Introduction
Cardiovascular diseases (CVDs) are a leading cause
of mortality and disability worldwide (1). Atherosclerosis (AS), a chronic
inflammatory condition, underlies a number of cardiovascular
pathologies and serves as their principal pathological basis. The
long-term sequelae of AS contribute substantially to the
persistently high mortality rates observed in both developed and
developing countries (2). Research
has identified endothelial cell injury and dysfunction as
initiating events in the pathogenesis of AS (3), with endothelial apoptosis serving a
central role in these processes. Accordingly, inhibition of
endothelial apoptosis has emerged as a promising therapeutic
strategy for AS (4). These
observations underscore the importance of elucidating the specific
molecular mechanisms governing endothelial cell apoptosis in AS to
develop effective preventive and therapeutic approaches.
Oxidized low-density lipoprotein (ox-LDL), a
necrotic lipid substrate, is considered a central pathogenic driver
in AS (5). It contributes to
disease progression by inducing endothelial cell injury, promoting
foam cell formation, exacerbating inflammatory responses and
increasing plaque instability (6,7).
Owing to these roles, ox-LDL-stimulated human umbilical vein
endothelial cells (HUVECs) are widely used as in vitro
models of AS.
Notably, long non-coding RNAs and microRNAs
(miRNAs/miRs) have emerged as important regulators in the
pathogenesis of AS (8–10). miRNAs are short, non-coding RNA
molecules 18–22 nucleotides in length that modulate gene expression
by suppressing mRNA translation or promoting mRNA degradation
(11). In addition, it has been
shown that miRNAs are highly conserved and stable across different
species (12), and that they
possess important physiological functions, including the modulation
of protein-coding gene expression (13). miRNAs have been investigated in the
context of various diseases, particularly cardiovascular conditions
such as AS, cardiomyopathy and heart failure (HF). For example,
serum levels of miR-21 in patients with HF exhibit high sensitivity
and specificity for diagnosis, indicating its potential as a
biomarker (14). Other miRNAs have
also been implicated in disease pathogenesis. For example,
miR-125b-1-3p mitigates AS progression via the Ras-related
GTP-binding protein D/mTOR/Unc-51-like autophagy activating kinase
1 signaling pathway (15), whereas
miR-199b-3p suppresses ovarian cancer progression by targeting zinc
finger E-box-binding homeobox 1 (16). Furthermore, miRNAs influence key
cellular processes, such as proliferation, migration, apoptosis and
differentiation, and contribute to the structural stability of
atherosclerotic plaques. For example, miR-129 is notably
downregulated in colorectal cancer tissues and cell lines, and its
upregulation markedly inhibits cell proliferation, migration,
invasion and epithelial-mesenchymal transition (17). Similarly, miR-125b targets the
vitamin D receptor in renal cell carcinoma to promote cell
migration and invasion (18),
while miR-378 suppresses cell proliferation and induces apoptosis
in myelodysplastic syndrome cells (19). Among these, the role of miRNAs in
regulating apoptosis has garnered increasing research
attention.
Previous studies have revealed that miR-222-5p
expression is significantly higher in the serum of atherosclerotic
mice compared with in age-matched healthy C57BL/6J mice (7), and in patients with AS compared with
in healthy individuals without AS or any cardiovascular
comorbidities (20), implicating
its role in AS pathogenesis. Although high expression of miR-222-5p
in AS has been reported, its specific function and molecular
mechanism in ox-LDL-induced endothelial cell apoptosis have yet to
be clarified.
Integrins are cell surface adhesion receptors that
regulate cytoskeletal organization, migration, proliferation and
survival (21,22). Integrin subunit α5 (ITGA5), a
member of the integrin family, has been shown to participate in the
regulation of both cell proliferation and apoptosis. For example,
ITGA5 mediates epidermal growth factor-induced proliferation and
migration in retinal pigment epithelial cells (23), and promotes both
epithelial-mesenchymal transition and progression in oral squamous
cell carcinoma (24). However,
whether a direct regulatory relationship exists between miR-222-5p
and ITGA5, and whether this regulatory axis is involved in
endothelial cell apoptosis and AS development lacks experimental
verification. It has been shown that ITGA5 expression is reduced in
AS injury models (25). These
findings suggest that miR-222-5p may represent a promising
therapeutic target in AS development. In the present study, the
role of miR-222-5p in the regulation of endothelial cell apoptosis
was, to the best of our knowledge, investigated for the first time
in an ox-LDL-induced HUVEC model. In addition, the current study
aimed to verify whether ITGA5 acts as a functional target gene for
miR-222-5p, thus elucidating the miR-222-5p/ITGA5 axis in AS
pathogenesis.
Materials and methods
Cell culture and establishment of the
apoptosis model
Immortalized HUVECs (cat. no. iCell-h110; iCell
Bioscience Inc.) were cultured in Dulbecco's Modified Eagle Medium
(DMEM; Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10%
fetal bovine serum (FBS; Biological Industries; Sartorius AG) and
1% streptomycin/penicillin (Beyotime Biotechnology). These
immortalized cells achieve long-term proliferation in vitro
while retaining the key phenotypic and functional characteristics
of primary HUVECs. Subsequently, the cells were incubated at 37°C
in a 5% CO2 atmosphere, with the medium replaced every
1–2 days and subcultured at 80–90% confluence using a 1:3 split
ratio. An endothelial cell apoptosis model was established by
treating HUVECs with different concentrations of ox-LDL (cat. no.
YB002; Yiyuan Biotechnology Co., Ltd.) following a 12-h serum
starvation period. The specific procedure was as follows: Cells
were treated with 25, 50 or 100 mg/l ox-LDL and cultured
continuously at 37°C in a 5% CO2 environment for 48 h.
This concentration gradient aimed to evaluate the dose-dependent
effects of ox-LDL on endothelial cell apoptosis. Based on the
results shown in Fig. 1, this dose
significantly induced apoptosis; ultimately, 100 mg/l ox-LDL was
selected as the optimal concentration for subsequent functional
experiments. Apoptotic effects were then detected immediately after
the 48-h ox-LDL treatment period.
Cell transfection and
co-transfection
Cells were seeded into 6-well plates, and
transfection was initiated once cell confluence reached 70–90%.
miR2225p mimic (final concentration: 50 nM; cat. no.
miR10004569-1-5), miR2225p inhibitor (final concentration: 100 nM;
cat. no. miR20004569-1-5) and their respective negative controls
(NCs; mimic NC; cat. no. miR1N0000001-1-5; final concentration: 100
nM; NC inhibitor; cat. no. miR2N0000001-1-5; final concentration:
100 nM) were obtained from Guangzhou RiboBio Co., Ltd. Similarly,
the small interfering RNAs (siRNAs) targeting ITGA5 (siITGA5; final
concentration: 100 nM) and its NC (siNC; final concentration: 100
nM), were acquired from Hanheng Biotechnology (Shanghai) Co., Ltd.
For co-transfection of miR-222-5p inhibitor and si-ITGA5, cells
were transfected with a mixture of miR-222-5p inhibitor (100 nM)
and si-ITGA5 (100 nM) at the aforementioned final concentrations.
The corresponding NC group was co-transfected with the NC inhibitor
(100 nM) and si-NC (100 nM) to exclude non-specific effects of the
nucleic acids. According to the manufacturer's instructions, the
liquid transfection reagent (Lipofectamine® 3000; Thermo
Fisher Scientific, Inc.) was briefly centrifuged (1,000 × g for 10
sec at 4°C) before use to collect any residual liquid reagent
adhering to the tube walls, thus ensuring an accurate sample
volume. Subsequently, RNA dilutions were prepared in sterile,
RNase-free microcentrifuge tubes and combined with the transfection
reagent, after which the resulting complexes were added dropwise to
the cells. The transfected cells were then placed in a 37°C, 5%
CO2 incubator and the medium was replaced with complete
medium (DMEM supplemented with 10% FBS) after 6–8 h. Transfection
efficiency was validated by reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) 24 h post-transfection.
Functional experiments were performed 48 h post-transfection; this
time window was selected to allow sufficient time for the
mimic/inhibitor/siRNA to regulate target gene expression and induce
detectable changes in cell functional phenotypes. The siRNA
sequences were as follows: si-ITGA5, sense 5′CAGCUACCUAGGAUACUCU3′,
antisense 5′AGAGUAUCCUAGGUAGCUG3′; and si-NC, sense
5′UUCUCCGAACGUGUCACGU3′, antisense 5′ACGUGACACGUUCGGAGAA3′. To
eliminate potential interference from ox-LDL on the basal
expression levels of miR-222-5p and the results of transfection
efficiency assays, thus ensuring that the miR-222-5p
mimic/miR-222-5p inhibitor alone was evaluated for its effects on
the target miRNA, HUVECs used in the transfection efficiency
validation experiments were not treated with ox-LDL.
Cell Counting Kit-8 (CCK-8)
Cell viability was assessed using the CCK8 [cat. no.
SC119; SevenBio (Beijing) Biotechnology Co., Ltd.]. For this assay,
HUVECs were subjected to the following treatments: To assess the
effects of transfection on cell viability, the cells were seeded
into 96-well plates at a density of 5×103 cells/well and
were cultured until 70–80% confluence. They were then transfected
as aforementioned. For the functional experiments evaluating
ox-LDL-induced cytotoxicity and the effects of miR-222-5p, the
cells were seeded into 96-well plates at 5×103
cells/well, underwent 12-h serum starvation, and were then treated
with ox-LDL at concentrations of 25, 50 and 100 mg/l for 24 h to
induce endothelial injury. After ox-LDL treatment, the cells were
transfected as aforementioned. Subsequently, the CCK-8 reagent was
added to each well (10/100 µl medium), and the plates were
incubated at 37°C in a 5% CO2 atmosphere for 2 h.
Absorbance was measured at 450 nm using a microplate reader
(Bio-Rad Laboratories, Inc.), with cell viability calculated
relative to the untreated control group. Cell viability was
calculated using the following formula: Cell viability
(%)=[(experimental OD value-blank OD value)/(control OD value-blank
OD value)] ×100.
Western blot analysis
All protein extraction procedures were conducted on
ice. Initially, a lysis buffer comprising RIPA buffer (cat. no.
P0013B; Beyotime Biotechnology) and PMSF protease inhibitor (cat.
no. ST506; Beyotime Biotechnology) was prepared at a ratio of
100:1, and total protein was extracted from the cells using the BCA
Protein Concentration Assay Kit (cat. no. P0010S; Beyotime
Biotechnology). The cells processed as aforementioned were lysed
with this solution, and the total protein was subsequently
quantified using the same BCA kit. Absorbance at 562 nm was
measured using a plate reader (Bio-Rad Laboratories, Inc.) and
protein concentration was calculated based on a standard curve and
the sample volume. Subsequently, a 5X loading buffer (cat. no.
P0015; Beyotime Biotechnology) was added to the supernatant, and
the proteins were denatured by heat treatment. The denatured
protein samples were stored at −20°C. Subsequently, equal amounts
of protein (20 µg) were separated by SDSPAGE on 7.5 and 12.5% gels,
depending on the protein size, and transferred to a PVDF membrane.
The membrane was then blocked with 1X protein-free rapid-blocking
solution (cat. no. PS108P; Shanghai Epizyme Biopharmaceutical
Technology Co., Ltd.; Ipsen Pharma) for 30 min at room temperature.
Primary antibody solutions were prepared in advance using a primary
antibody diluent (cat. no. P0023A; Beyotime Biotechnology) for the
following antibodies: Bcl2 rabbit anti-human monoclonal antibody
(cat. no. CY6717; 26 kDa; 1:1,500; Shanghai Abways Biotechnology
Co., Ltd.), Bax rabbit anti-human monoclonal antibody (cat. no.
CY5059; 21 kDa; 1:1,500; Shanghai Abways Biotechnology Co., Ltd.),
ITGA5 rabbit polyclonal antibody (cat. no. YT5589; 115 kDa;
1:1,500; ImmunoWay Biotechnology Company) and βactin rabbit
anti-human monoclonal antibody (cat. no. AB0035; 42 kDa; 1:25,000;
Shanghai Abways Biotechnology Co., Ltd.). After blocking, the
membranes were incubated with the primary antibody solution at 4°C
overnight. Following primary antibody incubation, the membranes
were washed three times with TBST Buffer (Powder) (cat. no.
SW142-01; SevenBio (Beijing) Biotechnology Co., Ltd.) (10 min each)
at room temperature to remove unbound primary antibody.
Subsequently, the membranes were incubated with the secondary
antibody [cat. no. RS0002; HRPconjugated Goat Anti-Rabbit IgG
(H+L); 1:15,000; ImmunoWay Biotechnology Company] for 2 h at room
temperature. Finally, the PVDF membrane was visualized using an ECL
reagent (cat. no. P0018S; Beyotime Biotechnology) and was imaged
using a gel documentation system; the resulting images were
analyzed with ImageJ 1.54g software (version 1.8.0; National
Institutes of Health).
RNA extraction and RT-qPCR
Total RNA was extracted from HUVECs following the
aforementioned transfection or treatment procedures using the
SevenFast® Total RNA Extraction Kit for Cells [cat. no.
SM130; SevenBio (Beijing) Biotechnology Co., Ltd.] in accordance
with the manufacturer's instructions. The extracted RNA was then
reverse transcribed into cDNA using the Allinone First Strand cDNA
Synthesis Kit II Reverse Transcription Kit [cat. no. SM134;
SevenBio (Beijing) Biotechnology Co., Ltd.], strictly following the
manufacturer's protocol to eliminate genomic DNA contamination and
synthesize cDNA. The RT reaction program specified by the
manufacturer was as follows: 42°C for 15 min, followed by 5-sec
denaturation at 95°C. A 2X SYBR Green qPCR MasterMix [Green qPCR
MasterMix II kit; cat. no. SM143; SevenBio (Beijing) Biotechnology
Co., Ltd.] was utilized for qPCR amplification of the cDNA under
the following cycling conditions: Predenaturation at 95°C for 30
sec, followed by denaturation at 95°C for 15 sec and
annealing/extension at 60°C for 25 sec for 40 cycles. Subsequently,
data were normalized to the expression levels of individual
samples, and relative expression changes were calculated using the
standard 2−ΔΔCq method (26). Corresponding graphs were generated
based on the recorded data. miR2225p and ITGA5 expression levels
were normalized to U6 and GAPDH, respectively. The primer sequences
were as follows: miR-222-5p (General Biosystems Corp. Ltd.), stem
loop RT primer
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAATCTA, forward
5′-GAATCACGCTCAGTAGTCAGTG-3′, reverse 5′-ATCCAGTGCAGGGTCCGAGG-3′;
ITGA5 [SevenBio (Beijing) Biotechnology Co., Ltd.], forward
5′-GCCGATTCACATCGCTCTCAAC-3′, reverse 5′-GTCTTCTCCACAGTCCAGCAAG-3′;
U6 [Saiwen Innovation (Beijing) Biotechnology Co., Ltd.], forward
5′-GCTTCGGCAGCACATATACTAAAAT-3′, reverse
5′-CGCTTCACGAATTTGCGTGTCAT-3′; and GAPDH [Saiwen Innovation
(Beijing) Biotechnology Co., Ltd.], forward
5′-TGTTGCCATCAATGACCCCTT-3′, reverse 5′-CTCCACGACGTACTCAGCG-3′.
Apoptosis assay
Apoptosis was assessed using the Annexin VFITC/PI
Assay Kit [cat. no. SC123; SevenBio (Beijing) Biotechnology Co.,
Ltd.]. Cells processed as aforementioned were initially collected
by digestion with EDTA-free trypsin (cat. no. C0205; Beyotime
Biotechnology) and were subsequently resuspended in 1X Annexin V
buffer. Thereafter, Annexin VFITC and PI were added, and the
mixture was thoroughly mixed. The cell pellet was then incubated in
the dark at room temperature for 5–10 min to facilitate apoptosis
analysis. Subsequently, the cells were assessed using a flow
cytometer (A50 universal; Apogee Flow Systems Ltd.). Fluorescence
compensation was adjusted using single-stained controls and
unstained controls to correct for spectral overlap and to set
gating parameters. The results were analyzed with FlowJo v10.9.0
software (BD Biosciences), and scatter plots were generated with
FITC on the horizontal axis and PI on the vertical axis.
Bioinformatics analysis
The online bioinformatics tools TargetScan
(https://www.targetscan.org/vert_80/),
miRDB (https://mirdb.org), miRWalk
(mirwalk.umm.uni-heidelberg.de) and miRTarBase (https://mirtarbase.cuhk.edu.cn/) were used to
screen potential miR-222-5p target genes. After target gene
prediction was performed using the aforementioned databases, the
Venny 2.1.0 tool (https://bioinfogp.cnb.csic.es/tools/venny/index.html)
was used to determine intersections and obtain high-confidence
candidate target genes.
Statistical analysis
All data were analyzed and plotted using GraphPad
Prism 10.0 software (Dotmatics). All data were first validated for
normality using the Shapiro-Wilk test; the test results indicated
that all data met the criteria for normal distribution, with no
instances of non-normal distribution observed. Data that met the
criteria for normal distribution were expressed as mean ± standard
deviation. Comparisons between two groups were performed using an
independent samples t-test. Comparisons among multiple groups were
performed using one-way analysis of variance (ANOVA). If the ANOVA
results indicated statistically significant differences between
groups, further comparisons were performed using Tukey's honestly
significant difference test. P<0.05 was considered to indicate a
statistically significant difference. All experiments were repeated
at least three times.
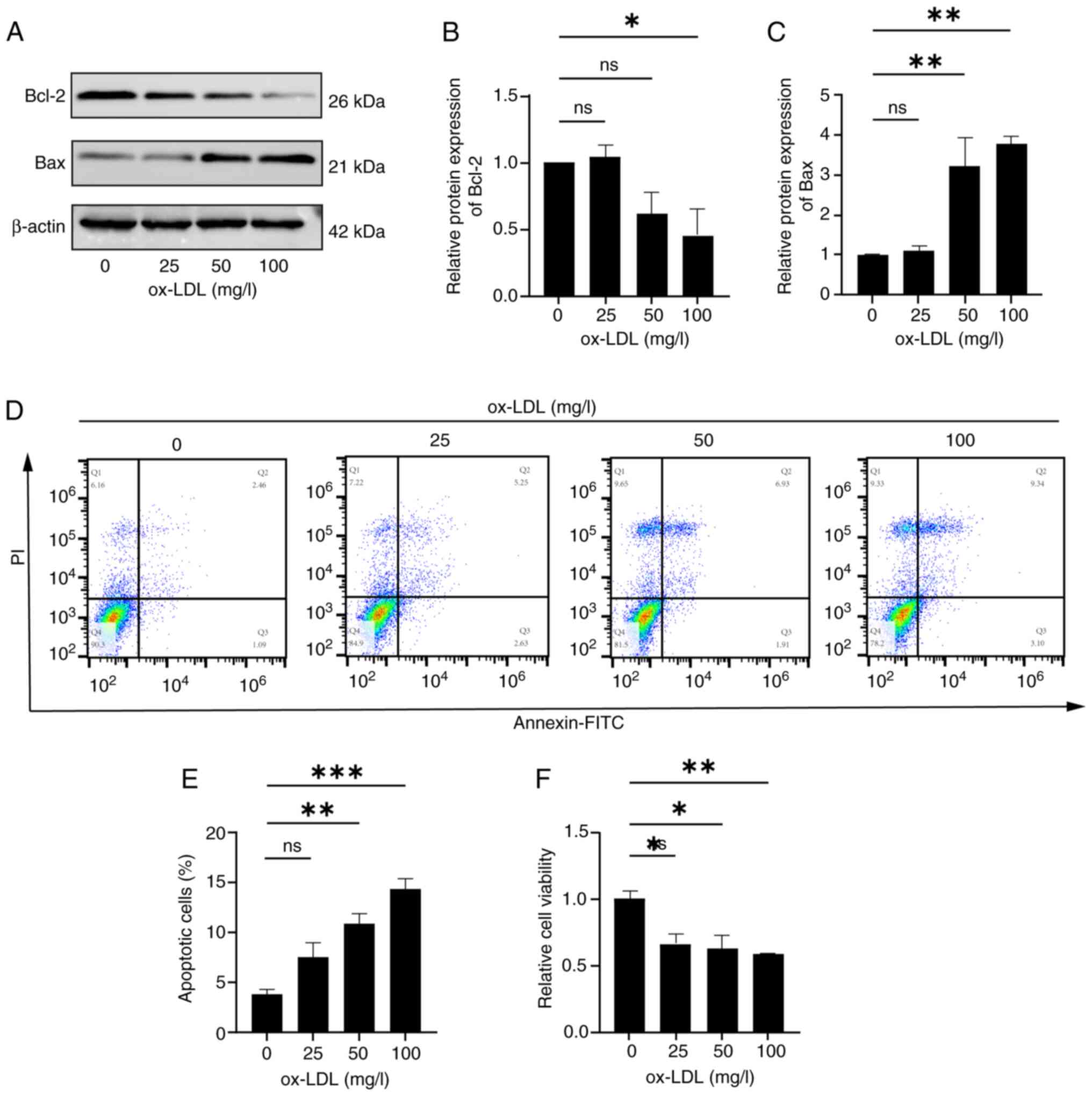
Results
Stimulation of endothelial cells by
ox-LDL establishes a model of endothelial cell apoptosis
To establish a model of endothelial cell apoptosis,
HUVECs were treated with various concentrations of ox-LDL. Prior to
stimulation, HUVECs were serum-starved for 12 h, then exposed to a
gradient of ox-LDL concentrations (0, 25, 50 and 100 mg/l). After
48 h of treatment, protein levels (via western blotting), apoptosis
(via flow cytometry) and cell viability (via CCK-8 assay) were
assessed to determine the optimal ox-LDL concentration. The
pro-apoptotic marker Bax and anti-apoptotic marker Bcl-2 were used
to evaluate apoptotic signaling. Western blot analysis revealed a
significant decrease in Bcl-2 expression and a significant increase
in Bax expression following 48 h of treatment with 100 mg/l ox-LDL
(Fig. 1A-C). Flow cytometric
analysis showed that 100 mg/l ox-LDL significantly increased the
apoptotic rate, defined as the sum of late apoptotic cells (second
quartile Q2: Annexin V+/PI+) and early
apoptotic cells (third quartile Q3: Annexin
V+/PI−), (Q2 + -Q3) from 2.46+1.09% in the 0
mg/l group to 9.34+3.10% (Fig. 1D and
E). Notably, Fig. 1D displays
the quantitative proportions of the four quadrants in the flow
cytometric apoptosis analysis, including the Q1 quadrant (necrotic
cells), Q2 quadrant (late apoptotic cells), Q3 quadrant (early
apoptotic cells) and Q4 quadrant (viable cells). Fig. 1E shows a quantitative histogram
depicting the total apoptosis rate across different treatment
groups (0, 25, 50 and 100 mg/l ox-LDL), defined as the sum of the
percentages of cells in the Q2 (late apoptosis) and Q3 (early
apoptosis) quadrants. Furthermore, 100 mg/l ox-LDL significantly
decreased cell viability compared with untreated cells (Fig. 1F). Based on the results shown in
Fig. 1, 100 mg/l ox-LDL induced a
strong pro-apoptotic effect in HUVECs. Therefore, this
concentration was selected for subsequent experiments.
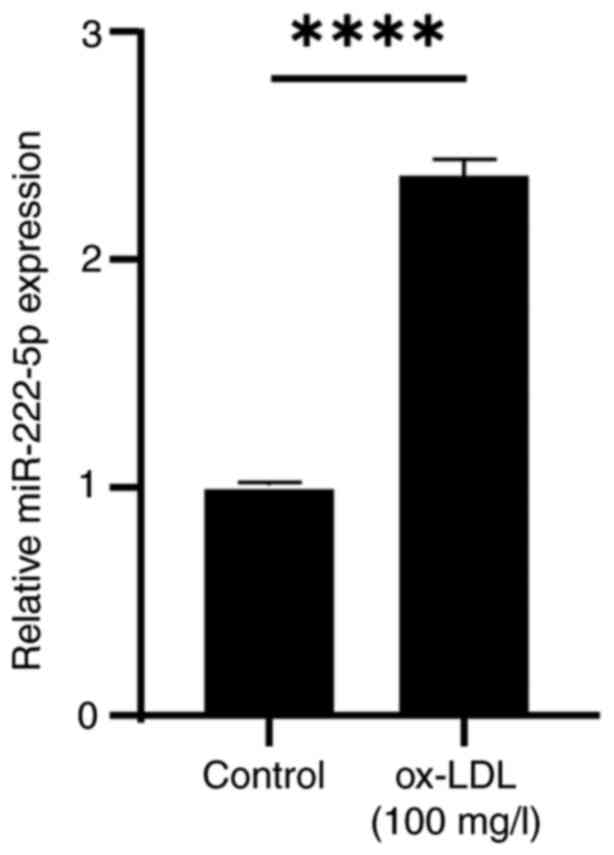
miR-222-5p is upregulated in
ox-LDL-stimulated endothelial cells
To examine the effect of ox-LDL on miR-222-5p
expression, HUVECs were treated with 100 mg/l ox-LDL for 48 h.
RT-qPCR analysis revealed a significant upregulation of miR-222-5p
expression in the ox-LDL group compared with that in the untreated
control group (Fig. 2), suggesting
a potential role for miR-222-5p in endothelial cell apoptosis.
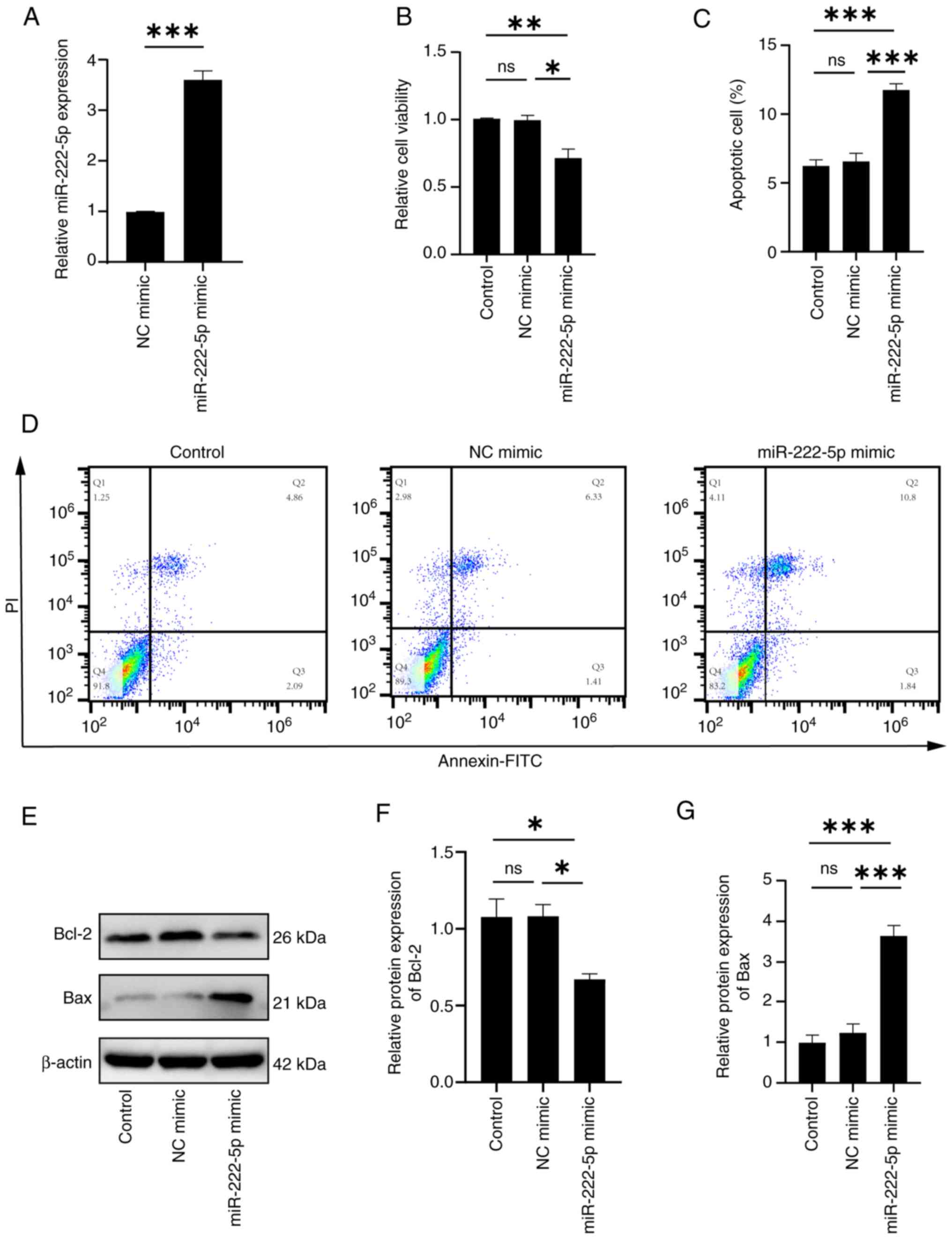
Effects of transfection with a
miR-222-5p mimic on endothelial cells
To further assess whether increased miR-222-5p
levels influenced endothelial apoptosis, HUVECs were transfected
with a miR-222-5p mimic using liposome-mediated delivery. Cells
were transfected with either a miR-222-5p mimic to induce
miR-222-5p overexpression or an NC mimic, and RT-qPCR was used to
quantify miR-222-5p expression. The results showed that miR-222-5p
expression in the overexpression group was ~361% higher than that
in the control group (Fig. 3A).
Subsequently, apoptosis-related parameters were evaluated,
including cell viability (via CCK-8 assay), apoptosis rate (via
flow cytometry) and Bax/Bcl-2 protein levels (via western
blotting). The CCK-8 assay demonstrated that transfection with the
miR-222-5p mimic significantly reduced cell viability compared with
that in the control groups (Fig.
3B), indicating impaired endothelial cell proliferation. Flow
cytometric analysis revealed that compared with in the NC group
(6.33+1.41%, Q2 + Q3), the miR-222-5p overexpression group
(10.80+1.84%, Q2 + Q3) exhibited significantly elevated total
apoptosis rates [(including late apoptotic cells (Q2: Annexin
V+/PI+) and early apoptotic cells (Q3:
Annexin V+/PI−] (Fig. 3C and D). Fig. 3D displays the scatter plot (Q1-Q4),
whereas Fig. 3C presents the
combined total apoptosis rate for Q2/Q3. Consistently, western blot
analysis showed that miR-222-5p mimic transfection significantly
increased Bax expression and significantly decreased Bcl-2
expression compared with those in the control groups (Fig. 3E-G). Taken together, these results
indicated that miR-222-5p promoted endothelial cell apoptosis.
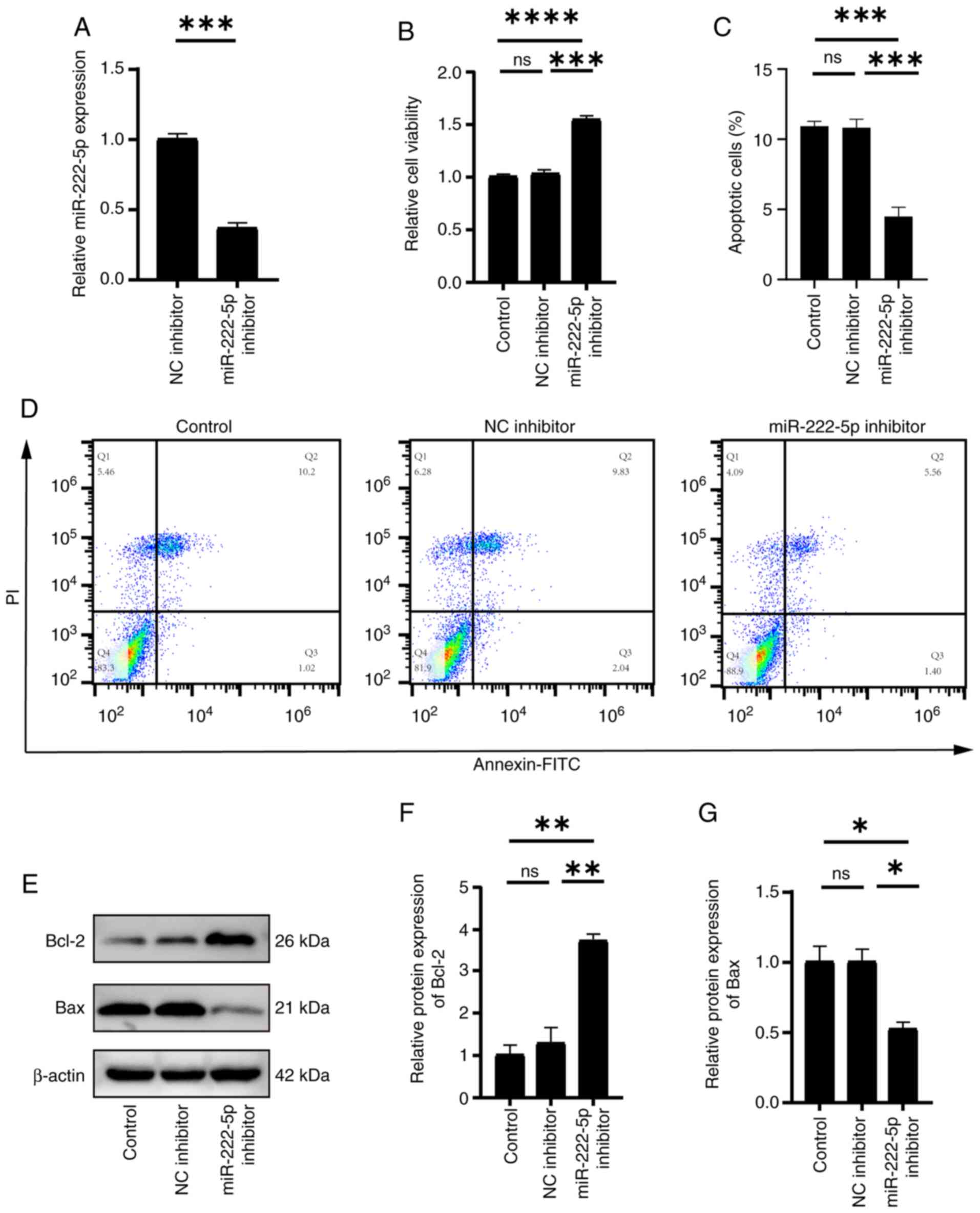
Effect of miR-222-5p inhibitor
transfection on endothelial cells
To further validate the role of miR-222-5p in
endothelial cell apoptosis, HUVECs were transfected with a
miR-222-5p inhibitor. RT-qPCR confirmed that the inhibitor
significantly suppressed ox-LDL-induced miR-222-5p expression
compared with the NC (Fig. 4A).
The CCK-8 assay revealed that miR-222-5p inhibition significantly
enhanced cell viability (Fig. 4B),
whereas flow cytometry analysis revealed that the total apoptosis
rate [sum of late apoptotic cells (Q2: Annexin
V+/PI+) and early apoptotic cells (Q3:
Annexin V+/PI−] in HUVECs significantly
decreased from 9.83+2.04% (Q2 + Q3) in the NC inhibitor group to
5.56+1.40% (Q2 + Q3) in the miR-222-5p inhibitor group (Fig. 4C and D). Fig. 4D displays a dot plot (Q1-Q4
distribution), while Fig. 4C
presents the combined total apoptosis rate (Q2 + Q3). Furthermore,
in endothelial cells treated with the miR-222-5p inhibitor, Bax
expression was significantly reduced, whereas Bcl-2 expression was
significantly increased compared with those in the control groups
(Fig. 4E-G). In summary, these
results indicated that the miR-222-5p inhibitor mitigated
ox-LDL-induced endothelial cell apoptosis.
miR-222-5p directly binds to the 3′
untranslated region (UTR) of ITGA5 and negatively regulates its
expression
To explore the regulatory mechanisms of miR-222-5p,
multiple bioinformatics databases, including TargetScan, miRDB,
miRWalk and miRTarBase, were searched to identify potential
downstream targets (Fig. 5B).
Cross-matching of predictions from these databases yielded 20
high-confidence candidate genes. Among them, ITGA5 contained a
predicted miR-222-5p binding site (Fig. 5A). Endothelial cells were
transfected with either a miR-222-5p mimic or inhibitor, before
RT-qPCR was used to quantify ITGA5 mRNA levels and western blotting
was employed to detect ITGA5 protein expression. The results showed
that transfection with the miR-222-5p mimic significantly
downregulated ITGA5 expression, whereas inhibitor transfection
significantly increased ITGA5 expression (Fig. 5C-H). These findings suggested that
ITGA5 was a downstream target of miR-222-5p and was negatively
regulated by it.
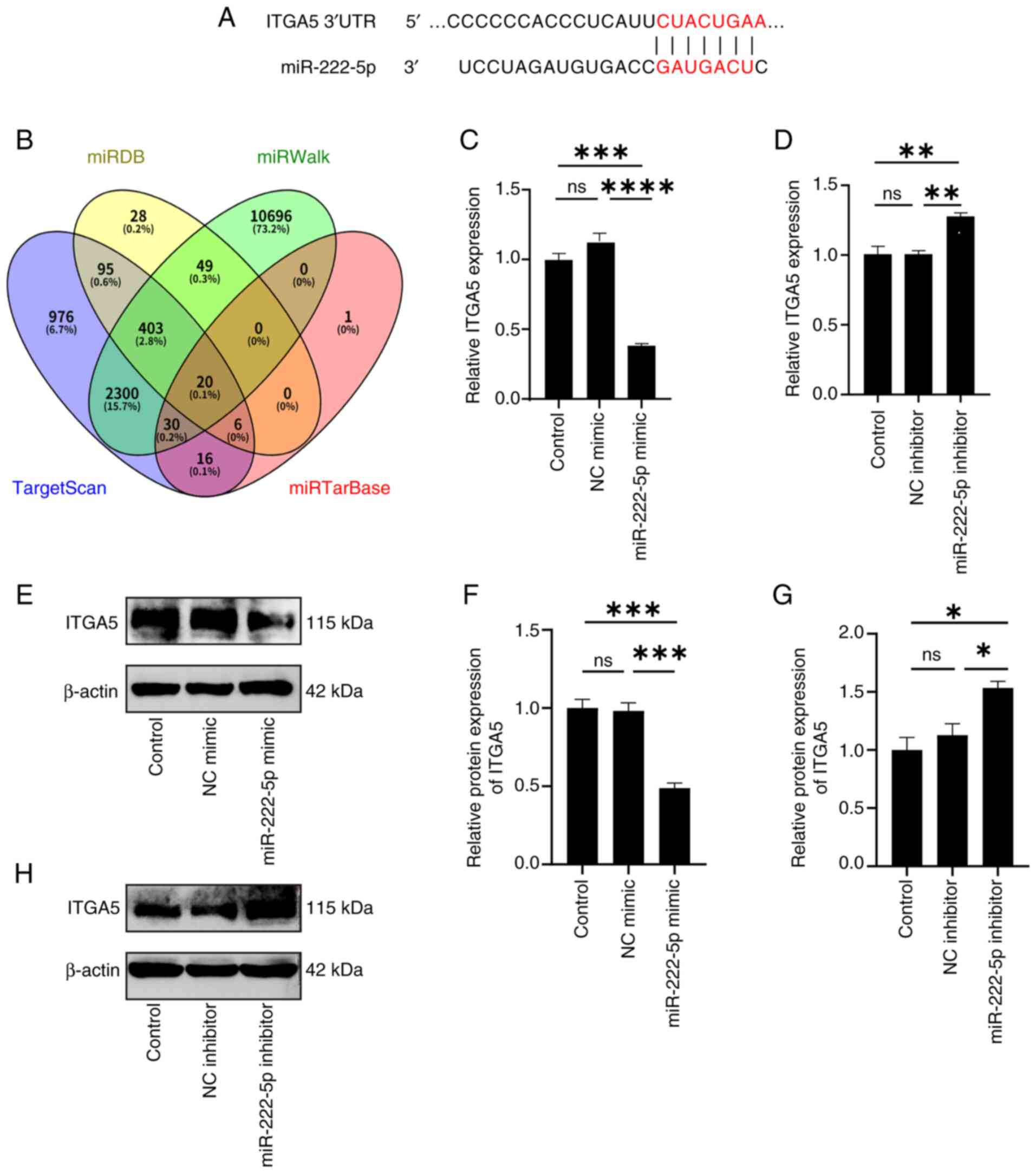 | Figure 5.miR-222-5p directly binds to the
3′UTR region of ITGA5 and negatively regulates its expression. (A)
Predicted miR-222-5p binding site in the ITGA5 3′UTR. (B)
Bioinformatics databases (TargetScan, miRDB, miRWalk and
miRTarBase) identified ITGA5 as a candidate target. Reverse
transcription-quantitative PCR to quantify ITGA5 expression
following (C) mimic (n=3) or (D) inhibitor transfection (n=3).
Western blotting was used to detect the expression levels of ITGA5
protein after transfection with (E) a miR-222-5p mimic and NC
mimic, (F) which was semi-quantified (n=3). (G) Western blotting
was used to detect the expression levels of ITGA5 protein after
transfection with a miR-222-5p inhibitor and NC, (H) which was
semi-quantified (n=3). *P<0.05, **P<0.01, ***P<0.001 and
****P<0.0001. ITGA5, integrin subunit α5; miR, microRNA; NC,
negative control; ns, not significant; UTR, untranslated
region. |
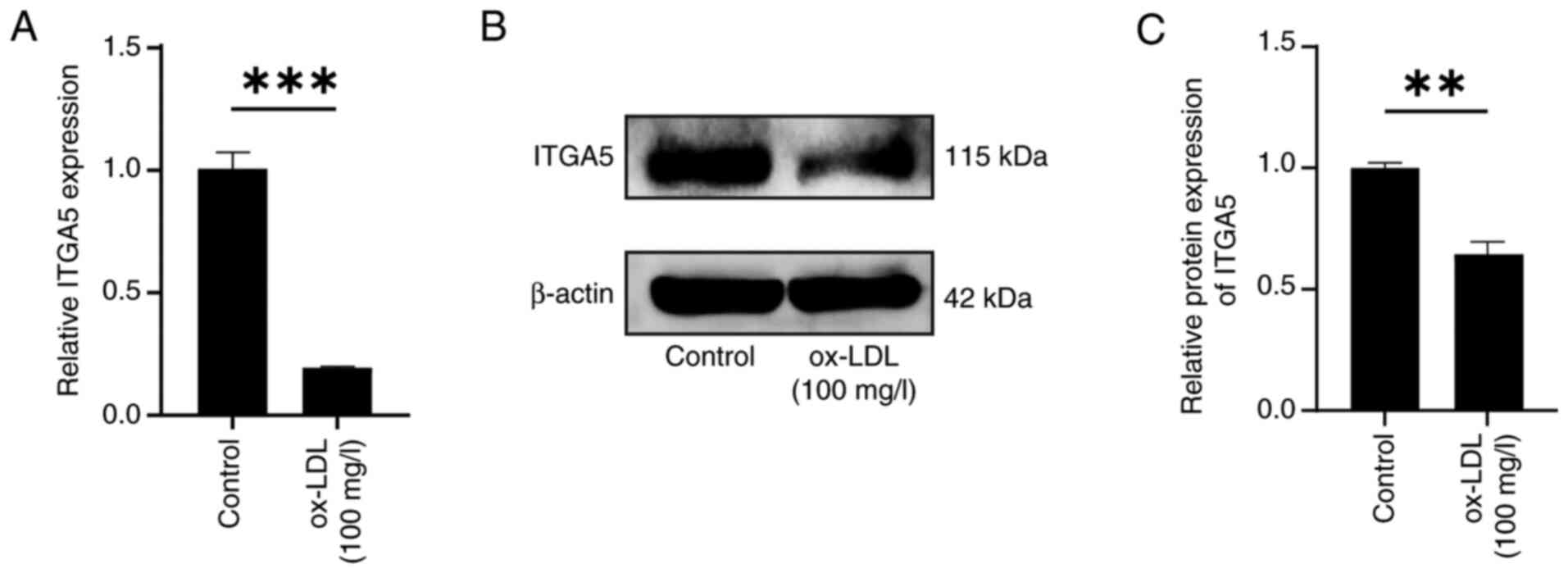
ITGA5 expression is decreased in
ox-LDL-treated endothelial cells
To assess ITGA5 expression in response to ox-LDL
treatment, HUVECs were treated with the optimal concentration of
ox-LDL (100 mg/l) for 48 h. Both RT-qPCR and western blot analysis
demonstrated a significant downregulation of ITGA5 expression in
treated cells compared with that in the untreated control cells
(Fig. 6A-C). These findings
implied a potential role for ITGA5 in ox-LDL-induced endothelial
cell apoptosis.
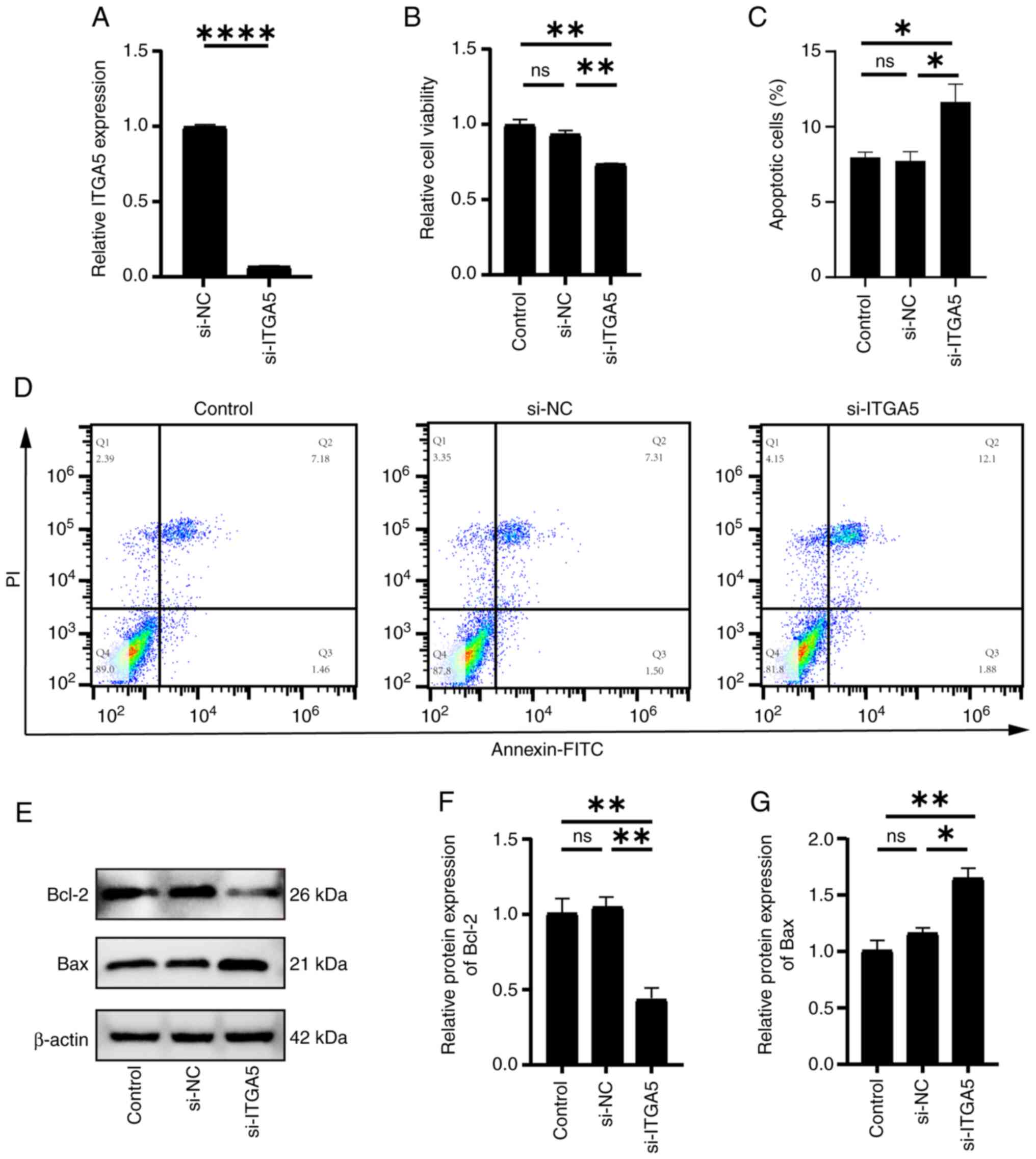
Effect of si-ITGA5 transfection on
endothelial cells
To further investigate the role of ITGA5 in
endothelial cell apoptosis, cells were transfected with si-ITGA5
and knockdown efficiency was assessed by RT-qPCR. The analysis
demonstrated a ~90% reduction in ITGA5 expression compared with
that in the si-NC group (Fig. 7A).
The CCK-8 assay revealed that ITGA5 knockdown significantly reduced
cell viability (Fig. 7B), and the
flow cytometric analysis revealed that a significant increase in
the total apoptosis rate [sum of late apoptotic cells (Q2: Annexin
V+/PI+) and early apoptotic cells (Q3:
Annexin V+/PI−)], rising from 7.31+1.50% (Q2
+ Q3) in the si-NC group to 12.10+1.88% (Q2 + Q3) in the
si-ITGA5-transfected HUVECs (Fig. 7C
and D). Fig. 7D displays a dot
plot (Q1-Q4 distribution), while Fig.
7C presents the combined total apoptosis rate (Q2 + Q3).
Western blot analysis revealed that si-ITGA5 transfection
significantly upregulated Bax and significantly downregulated Bcl-2
expression (Fig. 7E-G). These
findings suggested that ITGA5 exerted an anti-apoptotic effect in
endothelial cells.
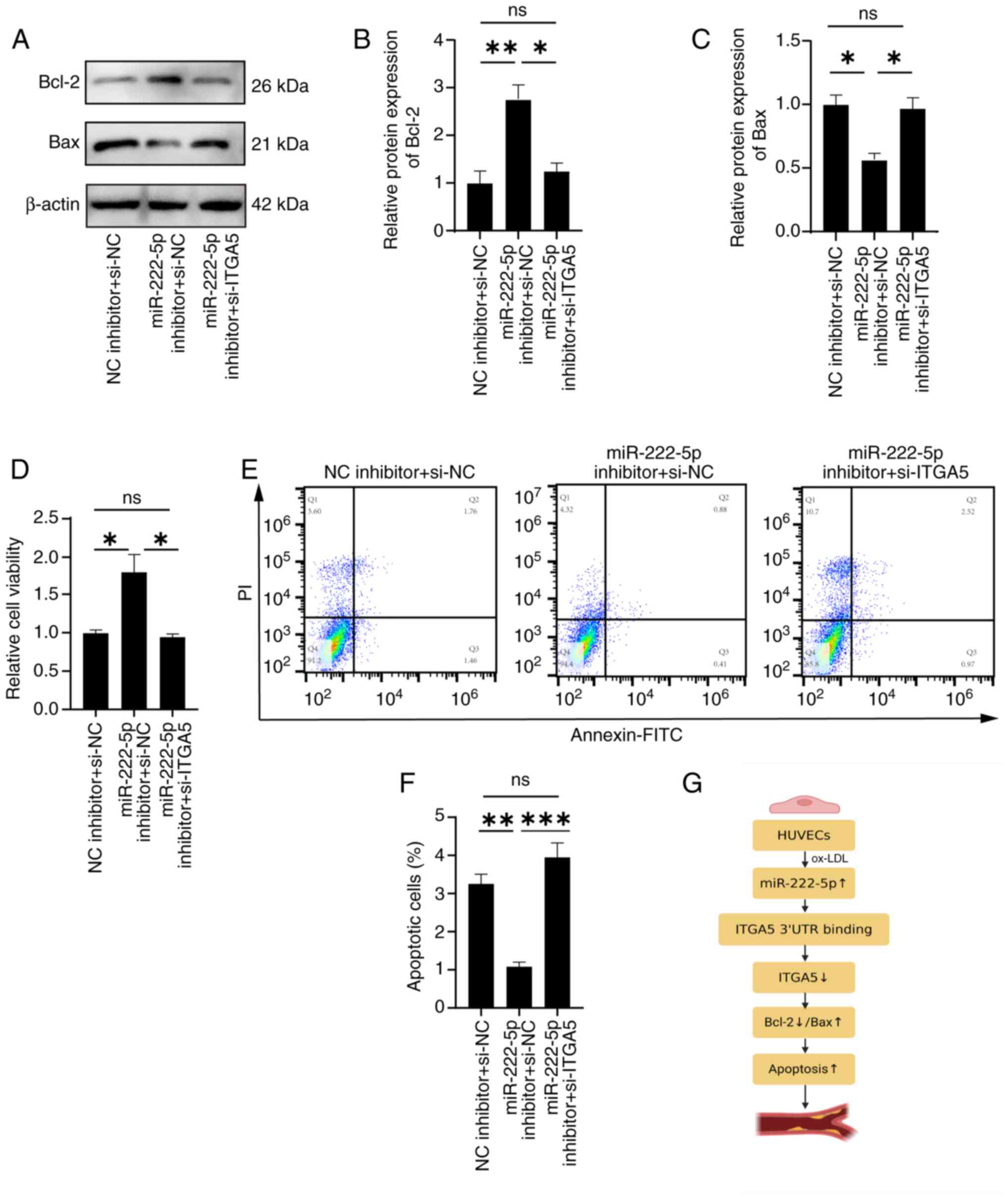
si-ITGA5 partially reverses the
anti-apoptotic effects of the miR-222-5p inhibitor
To validate the regulatory relationship between
miR-222-5p and ITGA5, HUVECs were co-transfected with a miR-222-5p
inhibitor and si-ITGA5. Western blotting revealed that miR-222-5p
inhibition significantly reduced Bax and significantly increased
Bcl-2 levels, whereas co-transfection with si-ITGA5 significantly
reversed these effects, increasing Bax and decreasing Bcl-2
expression (Fig. 8A-C), thereby
attenuating the protective effect of miR-222-5p inhibition. Flow
cytometry revealed a significant reduction in the total apoptosis
rate [sum of late apoptotic cells (Q2: Annexin
V+/PI+) and early apoptotic cells (Q3:
Annexin V+/PI−)], decreasing from 1.76+1.46%
(Q2 + Q3) in the NC inhibitor + si-NC group to 0.88+0.41% (Q2 + Q3)
in the miR-222-5p inhibitor + si-NC group, whereas co-transfection
with si-ITGA5 restored it to 2.52+0.97% (Q2 + Q3) in the miR-222-5p
inhibitor + si-ITGA5 group (Fig. 8E
and F). Fig. 8E displays a
scatter plot (Q1-Q4 distribution), while Fig. 8F presents the combined total
apoptosis rate (Q2 + Q3).
The CCK-8 assay also showed that si-ITGA5 reversed
the promoting effect of miR-222-5p inhibition on cell viability
(Fig. 8D). Taken together, these
findings supported that miR-222-5p promoted endothelial cell
apoptosis by targeting ITGA5.
Discussion
AS is a chronic inflammatory disease characterized
by lipid accumulation, endothelial injury, and superimposed
thrombus formation in medium and large arteries (27). These complications often contribute
to high mortality rates worldwide (28,29).
The progression of AS is largely driven by the deposition of oxLDL,
endothelial dysfunction and plaque accumulation within the vessel
wall (30). Endothelial cell
damage and dysfunction are important in the pathogenesis of AS;
therefore, elucidating their underlying mechanisms is important for
prevention, management and treatment (3,31–33).
Apoptosis is an important event contributing to endothelial injury
and dysfunction (34), and
understanding its regulatory mechanisms may reveal potential
therapeutic targets for AS.
Recently, miRNAs have emerged as promising
biomarkers with considerable therapeutic potential in CVDs
(35,36). A previous study focusing on
miRNA-mediated regulation of AS pathophysiology demonstrated that
miRNAs serve a notable role in modulating cardiovascular
pathophysiology, particularly in AS (37). Extensive research has confirmed
that abnormalities in various genes and signaling pathways are
closely associated with the onset and progression of AS, and that
specific miRNAs can modulate these dysregulated processes. By
regulating cell migration, differentiation, proliferation, lipid
metabolism and cytokine production, miRNAs offer novel insights
into the molecular mechanisms underlying AS and have attracted
interest. For example, miR-213p promotes the proliferation and
migration of smooth muscle cells by targeting PTEN, thereby
accelerating AS development (38).
Obesity-induced exosomal miR-27b3p directly binds to the coding DNA
sequence region of peroxisome proliferator-activated receptor α
mRNA, enhancing endothelial cell inflammation and contributing to
atherosclerotic progression (39).
In vivo animal experiments have shown that miR2225p
expression is upregulated in the serum of ApoE-knockout mice
(7), whereas clinical studies have
reported elevated miR2225p levels in the serum of patients with AS
(20). However, the role of
miR-222-5p in in vitro models of AS remains to be fully
elucidated. Therefore, the present study aimed to investigate the
expression and function of miR-222-5p in an endothelial cell
apoptosis model.
HUVECs are frequently used as cellular models in AS
research (40). To investigate the
role of miR-2225p in endothelial apoptosis, HUVECs were employed in
the experiments of the present study. Disease flare-ups are
initiated by the accumulation of oxLDL in the vascular endothelium,
which is a principal component of atherosclerotic lesions and a key
contributor to disease development (41–43).
As an independent pathogenic factor, oxLDL-induced endothelial cell
apoptosis serves an important role in the pathogenesis of AS
(44). Therefore, oxLDL-stimulated
HUVECs were used to mimic the atherosclerotic vascular
microenvironment and to establish a model of endothelial cell
apoptosis. Initially, HUVECs were treated with oxLDL at 0, 25, 50
and 100 mg/l. After 48 h, cell viability was significantly reduced
at the 100 mg/l concentration. RT-qPCR analysis revealed that
miR-2225p expression was significantly upregulated in cells treated
with 100 mg/l oxLDL, suggesting that aberrant miR-2225p expression
may be associated with endothelial apoptosis. To evaluate the
functional role of miR2225p, HUVECs were transfected with a
miR2225p mimic and inhibitor using a liposome-mediated method.
Overexpression of miR-2225p led to increased apoptosis, reduced
cell viability, upregulation of the pro-apoptotic protein Bax and
downregulation of the anti-apoptotic protein Bcl-2. Conversely,
transfection with the miR-2225p inhibitor resulted in decreased
apoptosis, enhanced viability, reduced Bax expression and elevated
Bcl-2 levels. In the present study, overexpression of miR-222-5p
was demonstrated to have significantly promoted endothelial cell
apoptosis in the ox-LDL-induced HUVEC model, whereas knockdown of
miR-222-5p inhibited apoptosis. Flow cytometry results demonstrated
that the proportion of necrotic cells remained consistently low
across most experimental groups. This low necrosis rate coupled
with significant apoptotic regulation directly and unequivocally
indicated that miR-222-5p primarily promotes cell death through the
apoptotic pathway. This conclusion provides more precise evidence
for the crucial role of miR-222-5p in AS.
miRNAs exert post-transcriptional gene regulation by
binding to the 3′UTR of target mRNAs (45). In the present study, bioinformatics
tools were used to predict ITGA5 as a downstream target of
miR-222-5p. ITGA5 typically forms a heterodimer, known as α5β1
integrin, with the integrin β1 subunit. In the extracellular matrix
(ECM), the primary ligand of ITGA5 is fibronectin (FN).
Specifically, FN contains an Arg-Gly-Asp sequence that is
recognized by ITGA5, which further mediates cell adhesion,
migration and signal transduction. ITGA5 is involved in
physiological processes, such as embryonic development and tissue
repair, and serves a key role in pathological processes, including
tumor invasion and fibrotic diseases (24,46,47).
In the context of apoptosis, ITGA5 transmits survival signals
through various mechanisms. Studies have shown that cell adhesion
to the ECM directly affects susceptibility to apoptosis, an
anchorage-dependent phenomenon, and that ITGA5FN binding activates
downstream FAK and PI3K/Akt signaling pathways. These pathways
inhibit apoptosis by downregulating pro-apoptotic proteins such as
Bax and upregulating anti-apoptotic members of the Bcl-2 family,
thereby counteracting anoikis (48). ITGA5 levels have been found to be
reduced in AS injury models (25).
Consistently, ITGA5 expression was significantly decreased in
HUVECs treated with 100 mg/l oxLDL. To further assess the role of
ITGA5 in endothelial apoptosis, ITGA5 was knocked down using
liposome-mediated transfection. The results indicated that si-ITGA5
promoted apoptosis, thereby supporting the involvement of ITGA5 in
regulating endothelial cell death. Whether miR-2225p modulates
apoptosis via ITGA5, however, requires further investigation.
To further evaluate whether miR2225p regulated
apoptosis via ITGA5 and contributed to AS, HUVECs were transfected
with a miR2225p mimic and inhibitor and ITGA5 expression was
examined. ITGA5 expression was significantly reduced following
mimic transfection and increased following inhibitor transfection.
In addition, co-transfection with the miR-222-5p inhibitor and
si-ITGA5 reversed the protective effect of the inhibitor on
apoptosis. Specifically, the level of apoptosis, initially reduced
by the inhibitor, was restored when ITGA5 was downregulated. These
findings provided functional evidence that miR-222-5p regulated
apoptosis by directly modulating ITGA5 expression. In the present
study, the reversal of the effect of the miR-222-5p inhibitor by
transfection with si-ITGA5 not only supported the importance of
ITGA5 in the anti-apoptotic signaling pathway of miR-222-5p but
also identified ITGA5 as an important biomolecule in
miR-222-5p-mediated cell survival signaling. This process
reinforced the central role of the miR-222-5p/ITGA5 axis in
regulating endothelial cell apoptosis. The molecular mechanism
mediating this regulatory cascade is visualized in Fig. 8G, which outlines the sequential
effects of the miR-222-5p-ITGA5 interaction on downstream signaling
and apoptotic responses in the context of AS.
In recent years, dysregulated miRNA expression
profiles in various diseases have highlighted their potential as
biomarkers (49). Previous studies
have only reported the abnormal expression of miR-222-5p in AS
(7). The present study is, to the
best of our knowledge, the first to provide evidence for the
functional role of miR-222-5p in ox-LDL-induced endothelial cell
apoptosis. To the best of our knowledge, the present study was also
the first to investigate the role of miR-222-5p in AS progression
via ITGA5. The findings of the present study indicated that
miR-222-5p regulated apoptosis and AS by targeting ITGA5. These
cellular findings provide a valuable foundation for future animal
model studies, and lay an important experimental foundation for
subsequent in vivo mechanism validation and translational
research.
From a translational medicine perspective, the
potential of miR-222-5p as a biomarker for AS warrants particular
attention: The present study found that miR-222-5p was upregulated
in endothelial cells induced by ox-LDL and positively associated
with the degree of endothelial cell apoptosis. Combined with
previous studies confirming the upregulation of miR-222-5p in serum
from patients with AS and animal models (7,20),
these results suggest that it may serve as a potential molecular
marker for the early diagnosis of AS. Compared with traditional
diagnostic markers, miRNAs offer advantages such as high stability
in bodily fluids and rapid quantitative detection via techniques
such as RT-qPCR (50), making them
promising candidates for early screening of AS, particularly in
asymptomatic high-risk populations, such as those with
hyperlipidemia (51,52). Dynamic monitoring of serum
miR-222-5p levels may reflect the extent of endothelial damage,
providing objective evidence for selecting optimal timing for
clinical intervention. Furthermore, if subsequent large-scale
clinical cohort studies confirm that high miR-222-5p expression is
positively associated with plaque instability and future
cardiovascular event risk, it may emerge as a novel molecular
indicator for assessing the prognosis of patients with AS.
In terms of treatment strategies, based on the core
mechanism revealed in the present study that miR-222-5p promotes
endothelial apoptosis by inhibiting ITGA5, the miR-222-5p/ITGA5
axis offers a new direction for targeted therapy in AS. For
example, miR-222-5p antagonists could be developed and delivered
via an intravascular local delivery system to inhibit its activity,
thereby restoring ITGA5 expression and activating the FAK/Akt
pathway to protect endothelial cells and slow plaque progression.
In addition, the application of ITGA5 agonists or recombinant
proteins may directly enhance endothelial cell survival signals,
which could be particularly suitable for patients with AS with high
miR-222-5p expression. Additionally, concurrent detection of serum
miR-222-5p and ITGA5 levels may optimize patient stratification,
providing a reference for the development of personalized treatment
regimens. Notably, the realization of these translational
applications requires further research support, including
large-scale clinical studies to clarify their diagnostic and
prognostic value, systematic evaluation of the safety and efficacy
of targeted drugs through animal models, and ultimately promotion
of the translation of basic research findings into clinical
applications.
Notably, the present study had certain limitations:
i) The in vitro experiments used only a single HUVEC line
and although it is a classic model for studying endothelial
function, umbilical vein endothelium differs from arterial
endothelium in anatomical location, physiological characteristics
and response to pathological stimuli. Therefore, the conclusions
drawn in the present study regarding the miR-222-5p/ITGA5 axis
regulating endothelial cell apoptosis require further validation in
arterial endothelial cells. ii) Although the findings of the
present study preliminarily supported the mediating role of ITGA5
in miR-222-5p regulation of endothelial cell apoptosis via ITGA5
knockdown experiments, ITGA5 overexpression rescue experiments were
not conducted, nor was the 3′UTR binding specificity between
miR-222-5p and ITGA5 directly validated through dual-luciferase
reporter gene experiments. To some extent this weakens the
discovery of miR-222-5p targeting and inhibiting ITGA5. iii) While
ITGA5 is known to exert its effects through pathways such as the
FAK/PI3K/Akt pathway (22), the
present study did not employ specific pathway inhibitors in rescue
experiments, meaning that the specific role of this pathway within
the regulatory axis was not clarified. Future studies should
perform other experiments at the signaling pathway level to
elucidate the specific mechanisms and pathway details underlying
the miR-222-5p-mediated regulation of ITGA5. iv) Finally, the
present study did not validate the in vivo effects of this
regulatory axis on AS plaque formation and progression in animal
models, which to some extent limits the physiological and
pathological significance of results, as well as their
translational value.
Furthermore, the present study distinguished
apoptosis from necrosis by analyzing the proportion of necrotic
cells, which is crucial for interpreting AS-related vascular cell
death. In certain cases, the proportion of cells in the Q1 quadrant
was higher than that in the Q2 (late apoptosis) or Q3 (early
apoptosis) quadrants. In the apoptosis analysis, the Q1 quadrant
represents necrotic cells (as PI can penetrate damaged cell
membranes, whereas Annexin V cannot bind to intact cell membranes
that have not undergone phosphatidylserine reversal). Elevated Q1
proportions in specific groups may be associated with the following
factors, consistent with the experimental context: i) Cytotoxic
effects of high-concentration ox-LDL: The present study employed
ox-LDL to establish an endothelial cell apoptosis model, which is
known to induce apoptosis and necrosis in a dose-dependent manner.
At higher ox-LDL concentrations, cumulative toxic effects may cause
acute necrosis in some cells, particularly affecting sensitive cell
subpopulations with inherently fragile membrane integrity. This
aligns with previous studies indicating that excessive ox-LDL can
trigger necrotic apoptosis or passive necrosis alongside inducing
endothelial cell apoptosis (53–56).
ii) Minor technical factors in sample processing: Gentle pipetting
was employed during cell collection and staining to minimize
mechanical damage. However, prolonged incubation or minor shear
force may still cause minor membrane rupture in a small number of
cells, potentially slightly increasing the proportion of the Q1
subpopulation. The inclusion of unstained and single-stained
controls ruled out fragment interference, ensuring the Q1
subpopulation primarily reflects genuine necrotic cells. iii)
Biological heterogeneity of cell populations: Even within the same
passage of HUVECs, subtle variations in response to stimuli exist.
Some cells may exhibit higher intrinsic sensitivity, making them
more prone to necrosis rather than apoptosis under ox-LDL
stimulation, a normal biological phenomenon within heterogeneous
cell populations.
The present study did not deny that miR-222-5p may
regulate cell phenotypes through other target genes, as the
multi-target nature of miRNAs is a common phenomenon (57). In the future, other potential
target genes may be identified through high-throughput screening
techniques, such as RNA-sequencing. In addition, luciferase
reporter gene experiments, using ITGA5 3′UTR wild-type and
binding-site mutant vectors, with co-transfection of a miR-222-5p
mimic to detect differences in fluorescence activity, are critical
for directly verifying the binding specificity between miR-222-5p
and ITGA5. While this experiment was not performed in the present
study, the conclusions regarding their regulatory relationship are
supported by complementary evidence, including consistent
bioinformatics predictions, inverse expression trends and
functional rescue assays. Regarding the negative regulation of
miR-222-5p on ITGA5, specifically, the expression levels of
miR-222-5p were detected in HUVECs treated with 100 mg/l ox-LDL via
RT-qPCR, and the expression level of ITGA5 were assessed using
western blotting and RT-qPCR. The results showed that miR-222-5p
expression was increased, while ITGA5 expression was decreased.
Similarly, miR-222-5p overexpression led to decreased ITGA5 mRNA
and protein levels, whereas miR-222-5p inhibition increased ITGA5
mRNA and protein expression. These findings indicated a negative
association between miR-222-5p and ITGA5 expression, supporting a
potential regulatory relationship between the two. This key
validation step will be addressed in follow-up studies to further
confirm the direct interaction. Furthermore, we aim to collect
clinical samples from patients with AS to detect the expression
levels of miR-222-5p and ITGA5 in serum and plaque tissue, to
analyze their association with disease severity and clinical
outcomes, and to elucidate their potential as biomarkers. Animal
models could also be generated to analyze and validate in
vivo regulatory effects, providing evidence for clinical
translation. The present study considers miR-222-5p to hold notable
promise as a biomarker for disease diagnosis, prognosis and
therapeutic intervention in AS, which may provide notable clinical
benefits.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Wu Jieping Medical
Foundation Scientific Research Special Grant Fund (grant no.
320.6750.2024-06-85).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
LH was involved in the conception and design of the
paper. SW wrote the paper and conducted the experiments. BZ and LH
confirm the authenticity of all the raw data. BZ, YC, LG and JF
carried out the data collection and analysis, as well as the
literature review and organization. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Guo Y, Lu C, Hu K, Cai C and Wang W:
Ferroptosis in cardiovascular diseases: Current status, challenges,
and future perspectives. Biomolecules. 12:3902022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bułdak Ł: Cardiovascular diseases-a focus
on atherosclerosis, its prophylaxis, complications and recent
advancements in therapies. Int J Mol Sci. 23:46952022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang R, Wang M, Ye J, Sun G and Sun X:
Mechanism overview and target mining of atherosclerosis:
Endothelial cell injury in atherosclerosis is regulated by
glycolysis (Review). Int J Mol Med. 47:65–76. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Duan H, Zhang Q, Liu J, Li R, Wang D, Peng
W and Wu C: Suppression of apoptosis in vascular endothelial cell,
the promising way for natural medicines to treat atherosclerosis.
Pharmacol Res. 168:1055992021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang Y, Yang Y, Zhang T, Jia S, Ma X,
Zhang M, Wang L and Ma A: LncRNA SNHG16 accelerates atherosclerosis
and promotes ox-LDL-induced VSMC growth via the miRNA-22-3p/HMGB2
axis. Eur J Pharmacol. 915:1746012022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pirillo A, Norata GD and Catapano AL:
LOX-1, OxLDL, and atherosclerosis. Mediators Inflamm.
2013:1527862013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu Y, Jiang G, Lv C and Yang C:
miR-222-5p promotes dysfunction of human vascular smooth muscle
cells by targeting RB1. Environ Toxicol. 37:683–694. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pan Z, Fan Z, Ma J, Liu H, Shen L, He B
and Zhang M: Profiling and functional characterization of
circulation LncRNAs that are associated with coronary
atherosclerotic plaque stability. Am J Transl Res. 11:3801–3815.
2019.PubMed/NCBI
|
|
9
|
Huang P: Potential new therapeutic
targets: Association of microRNA with atherosclerotic plaque
stability. Int J Immunopathol Pharmacol. 37:39463202311856572023.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Abulsoud AI, Elshaer SS, Rizk NI, Khaled
R, Abdelfatah AM, Aboelyazed AM, Waseem AM, Bashier D, Mohammed OA,
Elballal MS, et al: Unraveling the miRNA puzzle in atherosclerosis:
Revolutionizing diagnosis, prognosis, and therapeutic approaches.
Curr Atheroscler Rep. 26:395–410. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Piko N, Bevc S, Hojs R and Ekart R:
Atherosclerosis and epigenetic modifications in chronic kidney
disease. Nephron. 147:655–659. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gaál Z: Implication of microRNAs in
carcinogenesis with emphasis on hematological malignancies and
clinical translation. Int J Mol Sci. 23:58382022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Saetrom P, Snøve O Jr and Rossi JJ:
Epigenetics and microRNAs. Pediatr Res. 61((5 Pt 2)): 17R–23R.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang J, Xing Q, Zhou X, Li J, Li Y, Zhang
L, Zhou Q and Tang B: Circulating miRNA-21 is a promising biomarker
for heart failure. Mol Med Rep. 16:7766–7774. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen X, Cao Y, Guo Y, Liu J, Ye X, Li H,
Zhang L, Feng W, Xian S, Yang Z, et al: microRNA-125b-1-3p mediates
autophagy via the RRAGD/mTOR/ULK1 signaling pathway and mitigates
atherosclerosis progression. Cell Signal. 118:1111362024.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wei L, He Y, Bi S, Li X, Zhang J and Zhang
S: miRNA-199b-3p suppresses growth and progression of ovarian
cancer via the CHK1/E-cadherin/EMT signaling pathway by targeting
ZEB1. Oncol Rep. 45:569–581. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen Z, Zhong T, Zhong J, Tang Y, Ling B
and Wang L: MicroRNA-129 inhibits colorectal cancer cell
proliferation, invasion and epithelial-to-mesenchymal transition by
targeting SOX4. Oncol Rep. 45:612021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
He X, Liao S, Lu D, Zhang F, Sun Y and Wu
Y: MiR-125b promotes migration and invasion by targeting the
vitamin D receptor in renal cell carcinoma. Int J Med Sci.
18:150–156. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kuang X, Wei C, Zhang T, Yang Z, Chi J and
Wang L: miR-378 inhibits cell growth and enhances apoptosis in
human myelodysplastic syndromes. Int J Oncol. 49:1921–1930. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gorur A, Celik A, Yildirim DD, Gundes A
and Tamer L: Investigation of possible effects of microRNAs
involved in regulation of lipid metabolism in the pathogenesis of
atherosclerosis. Mol Biol Rep. 46:909–920. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu X, Shen L, Li W, Liu X, Yang P and Cai
J: ITGA5 promotes tumor angiogenesis in cervical cancer. Cancer
Med. 12:11983–11999. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang C, Yu Z, Yang S, Liu Y, Song J, Mao
J, Li M and Zhao Y: ZNF460-mediated circRPPH1 promotes TNBC
progression through ITGA5-induced FAK/PI3K/AKT activation in a
ceRNA manner. Mol Cancer. 23:332024. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen Z, Chen CZ, Gong WR, Li JP and Xing
YQ: Integrin-alpha5 mediates epidermal growth factor-induced
retinal pigment epithelial cell proliferation and migration.
Pathobiology. 77:88–95. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Deng Y, Wan Q and Yan W: Integrin α5/ITGA5
promotes the proliferation, migration, invasion and progression of
oral squamous carcinoma by epithelial-mesenchymal transition.
Cancer Manag Res. 11:9609–9620. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang X, Mao W and Ma X: TLN1 synergizes
with ITGA5 to ameliorate cardiac microvascular endothelial cell
dysfunction. Folia Morphol (Warsz). 83:92–101. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Falk E: Pathogenesis of atherosclerosis. J
Am Coll Cardiol. 47 (8 Suppl):C7–C12. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hansson GK and Hermansson A: The immune
system in atherosclerosis. Nat Immunol. 12:204–212. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cheng J, Huang H, Chen Y and Wu R:
Nanomedicine for diagnosis and treatment of atherosclerosis. Adv
Sci (Weinh). 10:e23042942023. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bian W, Jing X, Yang Z, Shi Z, Chen R, Xu
A, Wang N, Jiang J, Yang C, Zhang D, et al: Downregulation of
LncRNA NORAD promotes Ox-LDL-induced vascular endothelial cell
injury and atherosclerosis. Aging (Albany NY). 12:6385–6400. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lin F, Yang Y, Wei S, Huang X, Peng Z, Ke
X, Zeng Z and Song Y: Hydrogen sulfide protects against high
glucose-induced human umbilical vein endothelial cell injury
through activating PI3K/Akt/eNOS pathway. Drug Des Devel Ther.
14:621–633. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zheng D, Liu J, Piao H, Zhu Z, Wei R and
Liu K: ROS-triggered endothelial cell death mechanisms: Focus on
pyroptosis, parthanatos, and ferroptosis. Front Immunol.
13:10392412022. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mao J, Yang R, Yuan P, Wu F, Wei Y, Nie Y,
Zhang C and Zhou X: Different stimuli induce endothelial
dysfunction and promote atherosclerosis through the Piezo1/YAP
signaling axis. Arch Biochem Biophys. 747:1097552023. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Winn RK and Harlan JM: The role of
endothelial cell apoptosis in inflammatory and immune diseases. J
Thromb Haemost. 3:1815–1824. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sessa F, Salerno M, Esposito M, Cocimano G
and Pomara C: miRNA dysregulation in cardiovascular diseases:
Current opinion and future perspectives. Int J Mol Sci.
24:51922023. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lozano-Velasco E, Inácio JM, Sousa I,
Guimarães AR, Franco D, Moura G and Belo JA: miRNAs in heart
development and disease. Int J Mol Sci. 25:16732024. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Madrigal-Matute J, Rotllan N, Aranda JF
and Fernández-Hernando C: MicroRNAs and atherosclerosis. Curr
Atheroscler Rep. 15:3222013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhu J, Liu B, Wang Z, Wang D, Ni H, Zhang
L and Wang Y: Exosomes from nicotine-stimulated macrophages
accelerate atherosclerosis through miR-21-3p/PTEN-mediated VSMC
migration and proliferation. Theranostics. 9:6901–6919. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tang Y, Yang LJ, Liu H, Song YJ, Yang QQ,
Liu Y, Qian SW and Tang QQ: Exosomal miR-27b-3p secreted by
visceral adipocytes contributes to endothelial inflammation and
atherogenesis. Cell Rep. 42:1119482023. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang F, Ge J, Huang S, Zhou C, Sun Z, Song
Y, Xu Y and Ji Y: KLF5/LINC00346/miR-148a-3p axis regulates
inflammation and endothelial cell injury in atherosclerosis. Int J
Mol Med. 48:1522021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Peng N, Meng N, Wang S, Zhao F, Zhao J, Su
L, Zhang S, Zhang Y, Zhao B and Miao J: An activator of mTOR
inhibits oxLDL-induced autophagy and apoptosis in vascular
endothelial cells and restricts atherosclerosis in apolipoprotein
E−/− mice. Sci Rep. 4:55192014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jensen HA and Mehta JL: Endothelial cell
dysfunction as a novel therapeutic target in atherosclerosis.
Expert Rev Cardiovasc Ther. 14:1021–1033. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Suciu CF, Prete M, Ruscitti P, Favoino E,
Giacomelli R and Perosa F: Oxidized low density lipoproteins: The
bridge between atherosclerosis and autoimmunity. Possible
implications in accelerated atherosclerosis and for immune
intervention in autoimmune rheumatic disorders. Autoimmun Rev.
17:366–375. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yang K, Zhang H, Luo Y, Zhang J, Wang M,
Liao P, Cao L, Guo P, Sun G and Sun X: Gypenoside XVII prevents
atherosclerosis by attenuating endothelial apoptosis and oxidative
stress: Insight into the ERα-Mediated PI3K/Akt pathway. Int J Mol
Sci. 18:772017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ali Syeda Z, Langden SSS, Munkhzul C, Lee
M and Song SJ: Regulatory mechanism of MicroRNA expression in
cancer. Int J Mol Sci. 21:17232020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sun B, Ding B, Chen Y, Peng C and Chen X:
AFAP1L1 promotes gastric cancer progression by interacting with
VAV2 to facilitate CDC42-mediated activation of ITGA5 signaling
pathway. J Transl Med. 21:182023. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xiao Y, Tao P, Zhang K, Chen L, Lv J, Chen
Z, He L, Jia H, Sun J, Cao M, et al: Myofibroblast-derived
extracellular vesicles facilitate cancer stemness of hepatocellular
carcinoma via transferring ITGA5 to tumor cells. Mol Cancer.
23:2622024. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Desgrosellier JS and Cheresh DA: Integrins
in cancer: Biological implications and therapeutic opportunities.
Nat Rev Cancer. 10:9–22. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Elmoselhi AB, Seif Allah M, Bouzid A,
Ibrahim Z, Venkatachalam T, Siddiqui R, Khan NA and Hamoudi RA:
Circulating microRNAs as potential biomarkers of early vascular
damage in vitamin D deficiency, obese, and diabetic patients. PLoS
One. 18:e02836082023. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Mir R, Elfaki I, Khullar N, Waza AA, Jha
C, Mir MM, Nisa S, Mohammad B, Mir TA, Maqbool M, et al: Role of
selected miRNAs as diagnostic and prognostic biomarkers in
cardiovascular diseases, including coronary artery disease,
myocardial infarction and atherosclerosis. J Cardiovasc Dev Dis.
8:222021.PubMed/NCBI
|
|
51
|
Su X, Nie M, Zhang G and Wang B: MicroRNA
in cardio-metabolic disorders. Clin Chim Acta. 518:134–141. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Xiang Y, Mao L, Zuo ML, Song GL, Tan LM
and Yang ZB: The role of MicroRNAs in hyperlipidemia: From
pathogenesis to therapeutical application. Mediators Inflamm.
2022:31019002022. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Lin F, Pei L, Zhang Q, Han W, Jiang S, Lin
Y, Dong B, Cui L and Li M: Ox-LDL induces endothelial cell
apoptosis and macrophage migration by regulating caveolin-1
phosphorylation. J Cell Physiol. 233:6683–6692. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Colles SM, Maxson JM, Carlson SG and
Chisolm GM: Oxidized LDL-induced injury and apoptosis in
atherosclerosis. Potential roles for oxysterols. Trends Cardiovasc
Med. 11:131–138. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Escargueil-Blanc I, Meilhac O, Pieraggi
MT, Arnal JF, Salvayre R and Nègre-Salvayre A: Oxidized LDLs induce
massive apoptosis of cultured human endothelial cells through a
calcium-dependent pathway. Prevention by aurintricarboxylic acid.
Arterioscler Thromb Vasc Biol. 17:331–339. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhang YZ, Wang L, Zhang JJ, Xiong XM,
Zhang D, Tang XM, Luo XJ, Ma QL and Peng J: Vascular peroxide 1
promotes ox-LDL-induced programmed necrosis in endothelial cells
through a mechanism involving β-catenin signaling. Atherosclerosis.
274:128–138. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Tolouei S, Curi TZ, Klider LM and Junior
AG: MicroRNA-30 and 145 as targets for the treatment of
cardiovascular diseases: Therapeutic feasibility and challenges.
Curr Pharm Des. 27:3858–3870. 2021. View Article : Google Scholar : PubMed/NCBI
|