Kokkiligadda S,
Sharon T and Kantamaneni P: A review on causes of myocardial
infarction and its management. World J Pharmaceutical Res.
12:511–529. 2023.
|
|
2
|
Sagris M, Antonopoulos AS, Theofilis P,
Oikonomou E, Siasos G, Tsalamandris S, Antoniades C, Brilakis ES,
Kaski JC and Tousoulis D: Risk factors profile of young and older
patients with myocardial infarction. Cardiovasc Res. 118:2281–2292.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Khan IA, Karim HMR, Panda CK, Ahmed G and
Nayak S: Atypical presentations of myocardial infarction: A
systematic review of case reports. Cureus. 15:e354922023.PubMed/NCBI
|
|
4
|
Camacho X, Nedkoff L, Wright FL, Nghiem N,
Buajitti E, Goldacre R, Rosella LC, Seminog O, Tan EJ, Hayes A, et
al: Relative contribution of trends in myocardial infarction event
rates and case fatality to declines in mortality: An international
comparative study of 1 95 million events in 80 4 million people in
four countries. Lancet Public Health. 7:e229–e239. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fathima SN: An update on myocardial
infarction. Curr Res Trends Med Sci Technol. 1:152021.
|
|
6
|
Dos Santos CC, Matharoo AS, Cueva EP, Amin
U, Ramos AAP, Mann NK, Maheen S, Butchireddy J, Falki VB, Itrat A,
et al: The influence of sex, age, and race on coronary artery
disease: A narrative review. Cureus. 15:e477992023.PubMed/NCBI
|
|
7
|
Radu RI, Ben Gal T, Abdelhamid M, Antohi
EL, Adamo M, Ambrosy AP, Geavlete O, Lopatin Y, Lyon A, Miro O, et
al: Antithrombotic and anticoagulation therapies in cardiogenic
shock: A critical review of the published literature. ESC Heart
Fail. 8:4717–4736. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zangrillo A, Lomivorotov VV, Pasyuga VV,
Belletti A, Gazivoda G, Monaco F, Nigro Neto C, Likhvantsev VV,
Bradic N, Lozovskiy A, et al: Effect of volatile anesthetics on
myocardial infarction after coronary artery surgery: A post hoc
analysis of a randomized trial. J Cardiothorac Vasc Anesth.
36:2454–2462. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Körner J, Albani S, Sudha Bhagavath
Eswaran V, Roehl AB, Rossetti G and Lampert A: Sodium channels and
local anesthetics-old friends with new perspectives. Front
Pharmacol. 13:8370882022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gitman M, Fettiplace MR, Weinberg GL, Neal
JM and Barrington MJ: Local anesthetic systemic toxicity: A
narrative literature review and clinical update on prevention,
diagnosis, and management. Plast Reconstr Surg. 144:783–795. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dell'Olio F, Capodiferro S, Lorusso P,
Limongelli L, Tempesta A, Massaro M, Grasso S and Favia G: Light
conscious sedation in patients with previous acute myocardial
infarction needing exodontia: An observational study. Cureus.
11:e65082019.PubMed/NCBI
|
|
12
|
Mosqueira M, Aykut G and Fink RH:
Mepivacaine reduces calcium transients in isolated murine
ventricular cardiomyocytes. BMC Anesthesiol. 20:102020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
de Groot-van der Mooren M, Quint S, Knobbe
I, Cronie D and van Weissenbruch M: Severe cardiorespiratory and
neurologic symptoms in a neonate due to mepivacaine intoxication.
Case Rep Pediatr. 2019:40135642019.PubMed/NCBI
|
|
14
|
Andrade A, Brennecke A, Mallat S, Brown J,
Gomez-Rivadeneira J, Czepiel N and Londrigan L: Genetic
associations between voltage-gated calcium channels and psychiatric
disorders. Int J Mol Sci. 20:35372019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gilbert G, Demydenko K, Dries E, Puertas
RD, Jin X, Sipido K and Roderick HL: Calcium signaling in
cardiomyocyte function. Cold Spring Harb Perspect Biol.
12:a0354282020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shah K, Seeley S, Schulz C, Fisher J and
Gururaja Rao S: Calcium channels in the heart: Disease states and
drugs. Cells. 11:9432022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Erdogmus S, Concepcion AR, Yamashita M,
Sidhu I, Tao AY, Li W, Rocha PP, Huang B, Garippa R, Lee B, et al:
Cavβ1 regulates T cell expansion and apoptosis independently of
voltage-gated Ca2+ channel function. Nat Commun. 13:20332022.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xuan L, Zhu Y, Liu Y, Yang H, Wang S, Li
Q, Yang C, Jiao L, Zhang Y, Yang B and Sun L: Up-regulation of
miR-195 contributes to cardiac hypertrophy-induced arrhythmia by
targeting calcium and potassium channels. J Cell Mol Med.
24:7991–8005. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lu Y, Zhang Y, Wang N, Pan Z, Gao X, Zhang
F, Zhang Y, Shan H, Luo X, Bai Y, et al: MicroRNA-328 contributes
to adverse electrical remodeling in atrial fibrillation.
Circulation. 122:2378–2387. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Couch LS, Fiedler J, Chick G, Clayton R,
Dries E, Wienecke LM, Fu L, Fourre J, Pandey P, Derda AA, et al:
Circulating microRNAs predispose to takotsubo syndrome following
high-dose adrenaline exposure. Cardiovasc Res. 118:1758–1770. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Scarparo HC, Maia RN, Filho ED, Soares E,
Costa F, Fonteles C, Bezerra TP, Ribeiro TR and Romero NR: Plasma
mepivacaine concentrations in patients undergoing third molar
surgery. Aust Dent J. 61:446–454. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhou Q and Zhang L: MicroRNA-183-5p
protects human derived cell line SH-SY5Y cells from
mepivacaine-induced injury. Bioengineered. 12:3177–3187. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cheng L, Zhang H, Wu F, Liu Z, Cheng Y and
Wang C: Role of Nrf2 and its activators in cardiocerebral vascular
disease. Oxid Med Cell Longev. 2020:46839432020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhao E, Xie H and Zhang Y: Predicting
diagnostic gene biomarkers associated with immune infiltration in
patients with acute myocardial infarction. Front Cardiovasc Med.
7:5868712020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang Q, Wang L, Wang S, Cheng H, Xu L,
Pei G, Wang Y, Fu C, Jiang Y, He C and Wei Q: Signaling pathways
and targeted therapy for myocardial infarction. Signal Transduct
Target Ther. 7:782022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Paolisso P, Foà A, Bergamaschi L, Donati
F, Fabrizio M, Chiti C, Angeli F, Toniolo S, Stefanizzi A,
Armillotta M, et al: Hyperglycemia, inflammatory response and
infarct size in obstructive acute myocardial infarction and MINOCA.
Cardiovasc Diabetol. 20:332021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhao X, Ren Y, Ren H, Wu Y, Liu X, Chen H
and Ying C: The mechanism of myocardial fibrosis is ameliorated by
myocardial infarction-associated transcript through the PI3K/Akt
signaling pathway to relieve heart failure. J Int Med Res. Jul
18–2021. View Article : Google Scholar
|
|
29
|
Burgos EG, García-García LL,
Gómez-Serranillos MP and Oliver FG: Local Anesthetics. Adv
Neuropharmacol. 351–388. 2020. View Article : Google Scholar
|
|
30
|
Lottinger C: Local anesthetics in
dentistry. Evidence-Based Oral Surgery. Springer; London: pp.
129–150. 2019
|
|
31
|
Ozbay S, Ayan M and Karcioglu O: Local
anesthetics, clinical uses, and toxicity: Recognition and
management. Curr Pharm Des. 29:1414–1420. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Aladağ N, Asoğlu R, Ozdemir M, Asoğlu E,
Derin AR, Demir C and Demir H: Oxidants and antioxidants in
myocardial infarction (MI): Investigation of ischemia modified
albumin, malondialdehyde, superoxide dismutase and catalase in
individuals diagnosed with ST elevated myocardial infarction
(STEMI) and non-STEMI (NSTEMI). J Med Biochem. 40:2862021.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mitsis A, Kadoglou NP, Lambadiari V,
Alexiou S, Theodoropoulos KC, Avraamides P and Kassimis G:
Prognostic role of inflammatory cytokines and novel adipokines in
acute myocardial infarction: An updated and comprehensive review.
Cytokine. 153:1558482022. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang H and Dhalla NS: The role of
pro-inflammatory cytokines in the Pathogenesis of Cardiovascular
Disease. Int J Mol Sci. 25:10822024. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shao L, Shen Y, Ren C, Kobayashi S,
Asahara T and Yang J: Inflammation in myocardial infarction: Roles
of mesenchymal stem cells and their secretome. Cell Death Discov.
8:4522022. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tuluc P, Theiner T, Jacobo-Piqueras N and
Geisler SM: Role of high Voltage-Gated Ca2+ Channel subunits in
pancreatic β-cell insulin release. From structure to function.
Cells. 10:20042021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang S, Yan F, Luan F, Chai Y, Li N, Wang
YW, Chen ZL, Xu DQ and Tang YP: The pathological mechanisms and
potential therapeutic drugs for myocardial ischemia reperfusion
injury. Phytomedicine. 129:1556492024. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang D, Wu H, Liu D, Li Y and Zhou G:
Research progress on the mechanism and treatment of inflammatory
response in myocardial Ischemia-reperfusion injury. Heart Surg
Forum. 25:E462–E468. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Algoet M, Janssens S, Himmelreich U, Gsell
W, Pusovnik M, Van den Eynde J and Oosterlinck W: Myocardial
ischemia-reperfusion injury and the influence of inflammation.
Trends Cardiovasc Med. 33:357–366. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li YW, Liu Y, Luo SZ, Huang XJ, Shen Y,
Wang WS and Lang ZC: The significance of calcium ions in cerebral
ischemia-reperfusion injury: Mechanisms and intervention
strategies. Front Mol Biosci. 12:15857582025. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
He J, Liu D, Zhao L, Zhou D, Rong J, Zhang
L and Xia Z: Myocardial ischemia/reperfusion injury: Mechanisms of
injury and implications for management. Exp Ther Med. 23:4302022.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Trujillo-Rangel WÁ, García-Valdés L,
Méndez-del Villar M, Castañeda-Arellano R, Totsuka-Sutto SE and
García-Benavides L: Therapeutic targets for regulating oxidative
damage induced by ischemia-reperfusion injury: A study from a
pharmacological perspective. Oxid Med Cell Longev.
2022:86243182022. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Vairamani K, Prasad V, Wang Y, Huang W,
Chen Y, Medvedovic M, Lorenz JN and Shull GE: NBCe1
Na+-HCO3-cotransporter ablation causes reduced apoptosis following
cardiac ischemia-reperfusion injury in vivo. World J Cardiol.
10:972018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang Y, Zhang X, Wen Y, Li S, Lu X, Xu R
and Li C: Endoplasmic reticulum-mitochondria contacts: A potential
therapy target for cardiovascular remodeling-associated diseases.
Front Cell Dev Biol. 9:7749892021. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Luan Y, Luan Y, Yuan R-X, Feng Q, Chen X
and Yang Y: Structure and function of mitochondria-associated
endoplasmic reticulum membranes (MAMs) and their role in
cardiovascular diseases. Oxid Med Cell Longev. 2021:45788092021.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Li W, Cao T, Luo C, Cai J, Zhou X, Xiao X
and Liu S: Crosstalk between ER stress, NLRP3 inflammasome, and
inflammation. Appl Microbiol Biotechnol. 104:6129–6140. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Saha S, Buttari B, Panieri E, Profumo E
and Saso L: An overview of Nrf2 signaling pathway and its role in
inflammation. Molecules. 25:54742020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wang L and Hauenstein AV: The NLRP3
inflammasome: Mechanism of action, role in disease and therapies.
Mol Aspects Med. 76:1008892020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Tastan B, Arioz BI and Genc S: Targeting
NLRP3 inflammasome with Nrf2 inducers in central nervous system
disorders. Front Immunol. 13:8657722022. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Nie Q, Zhang J, He B, Wang F, Sun M, Wang
C, Sun W, Guo J, Wen J and Liu P: A novel mechanism of protection
against isoproterenol-induced cardiac inflammation via regulation
of the SIRT1/NRF2 signaling pathway with a natural SIRT1 agonist.
Eur J Pharmacol. 886:1733982020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Mauro AG, Bonaventura A, Mezzaroma E,
Quader M and Toldo S: NLRP3 inflammasome in acute myocardial
infarction. J Cardiovasc Pharmacol. 74:175–187. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Shen Y, Liu X, Shi J and Wu X: Involvement
of Nrf2 in myocardial ischemia and reperfusion injury. Int J Biol
Macromol. 125:496–502. 2019. View Article : Google Scholar : PubMed/NCBI
|