Introduction
Inflammatory bowel diseases (IBDs) comprise chronic
as well as recurrent gastrointestinal conditions. A recent study
has reported on the notable increase in the global prevalence of
IBD, predominantly characterized by Crohn's disease (CD) and
ulcerative colitis (UC) (1).
Impaired intestinal barrier integrity is a contributing factor in
the pathogenesis of IBD and is closely associated with its
pathophysiology (2,3). Despite this, there is a
characteristic lack of therapeutic options that specifically
enhance mucosal barrier function and promote its regeneration in
IBD, which is likely owing to a limited understanding of the
underlying biological pathways and regulatory molecules involved
(4). Although various potential
therapeutic approaches have been developed for improving mucosal
barrier function (4,5), to the best of our knowledge, no
regenerative strategies have yet been developed specifically for
IBD.
Glycosylation is a post-translational modification
affecting proteins, lipids and RNA, which can profoundly influence
the structural and functional characteristics of the modified
molecules (6–8). This modification is important for
regulating a variety of biological mechanisms, including protein
maturation, transport and immune modulation (9). Glycosylation is also implicated in
the pathophysiology of severe diseases, such as during
carcinogenesis, and inflammation; a previous study also
demonstrated its association with IBD (10). In healthy individuals, protein
glycoforms exhibit substantial stability over time; however, they
tend to undergo notable alterations in response to pathological
conditions, particularly during inflammatory states. For example,
patients diagnosed with IBD exhibit increased expression levels of
short-chain O-glycans in the intestine compared with healthy
individuals, accompanied by alterations in the expression of
terminal glycan structures (for example, increased sialylation,
enhanced fucosylation, reduced sulfation and dysregulated Lewis
antigens). These changes adversely affect the integrity of the
intestinal mucus layer, impede glycan-lectin interactions, disrupt
the dynamic balance of the intestinal microbiota and impair the
functionality of the intestinal mucosal immune system (11).
Different sialylation modifications are catalyzed by
various sialyltransferases. However, any alterations in the
expression levels of sialyltransferases change their physiological
functions. For example, a decrease in sialyltransferase ST6GalNAc1
(ST6)-catalyzed α2,6-sialylation in mice results in impaired mucus
barrier function, gut microbiota dysbiosis and increased
susceptibility to intestinal inflammation (12). The α2,6-sialylation catalyzed by
ST6Gal1 promotes the activation of CD4+ T cells and
induces the development of UC (13), whereas α2,8-sialylation catalyzed
by ST8Sia4 facilitates the metastasis of breast cancer cells
(14). However, the role of
ST3Gal1-catalyzed α2,3-sialylation in intestinal inflammation
remains to be fully elucidated.
The upregulation of ST3Gal1 augments the sialylation
of O-glycan Tn and catalyzes its conversion into sialyl-Tn. This
modification introduces an α2,3-linkage between Gal and
β1,3-GalNAc, which is frequently observed in various cancer types,
including breast cancer and hematological malignancies, where the
upregulation of sialyltransferases leads to premature termination
of aberrant mucin-type O-glycan chains (15,16).
Notably, mucin-type O-linked glycosylation is not restricted to
mucins; this modification also occurs in an array of cell-surface
glycoproteins (17). Emerging
evidence has highlighted that glycosylation in intestinal
epithelial cells (IECs) is associated with specific
glycosyltransferases; however, the functional role of
ST3Gal1-catalyzed α2,3-sialylation, a key glycosylation event
mediated by this glycosyltransferase, in the regulation of
intestinal barrier function has often been neglected in relevant
research. Dysregulation of glycosylation exacerbates intestinal
inflammation by damaging the intestinal barrier integrity,
interfering with glycan-lectin interactions, disrupting intestinal
microbiota dynamics and impairing mucosal immunity (18–20).
Given that ST3Gal1-catalyzed α2,3-linkage formation
at the end of N-glycan or O-glycan is associated with
carcinogenesis, as well as a host of other diseases, it is
necessary to evaluate prognostic factors, identify predictive
biomarkers and explore possible therapeutic targets associated with
ST3Gal1 activity, particularly in IBD (15,21).
However, there is limited research on ST3Gal1 and its impact on the
function of the intestinal barrier.
The present study aimed to explore the potential
role of ST3Gal1-catalyzed α2,3-linkage in IBD, along with its
underlying mechanisms. The present experimental study employed an
in vitro triple-cell co-culture system to replicate both the
healthy and inflamed states of the human intestine to investigate
the impact of ST3Gal1-mediated α2,3-sialylation on the integrity of
intestinal mucosal barrier function. Notably, in the present model,
each cell type embodied distinct roles: i) Caco-2 cells formed a
monolayer that mimicked IECs; ii) HT29-MTX-E12 cells secreted mucus
and acted as a surrogate for intestinal goblet cells; and iii)
THP-1 cells, upon induction with phorbol 12-myristate 13-acetate
(PMA), differentiated into macrophage-like cells, simulating
intestinal inflammatory cells (22–24).
The present model adhered to the principles of the 3R framework,
which are replacement, reduction and refinement of animal model
usage, thus underscoring the increasing importance of in
vitro models in intestinal health research (25).
Materials and methods
Cell culture
The colorectal cancer cell line HT-29-MTX was
sourced from Beijing Bohui Innovation Biotechnology Co., Ltd.,
whereas the THP-1 human monocytic leukemia and Caco-2 human
colorectal adenocarcinoma cell lines were obtained from The Cell
Bank of Type Culture Collection of The Chinese Academy of Sciences.
All cells were authenticated by short tandem repeat profiling and
were cultured in RPMI-1640 medium (Merck KGaA) supplemented with
10% heat-inactivated fetal bovine serum (HyClone™; Cytiva). Cell
cultures were maintained in a humidified incubator at 37°C with 5%
CO2.
Cell culture model
To establish an IEC monolayer, Caco-2 and
HT29-MTX-E12 cells were co-seeded at a total density of
8×104 cells/well into the apical compartment of 24-well
Transwell inserts (pore size, 0.4 µm; Wuxi NEST Biotechnology Co.,
Ltd.) at a ratio of 9:1, based on this ratio and total cell
density, the seeding number of Caco-2 cells was 7.2×104
cells/well, and that of HT29-MTX-E12 cells was 8×103
cells/well, followed by a differentiation period of 19 days.
Concurrently, the culture medium was replaced three times a week to
maintain optimal growth conditions. Additionally, THP-1 cells were
stimulated to differentiate into macrophage-like cells by applying
PMA (100 ng/ml; Beyotime Biotechnology) at 37°C with 5%
CO2 for 48 h in 24-well plates. Subsequently, the
differentiated macrophage-like THP-1 cells were cultured in the
basolateral compartment of the same Transwell plate. The IEC
monolayer and macrophage-like THP-1 cells were co-cultured for 24 h
under standard conditions (37°C, 5% CO2) to simulate the
intestinal mucosa in vitro.
ST3Gal1 overexpression (ST3Gal1-OE)
assay
Lentiviral vectors designed for the overexpression
of ST3Gal1 were procured from Shanghai GenePharma Co., Ltd. The
ST3Gal1-OE lentiviral vector was constructed using the
LV5(EF-1α/CopGFP&Puro) backbone, based on the 3rd-generation
lentiviral system, with a working concentration of
1×1010 plaque-forming units (PFU)/ml. The lentiviral
particles were produced by transfecting 293T cells (American Type
Culture Collection). For transfection, a total of 10 µg plasmids
was used, and the molar ratio of lentiviral expression plasmid
(ST3Gal1-OE)/packaging plasmid/envelope plasmid was 4:3:1. The
transfection was performed using Lipofectamine® 3000
Transfection Reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions, under the conditions
of 37°C and 5% CO2 for a duration of 48 h to facilitate
lentivirus production. The negative control for overexpression
experiments was an empty LV5 vector (Shanghai GenePharma Co.,
Ltd.), used at the same concentration (1×1010
PFU/ml).
Following the precise guidelines set forth by the
manufacturer, infection of the cells in the triple co-culture model
was performed within the Transwell system, with lentiviral vectors
added to both the upper and lower chambers. For lentiviral
transduction, polybrene (Shanghai GenePharma Co., Ltd.) was added
to the culture medium at a final concentration of 5 µg/ml to
enhance infection efficiency, and the lentiviral particles were
used at a multiplicity of infection of 10. The infected cells were
then incubated at 37°C for 24 h. Subsequently, 4 µg/ml puromycin
(MilliporeSigma; Merck KGaA) was used for the initial selection of
stably infected cell lines for 7 days, followed by maintenance with
2 µg/ml puromycin thereafter. These cell lines were subsequently
classified as OE-Ctr/IEC (negative control group) and
ST3Gal1-OE/IEC groups. Cells were collected on day 9 post-infection
(after the completion of the 1-day infection plus a 7-day selection
process), and the successful overexpression of ST3Gal1 was
confirmed through reverse transcription-quantitative PCR (RT-qPCR)
and western blot analysis.
ST3Gal1 interference assay
Adenoviral particles designed for interfering with
the ST3Gal1 enzyme were obtained from Shanghai GenePharma Co., Ltd.
The adenoviral constructs were based on the ADV1(U6/CMV-GFP) vector
(2nd-generation adenoviral expression system), which separately
harbored three distinct ST3Gal1-targeting short hairpin RNA (shRNA;
ST3Gal1-sh) inserts or a non-targeting scrambled negative control
shRNA (sh-Ctr). The specific interference target sequences and
full-length sense/antisense shRNA fragments were as follows:
ST3Gal1 sh1, target sequence 5′-GCACCATTTCCCACACCTACA-3′, sense
strand′-AATTCGCACCATTTCCCACACCTACATTCAAGAGATGTAGGTGTGGGAAATGGTGCTTTTTTG-3′,
antisense strand
5′-GATCCAAAAAAGCACCATTTCCCACACCTACATCTCTTGAATGTAGGTGTGGGAAATGGTGCG-3′;
ST3Gal1 sh2, target sequence 5′-GCATCCTCTCGGTCATCTTCT-3′, sense
strand
5′-AATTCGCATCCTCTCGGTCATCTTCTTTCAAGAGAAGAAGATGACCGAGAGGATGCTTTTTTG-3′,
antisense strand
5′-GATCCAAAAAAGCATCCTCTCGGTCATCTTCTTCTCTTGAAAGAAGATGACCGAGAGGATGCG-3′;
ST3Gal1 sh3, target sequence 5′-GGTTCGATGAGAGGTTCAACC-3′, sense
strand
5′-AATTCGGTTCGATGAGAGGTTCAACCTTCAAGAGAGGTTGAACCTCTCATCGAACCTTTTTTG-3′,
antisense strand
5′-GATCCAAAAAAGGTTCGATGAGAGGTTCAACCCTCTCTTGAAGGTTGAACCTCTCATCGAACCG-3′;
and sh-Ctr, target sequence 5′-GTTCTCCGAACGTGTCACGT-3′, sense
strand
5′-GATCCGTTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGAACTTTTTTG-3′,
antisense strand
5′-AATTCAAAAAAGTTCTCCGAACGTGTCACGTTCTCTTGAAACGTGACACGTTCGGAGAACG-3′.
The adenoviral particles were produced by transfecting 293T cells.
For transfection, 10 µg plasmids were used per 10-cm culture dish,
and the molar ratio of adenoviral expression plasmid (ST3Gal1-sh or
sh-Ctr)/packaging plasmid/envelope plasmid was 1:1:1. The
transfection was performed using Lipofectamine 3000 Transfection
Reagent, according to the manufacturer's instructions, at 37°C in a
humidified atmosphere containing 5% CO2 for 48 h before
collecting the adenoviral supernatant.
The triple co-culture cells cultured within the
Transwell system were infected with these adenoviral particles in
accordance with the manufacturer's guidelines, with adenoviral
particles added to both the upper and lower chambers of the
Transwell system. The infection procedure was consistent with that
used for the infection of overexpression lentiviral vectors
aforementioned. These cell lines were subsequently classified as
sh-Ctr/IEC and ST3Gal1-sh/IEC groups. The working concentration of
adenovirus particles carrying interfering fragments was
1×1010 PFU/ml, and that of negative control adenovirus
particles was 1×1010 PFU/ml. For adenoviral infection,
the MOI was set at 40. Specifically, cells were collected at 48 h
post-infection, and the efficiency of the interference was assessed
through RT-qPCR and western blot analysis.
Induction of inflammation in the IEC
monolayer
To establish an in vitro UC cell model,
ST3Gal1-sh/IEC or ST3Gal1-OE/IEC and their controls were treated
with 2% DSS (MilliporeSigma) and incubated in a cell culture
incubator at 37°C for 4 days (26,27).
The supernatants were collected by centrifuging the cell cultures
at 3,000 × g and 4°C for 10 min, and were employed for subsequent
experiments.
RT-qPCR
RNA was isolated and PCR was carried out following
the methodologies detailed in our earlier study (28). Briefly, total RNA was extracted
from triple co-culture cells, including ST3Gal1-sh, ST3Gal1-OE and
their respective negative control cells, using the RNA Isolation
Kit (MilliporeSigma; Merck KGaA). RNA purity was assessed by
evaluating the A260/A280 ratio (1.8–2.0) with a NanoDrop 2000
spectrophotometer (Thermo Fisher Scientific, Inc.). RNA integrity
was verified through 1% agarose gel electrophoresis and by using an
Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.), obtaining an
RNA integrity number >8.0. cDNA synthesis was performed using
the PrimeScript™ RT Reagent Kit with gDNA Eraser (Takara Bio Inc.,
Japan), according to the manufacturer's protocol. For qPCR, the
SYBR® Green Premix Pro Taq HS qPCR Kit (Hunan Accurate
Biotechnology Co., Ltd.) was used, with SYBR Green as the PCR
fluorophore. Thermocycling conditions were as follows:
Pre-denaturation at 95°C for 3 min, followed by 40 cycles at 95°C
for 30 sec and 60°C for 30 sec. β-actin was used as the internal
reference gene for normalization. Quantification of the qPCR
results was performed using the 2−ΔΔCq method (29), and result analysis was performed
using Bio-Rad CFX Manager software (version 3.1; Bio-Rad
Laboratories, Inc.). Table I
displays the sequences of PCR primers utilized in the present
experimental investigation.
 | Table I.Primer sequences used in reverse
transcription-quantitative PCR. |
Table I.
Primer sequences used in reverse
transcription-quantitative PCR.
| Primer | Sequence | Product length,
bp |
|---|
| ST3Gal1 |
| 74 |
|
Forward |
5′-TTCCGGGAGCTGGGAGATAA-3′ |
|
|
Reverse |
5′-CTCACCACCCACTCCAAGTC-3′ |
|
| MUC2 |
| 165 |
|
Forward |
5′-GGGGAGTGCTGTAAGAAGTGTGA-3′ |
|
|
Reverse |
5′-GTTGGAGACGGACGAGATGAG-3′ |
|
| TFF3 |
| 143 |
|
Forward |
5′-CTCCAGCTCTGCTGAGGAGT-3′ |
|
|
Reverse |
5′-CAGGGATCCTGGAGTCAAAG-3′ |
|
| CDX2 |
| 85 |
|
Forward |
5′-CAGGACGAAAGACAAATATC-3′ |
|
|
Reverse |
5′-GATGTAGCGACTGTAGTG-3′ |
|
| β-actin |
| 132 |
|
Forward |
5′-GTGATCTCCTTCTGCATCCTGT-3′ |
|
|
Reverse |
5′-CCACGAAACTACCTTCAACTCC-3′ |
|
| IL-1β |
| 153 |
|
Forward |
5′-CAGAAGTACCTGAGCTCGCC-3′ |
|
|
Reverse |
5′-AGATTCGTAGCTGGATGCCG-3′ |
|
| IL-6 |
| 166 |
|
Forward |
5′-TACCCCCAGGAGAAGATTCC-3′ |
|
|
Reverse |
5′-AGTGCCTCTTTGCTGCTTTC-3′ |
|
| IL-8 |
| 210 |
|
Forward |
5′-GTGCAGTTTTGCCAAGGAGT-3′ |
|
|
Reverse |
5′-ACTTCTCCACAACCCTCTGC-3′ |
|
| STAT3 |
| 237 |
|
Forward |
5′-CATCCTGAAGCTGACCCAGG-3′ |
|
|
Reverse |
5′-TCCTCACATGGGGGAGGTAG-3′ |
|
Western blotting
The procedures for protein extraction and detection
were carried out in accordance with the methodologies outlined in
our previous publication (26).
Briefly, total protein was isolated from the triple co-culture
cells, including ST3Gal1-sh cells, ST3Gal1-OE cells and their
corresponding negative control cells. Table II presents the information
regarding the manufacturer, cat. no. and dilution ratios for the
primary and secondary antibodies utilized in the present study.
 | Table II.Antibodies used for western
blotting. |
Table II.
Antibodies used for western
blotting.
| A, Primary
antibodies |
|---|
|
|---|
| Name | Company | Cat. no. | Dilution |
|---|
| Rabbit polyclonal
IgG to ST3Gal1 | Invitrogen; Thermo
Fisher | PA5-42726 | 1:1,000 |
|
| Scientific,
Inc. |
|
|
| Rabbit monoclonal
IgG to CDX2 | Abcam | ab76541 | 1:1,000 |
| Rabbit monoclonal
IgG to p-STAT3 | Abcam | ab76315 | 1:1,000 |
| Mouse monoclonal
IgG to MUC2 | Abcam | ab11197 | 1:1,000 |
| Rabbit Polyclonal
IgG to TFF3 | Proteintech Group,
Inc. | 23277-1-AP | 1:1,000 |
| Rabbit polyclonal
IgG to STAT3 | Proteintech Group,
Inc. | 10253-2-AP | 1:500 |
| Rabbit polyclonal
IgG to α-tubulin | Proteintech Group,
Inc. | 11224-1-AP | 1:1,000 |
|
| B, Secondary
antibodies |
|
| HRP-conjugated
Affinipure Goat | Proteintech Group,
Inc. | SA00001-2 | 1:10,000 |
| Anti-Rabbit IgG
(H+L) |
|
|
|
| HRP-conjugated
Affinipure Goat | Proteintech Group,
Inc. | SA00001-1 | 1:10,000 |
| Anti-Mouse IgG
(H+L) |
|
|
|
ELISA
Cell culture supernatants from each group were
collected by centrifuging the cell cultures at 3,000 × g and 4°C
for 10 min. The levels of multiple inflammatory mediators, such as
IL-1β, IL-6 and IL-8, present in the harvested cell supernatants
were measured in accordance with the instructions outlined in the
ELISA kits specific to these inflammatory factors (Beijing Solarbio
Science & Technology Co., Ltd.): IL-1β (cat. no. SEKH-0002),
IL-6 (cat. no. SEKH-0013), and IL-8 (cat. no. SEKH-0016).
Trans-epithelial electrical resistance
(TEER) assay
Cells from each group were plated at a density of
1×105 cells/well into the upper chambers of a 24-well
Transwell plate with a polyester membrane (pore size, 0.4 µm;
Costar; Corning, Inc.). For the Transwell co-culture assay, both
the upper and lower chambers were initially supplemented with DMEM
containing 10% FBS. Cells were co-cultured at 37°C with 5%
CO2 for 14 days, and 2 days before TEER measurement, the
media in both chambers were replaced with serum-free DMEM. The
resistance values of different cell models, including inflammatory
and non-inflammatory cell models, as well as their ST3Gal1
knockdown or overexpression cell models, were measured using a
resistance meter. The relative TEER values were calculated for each
group by comparison with the control.
FITC-dextran permeability assay
Subsequently, the permeability of different cell
models was studied. Notably, the setup of Transwell inserts used in
this permeability assay (other than the addition of FITC-dextran)
was consistent with that of the aforementioned protocol for TEER
measurement. FITC-dextran (4 kDa; MilliporeSigma) was added to the
apical compartment of Transwell inserts, with a final concentration
of 1 mg/ml. These cell models were incubated at 37°C for 2 h.
Subsequently, a 100-µl aliquot was aspirated from the lower
chamber, and the fluorescence value was measured using a microplate
reader (Bio-Rad Laboratories, Inc.) at an excitation wavelength of
492 nm and an emission wavelength of 520 nm. The relative
fluorescence values were calculated for each group by comparison
with the control.
Bioinformatics analysis
For ST3Gal1 expression analysis, datasets including
human normal samples and UC samples (GDS3119) (30), as well as UC lesional and
non-lesional samples (GSE107499) were obtained from the Gene
Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geoprofiles/?term=) and
analyzed. To investigate the relationship between ST3Gal1 and the
severity score of UC, relevant clinical data were extracted from
the GEO dataset GSE92415 (31) for
analysis. The GEO dataset GDS3859 (32) was used to analyze changes in the
mRNA levels of ST3Gal1 in DSS-treated mice. Additionally, the GEO
dataset GSE107499 was used to analyze the expression changes of
ST3Gal1, as well as the expression profiles of inflammatory
mediators, intestine-associated secretory proteins and signaling
pathway-related proteins in lesional and non-lesional tissues of
UC. Furthermore, the relationships among these factors and proteins
in the GSE107499 dataset, specifically within the lesional tissues
of UC, were investigated. Dataset processing and statistical
analysis were performed using GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/), an official
GEO tool. In the present study, this tool was specifically utilized
to extract and compare the expression levels of pre-specified
target genes (including ST3Gal1, MUC2, TFF3, CDX2, IL-1β, IL-6,
IL-8 and STAT3) between experimental groups (for example, UC
lesional mucosa vs. UC non-lesional mucosa, and healthy human colon
mucosa vs. UC colon mucosa).
Statistical analysis
GraphPad Prism 6.0 software (Dotmatics) was utilized
for the graphical representation and statistical evaluation of
diverse data obtained during the experiment. Data are presented as
box and whisker plots (displaying median and interquartile range),
scatter plots or as the mean ± SD from at least three independent
replicates, with unpaired two-tailed t-test used for comparisons
between two groups and Pearson's correlation test used for
assessing linear correlations. In addition, one-way ANOVA with
Tukey's HSD post hoc multiple comparisons test was applied for
multi-group analyses. P<0.05 was considered to indicate a
statistically significant difference.
Results
ST3Gal1 expression in intestinal
mucosa from patients with UC and the mouse DSS-induced colitis
model is related to the inflammatory status of colitis
The present study compared the mRNA levels of
ST3Gal1 in normal human colon mucosa and the mucosa from patients
with UC obtained from the GDS3119 dataset in the GEO. The mRNA
expression levels of ST3Gal1 in the mucosal tissue of individuals
diagnosed with UC were significantly higher than those in healthy
colon mucosa (derived from healthy control individuals) (P<0.05;
Fig. 1A). Furthermore, mRNA levels
of ST3Gal1 in the inflamed mucosal regions of patients with UC were
significantly higher than those observed in normal colon mucosa.
Additionally, ST3Gal1 expression levels were significantly higher
in inflamed UC intestinal mucosa than in non-inflamed UC intestinal
mucosa (P<0.01; Fig. 1B).
Furthermore, the mRNA expression levels of ST3Gal1 in non-lesional
mucosa were significantly lower than those in lesional mucosa
(GSE107499; P<0.01; Fig. 1C).
Additionally, in patients with UC, ST3Gal1 mRNA within the inflamed
mucosa exhibited progressive, mostly significant upregulation
associated with an increasing modified Mayo score (GSE92415;
P<0.01; Fig. 1D).
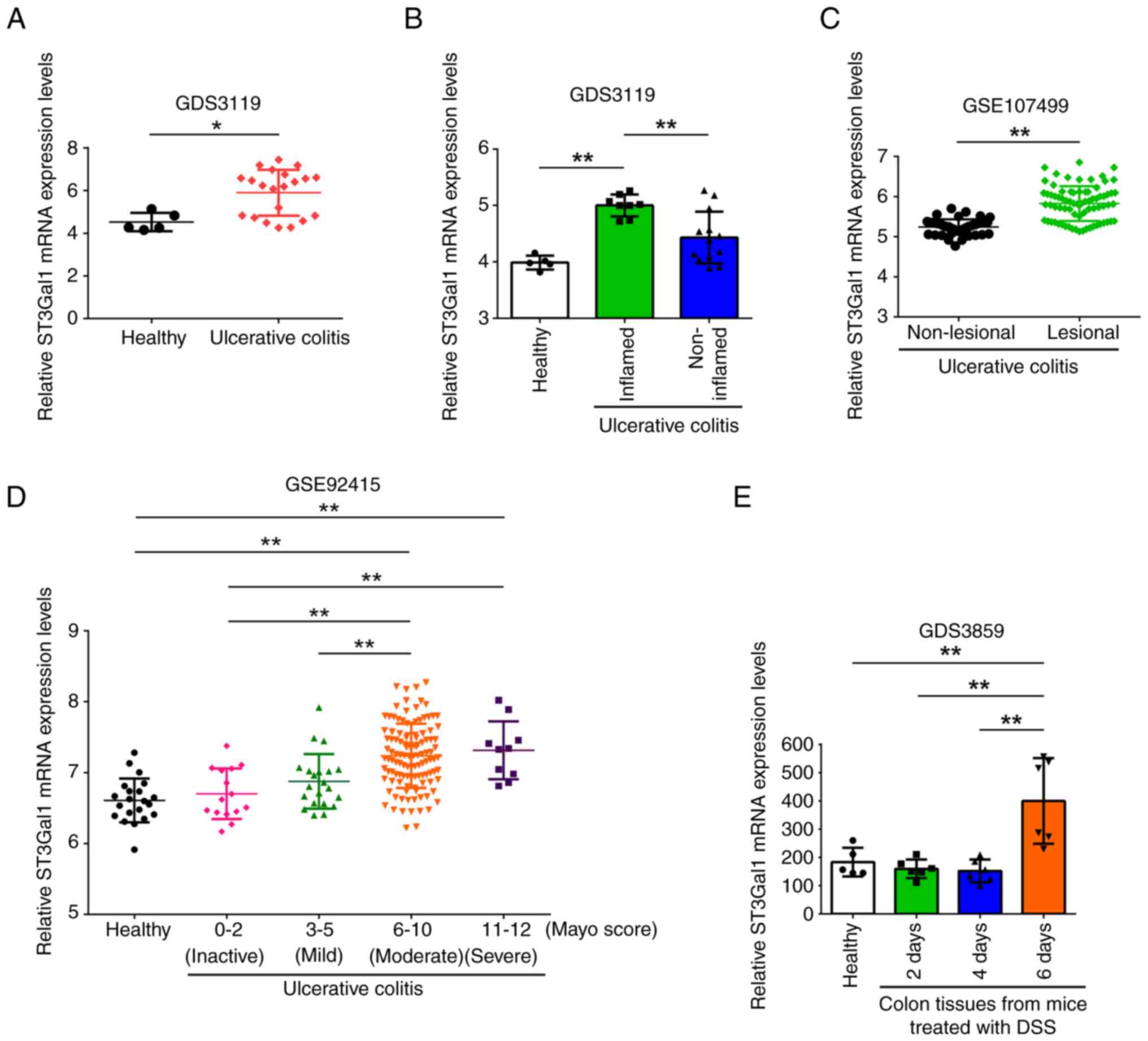 | Figure 1.ST3Gal1 mRNA levels in intestinal
mucosa from patients with UC and a mouse DSS-induced colitis model
are associated with inflammatory states. (A) Comparison of ST3Gal1
mRNA levels between human normal colon mucosa from healthy controls
(n=5) and colonic mucosa from patients diagnosed with UC (n=21) in
the GDS3119 dataset from the GEO database. (B) ST3Gal1 mRNA levels
in human normal colonic tissues from healthy controls (n=5),
inflamed colonic tissues from patients with UC and visible
macroscopic inflammation (n=8), and non-inflamed colonic mucosal
tissues from patients with UC without macroscopic signs of
inflammation (n=13) in the GEO dataset GDS3119. (C) ST3Gal1 mRNA
levels in non-lesional mucosa (n=44) and lesional mucosa (n=75)
from different patients with UC, obtained from the GEO dataset
GSE107499. (D) ST3Gal1 mRNA levels in the normal mucosa (n=21) from
healthy controls and the inflamed mucosa (n=162) from patients with
UC with varying modified Mayo scores in the GEO dataset GSE92415.
(E) ST3Gal1 mRNA levels in colon tissues from mice treated with DSS
to induce colitis (independent, non-overlapping mice) divided into
four groups: Before DSS administration (n=5), and at 2, 4 and 6
days post-DSS treatment (n=6/group). Data were obtained from the
GEO dataset GDS3859. Data in A, C and D are presented as box and
whisker plots, whereas data in B and E are presented as the mean ±
SD. Statistical significance was determined using specific tests
for each subpart based on group comparisons: (A and C) Unpaired
t-test and (B, D and E) one-way analysis of variance followed by
Tukey's post-hoc test. *P<0.05 and **P<0.01. DSS, dextran
sulfate sodium; GEO, Gene Expression Omnibus; UC, ulcerative
colitis. |
The present study further analyzed the mRNA levels
of ST3Gal1 in the mucosa of the DSS-induced mouse colitis model.
The obtained ST3Gal1 mRNA levels were classified into four groups
based on DSS usage: The normal group, classified as mRNA levels
obtained prior to DSS administration, and those obtained 2, 4 and 6
days after DSS feeding. ST3Gal1 mRNA expression in the mucosa of
mice 6 days after DSS feeding was significantly higher than that in
all other groups analyzed (GDS3859; P<0.01; Fig. 1E).
Establishment of an IEC monolayer and
ST3Gal1-knockdown or ST3Gal1-OE models
The present study first prepared an IEC monolayer
model through a co-culture of three distinct cell types, and then
obtained ST3Gal1-knockdown and ST3Gal1-OE IEC monolayer cell models
using either an ADV1(U6/CMV-GFP) vector and an ST3Gal1 interference
sequence insert or a ST3Gal1-OE lentiviral vector. The third
interference sequence (ST3Gal1-sh3) had the most efficient
inhibitory effect on ST3Gal1 expression in the IEC monolayer model,
named ST3Gal1-sh3/IEC for further experiments (P<0.01; Fig. 2A and C). A ST3Gal1-OE assay showed
that the ST3Gal1-OE group exhibited a significant increase in
ST3Gal1 expression at both mRNA and protein levels in the IEC
monolayer model compared with those in the untreated and
overexpression control groups (P<0.01; Fig. 2B and D). This model was designated
ST3Gal-OE/IEC for further experiments.
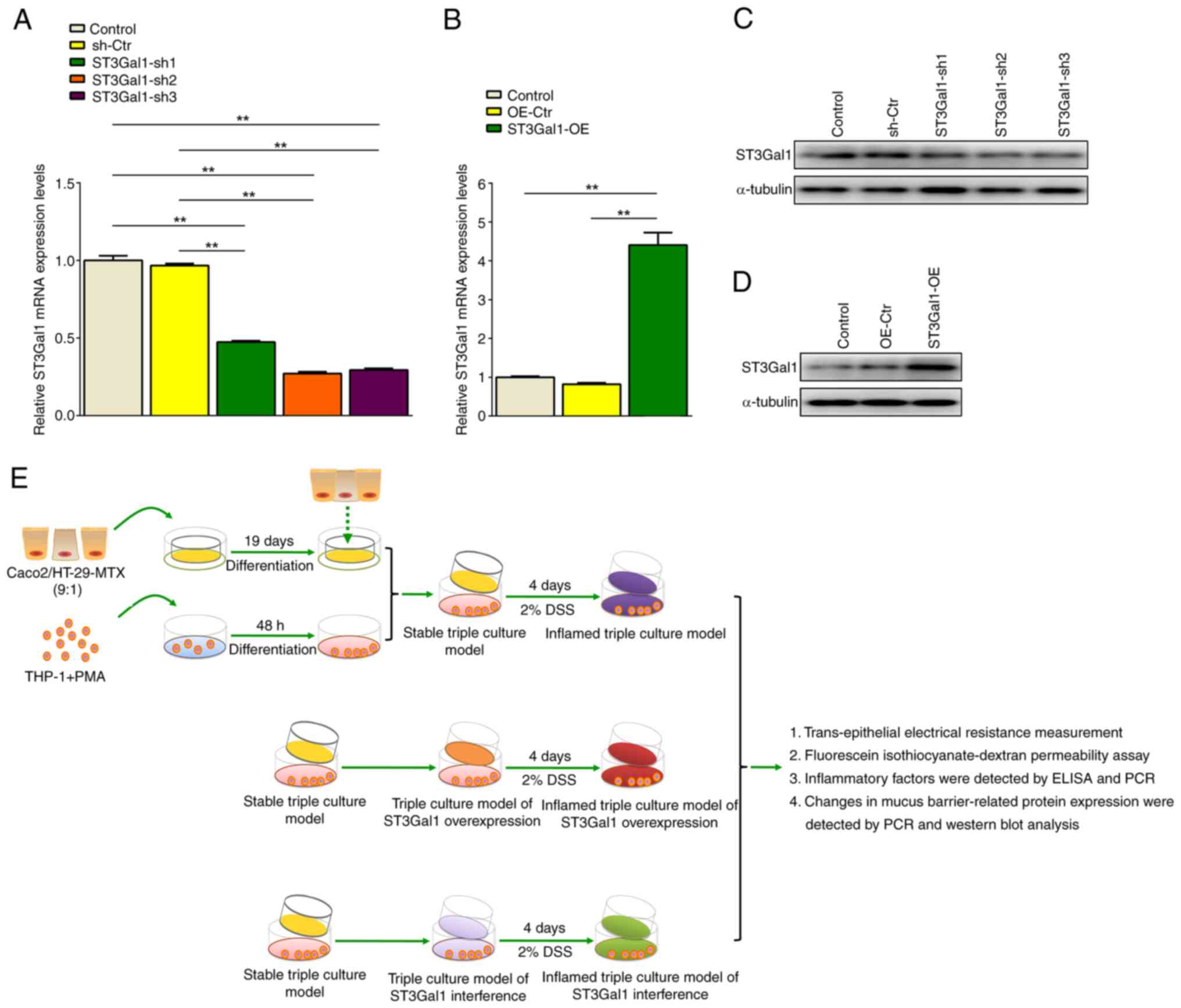 | Figure 2.Establishment of ST3Gal1-interfered
IEC and ST3Gal1-overexpressed IEC models, and the schematic
representation of IEC monolayer grouping. (A) ST3Gal1 mRNA levels
in IEC, sh-Ctr/IEC and ST3Gal1-interfered IEC groups 1–3. (B)
ST3Gal1 mRNA levels in IEC, OE-Ctr/IEC and ST3Gal1-OE/IEC groups.
(C) ST3Gal1 protein levels in IEC, sh-Ctr/IEC and
ST3Gal1-interfered IEC groups. (D) ST3Gal1 protein levels in IEC,
OE-Ctr/IEC and ST3Gal1-OE/IEC groups. (E) Establishment of IEC
monolayers containing sh-Ctr, ST3Gal1-sh3, OE-Ctr and ST3Gal1-OE.
The stable triple culture models were treated with fresh complete
medium supplemented with 2% DSS for 4 days to obtain inflamed
models. Data are presented as the mean ± SD. Statistical
significance was assessed using one-way analysis of variance
followed by Tukey's post-hoc test. **P<0.01. IEC, intestinal
epithelial cell; sh, short hairpin; Ctr, control; OE,
overexpression; PMA, phorbol 12-myristate 13-acetate; DSS, dextran
sulfate sodium. |
The present study used IEC monolayer models to
replicate both healthy and inflamed states of the human intestine.
To establish an inflamed IEC monolayer model, the aforementioned
infection models were administered fresh complete medium enriched
with 2% DSS for 4 days. The resulting inflamed IEC monolayer model
was employed for investigating intestinal barrier integrity and
inflammatory mediator secretion (Fig.
2E).
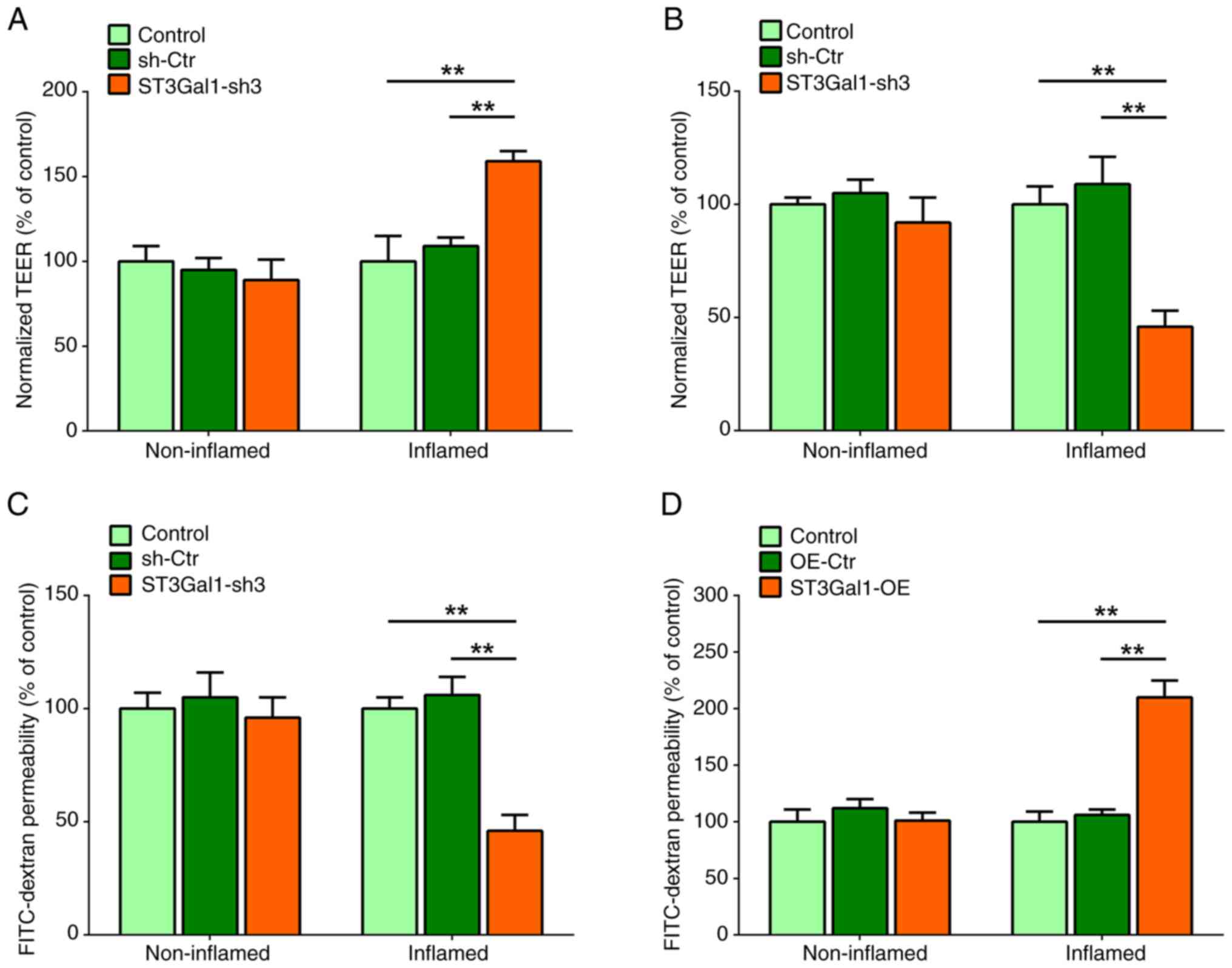
ST3Gal1 alters the barrier function of
the IEC monolayer
The IEC monolayer barrier was assessed by measuring
TEER. The TEER levels in the ST3Gal1-sh3/IEC group were
significantly higher than those in the IEC and sh-Ctr/IEC groups
after stimulation with 2% DSS (P<0.01; Fig. 3A). By contrast, no notable
differences in the TEER levels were observed among the non-inflamed
IEC monolayers of the stable triple culture models without 2% DSS
treatment (P>0.05; Fig. 3A).
The TEER levels of the inflamed ST3Gal1-OE/IEC group were
significantly lower compared with those in the IEC or Ctr/IEC
groups (P<0.01; Fig. 3B). By
contrast, no notable differences were observed among the
non-inflamed IEC monolayers of the stable triple culture models
(P>0.05; Fig. 3B). The impact
of modifications in ST3Gal1 expression on the permeability of IEC
monolayers was measured by performing a trans-epithelial
permeability assay using FITC-dextran. The assays demonstrated a
notable decrease in the FITC-dextran permeability of the inflamed
ST3Gal1-sh3/IEC population compared with that in the IEC and
sh-Ctr/IEC groups (P<0.01; Fig.
3C). However, no significant difference was observed among the
non-inflamed stable triple culture models (P>0.05; Fig. 3C). Notably, a significant increase
in the permeability of the inflamed ST3Gal1-OE/IEC group was noted
compared with that in the IEC and Ctr/IEC groups (P<0.01;
Fig. 3D). By contrast, the
non-inflamed triple culture models did not show any significant
differences in permeability (P>0.05; Fig. 3D). Therefore, it may be suggested
that ST3Gal1 interference in IECs enhanced the barrier function of
the IEC monolayer. However, enforced expression of the ST3Gal1 gene
in IECs damaged the barrier function of the IEC monolayer.
ST3Gal1 expression in the IEC
monolayer alters MUC2, TFF3 and CDX2 expression, as well as STAT3
phosphorylation
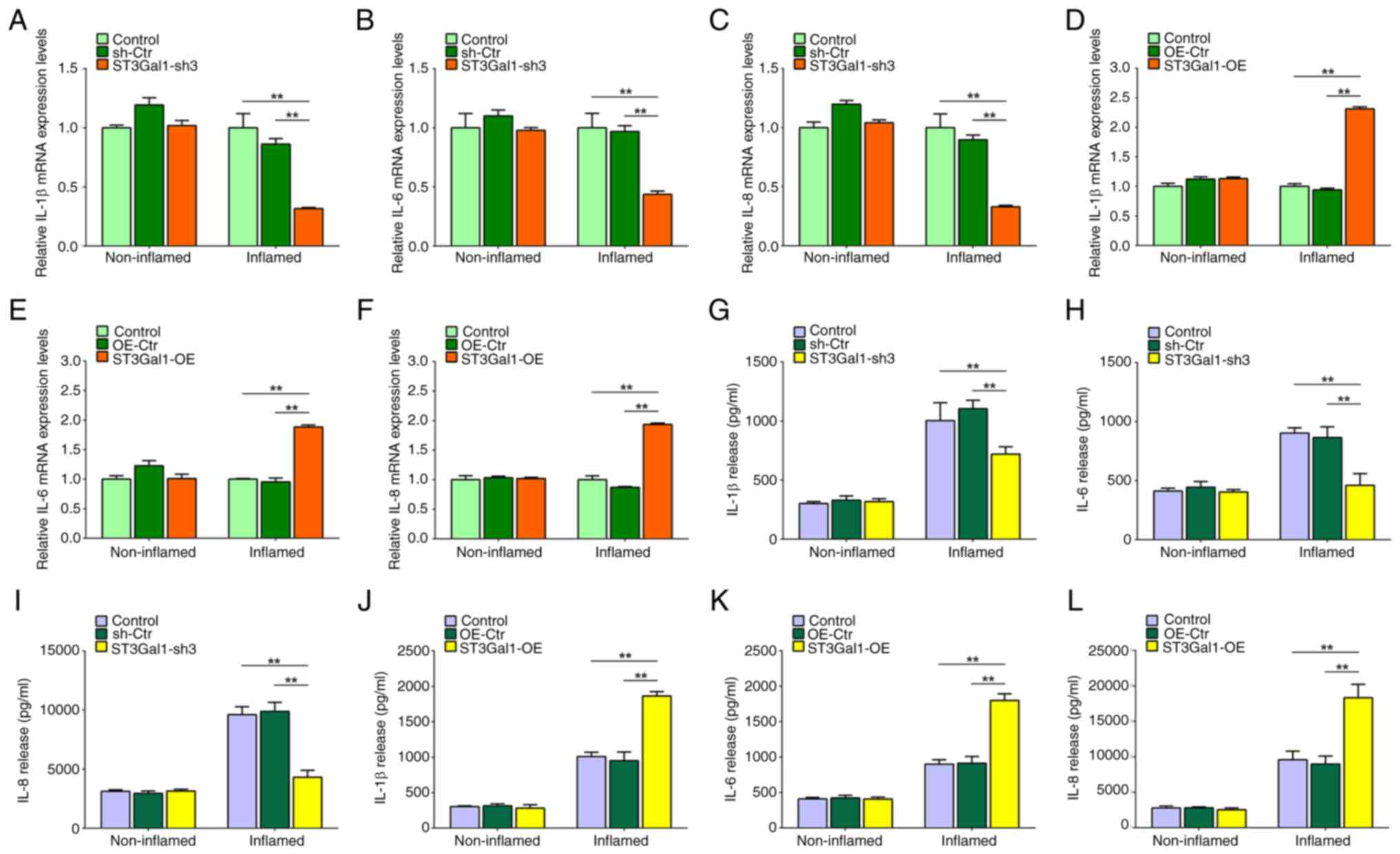
Owing to alterations in the intestinal epithelial
barrier caused by altered ST3Gal1 expression in the IEC monolayer,
the present study detected intestinal mucus barrier-associated MUC2
and TFF3, goblet cell differentiation-associated CDX2 expression
and inflammation-associated STAT3 phosphorylation in IEC
monolayers. The present study found that ST3Gal1 knockdown in IECs
significantly increased the mRNA levels of MUC2, TFF3 and CDX2, but
significantly decreased the mRNA levels of STAT3 in the inflamed
stable triple culture model compared with those in the control
groups (P<0.01; Fig. 4A-D). No
significant differences in MUC2, TFF3, CDX2 and STAT3 mRNA levels
were observed among the non-inflamed stable triple culture models
(P>0.05; Fig. 4A-D).
Overexpression of ST3Gal1 in IECs significantly reduced the mRNA
levels of MUC2, TFF3 and CDX2 in the inflamed stable triple culture
model compared with those in the control groups (P<0.05 or
P<0.01; Fig. 4E-H), and the
mRNA expression levels of STAT3 were decreased in the inflamed
stable triple culture model compared with those in the control
groups (P<0.05; Fig. 4H);
however, no significant difference was observed when compared with
the OE-Ctr group (P>0.05; Fig.
4H). No significant differences were observed in the mRNA
levels of MUC2, TFF3, CDX2 and STAT3 among the non-inflamed stable
triple-culture models (P>0.05; Fig.
4E-H).
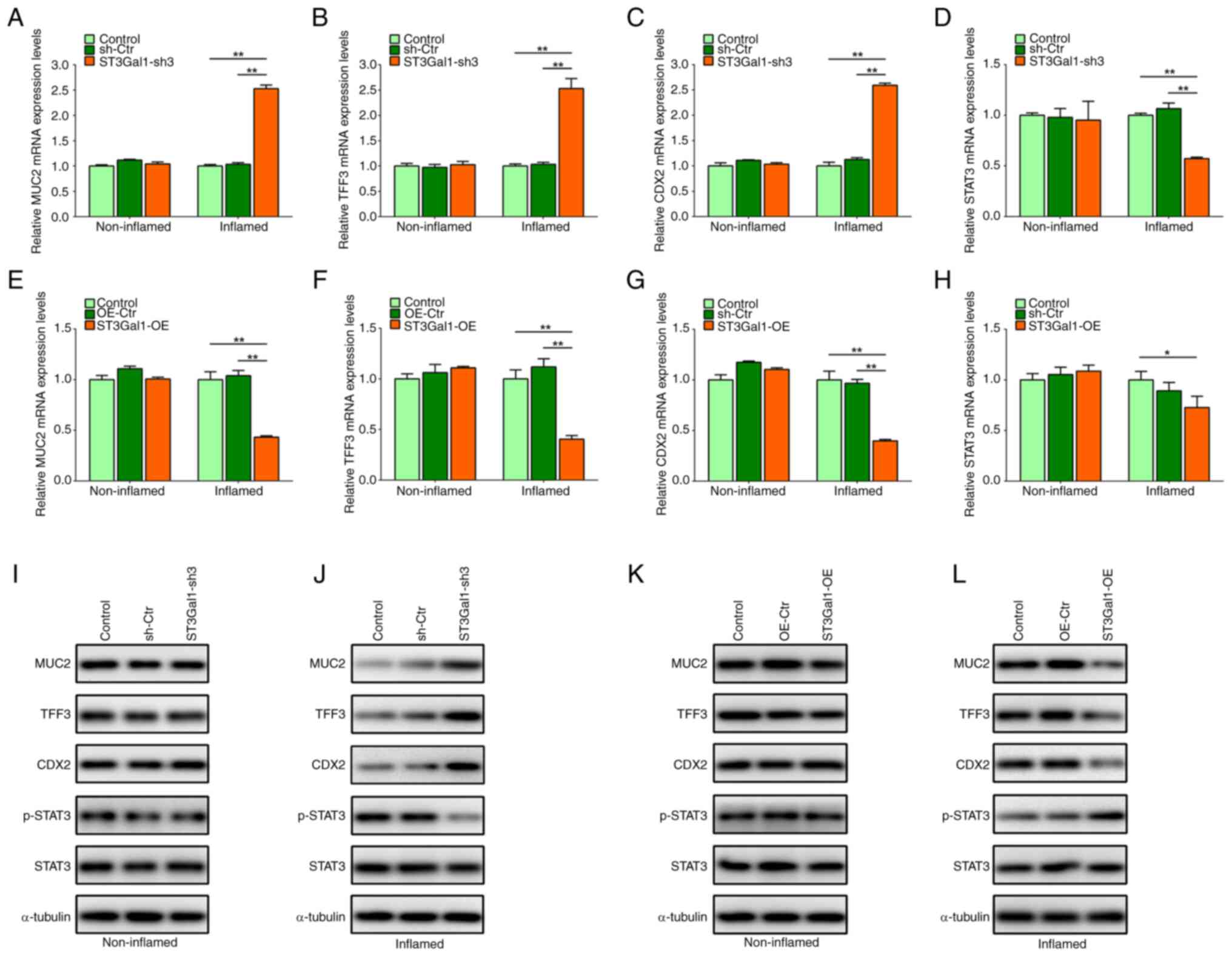 | Figure 4.ST3Gal1 expression in inflamed IEC
monolayers alters MUC2, TFF3, CDX2, STAT3 and p-STAT3 expression.
ST3Gal1 knockdown in ST3Gal1-sh3/IEC significantly increased (A)
MUC2, (B) TFF3 and (C) CDX2, but significantly reduced (D) STAT3
mRNA levels in the inflamed IEC monolayer. ST3Gal1 OE in
ST3Gal1-OE/IEC significantly reduced (E) MUC2, (F) TFF3 and (G)
CDX2 mRNA levels in the inflamed IEC monolayer. (H) ST3Gal1 OE in
ST3Gal1-OE/IEC slightly decreased STAT3 mRNA levels in the inflamed
IEC monolayer. (I) Western blotting showed no changes in protein
expression in non-inflamed knockdown samples. (J) Western blotting
showed that MUC2, TFF3 and CDX2 protein levels were increased, and
p-STAT3 expression was decreased in the inflamed IEC monolayer
comprising ST3Gal1-sh3/IEC. (K) Western blotting showed no notable
changes in protein expression in non-inflamed OE samples. (L)
Western blotting showed that MUC2, TFF3 and CDX2 protein levels
were decreased, and p-STAT3 expression was increased in the
inflamed IEC monolayer comprising ST3Gal1-OE/IEC. Data are
presented as the mean ± SD. Statistical significance was assessed
using one-way analysis of variance followed by Tukey's post-hoc
test. *P<0.05 and **P<0.01. IEC, intestinal epithelial cell;
sh, short hairpin; Ctr, control; OE, overexpression; MUC2, mucin 2;
TFF3, trefoil factor 3; CDX2, homeobox protein CDX-2; p-,
phosphorylated. |
The present study also detected the protein
expression levels of MUC2, TFF3, CDX2, phosphorylated (p)-STAT3 and
STAT3 in both the inflamed and non-inflamed stable triple culture
models. In the inflamed stable triple culture model comprising
ST3Gal1-sh3/IEC, the protein levels of MUC2, TFF3 and CDX2 were
increased, whereas p-STAT3 levels were notably decreased compared
with those in the control groups (Fig.
4J). However, no notable difference was noted in the MUC2,
TFF3, CDX2, STAT3 and p-STAT3 protein levels of the non-inflamed
stable triple culture models (Fig.
4I). Additionally, the protein levels of MUC2, TFF3 and CDX2
were notably decreased in the inflamed stable triple culture model
comprising ST3Gal1-OE/IEC, whereas p-STAT3 levels were increased
compared with those in the control groups (Fig. 4L). However, there were no marked
differences observed in the MUC2, TFF3, CDX2, STAT3 and p-STAT3
protein levels of the non-inflamed stable triple culture
overexpression model (Fig.
4K).
ST3Gal1 expression is correlated with
TFF3 and CDX2 mRNA levels in lesional mucosa obtained from patients
with UC
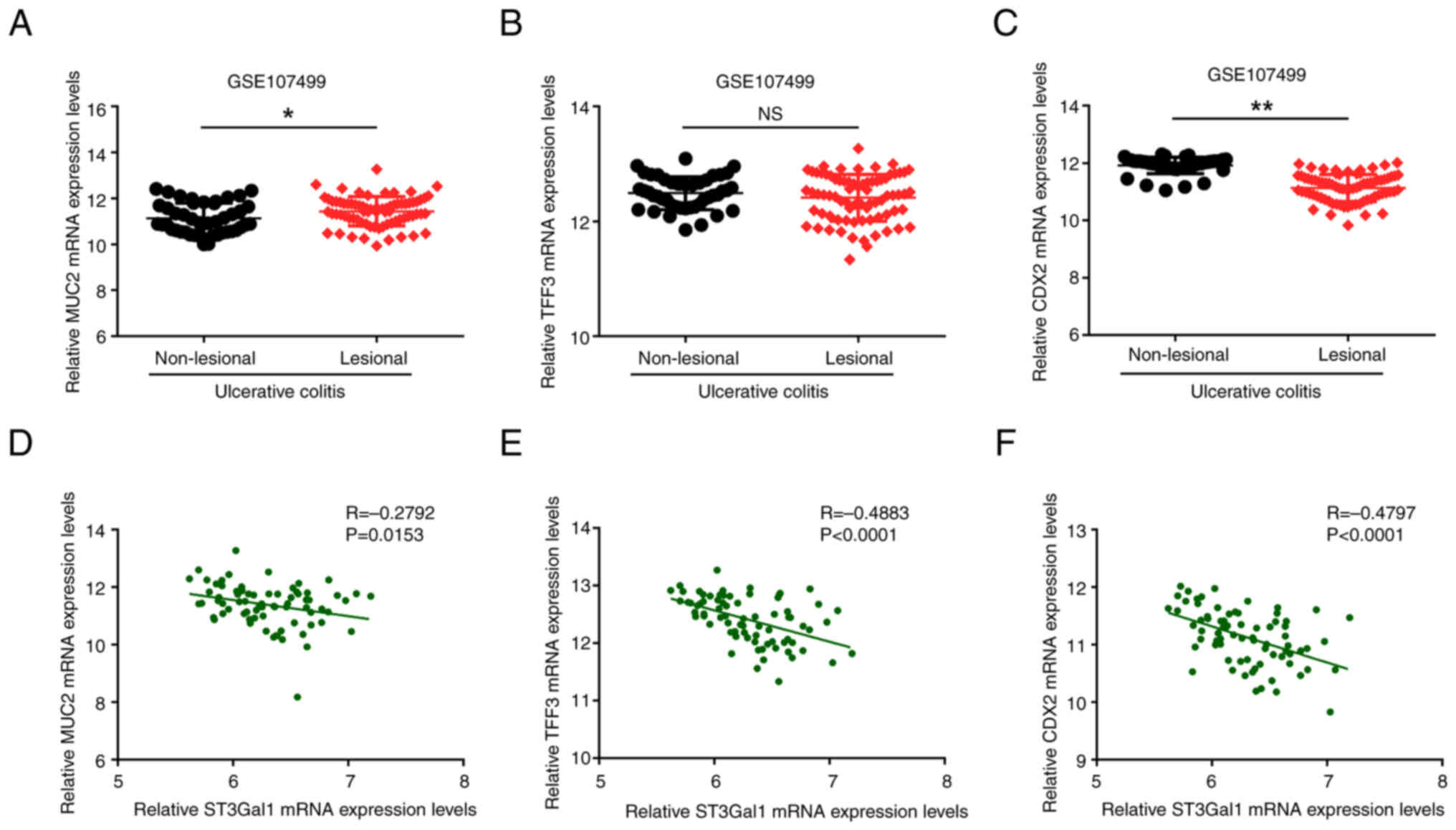
To support the relationship between ST3Gal1
expression and MUC2, TFF3 and CDX2 mRNA levels in patients with UC,
the expression dataset GSE107499 was downloaded, and the mRNA
levels of TFF3 and CDX2 in the non-lesional and lesional mucosa
from patients with UC, as well as their correlations with ST3Gal1
mRNA expression, were subsequently compared. As illustrated in
Fig. 5A, the mRNA expression
levels of MUC2 in the affected lesional mucosal tissue were
significantly elevated compared with those in the non-lesional
mucosa (P<0.05). By contrast, the TFF3 mRNA levels in the
lesional mucosa showed no significant difference when compared with
those in the non-lesional mucosa of patients with UC (P>0.05;
Fig. 5B). Notably, the mRNA levels
of CDX2 in the lesional mucosa were significantly lower than those
in non-lesional mucosa (P<0.01; Fig. 5C). The correlation analysis
revealed that the mRNA expression levels of ST3Gal1 in the lesional
mucosal tissue exhibited a negative correlation with the mRNA
levels of TFF3 (R=−0.4883; P<0.0001; Fig. 5E) and CDX2 (R=−0.4797; P<0.0001;
Fig. 5F) in patients diagnosed
with UC. However, no notable correlation was observed between
ST3Gal1 and of MUC2 expression in lesional tissues of patients with
UC (R=−0.2792; P<0.05; Fig.
5D).
ST3Gal1 expression in the IEC
monolayer alters the expression of inflammatory mediators
Supernatants from the inflamed and non-inflamed
stable triple culture models comprising ST3Gal1-sh3/IEC and
ST3Gal1-OE/IEC were collected to detect the levels of inflammatory
mediators. The protein concentrations of inflammatory mediators
were quantified via ELISA, whereas the mRNA levels of inflammatory
mediators in cell lysates were assessed via RT-qPCR. Notably,
knockdown of ST3Gal1 significantly reduced the mRNA expression
levels of IL-1β, IL-6 and IL-8 in the inflamed intestinal IEC
monolayer, compared with that in the control groups (P<0.01;
Fig. 6A-C). This analysis,
however, revealed no significant differences in the mRNA levels of
IL-1β, IL-6 and IL-8 among the non-inflamed IEC monolayers
(P>0.05; Fig. 6A-C). However,
the overexpression of ST3Gal1 in ST3Gal1-OE/IEC cells significantly
elevated the mRNA levels of IL-1β, IL-6 and IL-8 within the
inflamed IEC monolayer compared with the control groups (P<0.01;
Fig. 6D-F). By contrast, the
non-inflamed IEC monolayer exhibited no significant differences in
the expression of these cytokines (P>0.05; Fig. 6D-F). Additionally, a significant
decrease in the protein concentrations of IL-1β (720.51±61.01
pg/ml), IL-6 (461.20±98.06 pg/ml) and IL-8 (4,325.37±579.25 pg/ml)
was observed in supernatants derived from the inflamed IEC
monolayer containing ST3Gal1-sh3/IEC when compared with the control
groups (P<0.01; Fig. 6G-I). No
significant differences were noted in the protein concentrations of
IL-1β (318.43±24.54 pg/ml), IL-6 (404.52±20.55 pg/ml) and IL-8
(3,151.23±156.78 pg/ml) in ST3Gal1-sh3/IEC compared with in the
control groups in the supernatants obtained from the non-inflamed
IEC monolayer (P>0.05; Fig.
6G-I). In addition, the protein concentrations of IL-1β
(1,863.41±62.25 pg/ml), IL-6 (1,797.57±94.93 pg/ml) and IL-8
(18,319.25±1,893.01 pg/ml) were significantly higher in the
supernatants derived from the inflamed IEC monolayer containing
ST3Gal1-OE/IEC compared with those from the control groups
(P<0.01; Fig. 6J-L). Consistent
with previous observations in the present study, there were no
significant differences in the concentrations of IL-1β
(282.01±48.33 pg/ml), IL-6 (408.14±28.22 pg/ml) and IL-8
(4,203.54±433.62 pg/ml) the aforementioned cytokines in the
supernatants obtained from the non-inflamed IEC monolayer
(P>0.05; Fig. 6J-L). Therefore,
it may be inferred that the expression of ST3Gal1 in IECs markedly
modified the expression, and subsequent release of IL-1β, IL-6 and
IL-8 in inflamed triple culture models.
ST3Gal1 levels are correlated with
IL-1β, IL-6, IL-8 and STAT3 mRNA levels in lesional mucosa obtained
from patients with UC
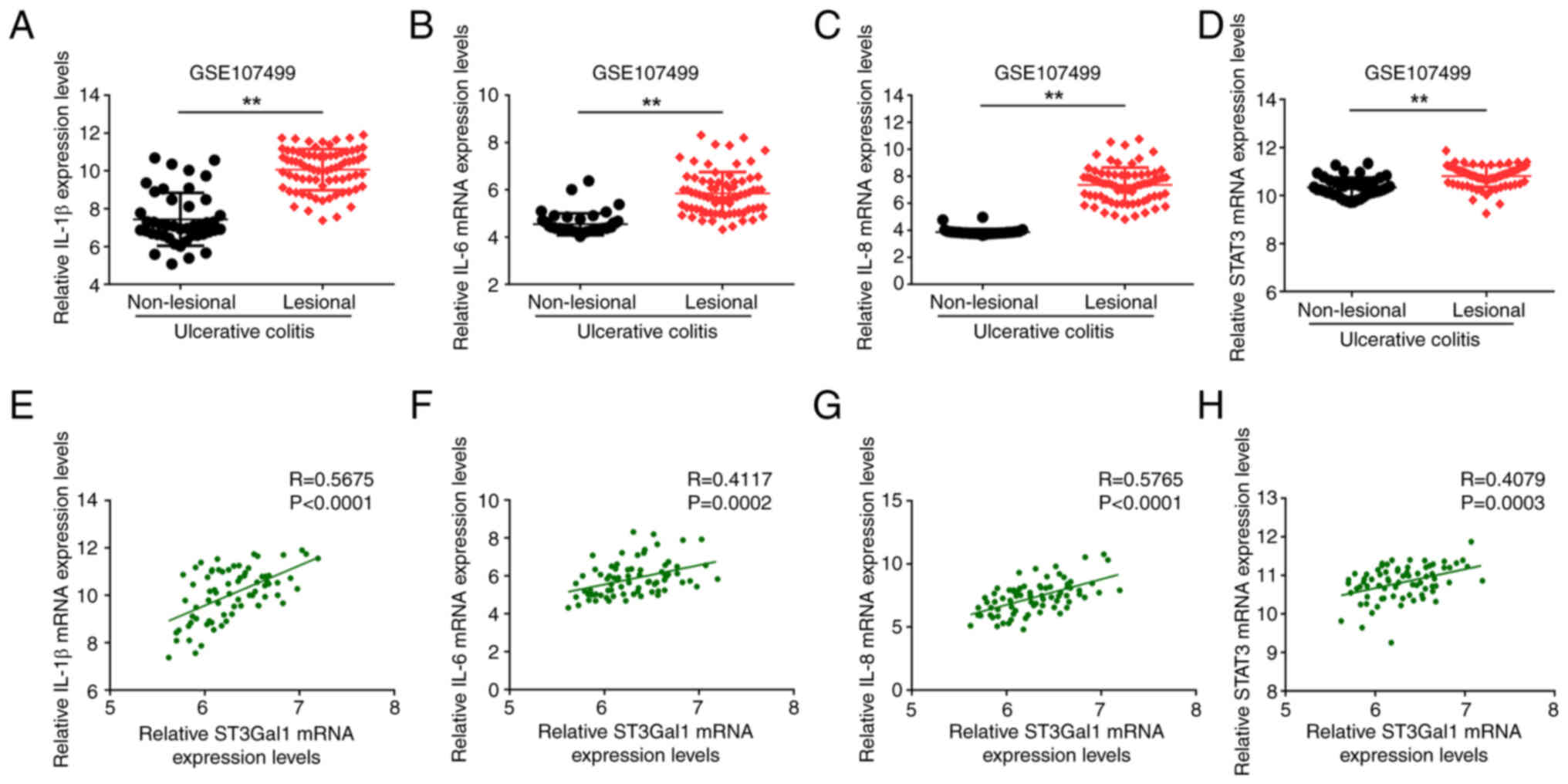
The present study conducted a comparative analysis
of the mRNA expression levels of IL-1β, IL-6, IL-8 and STAT3
between the non-lesional and lesional mucosal tissues of patients
diagnosed with UC, which was obtained from the GSE107499 dataset,
alongside the relationships of cytokine expression with ST3Gal1
mRNA expression levels. The present results demonstrated that the
mRNA levels of IL-1β (P<0.01; Fig.
7A), IL-6 (P<0.01; Fig.
7B), IL-8 (P<0.01; Fig. 7C)
and STAT3 (P<0.01; Fig. 7D) in
the lesional mucosa were significantly elevated compared with those
in the non-lesional mucosa. Furthermore, correlation analyses
revealed positive correlations between ST3Gal1 mRNA expression and
the expression levels of the cytokines IL-1β (R=0.5675;
P<0.0001; Fig. 7D), IL-6
(R=0.4117; P=0.0002; Fig. 7E),
IL-8 (R=0.5765; P<0.0001; Fig.
7F) and STAT3 (R=0.4079; P=0.0003; Fig. 7H) in the lesional mucosa of
patients with UC.
Discussion
The results of the present study indicated a strong
relationship between ST3Gal1 expression in the intestinal
epithelium and the barrier function of IECs, as well as with the
secretion of inflammatory mediators within the context of
intestinal inflammation. ST3Gal1 knockdown in the IEC monolayer
increased the barrier function, whereas its overexpression resulted
in intestinal barrier function deterioration. Overexpression of
ST3Gal1 in the IEC monolayer reduced the expression of intestinal
mucus barrier-associated MUC2, TFF3 and CDX2, as well as the
expression of inflammation-associated p-STAT3. By contrast,
overexpression of ST3Gal1 elevated the levels of inflammatory
mediators (IL-1β, IL-6, and IL-8), which are important for
maintaining intestinal mucosa homeostasis (33). The DSS-induced colitis model is
linked to the inflammatory status of the colonic tissue, as
evidenced by the dynamic association between disease progression
and the expression profiles of key molecules: Specifically,
inflammatory factors (IL-1β, IL-6, IL-8), goblet cell secretory
proteins (MUC2, TFF3) and signaling pathway-related proteins
(STAT3, p-STAT3). This is supported by the bioinformatics analysis
of the GEO dataset GDS3859, where ST3Gal1 mRNA expression in mouse
colonic mucosa was significantly upregulated with prolonged DSS
treatment (peaking at 6 days post-administration), concurrent with
the exacerbation of colonic inflammation. Additionally, in the
in vitro inflamed triple-culture model induced by 2% DSS,
the expression patterns of the aforementioned molecules were
consistently altered in response to ST3Gal1 knockdown or
overexpression, further confirming that the DSS-induced model
recapitulates the intrinsic association between colonic
inflammatory status and the expression of these functional proteins
in UC. ST3Gal1 facilitates the formation of α2,3-sialic acid
residues and contributes to the modification of glycan ends on
proteins that possess α2,3-sialic acid residues (34). Dysregulation of ST3Gal1 within IECs
can result in the abnormal expression of sialic acid, which is a
unique carbohydrate that anchors glycoproteins and glycolipids to
the cell membrane through α2,3-sialic acid residue modifications
(35). The present study
elucidated the role of ST3Gal1 in various IEC monolayers, including
inflamed and non-inflamed states, to identify knowledge gaps, and
explore its potential application as a novel diagnostic and
therapeutic target.
Sialyltransferases are key enzymes in the regulatory
mechanisms of various life processes, including cell signaling,
cellular recognition, interactions between cells and pathogens, and
cancer metastasis (36–38). Among these enzymes, the ST3Gal
family stands out as one of the most important enzyme families,
comprising six distinct members in both mice and humans: ST3Gal1-6
(39). The ST3Gal family catalyzes
the synthesis and attachment of α2,3-linked sialic acid residues to
the terminal galactose residues of glycans in proteins through a
process known as sialylation. The evolution of distinct human
ST3Gal family members, which establish specific α2,3-linkages,
suggests a degree of functional specialization; however, the roles
of α2,3-linkage in health and disease remain insufficiently
investigated (39). Prior research
has documented increased sialylation in IBD, as well as in other
autoimmune conditions and instances of acute inflammation (40). Notably, α2,6- and α2,3-sialylation
of glycans with extensively branched structures appear to escalate
during inflammatory responses in CD and UC, respectively, which are
mediated by α2,6-sialyltransferases and α2,3-sialyltransferases
(41,42). Similarly, ST6 mutations in animal
models have been shown to cause impairment of the intestinal mucus
barrier, dysbiosis of the intestinal microbiota and increased
susceptibility to intestinal inflammation (11,43).
Therefore, it can be suggested that ST6 is important for inhibiting
bacterial translocation and mitigating inflammation within the
intestinal mucus barrier (44).
Consequently, the role of α2,3-linkage, catalyzed by ST3Gal1
sialyltransferases, in the pathogenesis of patients with IBD and
its underlying mechanisms remain largely unexplored.
IBD refers to a persistent inflammatory disorder
affecting the intestines, which is influenced by a combination of
genetic predispositions, immune responses and environmental
elements (45). To facilitate
research on IBD, 66 distinct animal models have been developed at
present, which can be classified into chemical treatment models,
cell migration models, gene mutation models and gene transfer
models (46). However, in some
mouse models with few IBD-related genes, the specific pathogenic
mechanisms of IBD are more complex than previously predicted
(46). To investigate the
pathogenic mechanisms and therapeutic approaches of IBD under
healthy conditions and inflammatory microenvironments,
respectively, researchers have established tri-culture cell models
of healthy and inflamed states in vitro. Based on these
models, various complex in vitro culture systems and
derivative systems have been developed, including primary cell
organoid culture systems, multicellular co-culture systems and chip
device systems (24,47,48).
The present study established an in vitro intestinal barrier
simulation model by co-culturing three cell lines: i) HT29-MTX-E12
cells, which exhibit goblet cell characteristics and have a
mucus-secreting ability, can be used to simulate the intestinal
mucus layer in vitro; ii) Caco-2 cells with epithelial cell
properties, which were seeded in the upper layer; and iii)
differentiable THP-1 inflammatory cells, which were seeded in the
basal side. Subsequently, this model was treated with 2% DSS to
induce an inflammatory state. The effect of ST3Gal on the integrity
of the IEC monolayer barrier was then evaluated by measuring TEER
and FITC-dextran permeability. Additionally, as reported in
previous studies, the secretion levels of relevant inflammatory
factors and the expression of intestinal barrier-related proteins
were compared between inflammatory and non-inflammatory states
(49–51). The tri-culture system designed to
simulate intestinal conditions streamlined the present model,
enhancing its robustness and reproducibility (24). The epithelial Transwell cultures
incorporating Caco-2 and HT29-MTX-E12 cell lines exhibited traits
that closely resemble those of human intestinal mucosal epithelium.
Furthermore, the inclusion of immune-competent THP-1 cells
contributed to the immune mechanisms associated with the
development of IBD (21). Notably,
both the presence of these immune effector cells and the
persistence of inflammatory processes substantially influenced the
sensitivity of the IEC monolayer.
However, there remains a lack of
α2,3-sialylation-linked target molecules bridging ST3Gal1 and
intestinal mucus barrier-associated MUC2, TFF3 and CDX2, as well as
inflammation-associated molecules, such as IL-1β, IL-6, IL-8, STAT3
and p-STAT3. Further study is required to identify ST3Gal1-mediated
molecules with post-translation α2,3-sialylation at their glycan
termini. This may help identify novel target molecules to modulate
the intestinal epithelial barrier function important for the
pathogenesis of human IBD.
The present study provided a new direction for the
treatment of UC and other IBDs and showed that ST3Gal1 can act as a
potential therapeutic target. Overexpression of ST3Gal1 was
directly associated with impaired intestinal barrier function,
elevated expression of the pro-inflammatory cytokines IL-1β, IL-6
and IL-8, and increased disease activity in patients with UC,
supported by in vitro and clinical data: i) In vitro,
ST3Gal1 overexpression in the inflamed triple-culture model showed
significantly reduced TEER, increased FITC-dextran permeability,
downregulated expression of barrier-protective proteins MUC2, TFF3
and CDX2, and upregulated p-STAT3, as well as elevated mRNA and
protein levels of IL-1β, IL-6 and IL-8. ii) Clinically,
bioinformatics analysis of GEO datasets (GSE107499 and GSE92415)
revealed that ST3Gal1 mRNA expression was significantly higher in
lesional vs. non-lesional mucosa inpatients with UC, it was
positively associated with modified Mayo scores (a measure of UC
disease activity), and it was positively correlated with the mRNA
levels of IL-1β, IL-6, IL-8 and STAT3 in UC lesional tissues. By
contrast, ST3Gal1 knockdown upregulated the barrier-protective
molecules (MUC2, TFF3 and CDX2) and reduced p-STAT3 expression,
thereby repairing the mucosal barrier and alleviating inflammation.
Compared with traditional broad-spectrum immunosuppressive
therapies, ST3Gal1-targeted interventions may act precisely on the
glycosylation pathway, thereby potentially minimizing off-target
effects. Additionally, the characteristic high expression of
ST3Gal1 in inflamed mucosal tissues enables it to serve as a
potential biomarker for assessing disease activity or treatment
response. Future studies should validate the efficacy of ST3Gal1
inhibitors in animal models or explore their synergistic effects
with existing drugs, such as mesalamine, to provide more precise
therapeutic options for IBD.
While the Caco-2/HT29-MTX-E12/THP-1 triple-culture
model recapitulated key intestinal mucosal features and followed
the 3R principles, it failed to fully replicate the in vivo
intestinal microenvironment, lacking fibroblasts, endothelial cells
and diverse gut microbiota, which are all regulators of mucosal
barrier function and IBD inflammation (52). Furthermore, immortalized cell lines
such as those used in the present study may not reflect the
phenotypes or functions of primary cells of patients with IBD, thus
limiting translational relevance.
The clinical analyses of the present study, which
relied on retrospective GEO data, also had inherent constraints: i)
The lack of access to detailed patient metadata, such as age and
treatment history, or long-term paired samples, which would
preclude ST3Gal1-prognosis/treatment response analyses; and ii)
limited sample sizes of subgroups, such as the UC non-inflamed
mucosa group, reduced the power of the correlation analysis for
ST3Gal1 with MUC2, which as a key intestinal mucosal barrier
component and goblet cell marker, is closely implicated in UC
pathogenesis (53). To address
these limitations, future studies should: i) Validate the findings
of the present study by using primary IEC-organoid models with
microbiota or stromal cells for improved physiological relevance;
ii) use prospective, well-characterized clinical cohorts to clarify
the clinical utility of ST3Gal1; and iii) apply glycoproteomic
methods, such as lectin affinity chromatography-mass spectrometry,
to identify ST3Gal1-mediated α-2,3-sialylation target molecules to
strengthen the mechanistic and translational value of the present
results.
In conclusion, the present study provided evidence
that ST3Gal1-catalyzed α2,3-linkage formation in IEC may be closely
associated with intestinal barrier function. This relationship
could be mediated through the distinct influence of ST3Gal1 on the
expression of intestinal barrier-associated proteins, including
MUC2, TFF3 and CDX2, which are important for preserving the
integrity of the intestinal barrier. Additionally, ST3Gal1
modulated the expression of associated inflammatory mediators and
transcription factors, including the expression of IL-1β, IL-6,
IL-8 and STAT3, which serve important roles in regulating
intestinal mucosal inflammation, thereby further linking ST3Gal1 to
barrier function.
Acknowledgements
Not applicable.
Funding
The present work was supported by the Natural Science Foundation
of Chongqing (grant no. cstc2020jcyj-msxmX1094).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
YT, YL, YS, LR and LL were responsible for
performing the experiments, and collecting and analyzing the data;
specifically, they performed in vitro cell culture
(triple-culture model establishment, ST3Gal1
overexpression/knockdown), functional assays (TEER, FITC-dextran
permeability), molecular experiments (RT-qPCR, western blotting,
ELISA), and bioinformatics data processing (GEO dataset analysis).
JY substantially contributed to study conception and design,
provided critical project oversight (experimental direction, data
validation), and led manuscript drafting and critical revision for
key intellectual content. RW critically revised the manuscript by
interpreting the biological significance of core findings (for
example, linking ST3Gal1-mediated α2,3-sialylation to intestinal
barrier dysfunction in UC) and standardizing the academic
expression of results (refining data description and mechanistic
discussion logic). The manuscript was authored by JY and RW, with
JY providing project oversight. JY and YT confirm the authenticity
of all the raw data. All authors have read, critically revised, and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Furfaro F, Ragaini E, Peyrin-Biroulet L
and Danese S: Novel therapies and approaches to inflammatory bowel
disease (IBD). J Clin Med. 11:43742022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Selvakumar B and Samsudin R: Intestinal
barrier dysfunction in inflammatory bowel disease: Pathophysiology
to precision therapeutics. Inflamm Bowel Dis. 31:450–3464. 2025.
View Article : Google Scholar
|
|
3
|
Marincola Smith P, Choksi YA, Markham NO,
Hanna DN, Zi J, Weaver CJ, Hamaamen JA, Lewis KB, Yang J, Liu Q, et
al: Colon epithelial cell TGFβ signaling modulates the expression
of tight junction proteins and barrier function in mice. Am J
Physiol Gastrointest Liver Physiol. 320:G936–G957. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rader AG, Cloherty APM, Patel KS,
Almandawi DDA, Perez-Vargas J, Wildenberg ME, Muncan V, Schreurs R,
Jean F and Ribeiro CMS: Autophagy-enhancing strategies to promote
intestinal viral resistance and mucosal barrier function in
SARS-CoV-2 infection. Autophagy Rep. 4:25142322025. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang R, Xie L, Jiang P, Hou Y, Li D and
Wang W: Metformin may improve intestinal mucosal barrier function
and help prevent and reverse colorectal cancer in mice. J Cancer.
16:3703–3711. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Esmail S and Manolson MF: Advances in
understanding N-glycosylation structure, function, and regulation
in health and disease. Eur J Cell Biol. 100:1511862021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Montag N, Gousis P and Wittmann J: The
emerging role of GlycoRNAs in immune regulation and recognition.
Immunol Lett. 276:1070482025. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Flynn RA, Pedram K, Malaker SA, Batista
PJ, Smith BAH, Johnson AG, George BM, Majzoub K, Villalta PW,
Carette JE, et al: Small RNAs are modified with N-glycans and
displayed on the surface of living cells. Cell. 188:44702025.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
He M, Zhou X and Wang X: Glycosylation:
Mechanisms, biological functions and clinical implications. Signal
Transduct Target Ther. 9:1942024. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang G, Yuan J, Luo J, Ocansey DKW, Zhang
X, Qian H, Xu W and Mao F: Emerging role of protein modification in
inflammatory bowel disease. J Zhejiang Univ Sci B. 23:173–188.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kudelka MR, Stowell SR, Cummings RD and
Neish AS: Intestinal epithelial glycosylation in homeostasis and
gut microbiota interactions in IBD. Nat Rev Gastroenterol Hepatol.
17:597–617. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yao Y, Kim G, Shafer S, Chen Z, Kubo S, Ji
Y, Luo J, Yang W, Perner SP, Kanellopoulou C, et al: Mucus
sialylation determines intestinal host-commensal homeostasis. Cell.
185:1172–1188.e28. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fan Q, Li M, Zhao W, Zhang K, Li M and Li
W: Hyper α2,6-Sialylation promotes CD4+ T-Cell activation and
induces the occurrence of ulcerative colitis. Adv Sci (Weinh).
10:e23026072023. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shu X, Li J, Chan UI, Su SM, Shi C, Zhang
X, An T, Xu J, Mo L, Liu J, et al: BRCA1 insufficiency induces a
hypersialylated acidic tumor microenvironment that promotes
metastasis and immunotherapy resistance. Cancer Res. 83:2614–2633.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fan TC, Yeo HL, Hung TH, Chang NC, Tang
YH, Yu J, Chen SH and Yu AL: ST3GAL1 regulates cancer cell
migration through crosstalk between EGFR and neuropilin-1
signaling. J Biol Chem. 301:1083682025. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hong Y, Walling BL, Kim HR, Serratelli WS,
Lozada JR, Sailer CJ, Amitrano AM, Lim K, Mongre RK, Kim KD, et al:
ST3GAL1 and βII-spectrin pathways control CAR T cell migration to
target tumors. Nat Immunol. 24:1007–1019. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tajadura-Ortega V, Gambardella G, Skinner
A, Halim A, Van Coillie J, Schjoldager KTG, Beatson R, Graham R,
Achkova D, Taylor-Papadimitriou J, et al: O-linked mucin-type
glycosylation regulates the transcriptional programme downstream of
EGFR. Glycobiology. 31:200–210. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zaro BW, Bateman LA and Pratt MR: Robust
in-gel fluorescence detection of mucin-type O-linked glycosylation.
Bioorg Med Chem Lett. 21:5062–5066. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang Y, Wang L, Ocansey DKW, Wang B, Wang
L and Xu Z: Mucin-Type O-Glycans: Barrier, microbiota, and immune
anchors in inflammatory bowel disease. J Inflamm Res. 14:5939–5953.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bergstrom K, Shan X, Casero D, Batushansky
A, Lagishetty V, Jacobs JP, Hoover C, Kondo Y, Shao B, Gao L, et
al: Proximal colon-derived O-glycosylated mucus encapsulates and
modulates the microbiota. Science. 370:467–472. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhao Z, Zheng W, Zhang L, Song W and Wang
T: Sialyltransferase ST3GAL1 promotes malignant progression in
glioma. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 41:308–317. 2025.(In
Chinese). PubMed/NCBI
|
|
22
|
Busch M, Kämpfer AAM and Schins RPF: An
inverted in vitro triple culture model of the healthy and inflamed
intestine: Adverse effects of polyethylene particles. Chemosphere.
284:1313452021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Busch M, Ramachandran H, Wahle T, Rossi A
and Schins RPF: Investigating the role of the NLRP3 inflammasome
pathway in acute intestinal inflammation: Use of THP-1 knockout
cell lines in an advanced triple culture model. Front Immunol.
13:8980392022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kämpfer AAM, Busch M, Büttner V, Bredeck
G, Stahlmecke B, Hellack B, Masson I, Sofranko A, Albrecht C and
Schins RPF: Model complexity as determining factor for in vitro
nanosafety studies: Effects of silver and titanium dioxide
nanomaterials in intestinal models. Small. 17:e20042232021.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Aske KC and Waugh CA: Expanding the 3R
principles: More rigour and transparency in research using animals.
EMBO Rep. 18:1490–1492. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Deng X, Shang L, Du M, Yuan L, Xiong L and
Xie X: Mechanism underlying the significant role of the
miR-4262/SIRT1 axis in children with inflammatory bowel disease.
Exp Ther Med. 20:2227–2235. 2020.PubMed/NCBI
|
|
27
|
Samak G, Chaudhry KK, Gangwar R, Narayanan
D, Jaggar JH and Rao R: Calcium/Ask1/MKK7/JNK2/c-Src signalling
cascade mediates disruption of intestinal epithelial tight
junctions by dextran sulfate sodium. Biochem J. 465:503–515. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pan Q, Tian Y, Li X, Ye J, Liu Y, Song L,
Yang Y, Zhu R, He Y, Chen L, et al: Enhanced membrane-tethered
mucin 3 (MUC3) expression by a tetrameric branched peptide with a
conserved TFLK motif inhibits bacteria adherence. J Biol Chem.
288:5407–5416. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Olsen J, Gerds TA, Seidelin JB, Csillag C,
Bjerrum JT, Troelsen JT and Nielsen OH: Diagnosis of ulcerative
colitis before onset of inflammation by multivariate modeling of
genome-wide gene expression data. Inflamm Bowel Dis. 15:1032–1038.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sandborn WJ, Feagan BG, Marano C, Zhang H,
Strauss R, Johanns J, Adedokun OJ, Guzzo C, Colombel JF, Reinisch
W, et al: Subcutaneous golimumab induces clinical response and
remission in patients with moderate-to-severe ulcerative colitis.
Gastroenterology. 146:85–95. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fang K, Bruce M, Pattillo CB, Zhang S,
Stone R II, Clifford J and Kevil CG: Temporal genomewide expression
profiling of DSS colitis reveals novel inflammatory and
angiogenesis genes similar to ulcerative colitis. Physiol Genomics.
43:43–56. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xu W, Guo Y, Huang Z, Zhao H, Zhou M,
Huang Y, Wen D, Song J, Zhu Z, Sun M, et al: Small heat shock
protein CRYAB inhibits intestinal mucosal inflammatory responses
and protects barrier integrity through suppressing IKKβ activity.
Mucosal Immunol. 12:1291–1303. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fan J, Huang S, Cao C, Jin X and Su Y: The
roles of ST3Gal1-6 in cancer: Expression profiles and functional
implications. Carbohydr Res. 559:1097402025. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ma X, Li M, Wang X, Qi G, Wei L and Zhang
D: Sialylation in the gut: From mucosal protection to disease
pathogenesis. Carbohydr Polym. 343:1224712024. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Al Saoud R, Hamrouni A, Idris A, Mousa WK
and Abu Izneid T: Recent advances in the development of
sialyltransferase inhibitors to control cancer metastasis: A
comprehensive review. Biomed Pharmacother. 165:1150912023.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Perez S, Fu CW and Li WS:
Sialyltransferase inhibitors for the treatment of cancer
metastasis: Current challenges and future perspectives. Molecules.
26:56732021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Uslupehlivan M, Şener E and İzzetoğlu S:
Computational analysis of the structure, glycosylation and CMP
binding of human ST3GAL sialyltransferases. Carbohydr Res.
486:1078232019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hatano K, Miyamoto Y, Nonomura N and
Kaneda Y: Expression of gangliosides, GD1a, and sialyl
paragloboside is regulated by NF-κB-dependent transcriptional
control of α2,3-sialyltransferase I, II, and VI in human
castration-resistant prostate cancer cells. Int J Cancer.
129:1838–1847. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kelm M, Quiros M, Azcutia V, Boerner K,
Cummings RD, Nusrat A, Brazil JC and Parkos CA: Targeting
epithelium-expressed sialyl Lewis glycans improves colonic mucosal
wound healing and protects against colitis. JCI Insight.
5:e1358432020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Taniguchi M, Okumura R, Matsuzaki T,
Nakatani A, Sakaki K, Okamoto S, Ishibashi A, Tani H, Horikiri M,
Kobayashi N, et al: Sialylation shapes mucus architecture
inhibiting bacterial invasion in the colon. Mucosal Immunol.
16:624–641. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhao T, Liu S, Ma X, Shuai Y, He H, Guo T,
Huang W, Wang Q, Liu S, Wang Z, et al: Lycium barbarum
arabinogalactan alleviates intestinal mucosal damage in mice by
restoring intestinal microbes and mucin O-glycans. Carbohydr Polym.
330:1218822024. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kotlarz D: Mucus sialylation maintains the
peace in intestinal host microbe relations. Gastroenterology.
163:527–528. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sánchez-Martínez E, Garrido-Romero M and
Moreno FJ: Functional role of ST6GALNAC1-mediated sialylation of
mucins in preserving intestinal barrier integrity and ameliorating
inflammation. Allergy. 77:3697–3698. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Petit C, Rozières A, Boschetti G, Viret C,
Faure M, Nancey S and Duclaux-Loras R: Advances in understanding
intestinal homeostasis: Lessons from inflammatory bowel disease and
monogenic intestinal disorder pathogenesis. Int J Mol Sci.
26:61332025. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Mizoguchi A: Animal models of inflammatory
bowel disease. Prog Mol Biol Transl Sci. 105:263–320. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Le NPK, Altenburger MJ and Lamy E:
Development of an Inflammation-triggered in vitro ‘Leaky Gut’ Model
using Caco-2/HT29-MTX-E12 combined with Macrophage-like THP-1 cells
or primary Human-derived macrophages. Int J Mol Sci. 24:74272023.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Weber L, Kuck K, Jürgenliemk G, Heilmann
J, Lipowicz B and Vissiennon C: Anti-Inflammatory and
Barrier-stabilising effects of myrrh, coffee charcoal and chamomile
flower Extract in a Co-Culture cell model of the intestinal mucosa.
Biomolecules. 10:10332020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ma L, Zhang X, Zhang C, Hou B and Zhao H:
FOSL1 knockdown ameliorates DSS-induced inflammation and barrier
damage in ulcerative colitis via MMP13 downregulation. Exp Ther
Med. 24:5512022. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Roselli M, Maruszak A, Grimaldi R,
Harthoorn L and Finamore A: Galactooligosaccharide treatment
alleviates DSS-induced colonic inflammation in Caco-2 cell model.
Front Nutr. 9:8629742022. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang Y, Wen R, Liu D, Zhang C, Wang ZA and
Du Y: Exploring effects of chitosan oligosaccharides on the
DSS-Induced intestinal barrier impairment in vitro and in vivo.
Molecules. 26:21992021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
De Cecco F, Franceschelli S, Panella V,
Maggi MA, Bisti S, Bravo Nuevo A, D'Ardes D, Cipollone F and
Speranza L: Biological response of treatment with saffron petal
extract on Cytokine-induced oxidative stress and inflammation in
the Caco-2/Human leukemia monocytic Co-Culture model. Antioxidants
(Basel). 13:12572024. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yin S, Yang H, Tao Y, Wei S, Li L, Liu M
and Li J: Artesunate ameliorates DSS-induced ulcerative colitis by
protecting intestinal barrier and inhibiting inflammatory response.
Inflammation. 43:765–776. 2020. View Article : Google Scholar : PubMed/NCBI
|