Introduction
Systemic lupus erythematosus (SLE) is a chronic,
multisystem autoimmune disease, and renal involvement, which is
referred to as lupus nephritis (LN), occurs in >60% of patients
with SLE (1–3). LN is among the most severe
complications of SLE; if uncontrolled, it can progress to renal
failure and remains a leading cause of mortality in patients with
SLE (4–6). Therefore, elucidating the pathogenic
mechanisms of LN has important clinical significance.
Traditionally, research on LN has focused primarily
on glomerular injury, but growing evidence indicates that
tubulointerstitial pathology plays a key role in both the onset and
progression of LN (7).
Tubulointerstitial lesions are more notably associated with LN
severity than glomerular fibrosis, and markers of tubular injury
(such as tubular proteinuria) often precede microalbuminuria
(8–12). In addition, severe
tubulointerstitial damage has been identified as a major risk
factor for LN progression (13,14).
Mechanistically, epithelial-mesenchymal transition of renal tubular
epithelial cells promotes extracellular matrix deposition and
fibrosis, while infiltration of immune cells (such as macrophages
and lymphocytes) and release of proinflammatory mediators
exacerbate tubular and interstitial injury (14–16).
Multiple signaling pathways, including NF-κB, TGF-β and
Wnt/β-catenin, have been implicated in tubulointerstitial lesion
regulation in LN, underscoring the complex and multifactorial
immune-inflammatory mechanisms involved (17,18).
Fos-related antigen 1 (FRA1) is a member of the
activator protein 1 (AP-1) transcription factor family, which
comprises Fos-family proteins (c-Fos, FosB, FRA1 and FRA2) that
dimerize with Jun-family proteins (19–21).
AP-1 regulates key processes such as cell proliferation,
differentiation and inflammatory responses, and controls cytokine
expression in various immune and inflammatory disorders (22). FRA1 has been implicated not only in
tumorigenesis but also in modulation of inflammatory and autoimmune
diseases, including arthritis, pneumonia, psoriasis, myasthenia
gravis and cardiovascular disorders (22). In immune cells, FRA1 influences
B-cell fate; for example, FRA1 is upregulated in activated B cells
and negatively regulates follicular B-cell differentiation into
plasma cells by suppressing the expression of the key transcription
factor Blimp-1, thereby limiting antibody production (23). In epithelial cells, Li et al
(24) reported that FRA1 disrupts
inflammatory cytokine secretion by medullary thymic epithelial
cells. Thus, the role of FRA1 in immune regulation has received
increasing attention. Promoter regions of numerous inflammatory
cytokines and chemokines, such as TNF-α, IL-1β, IL-11, IL-8 and
MCP-1, contain AP-1 binding sites, suggesting that FRA1 may
directly regulate their expression (22).
In the context of renal injury, FRA1 also exerts
notable functions; for example, in an acute kidney injury model,
FRA1 expression is upregulated in proximal tubular cells and
mitigates tubular cell damage and inflammation by maintaining
expression of the anti-aging protein Klotho (25). Moreover, a gene-screening study in
IgA nephropathy identified FRA1/2 as novel prognostic biomarkers,
implicating TNF and MAPK signaling pathways in disease progression
(26). Notably, METTL3 has been
shown to exacerbate renal inflammation by enhancing m6A
modification of FRA1 transcripts (27). Nevertheless, the role of FRA1 in LN
and other autoimmune kidney diseases remains unexplored, and its
functions and molecular mechanisms in LN-associated
tubulointerstitial injury remain unclear.
Based on these observations, bioinformatic analysis
of public gene expression datasets was performed to identify FRA1
as a potential key candidate in LN. The present study aimed to
investigate the role of FRA1 in the pathogenesis of LN and to
elucidate its underlying molecular mechanisms. The bioinformatics
analyses were combined with validation in the MRL/lpr mouse model
and in vitro experiments using HK-2 cells to systematically
characterize FRA1 expression patterns and its regulatory effects on
the inflammatory cytokine network in LN. By constructing FRA1
overexpression and knockdown cell models, the impact of FRA1 on
tubular epithelial cell function and inflammatory responses, was
further examined aiming to provide new insights into LN
pathogenesis and to identify potential therapeutic targets.
Materials and methods
Data sources
Gene expression data for renal tubulointerstitial
samples from patients with LN and healthy controls were obtained
from the gene expression omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). A total of
three datasets were included: GSE113342 (47 LN and 10 control
tubulointerstitial samples), GSE200306 (14 LN and 10 control
samples) and GSE127797 (44 LN samples) (28–30).
Probe identifiers were annotated to gene symbols using the platform
annotation file; for genes represented by multiple probes, one
probe was randomly selected to avoid redundancy. Background
correction and normalization were performed with the ‘limma’
package (v3.6.2) (31) in R
(v4.2.1; RStudio, Inc.) to ensure data quality and comparability,
yielding the final gene expression matrices. All analyses adhered
to the principles of The Declaration of Helsinki (2013 revision)
(32).
Identification of differentially
expressed genes (DEGs)
Differential expression analysis between LN and
control tubulointerstitial samples was conducted separately for
GSE113342 and GSE200306 using the ‘limma’ package (31). Genes with |log2fold
change| (|log2FC|)>1 and adjusted P<0.05 were
defined as DEGs. The intersection of DEGs from both datasets was
taken to identify consistently dysregulated genes, and results were
visualized using ggplot2 (v3.3.6; http://ggplot2.tidyverse.org/) and VennDiagram
(v1.7.3).
Functional enrichment analysis
To explore the potential biological roles of DEGs,
gene identifiers were converted using the org.Hs.eg.db package
(v3.22; http://bioconductor.org/packages/org.Hs.eg.db), and
Gene Ontology (GO) (https://geneontology.org/) was used to determine
relevant biological processes (BPs) and molecular functions (MFs)
and Kyoto Encyclopedia of Genes and Genomes (KEGG) (www.genome.jp/kegg/) for pathway enrichment analyses
were performed with clusterProfiler, with adjusted P<0.05
considered significant (33).
Enrichment results were visualized using ggplot2 (v3.3.6), igraph
(v2.2.1; http://r.igraph.org/) and ggraph
(v2.2.2; http://ggraph.data-imaginist.com/).
Relationship between FRA1 and
immunity
To assess the association between FRA1 expression
and immune cell infiltration in LN tubulointerstitial samples, the
CIBERSORT algorithm was applied, which is a linear support vector
regression-based deconvolution tool that estimates immune cell
proportions from microarray or RNA-seq data (34). In GSE127797, LN samples were
stratified into high- and low-FRA1 expression groups based on the
median, and only samples with CIBERSORT P<0.05 were retained for
subsequent analyses. The correlations between FRA1 expression and
infiltrating immune cell fractions and correlations between FRA1
and tubular epithelial cell-related cytokines were examined; all
visualizations were generated with ggplot2 (v3.3.6).
Animal experiments in MRL/lpr
mice
A total of three female MRL/lpr mice and three
female MRL/MPJ control mice (age, 16 weeks; body weight, 38–46 g)
were obtained from Cavens Biogle (Suzhou) Model Animal Research Co.
Ltd. Mice were maintained under standard housing conditions, which
included a climate-controlled environment at 22±2°C and 50±10%
relative humidity, a 12-h light/dark cycle and free access to food
and water, for 4 weeks and then euthanized for kidney tissue
collection to assess protein expression. All procedures complied
with the AVMA Guidelines for the Euthanasia of Animals (2020) and
approved institutional animal care protocols. Specific humane
endpoints included: Weight loss ≥20% of body weight; inability to
eat or drink; severe dehydration; severe respiratory distress,
paralysis or continuous convulsions; uncontrolled progressive
infection; or unrelievable pain or clinical signs severely
compromised quality of life. Health and behaviour were routinely
monitored, with weekly weighing and observation after enrolment,
and immediate escalation to daily or more frequent monitoring if
abnormalities arose. Mice were deeply anesthetized with
pentobarbital sodium (150–200 mg/kg, intraperitoneally) until loss
of the pedal withdrawal reflex, followed by cervical dislocation to
ensure complete euthanasia. Death was confirmed by the absence of
heartbeat and respiration, dilated pupils and lack of reflex
response. No experimental intervention was performed; MRL/lpr mice,
which spontaneously develop LN (35), were used as the disease model,
whereas MRL/MPJ mice served as healthy controls and were used
solely for terminal tissue collection. The power calculation for
the mouse experiments is shown in Table SI, following the methodology
described by Festing and Altman (36).
Immunohistochemistry (IHC)
Kidney tissues were fixed in 10% neutral-buffered
formalin for 24–48 h at room temperature and were then processed
for paraffin embedding. Paraffin-embedded tissues were cut into
5-µm sections. The sections were dewaxed twice in xylene (15 min
each), rehydrated through a descending ethanol series (100% ethanol
twice, then 95, 85 and 75% ethanol, 5 min each), and rinsed in
phosphate-buffered saline (PBS, pH 7.4) three times for 5 min each.
Antigen retrieval was performed in citrate buffer (pH 6.0) in a
microwave: Medium power for 8 min, rest for 8 min, then low power
for 7 min. Subsequently, the slides were allowed to cool to room
temperature for 20–40 min and then rinsed in PBS (three times, 5
min each). Endogenous peroxidase activity was quenched by
incubation in 3% hydrogen peroxide at room temperature in the dark
for 25 min, followed by three PBS washes (5 min each). Blocking was
performed using the normal goat serum blocking solution supplied
with the Vectastain ABC Elite HRP Kit (cat. no. SAP-9100; Beijing
Zhongshan Jinqiao Biotechnology Co., Ltd.) according to the
manufacturer's instructions (incubation at room temperature for
10–15 min). For primary antibody incubation, the sections were
incubated with rabbit polyclonal anti-FRA1 (cat. no. A5372;
ABclonal Biotech Co., Ltd.) at a dilution of 1:100 (diluted in PBS)
and incubated overnight at 4°C. After three PBS washes (5 min
each), the sections were incubated with the biotinylated goat
anti-rabbit IgG provided in the Vectastain ABC Elite HRP Kit at a
working dilution of 1:500 for 10–15 min at 37°C, followed by PBS
rinses (3×3 min). The HRP-labeled streptavidin working solution
(provided in the Vectastain ABC Elite HRP Kit) was applied and
incubated for 10–15 min at 37°C according to the manufacturer's
instructions, followed by PBS washes (3×3 min). Chromogenic
detection was performed with DAB substrate (cat. no. ZLI-9017;
Beijing Zhongshan Jinqiao Biotechnology Co., Ltd.); color
development was monitored under the microscope and stopped with
distilled water once an optimal signal was achieved. The sections
were then counterstained with hematoxylin at room temperature for
1–2 min, differentiated in 1% hydrochloric acid-ethanol (~1 sec),
rinsed in running tap water, blued in ammonia water and rinsed
again. Finally, the slides were dehydrated through an ascending
ethanol series (75, 85, 95 and 100%), cleared in xylene and mounted
with neutral resin. The percentage of FRA1-positive area/unit
tissue area was quantified using ImageJ (v2.3.0; National
Institutes of Health). All stained sections were examined and
images were captured using an Olympus BX40 upright light microscope
(Olympus Corporation).
Culture of HK-2 cells and retroviral
infection
HK-2 cells (Pricella; Elabscience Bionovation Inc.)
were maintained in DMEM/F12 (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% heat-inactivated fetal bovine serum (Beijing
Baiao Leibo Technology Co., Ltd.). A total of four groups were
established: FRA1 overexpression (FRA1-OE), FRA1 knockdown
(FRA1-shRNA) and their respective empty-vector controls (control-OE
and control-shRNA). The FRA1 overexpression vector
(pCDH_CMV_MCS_EF1_copGFP-FRA1) and shRNA vector
(pMAGic7.1-FRA1-shRNA) were constructed at the Clinical Laboratory,
Boai Hospital of Zhongshan (Zhongshan, China); plasmid maps and
sequences are provided in Fig. S1
and Table SII. For retroviral
packaging, 293T cells (Pricella; Elabscience Bionovation Inc.) were
co-transfected with 4 µg total plasmids using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37°C for 48 h. Specifically, the packaging
system consisted of the packaging plasmid (psPAX2), the envelope
plasmid (pMD2.G) and one of the four aforementioned transfer
plasmids (FRA1-OE, FRA1-shRNA, control-OE or control-shRNA) at a
mass ratio of 4:3:2. For gene manipulation, 2×105 HK-2
cells were infected with retroviral particles [2×107
transducing units/ml; multiplicity of infection=30; supplemented
with 6 µg/ml polybrene (MedChemExpress); infection efficiency
≥85%]. The infection was performed at 37°C for 24 h, after which
the medium was replaced with fresh complete medium. For the
FRA1-shRNA and control-shRNA groups, stably transduced cells were
selected using 3.0 µg/ml puromycin (MedChemExpress) starting 48 h
post-infection for 4 days. No additional selection was performed
for the FRA1-OE and control-OE groups, as the high transduction
efficiency was sufficient for subsequent experiments. All groups
were harvested for further analysis at 144 h post-transduction.
Western blotting
Kidney tissues or HK-2 cells were homogenized in
RIPA buffer (Biosharp Life Sciences), and protein concentrations
were determined using a BCA Protein Assay Kit (Wuhan Servicebio
Technology Co., Ltd.). For HK-2 experiments, cells were harvested
at 144 h post-transduction to allow time for lentiviral reverse
transcription, genomic integration and establishment of stable
transgene expression and for accumulation of downstream protein and
secreted cytokine changes. Lentiviral-mediated expression commonly
requires on the order of 5–7 days to reach stable levels, and
molecular-kinetic studies indicate that reverse transcription and
integration occur during the first few days post-transduction (~3
days) (37,38). Prior studies have documented
progressively increased reporter expression at 48-, 96- and 144-h
post-transduction, and have selected 144 h post-transduction as the
endpoint for subsequent mRNA and protein analyses of FRA1 effects
(24,39,40);
inflammatory mediators in renal cell models are also commonly
measured at ~5 days or later following perturbation (41). Equal amounts of protein (40
µg/lane) were separated by SDS-PAGE on 10% gels and were
transferred to PVDF membranes (MilliporeSigma). Protein molecular
weight markers (10–180 kDa; cat. no. 20350ES90; Shanghai Yeasen
Biotechnology Co., Ltd.) were run alongside samples to confirm
target sizes. Membranes were blocked in 5% skim milk at 37°C for 1
h, incubated with primary antibodies at 4°C overnight, then washed
with TBS-0.1% Tween-20 (TBST). The primary antibodies used in the
present study included: β-actin (1:5,000; cat. no. bs-0061R;
BIOSS), FRA1 (1:1,000; cat. no. A5372; ABclonal Biotech Co., Ltd.),
IL-1β (1:1,000; cat. no. HA601002; clone no. A7F9; HUABIO), IL-8
(1:1,000; cat. no. ab289967; clone no. EPR26511-74; Abcam), RANTES
(1:1,000; cat. no. 36467; CST Biological Reagents Co., Ltd.), IL-6
(1:2,000; cat. no. DF6087; Affinity Biosciences, Ltd.), MCP-1
(1:2,000; cat. no. A7277; ABclonal Biotech Co., Ltd.), TNF-α
(1:2,000; cat. no. 17590-1-AP; Proteintech Group, Inc.), and TGF-β
(1:2,000; cat. no. 81746-2-RR; clone no. 230544B7; Proteintech
Group, Inc.). The membranes were then incubated with HRP-conjugated
secondary antibodies at a 1:50,000 dilution for 30 min at room
temperature. The secondary antibodies use dincluded HRP-Goat anti
Rabbit (cat. no. 5220-0336) and HRP-Goat anti Mouse (cat. no.
5220-0341; SeraCare; LGC Clinical Diagnostics). After further
washing with TBST, bands were visualized using an ECL
Chemiluminescence Detection Kit (Dalian Meilun Biology Technology
Co., Ltd.) with the JS-1070P imaging system (Shanghai Peiqing
Technology Co., Ltd.) and densitometric analysis was performed with
IPWIN60 software (v6.0; Media Cybernetics, Inc.).
Statistical analysis
All analyses were performed in R (v4.0.3). For
two-group comparisons of continuous variables the following tests
were used: Unpaired Student's t-test for normally distributed data
with equal variances; Welch's t-test for normal data with unequal
variances; and the Wilcoxon rank-sum test for non-normal data. For
multi-group comparisons, one-way ANOVA followed by Tukey's Honestly
Significant Difference was applied to normal data with equal
variances and the Kruskal-Wallis test followed by Dunn's post hoc
test was applied for non-normal data. Pearson's correlation was
used for normally distributed variables, and Spearman's correlation
otherwise. Receiver operating characteristic curve analysis was
employed to evaluate the predictive performance of upregulated DEGs
for the disease. P<0.05 was considered to indicate a
statistically significant difference.
Results
Identification of DEGs in LN
tubulointerstitium
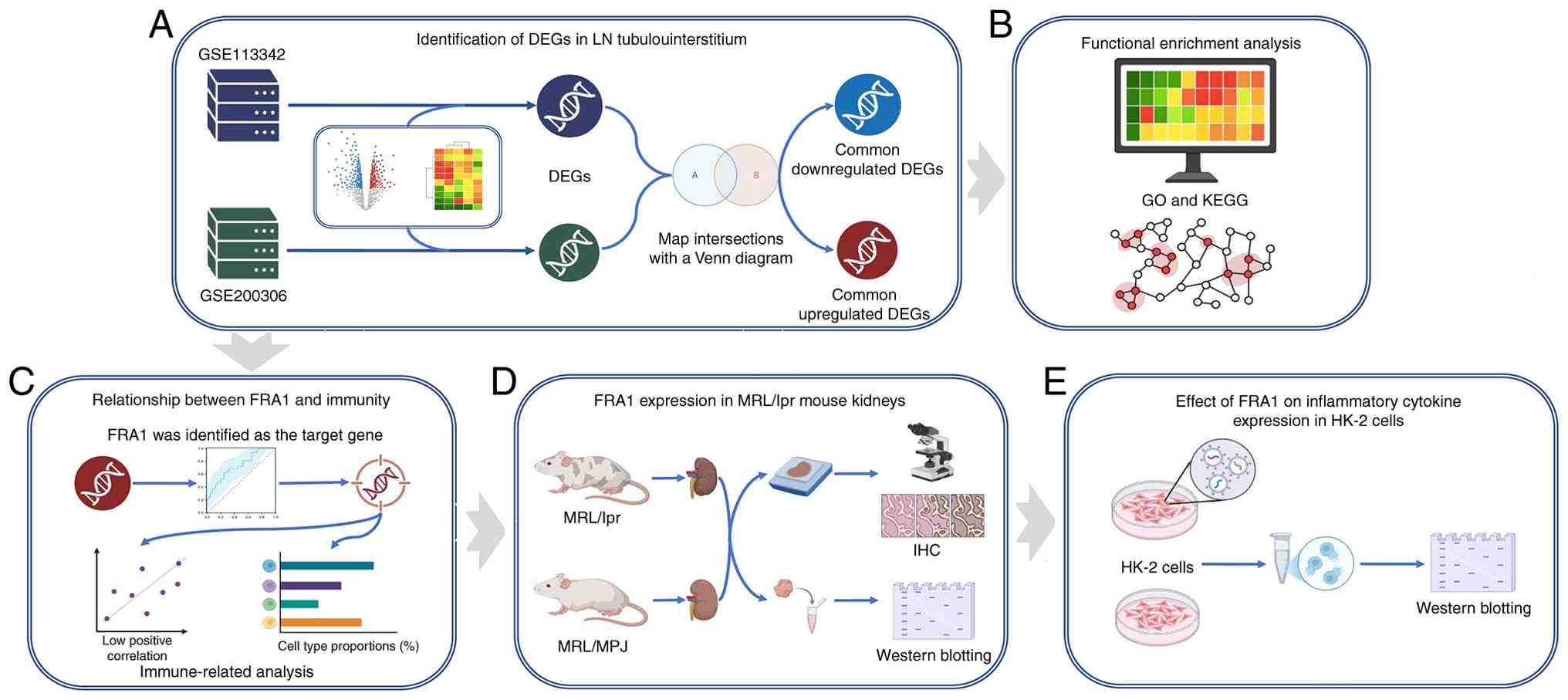
Design of the present study is illustrated in
Fig. 1. To determine gene
expression differences between LN and healthy controls
tubulointerstitial samples, the datasets GSE113342 and GSE200306
from GEO were analyzed. Using |log2FC|>1 and adjusted
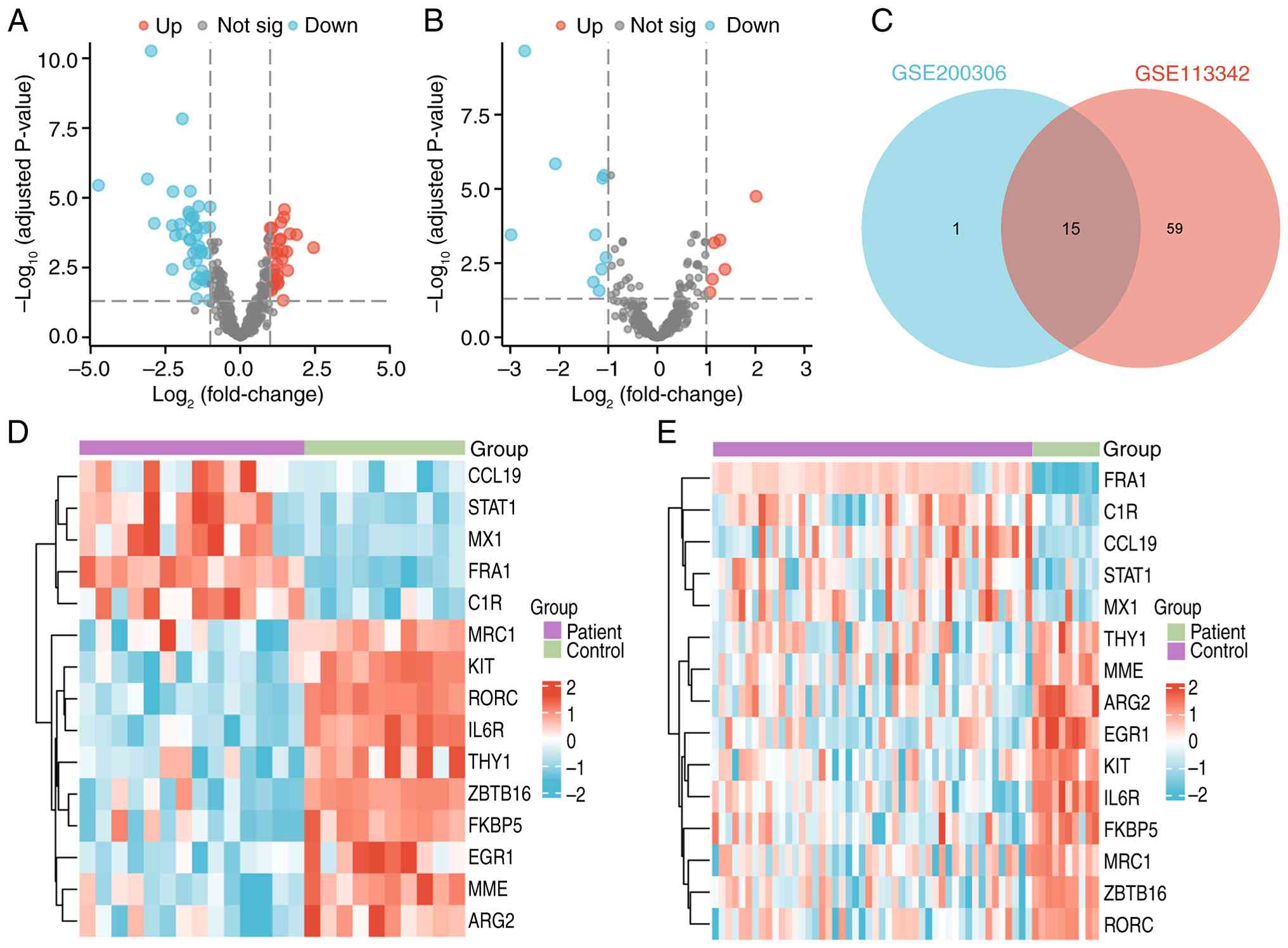
P<0.05, 74 DEGs were identified in GSE113342 (Fig. 2A) and 16 DEGs were identified in
GSE200306 (Fig. 2B). The
intersection of these yielded 15 common DEGs (Fig. 2C), of which FRA1, C1R, CCL19, STAT1
and MX1 were upregulated in LN, whereas THY1, MME, ARG2, EGR1, KIT,
IL6R, FKBP5, MRC1, ZBTB1 and RORC were downregulated. A heatmap
illustrates their expression patterns across both datasets
(Fig. 2D and E).
Functional enrichment of DEGs
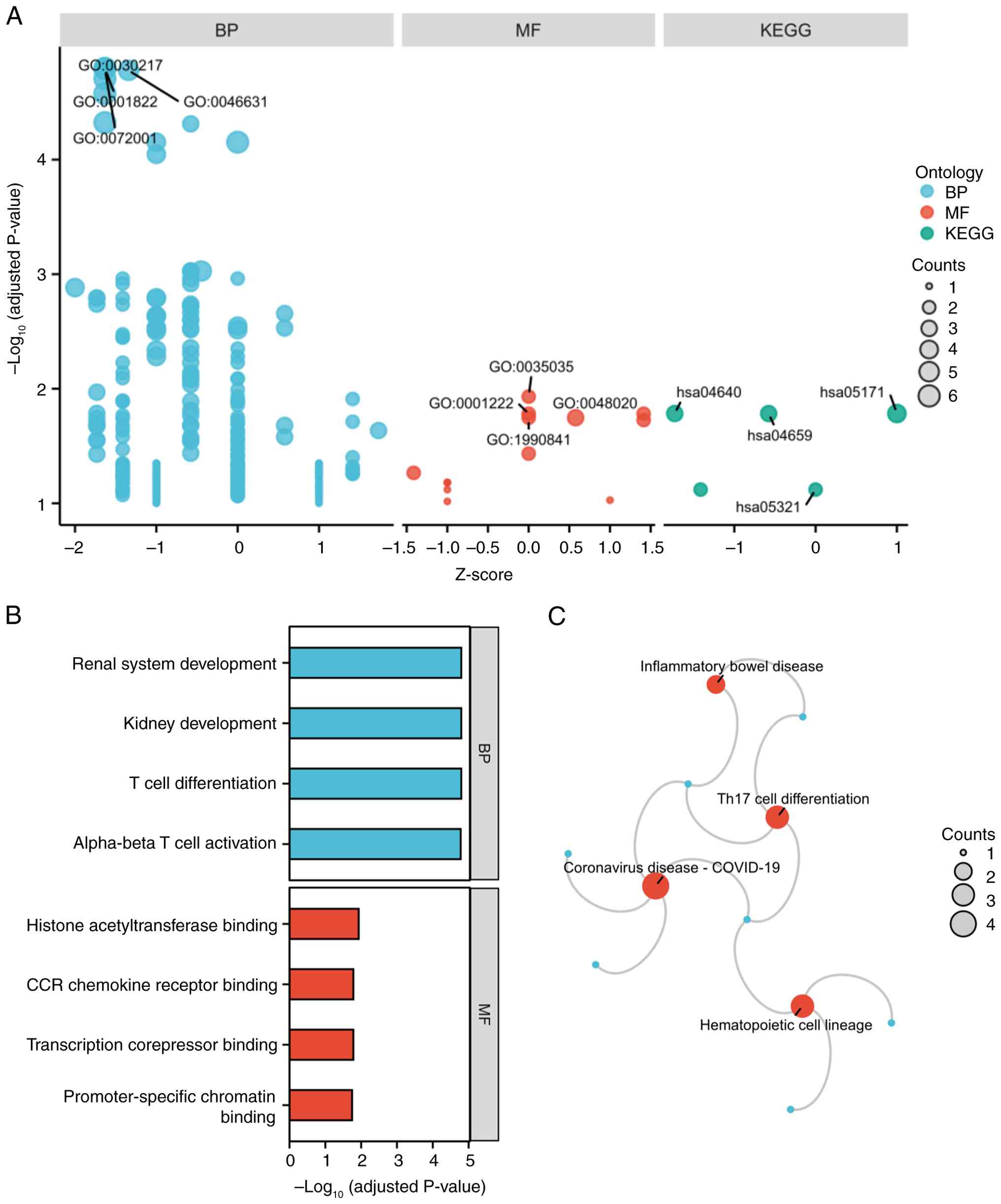
GO and KEGG enrichment analyses of the 15 shared
DEGs revealed significant associations with immune and
developmental processes (Fig. 3A).
In GO-BP, DEGs were enriched in ‘T cell differentiation’, ‘kidney
development’, ‘renal system development’ and ‘alpha-beta T cell
activation’ (Fig. 3B). In GO-MF,
terms included ‘histone acetyltransferase binding’, ‘transcription
corepressor binding’, ‘CCR chemokine receptor binding’,
‘promoter-specific chromatin binding’ (Fig. 3B). KEGG pathways highlighted
‘Coronavirus disease-COVID-19’, ‘Hematopoietic cell lineage’, ‘Th17
cell differentiation’ and ‘Inflammatory bowel disease’ as pathways
significantly associated with the DEGs (Fig. 3C).
Association between FRA1 and immune
cell infiltration
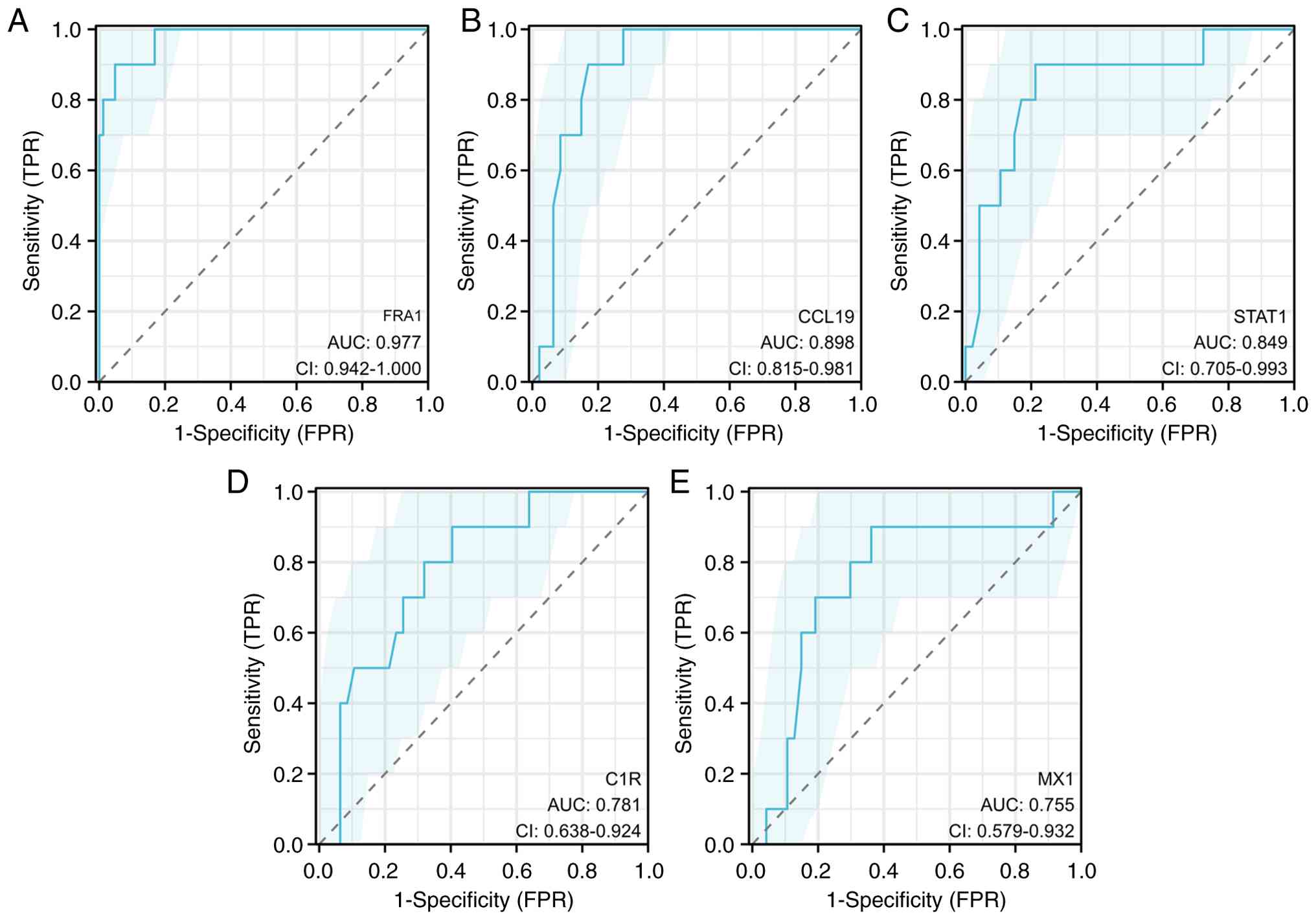
To prioritize candidates for functional follow-up,
the diagnostic performance of five upregulated DEGs was assessed,
and FRA1 was identified as the top performer (AUC=0.977; Fig. 4); therefore, downstream analyses
focused on FRA1. In addition, further analysis of the datasets
showed that the trend of FRA1 remained stable across different
subgroups and after exclusion of outliers, indicating that the
upregulation of FRA1 in LN is robust and minimally affected by
dataset heterogeneity (Fig. S2).
Next, the relationship between FRA1 expression and immune cell
infiltration in LN tubulointerstitial samples was explored using
CIBERSORT on dataset GSE127797.
The results showed statistically increased
proportions of CD8+ T cells, regulatory T cells (Tregs),
monocytes, M1 macrophages and activated mast cells in the high-FRA1
group, whereas plasma cells, resting CD4+ memory T
cells, M0/M2 macrophages, resting dendritic cells and resting mast
cells were reduced (Fig. 5A-C).
Correlation analyses further confirmed these associations.
Specifically, Fig. 5D illustrates
that FRA1 expression was significantly and positively correlated
with several immune cell types, most notably naive CD4+
T cells, monocytes and Tregs. Conversely, FRA1 showed strong
negative correlations with M0/M2 macrophages, resting
CD4+ memory T cells and plasma cells. Furthermore, FRA1
expression was positively correlated with key tubular
epithelial-related cytokines, including IL-6, IL-1β, IL-8, MCP-1,
RANTES, TGF-β and TNF (Fig. 5E-K).
Collectively, these findings underscore the diagnostic potential of
FRA1 among upregulated DEGs and also suggest a key immunomodulatory
role for FRA1 in the tubulointerstitial microenvironment of LN.
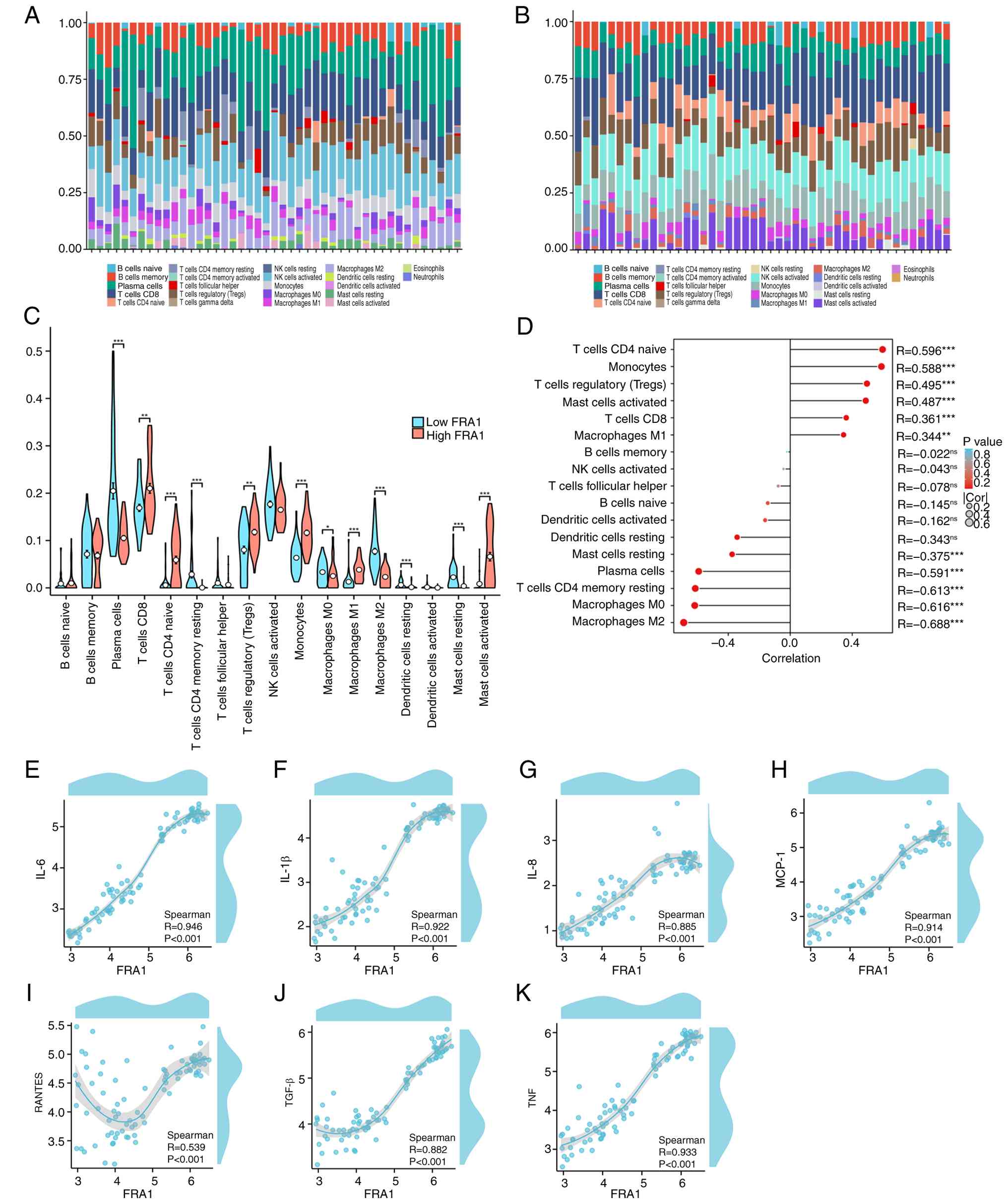 | Figure 5.Association between FRA1 and immune
cell infiltration. CIBERSORT-estimated immune cell proportions in
each sample stratified by FRA1 expression (median split): (A)
Low-Fra1 group; (B) high-Fra1 group. (C) Bar plots comparing the
proportions of immune cell types between high- and low-FRA1 groups.
Cells significantly increased in the high-FRA1 group included
CD8+ T cells, T regulatory cells, monocytes, M1
macrophages and activated mast cells; cells reduced in the
high-FRA1 group included plasma cells, resting CD4+
memory T cells, M0/M2 macrophages, resting dendritic cells and
resting mast cells (mean ± SEM; n=44; group comparisons were
performed using two-sided Wilcoxon rank-sum tests; *P<0.05;
**P<0.01, ***P<0.001). (D) Spearman correlation analyses
showing associations between FRA1 expression and immune cell
fractions, as well as tubular epithelial-related cytokines,
including (E) IL-6, (F) IL-1β, (G) IL-8, (H) MCP-1 (CCL2), (I)
RANTES (CCL5), (J) TGF-β and (K) TNF. All correlation tests are
two-sided Spearman rank tests; *P<0.05, **P<0.01 and
***P<0.001. |
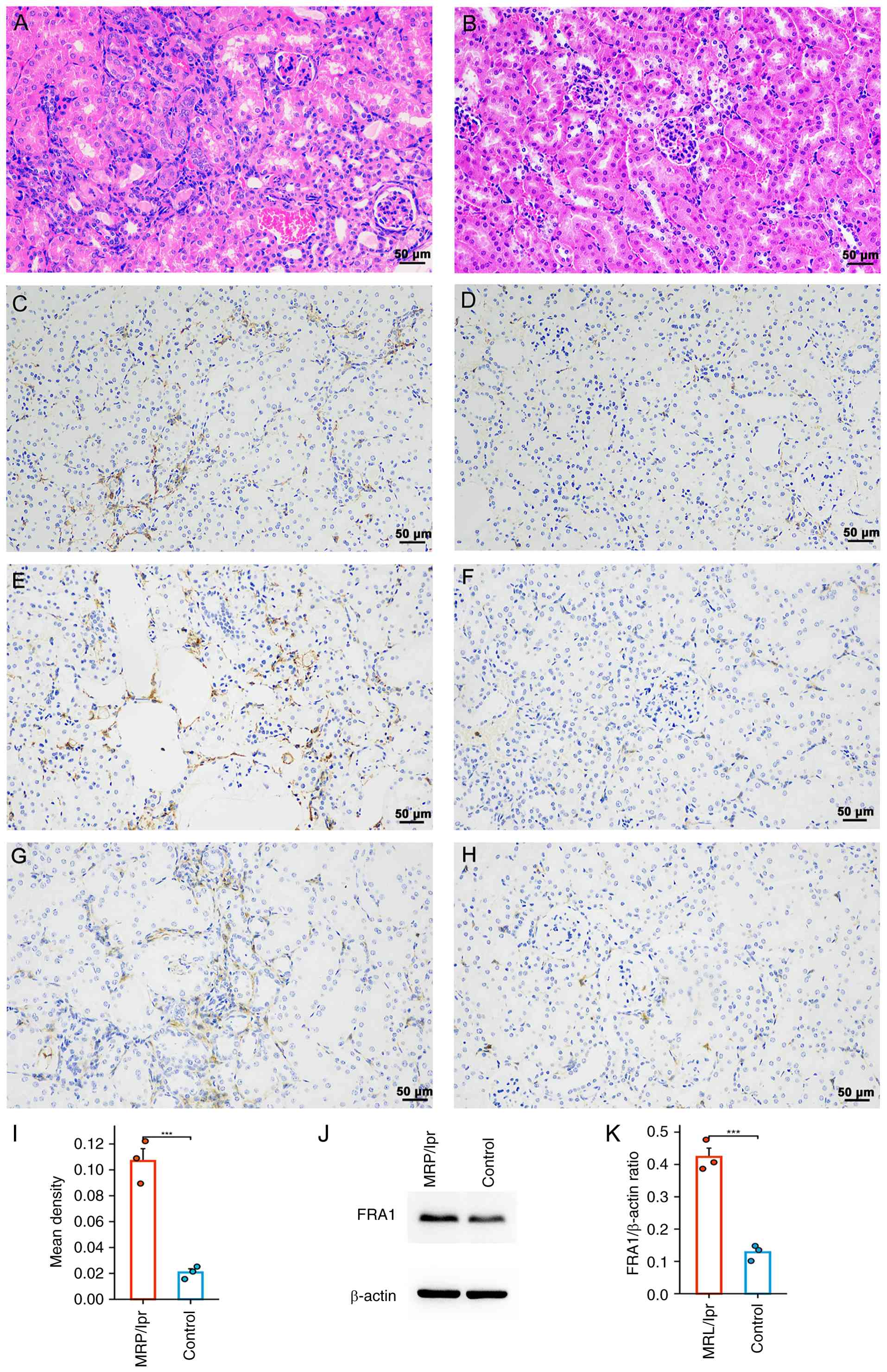
FRA1 expression in MRL/lpr mouse
kidneys
To verify the expression of FRA1 in LN kidneys,
kidneys from 20-week-old MRL/lpr mice were used. Compared with the
control group, H&E staining of MRL/lpr mouse kidneys showed
marked inflammatory infiltration (Fig.
6A and B), and tubulointerstitial injury was a distinctive and
prominent feature of the diseased kidneys. Moreover, IHC revealed
significantly increased FRA1 expression in tubular epithelial cells
of MRL/lpr kidneys compared with the control (Fig. 6C-I), which was corroborated by
elevated protein levels in western blot analysis (Fig. 6J and K).
Effect of FRA1 on inflammatory
cytokine expression in HK-2 cells
To investigate the functional role of FRA1 in renal
tubular epithelial cells, the expression of key cytokines were
measured in HK-2 cells following lentivirus-mediated FRA1
overexpression (FRA1-OE) or knockdown (FRA1-shRNA). Western blot
analysis at 144 h post-infection showed that IL-6, IL-1β, IL-8,
MCP-1 and RANTES were significantly upregulated in the FRA1-OE
group, whereas IL-6 and RANTES levels were decreased in the
FRA1-shRNA group (Fig. 7). TNF-α
and TGF-β expression remained unchanged in both FRA1-OE and
FRA1-shRNA cells. These results demonstrate that FRA1 modulates
multiple proinflammatory cytokines in human renal tubular
epithelial cells, suggesting its potential role as a regulator of
the tubulointerstitial immune microenvironment in LN.
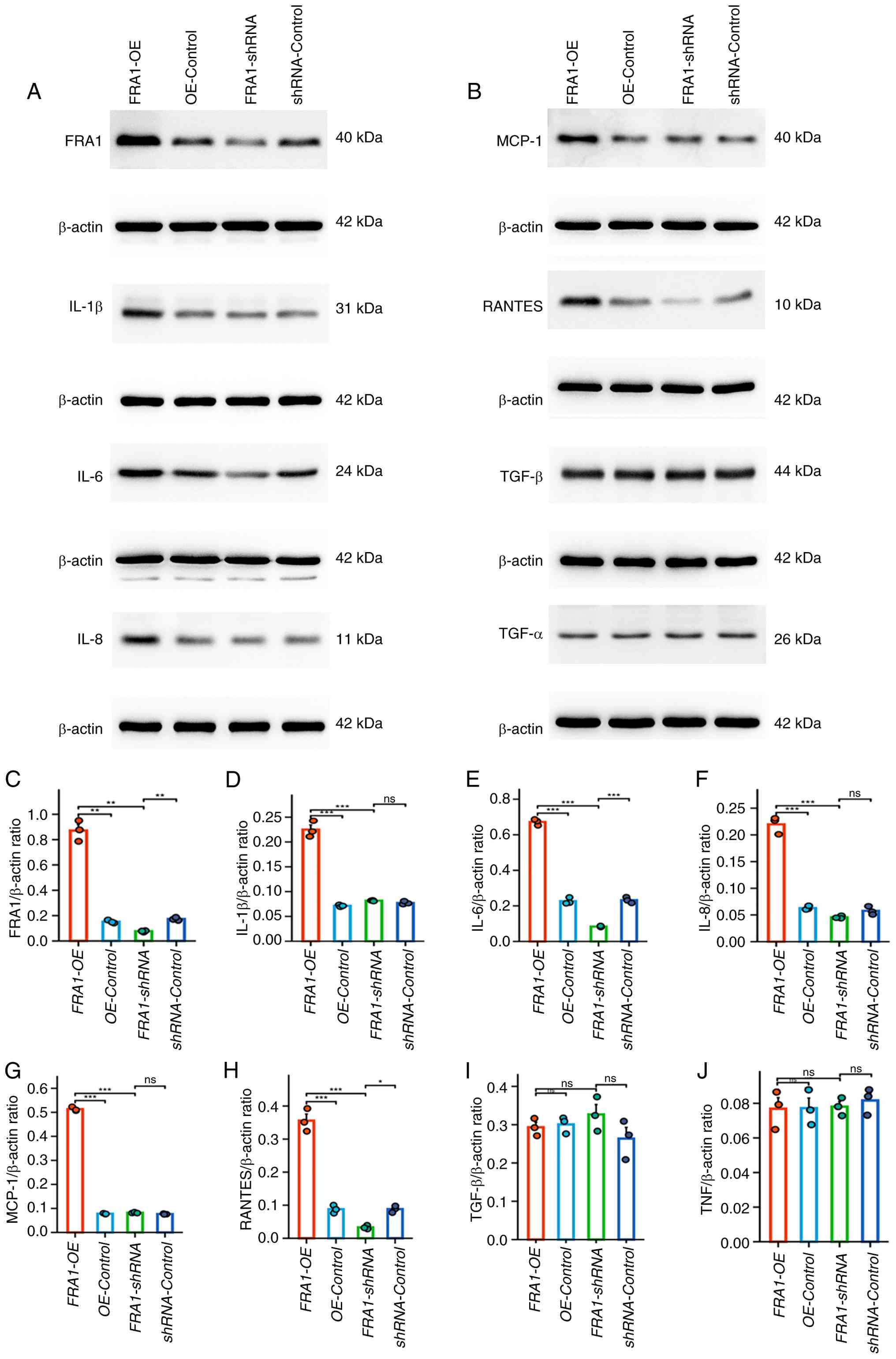 | Figure 7.Effect of FRA1 on inflammatory
cytokine expression in HK-2 cells. (A) Representative western blots
of FRA1 and inflammatory cytokines (IL-1β, IL-6, and IL-8) in HK-2
cells 144 h after transduction with FRA1-OE, FRA1-shRNA or their
corresponding controls. (B) Representative western blots of MCP-1,
RANTES, TGF-β and TNF-α in HK-2 cells following the same
transduction conditions. Densitometric semi-quantification of
protein bands normalized to β-actin for: (C) FRA1, (D) IL-1β, (E)
IL-6, (F) IL-8, (G) MCP-1, (H) RANTES, (I) TGF-β and (J) TNF-α.
Data are presented as the mean ± SEM; n=3 per group; comparisons
among subgroups were assessed using the two-sided Kruskal-Wallis
test; *P<0.05, **P<0.01 and ***P<0.001. OE, over
expression; ns, not significant; sh, short hairpin. |
Discussion
In the present study, an integrated analysis of FRA1
expression and its association with the immune microenvironment in
the renal tubulointerstitium of patients with LN was performed.
Bioinformatic analyses revealed significant upregulation of FRA1 in
LN tubulointerstitial samples, and CIBERSORT deconvolution
demonstrated that high FRA1 expression coincided with increased
infiltration of CD8+ T cells, Tregs, monocytes, M1
macrophages and activated mast cells, alongside decreased
proportions of plasma cells, resting CD4+ memory T
cells, M0/M2 macrophages, resting dendritic cells and resting mast
cells. These correlations suggest that FRA1 contributes to shaping
a proinflammatory microenvironment in LN. Consistent with the
bioinformatics analysis of GEO datasets, kidneys from MRL/lpr mice
exhibited elevated tubular epithelial FRA1 expression. In
vitro, FRA1 overexpression in HK-2 cells markedly increased
levels of IL-6, IL-1β, IL-8, MCP-1 and RANTES, whereas FRA1
knockdown resulted in reduced expression of IL-6 and RANTES.
Collectively, these data indicate that FRA1 may promote
tubulointerstitial inflammation in LN by modulating key
proinflammatory cytokines.
Prior research has indicated that LN renal tissue
exhibits abundant immune cell infiltration and a pro-inflammatory
cytokine environment, and that the renal microenvironment can
bidirectionally regulate immune cell recruitment, survival and
function (42). M1 macrophages in
LN are considered to be associated with disease activity, whereas
M2 macrophages are more commonly associated with the remission
phase (43). In the present study,
an increase in M1 macrophages and a decrease in M2 macrophages in
the FRA1-high group was observed, consistent with the
proinflammatory and alleviating roles attributed to these subsets
in LN (43). Additionally, the
increased monocyte infiltration may also promote macrophage
proliferation and activation (44). Notably, Tregs were also increased
in the FRA1-high group, despite their canonical role as suppressors
of SLE/LN inflammation (43); this
apparent paradox can be explained by several non-exclusive
considerations: i) Increased Foxp3+Treg infiltration has
been reported in active/proliferative LN and lupus-prone mouse
models and may reflect compensatory recruitment or expansion in
response to intense local inflammation rather than restored
suppressive function (45,46); ii) inflammatory milieus,
characterized by cytokines such as IL-6, can impair Treg stability
and suppressive capacity and promote phenotypic plasticity toward
exhausted or effector-like states (such as IL-17 production)
(46,47); and iii) CIBERSORT estimates only
relative cell proportions from bulk transcriptomes and cannot
determine Treg functional status. Thus, the increased Treg
frequency observed in the FRA1-high group may represent numerically
expanded but functionally compromised Tregs that fail to restrain
local LN inflammation (46,48).
The reduction in plasma cells in the FRA1 high-expression group may
reflect the fact that plasma cell aggregation and antibody
production mainly occur in the glomerular region (23,49),
whereas the present analysis focused on interstitial infiltration.
Overall, upregulation of FRA1 is consistent with increased
proinflammatory cell infiltration and aligns with the inflammatory
phenotype of LN renal tissue.
FRA1, as a member of the AP-1 transcription factor
family, regulates the expression of various inflammation-related
genes, including pro-inflammatory cytokines (such as IL-6 and TNF),
chemokines (such as MCP-1 and CXCL1) and matrix metalloproteinases
(22). Prior research has
indicated that the promoter regions of several pro-inflammatory
cytokines contain AP-1 binding sites, suggesting that AP-1 can
directly participate in the transcriptional regulation of these
genes (22). FRA1 can both form
heterodimers with the Jun family to activate gene expression and
may also suppress target genes under specific contexts (50–52).
The present data demonstrated that FRA1 overexpression upregulates
IL-6, a cytokine implicated in promoting antibody-mediated renal
inflammation; IL-6 deficiency delays LN onset and reduces renal
macrophage and T-cell infiltration in MRL/lpr mice (53). Therefore, FRA1-mediated
upregulation of IL-6 may exacerbate LN pathogenesis. MCP-1 is a key
chemokine in LN, produced by renal intrinsic cells and extensively
attracting monocytes/macrophages to inflammatory sites (54); the present study revealed that FRA1
overexpression induced MCP-1 upregulation, which may explain the
increased monocyte and M1 macrophage infiltration in the high-FRA1
group. Similarly, RANTES recruits immune cells such as T cells and
natural killer cells, and research has reported elevated urinary
RANTES levels in patients during active LN (55); FRA1-driven upregulation of RANTES
may also promote T cell (including CD8+ T cells and Tregs)
chemotaxis to the kidney (56–58).
IL-8 is a known neutrophil chemokine that is upregulated in various
nephritis conditions, and its pro-inflammatory effects may
indirectly affect other cell types through interactions (59). FRA1 can exert context-dependent
regulatory effects on IL-6 and other cytokines, influenced by cell
type, microenvironmental stimuli and dimer composition (60–62).
Future work should investigate whether the selective effects of
FRA1 knockdown on IL-6 and RANTES involve distinct AP-1 dimer
configurations or specific co-factor interactions.
FRA1 appears to carry out context-dependent roles
across different forms of renal injury. In an acute kidney injury
model, FRA1 was reported to be protective by preserving expression
of the anti-aging protein Klotho, thereby mitigating tubular damage
and inflammation (25). By
contrast, a bioinformatic screen in IgA nephropathy identified
FRA1/2 among prognostic candidates associated with TNF and MAPK
signaling, consistent with an inflammatory/prognostic association
(26); the present data in LN
align more closely with a proinflammatory role. These apparently
divergent roles may reflect several non-exclusive,
context-dependent factors, for example, acute vs. chronic injury
dynamics, differences in the principal affected cell types,
variation in upstream signaling milieus that influence AP-1
dimerization and post-transcriptional or epigenetic modulation
(such as m6A) of FRA1 expression or activity (27). These factors can alter the partner
selection, co-factor recruitment and promoter occupancy of FRA1,
thereby producing disease-specific transcriptional outputs.
The present study has several limitations; first,
the present data are primarily correlative and derived from in
vitro and ex vivo analyses. Definitive causal roles for
FRA1 in LN pathogenesis will require in vivo manipulation
(such as tubular epithelial-specific FRA1 knockout or
pharmacological inhibition) with larger sample sizes to assess its
effects on renal inflammation and function. Although empty-vector
controls were used to minimize non-specific viral effects,
potential vector integration and shRNA off-target effects cannot be
completely excluded. Second, although the present study
demonstrated that FRA1-dependent changes in selected cytokines, the
direct transcriptional targets of FRA1 in tubular cells remain to
be identified; chromatin immunoprecipitation or promoter-reporter
assays were not performed to confirm FRA1 binding at putative AP-1
sites, and these experiments constitute a clear next step. Third,
the different AP-1 dimer combinations were not distinguished or the
contributions of other Fos/Jun family members were not evaluated,
which may modulate FRA1 activity and selectivity. Fourth, the
present focus on the tubulointerstitium warrants complementary
studies in glomerular compartments and in additional cell types; in
particular, functional interrogation of FRA1 in LN-relevant immune
cells (such as macrophages and T cells) and in glomerular cells
would help determine cell-type-specific roles. Moreover,
species-specific differences between rodent models and human LN
necessitate analyses of human clinical samples to confirm the
translational relevance of FRA1 as a potential biomarker or
therapeutic target. Further investigation of classical profibrotic
and epithelial-mesenchymal transition markers (such as fibronectin,
collagen I and α-SMA) in FRA1 overexpression and knockdown models
would be valuable. Finally, it is worth considering the
translational potential of targeting FRA1/AP-1; for example,
small-molecule AP-1 inhibitors have shown efficacy in preclinical
kidney disease models: The selective c-Fos/AP-1 inhibitor T-5224
ameliorates renal inflammation and fibrosis in murine injury
models, and other AP-1 pathway blockers (such as the JNK inhibitor
SP600125) exhibit protective effects in inflammatory disease
(63). Thus, FRA1-directed
therapies or biomarker strategies, leveraging existing AP-1
modulators, could be explored in future studies of LN.
In conclusion, the present integrative analysis and
experimental validation revealed that FRA1 was upregulated in LN
tubulointerstitium and likely contributes to a proinflammatory
immune microenvironment by driving expression of key cytokines.
These findings position the FRA1/AP-1 axis as a novel regulator of
renal inflammation in LN and highlight its potential as a target
for therapeutic intervention. Further mechanistic and in
vivo studies are warranted to exploit FRA1 modulation for LN
treatment.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was funded by the Social Welfare and Basic
Research Project Fund of Zhongshan City (grant no. 2021B1088).
Availability of data and materials
The gene expression datasets analyzed in the present
study are publicly available from the Gene Expression Omnibus
(https://www.ncbi.nlm.nih.gov/geo/)
under accession numbers GSE113342, GSE200306 and GSE127797. All
other data generated during the present study may be requested from
the corresponding author upon reasonable request.
Authors' contributions
WN, JH, ZZ, JK, RL and GG contributed to the
research conception and design. WN, JH, ZZ, JK and RL performed the
experiments and drafted the manuscript. LW, YC, CZ, YL, JP and KD
analyzed and interpreted the raw data. WN, JH, ZZ, JK, RL, GG, JP
and KD confirm the authenticity of all the raw data. All authors
read and approved the final version of the manuscript.
Ethics approval and consent to
participate
All animal studies were approved by the ethics
committee of Boai Hospital of Zhongshan (approval no.
WHMYS-20250096). All animal operations were performed in compliance
with the National Institutes of Health Guide for the Care and Use
of Laboratory Animals.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
LN
|
lupus nephritis
|
|
AP-1
|
Activator Protein 1
|
|
FRA1
|
Fos-related antigen 1
|
|
GEO
|
Gene Expression Omnibus
|
|
Tregs
|
regulatory T cells
|
|
SLE
|
systemic lupus erythematosus
|
|
DEGs
|
differentially expressed genes
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
IHC
|
immunohistochemistry
|
References
|
1
|
Singh S, Saxena R and Palmer BF: Lupus
nephritis. Am J Med Sci. 337:451–460. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pons-Estel GJ, Serrano R, Plasín MA,
Espinosa G and Cervera R: Epidemiology and management of refractory
lupus nephritis. Autoimmun Rev. 10:655–663. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Guimarães JAR, Furtado SDC, Lucas ACDS,
Mori B and Barcellos JFM: Diagnostic test accuracy of novel
biomarkers for lupus nephritis-An overview of systematic reviews.
PLoS One. 17:e02750162022. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Qi J, Wu T, Wang J, Zhang J, Chen L, Jiang
Z, Li Y, Jiang H, Sun Q, Gu Q and Ying Z: Research trends and
frontiers in lupus nephritis: A bibliometric analysis from 2012 to
2022. Int Urol Nephrol. 56:781–794. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kostopoulou M, Pitsigavdaki S and Bertsias
G: Lupus nephritis: Improving treatment options. Drugs. 82:735–748.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Parodis I, Rovin BH, Tektonidou MG, Anders
HJ, Malvar A, Mok CC and Mohan C: Lupus nephritis. Nat Rev Dis
Primers. 11:692025. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cen B, Liao W, Wang Z, Gao L, Wei Y, Huang
W, He S, Wang W, Liu X, Pan X and Ji A: Gelofusine attenuates
tubulointerstitial injury induced by cRGD-conjugated siRNA by
regulating the TLR3 signaling pathway. Mol Ther Nucleic Acids.
11:300–311. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Parikh SV, Madhavan S, Shapiro J, Knight
R, Rosenberg AZ, Parikh CR, Rovin B and Menez S; Kidney Precision
Medicine Project, : Characterization of glomerular and
tubulointerstitial proteomes in a case of nonsteroidal
anti-inflammatory drug-attributed acute kidney injury: A clinical
pathologic molecular correlation. Clin J Am Soc Nephrol.
18:402–410. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hong S, Healy H and Kassianos AJ: The
emerging role of renal tubular epithelial cells in the
immunological pathophysiology of lupus nephritis. Front Immunol.
11:5789522020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gomes MF, Mardones C, Xipell M, Blasco M,
Solé M, Espinosa G, García-Herrera A, Cervera R and Quintana LF:
The extent of tubulointerstitial inflammation is an independent
predictor of renal survival in lupus nephritis. J Nephrol.
34:1897–1905. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rijnink EC, Teng YKO, Wilhelmus S,
Almekinders M, Wolterbeek R, Cransberg K, Bruijn JA and Bajema IM:
Clinical and histopathologic characteristics associated with renal
outcomes in lupus nephritis. Clin J Am Soc Nephrol. 12:734–743.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tesch GH: Review: Serum and urine
biomarkers of kidney disease: A pathophysiological perspective.
Nephrology (Carlton). 15:609–616. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hill GS, Delahousse M, Nochy D, Mandet C
and Bariéty J: Proteinuria and tubulointerstitial lesions in lupus
nephritis. Kidney Int. 60:1893–1903. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pamfil C, Makowska Z, De Groof A, Tilman
G, Babaei S, Galant C, Montigny P, Demoulin N, Jadoul M, Aydin S,
et al: Intrarenal activation of adaptive immune effectors is
associated with tubular damage and impaired renal function in lupus
nephritis. Ann Rheum Dis. 77:1782–1789. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nikolic-Paterson DJ, Wang S and Lan HY:
Macrophages promote renal fibrosis through direct and indirect
mechanisms. Kidney Int Suppl (2011). 4:34–38. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen H, Liu N and Zhuang S: Macrophages in
renal injury, repair, fibrosis following acute kidney injury and
targeted therapy. Front Immunol. 13:9342992022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang XD, Huang XF, Yan QR and Bao CD:
Aberrant activation of the WNT/β-catenin signaling pathway in lupus
nephritis. PLoS One. 9:e848522014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gröne EF, Federico G, Nelson PJ, Arnold B
and Gröne HJ: The hormetic functions of Wnt pathways in tubular
injury. Pflugers Arch. 469:899–906. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shaulian E and Karin M: AP-1 as a
regulator of cell life and death. Nat Cell Biol. 4:E131–E136. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Madrigal P and Alasoo K: AP-1 takes centre
stage in enhancer chromatin dynamics. Trends Cell Biol. 28:509–511.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gozdecka M and Breitwieser W: The roles of
ATF2 (activating transcription factor 2) in tumorigenesis. Biochem
Soc Trans. 40:230–234. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
He YY, Zhou HF, Chen L, Wang YT, Xie WL,
Xu ZZ, Xiong Y, Feng YQ, Liu GY, Li X, et al: The Fra-1: Novel role
in regulating extensive immune cell states and affecting
inflammatory diseases. Front Immunol. 13:9547442022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Grötsch B, Brachs S, Lang C, Luther J,
Derer A, Schlötzer-Schrehardt U, Bozec A, Fillatreau S, Berberich
I, Hobeika E, et al: The AP-1 transcription factor Fra1 inhibits
follicular B cell differentiation into plasma cells. J Exp Med.
211:2199–2212. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li QR, Ni WP, Lei NJ, Yang JY, Xuan XY,
Liu PP, Gong GM, Yan F, Feng YS, Zhao R and Du Y: The
overexpression of Fra1 disorders the inflammatory cytokine
secretion by mTEC of myasthenia gravis thymus. Scand J Immunol.
88:e126762018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cuarental L, Ribagorda M, Ceballos MI,
Pintor-Chocano A, Carriazo SM, Dopazo A, Vazquez E, Suarez-Alvarez
B, Cannata-Ortiz P, Sanz AB, et al: The transcription factor Fosl1
preserves Klotho expression and protects from acute kidney injury.
Kidney Int. 103:686–701. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Qian W, Wang X and Zi Y: Screening and
bioinformatics analysis of IgA nephropathy gene based on GEO
databases. Biomed Res Int. 2019:87940132019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu T, Zhuang XX, Li Zhu X, Wu X, Juan Qin
X, Bing Wei L, Chen Gao Y and Rong Gao J: Inhibition of METTL3
promotes mesangial cell mitophagy and attenuates glomerular damage
by alleviating FOSL1 m6A modifications via IGF2BP2-dependent
mechanisms. Biochem Pharmacol. 236:1168672025. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Parikh SV, Malvar A, Song H, Shapiro J,
Mejia-Vilet JM, Ayoub I, Almaani S, Madhavan S, Alberton V, Besso
C, et al: Molecular profiling of kidney compartments from serial
biopsies differentiate treatment responders from non-responders in
lupus nephritis. Kidney Int. 102:845–865. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mejia-Vilet JM, Parikh SV, Song H, Fadda
P, Shapiro JP, Ayoub I, Yu L, Zhang J, Uribe-Uribe N and Rovin BH:
Immune gene expression in kidney biopsies of lupus nephritis
patients at diagnosis and at renal flare. Nephrol Dial Transplant.
34:1197–1206. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Almaani S, Prokopec SD, Zhang J, Yu L,
Avila-Casado C, Wither J, Scholey JW, Alberton V, Malvar A, Parikh
SV, et al: Rethinking lupus nephritis classification on a molecular
level. J Clin Med. 8:15242019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
World Medical Association, . World medical
association declaration of Helsinki: Ethical principles for medical
research involving human subjects. JAMA. 310:2191–2194. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Newman AM, Liu CL, Green MR, Gentles AJ,
Feng W, Xu Y, Hoang CD, Diehn M and Alizadeh AA: Robust enumeration
of cell subsets from tissue expression profiles. Nat Methods.
12:453–457. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li Y, Ding T, Chen J, Ji J, Wang W, Ding
B, Ge W, Fan Y and Xu L: The protective capability of Hedyotis
diffusa Willd on lupus nephritis by attenuating the IL-17
expression in MRL/lpr mice. Front Immunol. 13:9438272022.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Festing MFW and Altman DG: Guidelines for
the design and statistical analysis of experiments using laboratory
animals. ILAR J. 43:244–258. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang RN, Wen Q, He WT, Yang JH, Zhou CY,
Xiong WJ and Ma L: Optimized protocols for γδ T cell expansion and
lentiviral transduction. Mol Med Rep. 19:1471–1480. 2019.PubMed/NCBI
|
|
38
|
Uchida N, Green R, Ballantine J, Skala LP,
Hsieh MM and Tisdale JF: Kinetics of lentiviral vector transduction
in human CD34(+) cells. Exp Hematol. 44:106–115. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sanber KS, Knight SB, Stephen SL, Bailey
R, Escors D, Minshull J, Santilli G, Thrasher AJ, Collins MK and
Takeuchi Y: Construction of stable packaging cell lines for
clinical lentiviral vector production. Sci Rep. 5:90212015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Malhotra D: Targeting epitope-specific T
cells and modelling protein hypercitrullination in rheumatoid
arthritis. [Master's thesis]. Hamilton (ON): McMaster University;
2024, Available from:. https://macsphere.mcmaster.ca/bitstream/11375/30359/2/Malhotra_Devon_2024August_MedicalSciences.pdf
|
|
41
|
Jian J, Liu Y, Zheng Q, Wang J, Jiang Z,
Liu X, Chen Z, Wan S, Liu H and Wang L: The E3 ubiquitin ligase
TRIM39 modulates renal fibrosis induced by unilateral ureteral
obstruction through regulating proteasomal degradation of PRDX3.
Cell Death Discov. 10:172024. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chernova I: Lupus nephritis: Immune cells
and the kidney microenvironment. Kidney360. 5:1394–1401. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cheng Y, Liu L, Ye Y, He Y, Hu W, Ke H,
Guo ZY and Shao G: Roles of macrophages in lupus nephritis. Front
Pharmacol. 15:14777082024. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sun L, Rautela J, Delconte RB,
Souza-Fonseca-Guimaraes F, Carrington EM, Schenk RL, Herold MJ,
Huntington ND, Lew AM, Xu Y and Zhan Y: GM-CSF quantity has a
selective effect on granulocytic vs. monocytic myeloid development
and function. Front Immunol. 9:19222018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Shakweer MM, Behairy M, Elhefnawy NG and
Elsaid TW: Value of Foxp3 expressing T-regulatory cells in renal
tissue in lupus nephritis; an immunohistochemical study. J
Nephropathol. 5:105–110. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kalim H, Pratama MZ, Nugraha AS,
Prihartini M, Chandra A, Sholihah AI, Qonita F and Handono K:
Regulatory T cells compensation failure cause the dysregulation of
immune response in pristane induced lupus mice model. Malays J Med
Sci. 25:17–26. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wu Y, Zhang W, Liao Y, Sun T and Liu Y and
Liu Y: Immune cell aberrations in systemic lupus erythematosus:
Navigating the targeted therapies toward precision management. Cell
Mol Biol Lett. 30:732025. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Jiang C, Wang H, Xue M, Lin L, Wang J, Cai
G and Shen Q: Reprograming of peripheral Foxp3+
regulatory T cell towards Th17-like cell in patients with active
systemic lupus erythematosus. Clin Immunol. 209:1082672019.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Espeli M, Bökers S, Giannico G, Dickinson
HA, Bardsley V, Fogo AB and Smith KG: Local renal autoantibody
production in lupus nephritis. J Am Soc Nephrol. 22:296–305. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Cao S, Schnelzer A, Hannemann N, Schett G,
Soulat D and Bozec A: The transcription factor FRA-1/AP-1 controls
lipocalin-2 expression and inflammation in sepsis model. Front
Immunol. 12:7016752021. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Hannemann N, Cao S, Eriksson D, Schnelzer
A, Jordan J, Eberhardt M, Schleicher U, Rech J, Ramming A, Uebe S,
et al: Transcription factor Fra-1 targets arginase-1 to enhance
macrophage-mediated inflammation in arthritis. J Clin Invest.
129:2669–2684. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Mittelstadt ML and Patel RC: AP-1 mediated
transcriptional repression of matrix metalloproteinase-9 by
recruitment of histone deacetylase 1 in response to interferon β.
PLoS One. 7:e421522012. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Cash H, Relle M, Menke J, Brochhausen C,
Jones SA, Topley N, Galle PR and Schwarting A: Interleukin 6 (IL-6)
deficiency delays lupus nephritis in MRL-Faslpr mice: the IL-6
pathway as a new therapeutic target in treatment of autoimmune
kidney disease in systemic lupus erythematosus. J Rheumatol.
37:60–70. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Alharazy S, Kong NC, Mohd M, Shah SA,
Ba'in A and Abdul Gafor AH: Urine monocyte chemoattractant
protein-1 and lupus nephritis disease activity: Preliminary report
of a prospective longitudinal study. Autoimmune Dis.
2015:9620462015.PubMed/NCBI
|
|
55
|
Chan RWY, Lai FMM, Li EKM, Tam LS, Chow
KM, Li PKT and Szeto CC: Messenger RNA expression of RANTES in the
urinary sediment of patients with lupus nephritis. Nephrology
(Carlton). 11:219–225. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Roh YS, Park S, Lim CW and Kim B:
Depletion of Foxp3+ Regulatory T cells promotes profibrogenic
milieu of cholestasis-induced liver injury. Dig Dis Sci.
60:2009–2018. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ahnstedt H, Roy-O'Reilly M, Spychala MS,
Mobley AS, Bravo-Alegria J, Chauhan A, Aronowski J, Marrelli SP and
McCullough LD: Sex differences in adipose tissue CD8+ T
cells and regulatory T cells in middle-aged mice. Front Immunol.
9:6592018. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Rovin BH, Lu L and Zhang X: A novel
interleukin-8 polymorphism is associated with severe systemic lupus
erythematosus nephritis. Kidney Int. 62:261–265. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Scheller J, Chalaris A, Schmidt-Arras D
and Rose-John S: the pro- and anti-inflammatory properties of the
cytokine interleukin-6. Biochim Biophys Acta. 1813:878–888. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Mishra RK, Potteti HR, Tamatam CR,
Elangovan I and Reddy SP: c-Jun is required for nuclear
factor-κB-dependent, LPS-stimulated fos-related antigen-1
transcription in alveolar macrophages. Am J Respir Cell Mol Biol.
55:667–674. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wang Q, Ni H, Lan L, Wei X, Xiang R and
Wang Y: Fra-1 protooncogene regulates IL-6 expression in
macrophages and promotes the generation of M2d macrophages. Cell
Res. 20:701–712. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Reck M, Baird DP, Veizades S, Sutherland
C, Bell RMB, Hur H, Cairns C, Janas PP, Campbell R, Nam A, et al:
Multiomic analysis of human kidney disease identifies a tractable
inflammatory and pro-fibrotic tubular cell phenotype. Nat Commun.
16:47452025. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Song D, Lian Y and Zhang L: The potential
of activator protein 1 (AP-1) in cancer targeted therapy. Front
Immunol. 14:12248922023. View Article : Google Scholar : PubMed/NCBI
|