Introduction
Small RNAs (microRNAs, miRNAs) are recently
discovered non-coding RNA molecules comprising approximately 18–25
nucleotides. The biological function of miRNAs remains to be
clarified. miRNAs are believed to play an important role in the
post-transcriptional regulation of mRNA (1). miRNAs have been shown to be closely
associated with the pathogenesis and differentiation of non-small
cell lung cancer (NSCLC) (9). In
the human genome, approximately 50% of all miRNAs are located on
chromosomes at sites associated with tumors, and their location
suggests that miRNAs play an important role in the pathogenesis of
tumors (2). Lethal-7
(let-7) miRNAs are a family of miRNAs whose expression has
been reported to be decreased in patients with lung cancer
(3). Ras is a major
oncogene, and the overexpression of ras proteins suppresses
apoptosis, promoting the pathogenesis and development of tumors. In
this study, the expression of the let-7 and K-ras
genes in NSCLC was examined, and the expression levels of these
genes in NSCLC patients were determined by performing reverse
transcription-polymerase chain reaction (RT-PCR).
Materials and methods
Materials
A total of 31 patients with NSCLC [22 males and 9
females; mean age, 61.3 (5.1) years; range, 45–68] who underwent
radical resection at our hospital between January 2007 and June
2007 were enrolled in the study. Fresh lung cancer and normal lung
tissues were harvested away from the tumor (pericancerous tissues
which served as the control) from specimens excised from the 31
patients within 30 min. The specimens were stored in Cryule vials
and were immediately frozen by placing them in liquid nitrogen.
Histopathological examinations performed after the operation using
sections prepared from paraffin-embedded slices confirmed that all
of the specimens were lung cancers, including 12 cases of squamous
cell carcinoma and 19 cases of adenocarcinoma. Following the
operation, the cancer specimens were staged according to the TNM
staging system: 9 cases were in stage IIa, 7 in stage IIb, 14 in
stage IIIa and 1 was in stage IIIb.
Reagents
TRIzol was purchased from Life Technologies, USA.
RNase inhibitor, MMLV reverse transcriptase and Taq polymerase were
from Promega. Oligo (dT)18 was from Shanghai Sangon,
China, and 10,000X SYBR-Green was purchased from Molecular Probes,
USA. For the primer design, software Primer 5.0 and Rotor-gene 6.0
were used, which were provided by Shanghai Sangon and Corbett
Research, respectively.
Real-time quantitative PCR
The TRIzol method was used to extract total RNA.
Subsequently, the ultraviolet absorption spectrum was examined and
denaturing RNA agarose gel electrophoresis was performed to
determine the purity and integrity of the RNA. RT-PCR was performed
to synthesize cDNA. Real-time Q-PCR was performed using the DK-8D
Electro-Thermostatic Water Cabinet (Shanghai Sibas Biotechnology
Development Co., Ltd., China), FeroTec Gradient PCR thermal cycler
(Ferotec, Germany), DYY-8 electrophoresis system (Shanghai Qite
Analytical Instruments Co., Ltd., China), DY-32
mini-electrophoresis chamber (Xinghua Analytical Instrument
Factory, China) and Rotor-Gene 3000 real-time PCR amplifier
(Corbett Research, Australia). Primer sequences (Table I) were obtained from Integrated DNA
Technologies, Inc. (Coralville, IA, USA).
 | Table IPrimers used for real-time Q-PCR. |
Table I
Primers used for real-time Q-PCR.
| Gene name | Bidirectional primer
sequences |
|---|
| β-actin | F:
5′CCTGTACGCCAACACAGTGC3′ |
| R:
5′ATACTCCTGCTTGCTGATCC3′ |
| K-ras | F:
5′TTGCCTCCCTACCTTCCACA3′ |
| R:
5′GTTCAAAGCATCAGCCACCAC3′ |
| Let-7 | F:
5′TCTTATGAATGGCCCAAA3′ |
| R:
5′CAGTTATCTCCCTTGATGTAA3′ |
Calculation of relative expression
Using real-time Q-PCR the let-7, K-ras
and β-actin genes were amplified from each sample. The
corrected value of expression was obtained by dividing the value of
let-7 and K-ras expression by the value of β-actin
(internal control). The value of relative expression of these genes
was obtained by dividing the corrected values of let-7 and
K-ras expression in lung tumors by those of their expression
in normal lung tissues.
Statistical analysis
SPSS13.0 was used for statistical analysis. Data
were expressed as the mean (standard deviation, SD). The t-test was
performed using the corrected values of the expression levels of
let-7 and K-ras in the lung cancer and normal lung
tissues. Pearson's correlation analysis was performed for the
corrected values of the expression levels of let-7 and
K-ras in the lung cancers. P<0.05 was considered to
indicate a statistically significant difference.
The relative expression was considered to be low
when its value was <0.5, as high expression when its value was
>2, and as moderate when its value was between 0.5 and 2. On the
basis of the values of the relative expression of let-7 and
K-ras, the 31 patients were assigned into low-, median- and
high-expression groups. Follow-up studies were conducted for 1 to
37 months to determine the survival status of each group.
Kaplan-Meier survival curves were plotted using these results and a
log-rank test was carried out to determine differential survival
between the two groups.
Results
Expression of let-7 and K-ras in lung
cancer
Let-7 expression was low in 21 cases (67.74%)
and moderate in 10 cases. However, a high expression of
let-7 was not detected in any of the lung cancer specimens.
K-ras expression was high in 20 cases (64.52%) and moderate
in 11 cases. However, a low expression of K-ras was not
detected in any of the lung cancer specimens.
Corrected values of let-7 and K-ras in
cancer and pericancerous tissues
The corrected values of let-7 and
K-ras expression in the 31 specimens were expressed as the
mean (SD) (Table II). The results
indicate that the corrected values of let-7 or K-ras
were significantly different between the cancer and normal tissues
(|t|let 7=6.658 or
tK-ras=6.617, P<0.05, respectively).
 | Table IICorrection of let-7 and
K-ras values in the cancer and pericancerous tissues. |
Table II
Correction of let-7 and
K-ras values in the cancer and pericancerous tissues.
| Let-7 | K-ras |
|---|
|
|
|---|
| Cancer tissue | Pericancerous
tissues | Cancer tissue | Pericancerous
tissues |
|---|
| 0.772 (0.383) | 2.235 (1.162) | 3.117 (1.253) | 1.430 (0.671) |
Correlation analysis
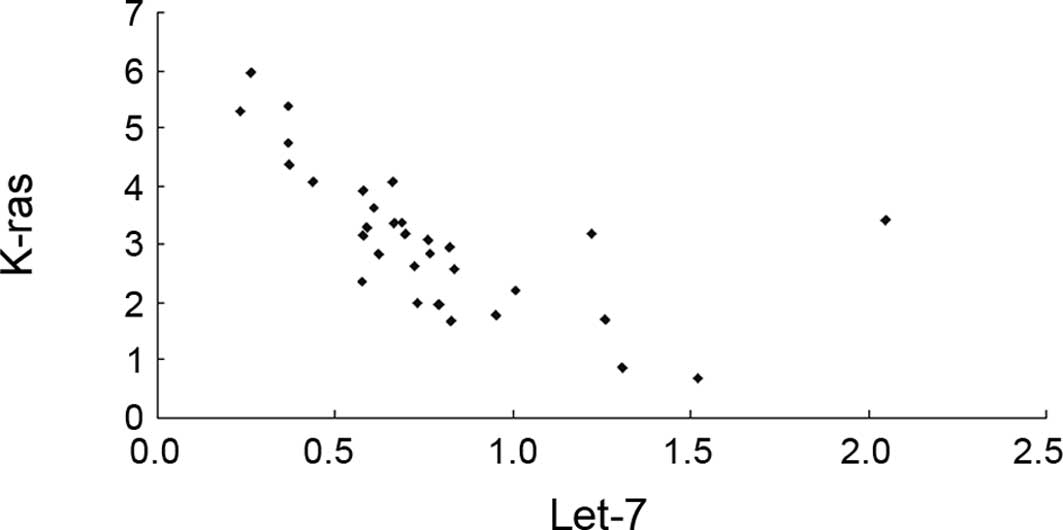
Fig. 1 shows the
importance of the corrected values of let-7 and K-ras
in the cancer tissues. The results show that the expression of
let-7 and K-ras in tumor tissues was closely
correlated (r=−0.6336, P<0.05).
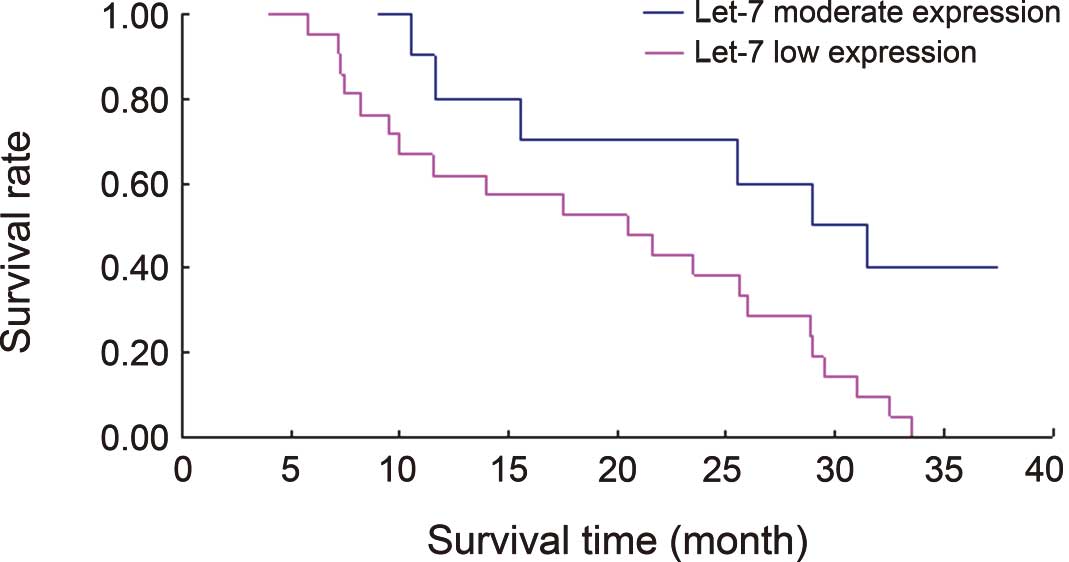
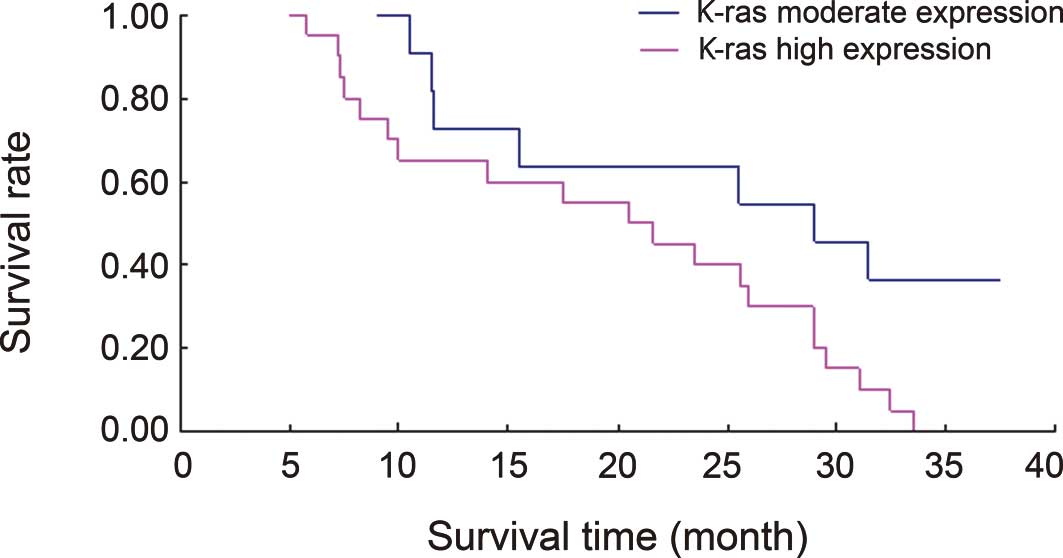
Survival curves associated with let-7 and
K-ras expression
Figs. 2 and 3 show Kaplan Meier survival curves
associated with let-7 and K-ras expression in the 31
lung cancer cases. The results from the log-rank differential
survival test indicated statistically significant differences in
the survival rates between the groups with a low and moderate
let-7 expression (χ2=6.1577) and between those
with a moderate and high K-ras expression
(χ2=5.0152); P<0.05.
Discussion
miRNAs are ubiquitous in eukaryotic genomes. These
small RNA molecules bind to specific target mRNAs through base
pairing and inhibit translation or negatively regulate gene
expression by the degradation of target mRNA. This method of gene
expression regulation plays an essential role in development, cell
differentiation and apoptosis (4).
Let-7 is a member of the miRNA family. It was first found in
nematodes and serves as a sequential control factor for cell fate
determination (3).
let-7 expression was found to be decreased in
human lung cancer, and a low expression was correlated with the
postoperative survival time of patients (3,5,6).
Takamizawa et al (3) studied
143 NSCLC patients undergoing radical resection, and a COX
proportional hazard model analysis was performed to determine the
factors that may affect prognosis, such as age, gender,
histological type, smoking history, TNM stage and the let-7
expression level. Results of these authors showed that the
let-7 expression level was an independent postoperative
prognostic factor for NSCLC.
Our results revealed that let-7 expression
was significantly lower than normal in 67.74% of the 31 lung cancer
patients. Follow-up studies of these patients showed that patients
in the low-expression group had a significantly lower survival rate
than those in the median-expression group. The results show that
let-7 expression is low in lung cancer and that patients
with a low let-7 expression have a short survival time.
Ras is an important human oncogene.
H-ras, K-ras and N-ras, three closely related members
of the ras family, are the most common oncogenes in human cancer.
K-ras is associated with lung cancer. Slebos et al
(7) found a marked decrease in the
survival rate of patients with a K-ras mutation. Nemunaitis
et al (8) studied
K-ras mutations and the expression of ras and c-erbB-2
proteins, and found that K-ras is a significant prognostic
factor for lung adenocarcinoma. In our study, K-ras
expression was high in 64.52% of the lung cancer patients, and the
postoperative survival rate of these patients was significantly
lower than that of the patients in the median-expression group.
These results show that K-ras plays an important role in the
pathogenesis of lung cancer.
Eder and Scherr (9)
found that the let-7 expression declined as the K-ras
expression increased in NSCLC, suggesting their significance.
Johnson et al (10) found
that the 3′-UTR of ras mRNA contains a number of
complementary binding sites for let-7 and inferred that
let-7 may regulate the expression of ras. These
authors reported that the target of let-7 is the
K-ras oncogene and that a decrease in let-7
expression resulted in an increase in ras expression or the
promotion of tumor growth. Tam (11) reported that a decrease in
let-7 expression caused an approximately 70% increase in the
level of the ras protein expression in transfected HeLa cells.
In the present study, Pearson's correlation analysis
revealed a negative correlation between the corrected values of
let-7 and K-ras in the NSCLC tissues (r=−0.6336).
This result suggests that during the pathogenesis of NSCLC, a
decrease in the level of let-7 expression may lead to an
enhanced expression of K-ras. Furthermore, our results
showed that a polygene was involved in the pathogenesis and
progression of NSCLC and that these genes act in synergy with each
other, thus promoting the pathogenesis and progression of lung
cancer and worsening patient prognosis.
References
|
1
|
Ambros V: microRNAs: tiny regulators with
great potential. Cell. 107:823–826. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Calin GA, Sevignani C, Dumitru CD, et al:
Human microRNA genes are frequently located at fragile sites and
genomic regions involved in cancers. Proc Natl Acad Sci USA.
101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Takamizawa J, Konishi H, Yanagisawa K, et
al: Reduced expression of the let-7 microRNAs in human lung cancers
in association with shortened postoperative survival. Cancer Res.
64:3753–3756. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Abrahante JE, Daul AL, Li M, Volk ML,
Tennessen JM, Miller EA and Rougvie AE: The Caenorhabditis
elegans hunchback-like gene lin-57/hbl-1 controls developmental
time and is regulated by microRNAs. Dev Cell. 4:625–637. 2003.
|
|
5
|
Yanaihara N, Caplen N, Bowman E, et al:
Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shell S, Park SM, Radjabi AR, et al: Let-7
expression defines two differentiation stages of cancer. Proc Natl
Acad Sci USA. 104:11400–11405. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Slebos RJ, Kibbelaar RE, Dalesio O, et al:
K-ras oncogene activation as a prognostic marker in adenocarcinoma
of the lung. N Engl J Med. 323:561–565. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nemunaitis J, Klemow S, Tong A, et al:
Prognostic value of K-ras mutations, ras oncoprotein, and c-erb B-2
oncoprotein expression in adenocarcinoma of the lung. Am J Clin
Oncol. 21:155–160. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Eder M and Scherr M: MicroRNA and lung
cancer. N Engl J Med. 352:2446–2448. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Johnson SM, Grosshans H, Shingara J, et
al: RAS is regulated by the let-7 microRNA family. Cell.
120:635–647. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tam W: Identification and characterization
of human BIC, a gene on chromosome 21 that encodes a noncoding RNA.
Gene. 274:157–167. 2001. View Article : Google Scholar : PubMed/NCBI
|