Introduction
Colorectal cancer (CRC) is one of the most common
types of cancer. Approximately 1 million new cases are diagnosed
and approximately 529,000 patients succumb to this type of cancer
worldwide each year (1). When this
cancer is diagnosed with localized disease, the five-year survival
rate following curative surgery is approximately 90% (2). However, the prognosis worsens with
advancing stage, and only 5% of patients diagnosed with distant
metastasis survive for five years. Detection of CRC in the early
stage is therefore a key factor for reducing CRC mortality
rates.
Among the various screening tests for CRC, fecal
occult blood testing (FOBT) is considered to be the most effective
non-invasive screening test. FOBT is convenient and relatively
cost-effective (3–6). Guaiac-based FOBT reduces incidence and
mortality (3,7,8), but
does not exhibit high sensitivity (9–11).
Immunochemical FOBT was reported to have a sensitivity of 65.8% and
a specificity of 95% for detecting CRC (12). Immunochemical FOBT exhibits a higher
sensitivity than that of guaiac-based FOBT. However, improvement in
the sensitivity of the fecal test for CRC screening is required to
reduce mortality rates from this type of cancer.
Numerous screening methods for CRC using fecal DNA
are available. Methods using fecal DNA allow for the detection of
mutated (13–16), methylated (17–22) or
long DNA (21,23,24).
Results from various studies on fecal RNA-based analysis by
reverse-transcription polymerase chain reaction (RT-PCR) for CRC
screening have been reported (25–31).
Altered messenger RNA (mRNA) expression of numerous genes has been
noted in CRC, but only a few genes have been studied to investigate
the usefulness of fecal RNA analysis in CRC detection.
Interferon-induced transmembrane protein (IFITM)
mRNA has been found to be overexpressed in CRC tissues compared
with expression levels in normal tissues by cDNA microarrays
(32). Three homologues (IFITM1,
IFITM2 and IFITM3) of the human IFITM gene exist. Frequent up-
regulation of the IFITM gene expression has been reported to be
highly specific to human colorectal carcinogenesis (33).
Quantification by real-time PCR is considered to be
useful for determining the optimal cut-off point for discriminating
between patients with and without CRC. However, the usefulness of
detecting fecal mRNA by real-time RT-PCR for CRC screening has yet
to be studied sufficiently (29).
The usefulness of detecting the fecal mRNA of IFITM1, IFITM2 and
IFITM3 by real-time RT-PCR for CRC screening was therefore
examined.
Materials and methods
Study design
This study consisted of 21 CRC and 23 control
patients (Table I), all of whom
underwent colonoscopy. The reasons for performing colonoscopy in
the CRC and control patients included positive results of a FOBT
test, abdominal pain, anemia, constipation and CRC screening. Stool
samples of CRC patients diagnosed with both colonoscopy and
histologically were collected prior to surgical resection. The
median age of the patients with CRC was 74 years (range 56–94).
There were 14 male and 7 female CRC patients. The primary tumor
sites were: rectum, 5 patients; sigmoid colon, 7 patients;
descending colon, 0 patients; transverse colon, 2 patients;
ascending colon, 4 patients; and cecum, 3 patients. The median size
of the primary tumors of 14 patients with CRC was 34 mm (range
10–70) and the size of the tumors of the remaining 7 patients with
CRC was unknown. The tumors were classified according to Dukes’
staging, yielding stage A (n=9), and stages B (n=3), C (n=7) and D
(n=2) (Table II). A total of 23
control patients (14 male and 9 female) who did not exhibit
neoplastic lesions colonoscopically were also included in this
study. The median age of the control patients was 61 years (range
40–80). This study was approved by the ethics committees at the
institutions in which fecal samples were collected. Oral and
written informed consent was obtained from the patients.
 | Table IThe characteristics of the CRC and
control patients. |
Table I
The characteristics of the CRC and
control patients.
| CRC patients | Control patients |
|---|
| No. | 21 | 23 |
| Gender
(male/female) | 14/7 | 14/9 |
| Median age
(range) | 74 (56–94) | 61 (40–80) |
 | Table IIThe characteristics of the CRC
patients. |
Table II
The characteristics of the CRC
patients.
| Patient no. | Age (years) | Gender
(male/female) | Location | Size (cm) | Dukes’ stage |
|---|
| 1 | 63 | M | R | un | C |
| 2 | 94 | M | R | un | B |
| 3 | 57 | M | S | 3.3 | B |
| 4 | 74 | F | Ce | un | D |
| 5 | 75 | M | R | 5.5 | A |
| 6 | 56 | M | S | un | A |
| 7 | 71 | M | R | 4.0 | C |
| 8 | 60 | M | R | 7.0 | A |
| 9 | 70 | F | S | 1.3 | A |
| 10 | 85 | M | T | un | C |
| 11 | 75 | M | Ce | un | C |
| 12 | 85 | M | A | un | C |
| 13 | 86 | M | Ce | 1.0 | A |
| 14 | 82 | F | S | 1.5 | A |
| 15 | 81 | M | A | 4.0 | A |
| 16 | 70 | M | S | 1.5 | A |
| 17 | 70 | M | S | 2.0 | A |
| 18 | 65 | F | A | 4.0 | C |
| 19 | 72 | F | T | 2.5 | C |
| 20 | 88 | F | S | 3.5 | B |
| 21 | 80 | F | A | 4.0 | D |
Fecal sample collection and RNA
isolation
Fecal samples were initially preserved at −80°C
within 24 h following evacuation. Total RNA was extracted from 500
mg of feces without isolating colonocytes using a combination of
Isogene (Nippon Gene, Toyama, Japan) and RNeasy kit (Qiagen, Tokyo,
Japan) as previously described (26).
cDNA synthesis
The concentration of isolated RNA was measured by
NanoDrop 1000 (Thermo Fisher Scientific, Yokohama, Japan). cDNA was
synthesized using SuperScript III RNase H− reverse
transcriptase (Invitrogen, Tokyo, Japan) with 1000 ng fecal total
RNA and 150 ng random primers in a total reaction volume of 18 μl
according to the manufacturer’s instructions.
Real-time polymerase chain reaction
Amplification and detection were performed by
real-time PCR with a Taq Man probe. The expression levels of
glyceraldehyde-3-phosphate dehydrogenase (GAPDH), β-actin (ACTB),
IFITM1, IFITM2 and IFITM3 were measured. The sequences of the PCR
primers and probes are listed in Table III. cDNA (2 μl) was used as
templates in each reaction. The reaction mixture consisted of
templates, 10 μl of QuantiTect Multiplex PCR kit (Qiagen, Tokyo,
Japan), 0.40 μM of forward and reverse primers and 0.20 μM of the
probe in a total reaction volume of 20 μl. The real-time PCR
reaction was performed with precycling heat activation at 95°C for
10 min, followed by 50 cycles of denaturation at 95°C for 15 sec
and annealing/extension at 60°C for 60 sec in an Applied Biosystems
7500 sequence detection system (Life Technologies, Tokyo,
Japan).
 | Table IIIThe primer and probe oligo
sequences. |
Table III
The primer and probe oligo
sequences.
| Gene | |
|---|
| GAPDH | Forward:
GAACGGGAAGCTTGTCATCA |
| GAPDH | Reverse:
ATCGCCCCACTTGATTTTG |
| GAPDH | Probe:
FAM-CCCATCACCATCTTCCAGGAGCGAGA-TAMRA |
| ACTB | Forward:
CCTCGCCTTTGCCGATCC |
| ACTB | Reverse:
CATGCCGGAGCCGTTGTC |
| ACTB | Probe:
FAM-CGTCCACACCCGCCGCCAGC-TAMRA |
| IFITM1 | Forward:
TCGCCTACTCCGTGAAGTCT |
| IFITM1 | Reverse:
TGTCACAGAGCCGAATACCA |
| IFITM1 | Probe:
FAM-ATGCCTCCACCGCCAAGTGCCT-TAMRA |
| IFITM2 | Forward:
TGTATCCCACGTACTCTATCTTCC |
| IFITM2 | Reverse:
GGACAGGGCGAGGAATGG |
| IFITM2 | Probe:
FAM-TGGAGTAAGTGGAATACAGGTCAAGGGCAG-TAMRA |
| IFITM3 | Forward:
CTGAGAACCATCCCAGTAACCC |
| IFITM3 | Reverse:
ACTGTTGACAGGAGAGAAGAAGG |
| IFITM3 | Probe:
FAM-CATGGTGTCCAGCGAAGACCAGCGG-TAMRA |
Statistical analysis
In the fecal RNA analysis used for detecting CRC,
when an amplification curve crossed the threshold line within the
end of definite cycles, the expression of the gene was interpreted
as positive, and multiple pairs of sensitivities and specificities
were determined for each gene. The receiver operating
characteristic (ROC) curve was created from pairs of sensitivities
and specificities, and the area under the curve (AUC) was
calculated (34). The optimal
sensitivity and specificity were determined using the Youden index:
(Youden index) = maximum (sensitivity + specificity − 1). The
sensitivities and specificities were estimated relative to the
results of the colonoscopy in the usual manner; 95% confidence
interval (CI) for each of the estimated parameters was based on the
exact binominal distribution. P<0.05 was considered to be
statistically significant. The reported P-values were evaluated by
a two-sided test.
Results
RNA concentration
The mean concentrations of RNA extracted from the
feces of the 21 CRC and 23 control patients were 876 ng/μl (range
105–3,333) and 1,044 ng/μl (range 105–2,000), respectively. No
significant difference was found between RNA concentrations in the
group of CRC patients and the group of control patients
(P=0.40).
Detection of GAPDH and ACTB mRNA
The detection rates of GAPDH were 91% (19/21) in the
CRC patients and 100% (23/23) in the control patients. No
significant difference was noted between the detection rates of
GAPDH in the CRC and control patients (P=0.2). The cycle threshold
(Ct) values of GAPDH in the CRC patients were significantly smaller
than those in the control patients (P=0.02). The detection rates of
ACTB were 76% (16/21) in the CRC patients and 91% (21/23) in the
control patients. No significant difference was found between the
detection rates of ACTB in the CRC and control patients (P=0.23).
The Ct values of ACTB in CRC patients were significantly smaller
than those in the control patients (P=0.003).
Sensitivities and specificities of the
gene expression analysis
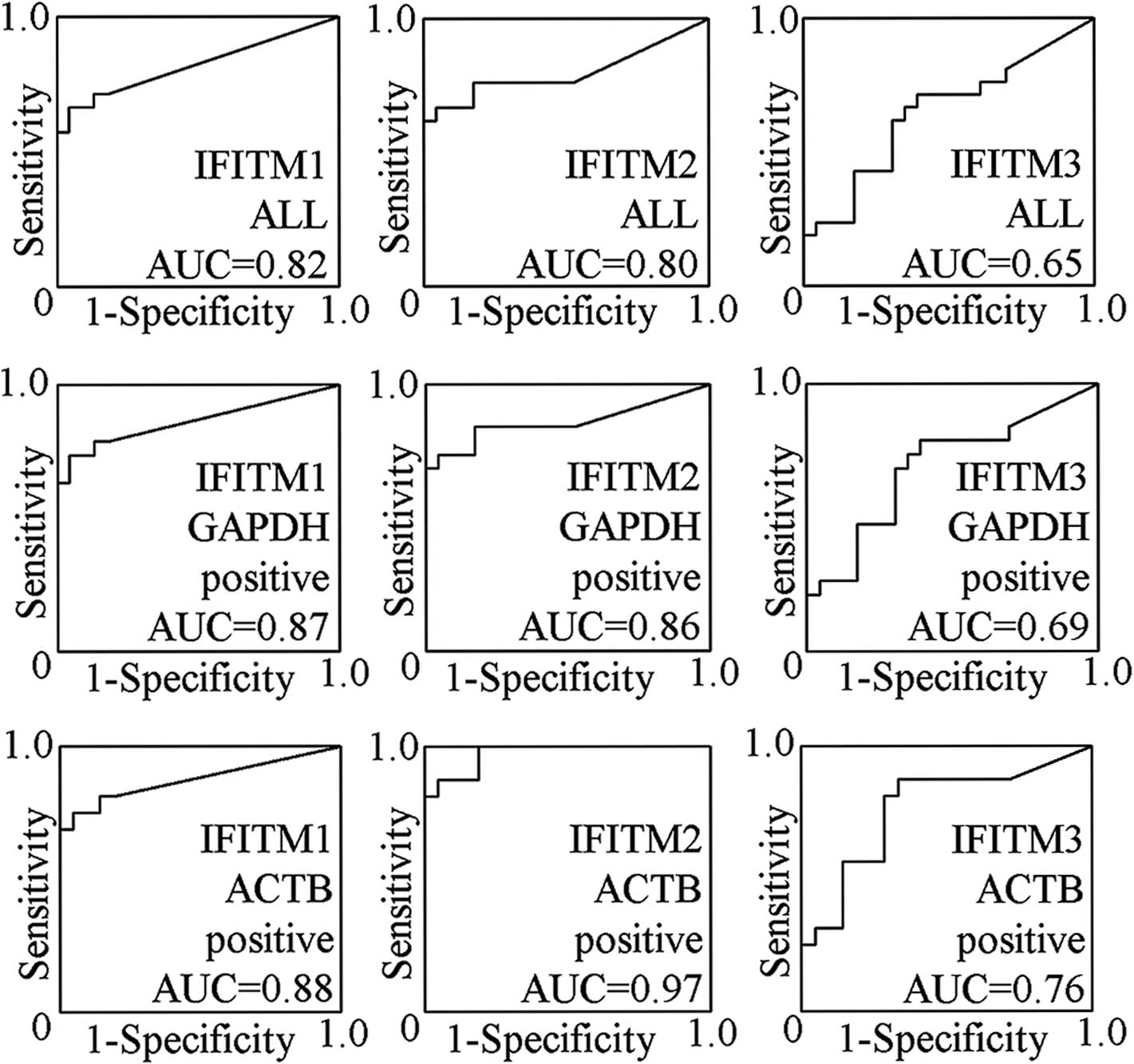
When the 44 cases were analyzed, the AUCs of fecal
IFITM1, IFITM2 and IFITM3 expression analysis for CRC were 0.82,
0.80 and 0.65, respectively (Fig.
1). The sensitivities were 67% (14/21; 95% CI 43–85%), 67%
(14/21; 95% CI 43–85%) and 71% (15/21; 95% CI 48–89%),
respectively. The specificities were 96% (1/23; 95% CI 78–100%),
96% (1/23; 95% CI 78–100%) and 61% (9/23; 95% CI 39–80%),
respectively. The Youden index values were 0.62, 0.62 and 0.32,
respectively (Table IV).
 | Table IVThe sensitivities and specificities
of fecal RNA expression analysis. |
Table IV
The sensitivities and specificities
of fecal RNA expression analysis.
| Expression | Gene | Sensitivity | Specificity | Threshold
(cycles) | Youden index |
|---|
| No. | % (95% CI) | No. | % (95% CI) |
|---|
| ALL |
| IFITM1 | 14/21 | 67 (43–85) | 1/23 | 96 (78–100) | 45 | 0.62 |
| IFITM2 | 14/21 | 67 (43–85) | 1/23 | 96 (78–100) | 39 | 0.62 |
| IFITM3 | 15/21 | 71 (48–89) | 9/23 | 61 (39–80) | 38 | 0.32 |
| IFITM1+2 | 18/21 | 86 (64–97) | 1/23 | 96 (78–100) | IFITM1:45
IFITM2:35 | 0.81 |
| GAPDH (+) |
| IFITM1 | 14/19 | 74 (49–91) | 1/23 | 96 (78–100) | 45 | 0.69 |
| IFITM2 | 14/19 | 74 (49–91) | 1/23 | 96 (78–100) | 39 | 0.69 |
| IFITM3 | 15/19 | 79 (54–94) | 9/23 | 61 (39–80) | 38 | 0.40 |
| IFITM1+2 | 18/19 | 95 (74–100) | 1/23 | 96 (78–100) | IFITM1:45
IFITM2:35 | 0.90 |
| ACTB (+) |
| IFITM1 | 12/16 | 75 (48–93) | 1/21 | 95 (76–100) | 44 | 0.70 |
| IFITM2 | 14/16 | 88 (62–99) | 1/21 | 95 (76–100) | 39 | 0.83 |
| IFITM3 | 14/16 | 88 (62–99) | 7/21 | 67 (43–85) | 38 | 0.54 |
| IFITM1+2 | 16/16 | 100 (79–100) | 0/21 | 100 (84–100) | IFITM1:44
IFITM2:35 | 1.00 |
When 42 cases were analyzed in which GAPDH mRNA was
detected, the AUCs of fecal IFITM1, IFITM2 and IFITM3 expression
analysis for CRC were 0.87, 0.86 and 0.69, respectively (Fig. 1). The sensitivities were 74% (14/19;
95% CI 49–91%), 74% (14/19; 95% CI 49–91%) and 79% (15/19; 95% CI
54–94%), respectively. The specificities were 96% (1/23; 95% CI
78–100%), 96% (1/23; 95% CI 78–100%) and 61% (9/23; 95% CI 39–80%),
respectively, and the Youden index values were 0.69, 0.69 and 0.40,
respectively (Table IV).
When 37 cases were analyzed in which ACTB mRNA was
detected, the AUCs of fecal IFITM1, IFITM2 and IFITM3 expression
analysis for CRC were 0.88, 0.97 and 0.76, respectively (Fig. 1). The sensitivities were 75% (12/16;
95% CI 48–93%), 88% (14/16; 95% CI 62–99%) and 88% (14/16; 95% CI
62–99%), respectively. The specificities were 95% (1/21; 95% CI
76–100%), 95% (1/21; 95% CI 76–100%) and 67% (7/21; 95% CI 43–85%),
respectively, and the Youden index values were 0.70, 0.83 and 0.54,
respectively (Table IV).
The sensitivities of fecal IFITM1, IFITM2 and IFITM3
expression analysis of the patients with Dukes’ A+B were 58% (7/12;
95% CI 30–87%), 67% (8/12; 95% CI 38–95%) and 67% (8/12; 95% CI
38–95%), respectively, and the patients with Dukes’ C+D exhibited
78% (7/9; 95% CI 45–100%), 67% (6/9; 95% CI 34–99%) and 78% (7/9;
95% CI 45–100%), respectively. Therefore, no significant difference
was found in the sensitivities between the two groups. There was
also no significant difference in the sensitivities of fecal
IFITM1, IFITM2 and IFITM3 expression analysis regarding gender,
tumor location, tumor size, RNA concentration or GAPDH expression.
The sensitivities of fecal IFITM2 and IFITM3 expression analysis of
the group in which ACTB expression was positive were significantly
higher than those of the group in which ACTB expression was
negative (P=0.001 and P=0.01, respectively), although no
significant difference was found in the sensitivity of fecal IFITM1
expression analysis between the two groups.
Combination of IFITM1 and IFITM2
The AUCs of IFITM1 and IFITM2 were larger than that
of IFITM3. Therefore, we calculated the sensitivities and
specificities for the combination of IFITM1 and IFITM2 (fecal
IFITM1+2 expression analysis). When analyzed for all 44 cases, the
sensitivity and specificity of fecal IFITM1+2 expression analysis
were found to be 86% (18/21; 95% CI 64–97%) and 96% (1/23; 95% CI
78–100%). When analyzed for cases in which GAPDH mRNA was detected,
the sensitivity and specificity were 95% (18/19; 95% CI 74–100%)
and 96% (1/23; 95% CI 78–100%). When analyzed for cases in which
ACTB mRNA was detected, the sensitivity and specificity were 100%
(16/16; 95% CI 79–100%) and 100% (0/21; 95% CI 84–100%) (Table IV).
The sensitivity of fecal IFITM1+2 analysis of
patients with Dukes’ A+B and that of patients with Dukes’ C+D were
83% (10/12; 95% CI 55–100%) and 89% (8/9; 95% CI 56–100%),
respectively, and no significant difference was noted in the
sensitivities between the two groups. There was also no significant
difference in the sensitivities of fecal IFITM1+2 expression
analysis with regards to gender, tumor location, tumor size or RNA
concentration. The sensitivity of fecal IFITM1+2 expression
analysis of the group in which GAPDH expression was positive was
significantly higher than that of the group in which GAPDH
expression was negative (P=0.01), and the sensitivity of the group
in which ACTB expression was positive was significantly higher than
that of the group in which ACTB expression was negative
(P=0.01).
Discussion
Results of numerous studies on fecal DNA-based
analysis for CRC screening have been reported. A fecal DNA panel
consisting of 21 mutations exhibited 51.6% sensitivity and 94.4%
specificity (11). Since CRC cells
undergo diverse genetic changes, it is difficult to detect CRC with
a high sensitivity by using only fecal DNA-based mutational
analysis.
Fecal methylation analysis of the vimentin gene
provided sensitivity and specificity of 72.5 and 86.9%,
respectively, and the combination of vimentin methylation plus a
DNA integrity assay resulted in 87.5% sensitivity and 82%
specificity (21). In another
study, a fecal SFRP2 methylation assay for CRC screening exhibited
77–90% sensitivity and 77% specificity (20). In the fecal methylation analysis, it
is crucial to decrease the effect of methylation with aging to
improve the specificity.
The number of studies on fecal RNA-based analysis
for CRC screening that are currently available are fewer than those
on fecal DNA-based analysis. The fecal RNA expression analysis of
PTGS2 by semi-quantitative RT-PCR was reported to have
sensitivities of 50–90% and specificities of 93–100% in previous
studies (22,26,30).
Semi-quantitative RT-PCR was used in the majority of previous
studies on fecal RNA expression analysis for CRC screening, while
real-time RT-PCR was used in only a few studies (29). Real-time PCR has a number of
advantages over semi-quantitative PCR, including high speed,
reduction of contamination and a high level of reproducibility,
from the viewpoint of a laboratory test. In the present study, the
sensitivity and specificity of fecal IFITM1+2 expression analysis
determined in all 44 cases were 86 and 96%, respectively. We showed
that CRC is potentially detected with a high sensitivity and
specificity by fecal RNA expression analysis using real-time
RT-PCR.
The quantitation of templates by real-time RT-PCR
allowed us to generate ROC curves for the fecal mRNA expression
analysis, compare the AUCs, and determine the cut-off points at
which optimal sensitivities and specificities are achieved.
Real-time RT-PCR is considered to be effective in determining the
optimal cut-off points efficiently when studying the usefulness of
fecal mRNA expression analysis by RT-PCR for CRC screening.
Up-regulation of the IFITM gene was considered to be
an early event in β-catenin intestinal tumorigenesis (33). No significant difference was noted
in the sensitivities of fecal IFITM1, IFITM2, IFITM3 and IFITM1+2
expression analysis between the group of CRC patients without
metastasis (Dukes’ A+B) and the group of CRC patients with
metastasis (Dukes’ C+D). Therefore, the fecal IFITM expression
analysis appears to be useful in the early detection of CRC.
No significant difference was found in sensitivities
regarding tumor size and location. However, the sensitivity for
tumors of less than 34 mm in diameter were lower than that for
tumors of more than 34 mm in diameter. The sensitivity for tumors
located on the right side of the colon were lower than that for
tumors located on the left side of the colon. A larger study is
therefore required to clarify the differences in sensitivities of
fecal mRNA expression analysis for CRC with regards to tumor size
and/or location.
When analyzed for cases in which the mRNA of the
housekeeping gene was detected, AUCs were found to be larger than
AUCs when analyzed for all 44 cases. Moreover, the sensitivity of
fecal IFITM1+2 expression analysis in the group in which the mRNA
of the housekeeping gene was not detected was significantly lower
than that in the group in which the mRNA of the housekeeping gene
was detected. Fecal IFITM mRNA-negative CRC cases appear to include
not only cases in which IFITM mRNA was not expressed in their
tumors but also ones in which human RNA as templates of RT-PCR was
insufficient in their fecal samples.
In conclusion, detection of fecal mRNA of IFITM1 and
IFITM2 had larger AUCs than that of IFITM3, and the sensitivity was
improved by combining IFITM1 and IFITM2. Since the number of cases
analyzed in the present study was limited, a larger study is
imperative in order to assess sensitivity and specificity. However,
the results of the present study suggest the usefulness of
detecting the fecal mRNA of IFITM1 and IFITM2 by real-time RT-PCR
for CRC screening.
Acknowledgements
Support by Grants-in-Aid for Scientific Research
from the Ministry of Education, Culture, Sports, Science and
Technology of Japan (to H.Y., K.I. and Y.S.)
Abbreviations:
|
ACTB
|
β-actin
|
|
AUC
|
area under the curve
|
|
CI
|
confidence interval
|
|
CRC
|
colorectal cancer
|
|
Ct
|
cycle threshold
|
|
FOBT
|
fecal occult blood testing
|
|
GAPDH
|
glyceraldehyde-3-phosphate
dehydrogenase
|
|
IFITM
|
interferon-induced transmembrane
protein
|
|
mRNA
|
messenger RNA
|
|
PCR
|
polymerase chain reaction
|
|
ROC
|
receiver operating characteristic
|
|
RT
|
reverse transcription
|
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar
|
|
2
|
Pfister DG, Benson AB III and Somerfield
MR: Clinical practice. Surveillance strategies after curative
treatment of colorectal cancer. N Engl J Med. 350:2375–2382. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mandel JS, Church TR, Bond JH, et al: The
effect of fecal occult-blood screening on the incidence of
colorectal cancer. N Engl J Med. 343:1603–1607. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Winawer S, Fletcher R, Rex D, et al:
Colorectal cancer screening and surveillance: clinical guidelines
and rationale-update based on new evidence. Gastroenterology.
124:544–560. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Smith A, Young GP, Cole SR and Bampton P:
Comparison of a brush-sampling fecal immunochemical test for
hemoglobin with a sensitive guaiac-based fecal occult blood test in
detection of colorectal neoplasia. Cancer. 107:2152–2159. 2006.
View Article : Google Scholar
|
|
6
|
Levi Z, Rozen P, Hazazi R, et al: A
quantitative immunochemical fecal occult blood test for colorectal
neoplasia. Ann Intern Med. 146:244–255. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kronborg O, Fenger C, Olsen J, Jørgensen
OD and Søndergaard O: Randomised study of screening for colorectal
cancer with faecal-occult-blood test. Lancet. 348:1467–1471. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hardcastle JD, Chamberlain JO, Robinson
MH, et al: Randomised controlled trial of faecal-occult-blood
screening for colorectal cancer. Lancet. 348:1472–1477. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lieberman DA and Weiss DG: One-time
screening for colorectal cancer with combined fecal occult-blood
testing and examination of the distal colon. N Engl J Med.
345:555–560. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sung JJ, Chan FK, Leung WK, et al:
Screening for colorectal cancer in Chinese: comparison of fecal
occult blood test, flexible sigmoidoscopy, and colonoscopy.
Gastroenterology. 124:608–614. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Imperiale TF, Ransohoff DF, Itzkowitz SH,
Turnbull BA and Ross ME: Fecal DNA versus fecal occult blood for
colorectal-cancer screening in an average-risk population. N Engl J
Med. 351:2704–2714. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Morikawa T, Kato J, Yamaji Y, Wada R,
Mitsushima T and Shiratori Y: A comparison of the immunochemical
fecal occult blood test and total colonoscopy in the asymptomatic
population. Gastroenterology. 129:422–428. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ahlquist DA, Skoletsky JE, Boynton KA, et
al: Colorectal cancer screening by detection of altered human DNA
in stool: feasibility of a multitarget assay panel.
Gastroenterology. 119:1219–1227. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dong SM, Traverso G, Johnson C, et al:
Detecting colorectal cancer in stool with the use of multiple
genetic targets. J Natl Cancer Inst. 93:858–865. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Traverso G, Shuber A, Levin B, et al:
Detection of APC mutations in fecal DNA from patients with
colorectal tumors. N Engl J Med. 346:311–320. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Diehl F, Schmidt K, Durkee KH, et al:
Analysis of mutations in DNA isolated from plasma and stool of
colorectal cancer patients. Gastroenterology. 135:489–498. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lenhard K, Bommer GT, Asutay S, et al:
Analysis of promoter methylation in stool: a novel method for the
detection of colorectal cancer. Clin Gastroenterol Hepatol.
3:142–149. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen WD, Han ZJ, Skoletsky J, et al:
Detection in fecal DNA of colon cancer-specific methylation of the
nonexpressed vimentin gene. J Natl Cancer Inst. 97:1124–1132. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zou H, Harrington J, Rego RL and Ahlquist
DA: A novel method to capture methylated human DNA from stool:
implications for colorectal cancer screening. Clin Chem.
53:1646–1651. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Müller HM, Oberwalder M, Fiegl H, et al:
Methylation changes in faecal DNA: a marker for colorectal cancer
screening? Lancet. 363:1283–1285. 2004.PubMed/NCBI
|
|
21
|
Itzkowitz SH, Jandorf L, Brand R, et al:
Improved fecal DNA test for colorectal cancer screening. Clin
Gastroenterol Hepatol. 5:111–117. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Leung WK, To KF, Man EP, et al: Detection
of hypermethylated DNA or cyclooxygenase-2 messenger RNA in fecal
samples of patients with colorectal cancer or polyps. Am J
Gastroenterol. 102:1070–1076. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Boynton KA, Summerhayes IC, Ahlquist DA
and Shuber AP: DNA integrity as a potential marker for stool-based
detection of colorectal cancer. Clin Chem. 49:1058–1065. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zou H, Harrington JJ, Klatt KK and
Ahlquist DA: A sensitive method to quantify human long DNA in
stool: relevance to colorectal cancer screening. Cancer Epidemiol
Biomarkers Prev. 15:1115–1119. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yamao T, Matsumura Y, Shimada Y, et al:
Abnormal expression of CD44 variants in the exfoliated cells in the
feces of patients with colorectal cancer. Gastroenterology.
114:1196–1205. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kanaoka S, Yoshida K, Miura N, Sugimura H
and Kajimura M: Potential usefulness of detecting cyclooxygenase 2
messenger RNA in feces for colorectal cancer screening.
Gastroenterology. 127:422–427. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lagerholm S, Lagerholm S, Dutta S and Nair
P: Non-invasive detection of c-myc p64, c-myc p67 and c-erbb-2 in
colorectal cancer. Scand J Gastroenterol. 40:1343–1350. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yajima S, Ishii M, Matsushita H, et al:
Expression profiling of fecal colonocytes for RNA-based screening
of colorectal cancer. Int J Oncol. 31:1029–1037. 2007.PubMed/NCBI
|
|
29
|
Koga Y, Yasunaga M, Moriya Y, et al:
Detection of colorectal cancer cells from feces using quantitative
real-time RT-PCR for colorectal cancer diagnosis. Cancer Sci.
99:1977–1983. 2008.PubMed/NCBI
|
|
30
|
Takai T, Kanaoka S, Yoshida K, et al:
Fecal cyclooxygenase 2 plus matrix metalloproteinase 7 mRNA assay
as a marker for colorectal cancer screening. Cancer Epidemiol
Biomarkers Prev. 18:1888–1893. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yu YJ, Majumdar AP, Nechvatal JM, et al:
Exfoliated cells in stool: a source for reverse
transcription-PCR-based analysis of biomarkers of gastrointestinal
cancer. Cancer Epidemiol Biomarkers Prev. 17:455–458.
2008.PubMed/NCBI
|
|
32
|
Kitahara O, Furukawa Y, Tanaka T, et al:
Alterations of gene expression during colorectal carcinogenesis
revealed by cDNA microarrays after laser-capture microdissection of
tumor tissues and normal epithelia. Cancer Res. 61:3544–3549.
2001.
|
|
33
|
Andreu P, Colnot S, Godard C, et al:
Identification of the IFITM family as a new molecular marker in
human colorectal tumors. Cancer Res. 66:1949–1955. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Akobeng AK: Understanding diagnostic tests
3: receiver operating characteristic curves. Acta Pediatrica.
96:644–647. 2007. View Article : Google Scholar : PubMed/NCBI
|