Introduction
Oral cancer accounts for approximately 2% of
systemic malignant tumors, of which 90% are squamous cell
carcinomas (1). Buccal cancer is
one of the most common oral cancers. Although great progress has
been made in a variety of oral cancer treatment methods, the 5-year
survival rate for buccal and oral cancer following treatment is
only 55–60% (1,2). Previous studies confirmed that oral
cancer is a complex, multiphase, multi-step pathological process
involving multiple genetic changes (3,4), where
a number of genes play various roles at different stages of cancer
development. The majority (80%) of oral cancers originate from
precancerous lesions (5). At
present, no effective treatment method for the prevention of the
occurrence of precancerous lesions exists. If effective
intervention to prevent the occurrence of precancerous lesions at
the molecular level is achieved, the early and effective prevention
of oral cancer may also be achieved.
The development of microarray technology allows for
whole-genome microarray technology to provide a comprehensive
understanding of genetic changes during carcinogenesis and screen
cancer-related genes to determine cellular gene expression
profiles. Microarray technology is currently used in studying oral
tumor markers (6,7), tumor molecular typing (8,9), drug
screening (10) and the comparative
analysis of discrepancies in gene expression patterns between
cancer tissues and adjacent or normal tissue (11,12).
However, no reports are currently available regarding the analysis
of disease-related genes during the transformation of normal buccal
mucosa to precancerous lesions on a whole-genome level.
An experimental animal model of
7,12-dimethylbenz(a)-anthracene (DMBA)-induced oral buccal mucosa
squamous cell carcinoma in Syrian golden hamsters was first
reported in 1954 by Salley (13).
The animal model was later shown to exhibit carcinogenesis, growth
characteristics and biological behavior similar to that of human
oral mucosal epithelial cells (14,15).
Thus, it is considered to be an ideal animal model for studying
oral mucosal carcinogenesis.
In this study, the DMBA-induced oral buccal mucosa
squamous cell carcinoma in Syrian golden hamsters was used to
establish precancerous lesions. Agilent whole-genome microarray
containing 41,000 genes/EST sequences and bioinformatics analysis
were used to screen and analyze precancerous disease-related genes
during the transformation of normal buccal mucosa to precancerous
lesions in order to identify cellular gene expression profiles.
This procedure may provide data and methodology for the exploration
of the molecular mechanisms underlying precancerous lesions and
their prevention.
Materials and methods
Main reagents and instruments
The following reagents and instruments were
utilized: DMBA (Sigma Corporation, USA), 20 Syrian golden hamsters
(Chengdu Institute of Biological Products, China), PCR instrument
(PTC-100, MJ Company), hybrid furnace (G-2545A, Agilent
Technologies, Palo Alto, CA, USA), agilent scanner (G2565AA,
Agilent Technologies), single-labeled agilent rat cDNA microarray
(Agilent Technologies), Cy3 NHS ester (GE Healthcare, PA13105),
aaUTP (Ambion, AM8436), low RNA input linear amplication kit
(Agilent, 5184-3523), gene expression hybridization kit (Agilent,
5188–5242), gene expression wash buffer kit (Agilent, 5188–5327),
stabilization and drying solution (Agilent, 5185–5979), gasket
slide (Agilent, G2534-60003), hybridization chamber (Agilent,
G2534A), RNeasy mini kit (Qiagen, 74106) and RT-PCR kit (Bioflux).
The animals and experimental procedures were approved by the
Management Committee of Laboratory Animals Use, Institute of
Laboratory Animal, Chongqing Medical University, China.
Establishment of hamster model with cheek
pouch precancerous lesions
A total of 20 Syrian golden hamsters (10 males and
10 females; weight, 90–120 g; age, 6–8 weeks) were randomly divided
into 2 groups (10 per group). Group I was considered the normal
control group and group II the experimental group. The hamsters in
group II were fixed on a self-made mouse shelf and the cheek
pouches were opened. The bilateral hamster cheek pouches were
painted with 0.5% DMBA acetone solution using a No. 1 oil pen every
Monday, Wednesday and Friday afternoon for 6 weeks. The animals
were then starved for 2 h. Group I was not treated. The animals
were sacrificed in the 6th week and cheek pouch tissue samples were
removed. Half of the tissue was immediately placed in liquid
nitrogen for preservation and the remaining half was fixed using a
10% formalin solution. The fixed tissue samples were then embedded
in paraffin, sectioned, stained with hematoxylin and eosin
(H&E) and mounted. The pathological examination was performed
via observation using light microscopy.
Diagnostic criteria for precancerous
lesions
The precancerous lesions were diagnosed according to
the 12 criteria set by the World Health Organization (WHO)
(16). These criteria included: i)
the disappearance of basal cell polarity, ii) stratified-like
changes in basal cells, iii) increased ratio of nucleus to
cytoplasm, iv) drop-shaped epithelial nail protrusion, v) disorder
of the epithelial layer, vi) increased and abnormal mitosis, vii)
increased mitosis (1/2 epithelium undergoing mitosis), viii)
pleomorphic cells, ix) hyperchromatic nuclei, x) enlarged
nucleolus, xi) diminished or lack of intercellular adhesion and
xii) dyskeratosis. The diagnostic classification was based on the
observation of the aforementioned criteria: 1–2 criteria, mild
dysplasia; 3–4, moderate epithelial dysplasia; and ≥5, severe
dysplasia or carcinoma in situ. In this study, moderate
dysplasial tissues were used for the experiments.
Extraction and purification of RNA from
normal hamster cheek pouch mucosa and precancerous lesion
samples
Total RNA from samples from groups I and II was
extracted using the TRIzol extraction kit (Invitrogen, Carlsbad,
CA, USA). The Qiagen RNeasy kit was used to purify total RNA,
according to the manufacturer's instructions. Equivalent amounts of
total RNA from groups I and II were dissolved in Milli-Q water and
preserved at −80°C.
cRNA labeling and synthesis
Extracted RNA samples were single-labeled with Cy3
using reverse transcription. The concrete steps were: 2 μg of RNA
from the samples obtained from the 2 groups was added to two test
tubes (1.5 ml) and 5 μl of primer was added. RNase-free water was
added for a total volume of 11.5 μl and the contents were combined.
The solution was incubated in a 60°C water bath for 10 min and
cooled in ice water for 5 min. Then, 2 μl DDT (100 mmol/l), 1 μl
dNTP, 4 μl 5 first strand buffer, 0.5 μl RNaseOUT and 1 μl MMLV RT
were added to the RNA samples and combined. cRNA was synthesized in
a 2-h reaction performed at 40°C, with a thermal cover heated to
65°C. The synthetic cRNA was labeled using aaUTP and purified using
the Qiagen RNeasy mini kit. The cRNA concentration was measured
using a spectrophotometer. The cRNA was then labeled and
repurified.
cRNA fragmentation and microarray
hybridization
Approximately 875 ng of Cy3-labeled cRNA extracted
from the normal mucosa and precancerous lesions was added to 11 μl
blocking agent and 2.2 μl fragmentation buffer. Nuclease-free water
was added to make the total volume 55 μl. The mixture was incubated
at 60°C for 30 min for fragmentation, following which 55 μl GEX
hybridization buffer was added. Then, 100 μl of the above solution
was added to the microarray for rolling hybridization at 65°C for
17 h at a speed of 10 rpm. The microarray was removed and washed in
lotion 1 for 1 min and then washed with lotion 2 for 1 min at
37°C.
Microarray scanning and data
processing
Microarray hybridization results were scanned using
an Agilent G2565AA fluorescence scanner and read by feature
extraction software. The scanner resolution was 5 μm, and the
photomultiplier tubes were set to 100 and 10% for 2 scans. The scan
results were automatically merged by Agilent software and feature
extraction was used for uniform treatment. The homogenization
coefficient in this experiment was 0.853. The ratio of average
signal intensity and average background of the microarray was
>3, which was consistent with the manufacturer's standards.
Bioinformatics analysis
Ratios of ≥2 and ≤0.5 were the threshold values for
screening differentially expressed genes. Differentially expressed
genes underwent functional classification according to the Gene
Ontology (GO) classification standard. The databases KECG, GENMAPP
and BIOCARTA (www.biorag.com) were used for signal
pathway analysis and identification of the signal pathways for
abnormally expressed genes. P<0.05 was used both to determine
the significance of pathways and screen for target pathways.
Verification of microarray results by
RT-PCR
From the microarray results, the Atp6v0d2
up-regulation and Sfrp2 down-regulation were randomly selected for
verification by RT-PCR. In accordance with the instructions, the
TRIzol kit (Invitrogen) was used for total RNA extraction from
samples from groups I and II. RNA (1 μg) was reverse transcribed to
cDNA in a 20 μl reaction using a thermocycler. The process involved
3 min pre-denaturation at 94°C; 30 cycles of 1 min denaturation at
94°C, 30 sec annealing at 52°C and 1 min extension at 72°C; and
after 10 min at 72°C it was terminated. The PCR primers were
designed by Primer 3.0 (Table
I).
 | Table IPrimers for RT-PCR used to verify the
microarray results. |
Table I
Primers for RT-PCR used to verify the
microarray results.
| Gene name | Gene library
number | Primer
sequences | Length of product
(bp) |
|---|
| Atp6v0d2 | NM_001011972 | F:
5′-CGAGGGTGCAAAGCCAGCCT-3′ | 210 |
| R:
5′-AGCCGCAGTCCCTCCGGATA-3′ |
| Sfrp2 | NM_001100700.1 | F:
5′-CTCCTGCCGCCCACAGAGGA-3′ | 180 |
| R:
5′-GATGCTGCGGGAGATGCGCT-3′ |
| β-actin | NM_001101.2 | F:
5′-CCCGCCACCAGTTCGCCAT-3′ | 240 |
| R:
5′-TGTGGGTGACCCCGTCTCCG-3′ |
Results
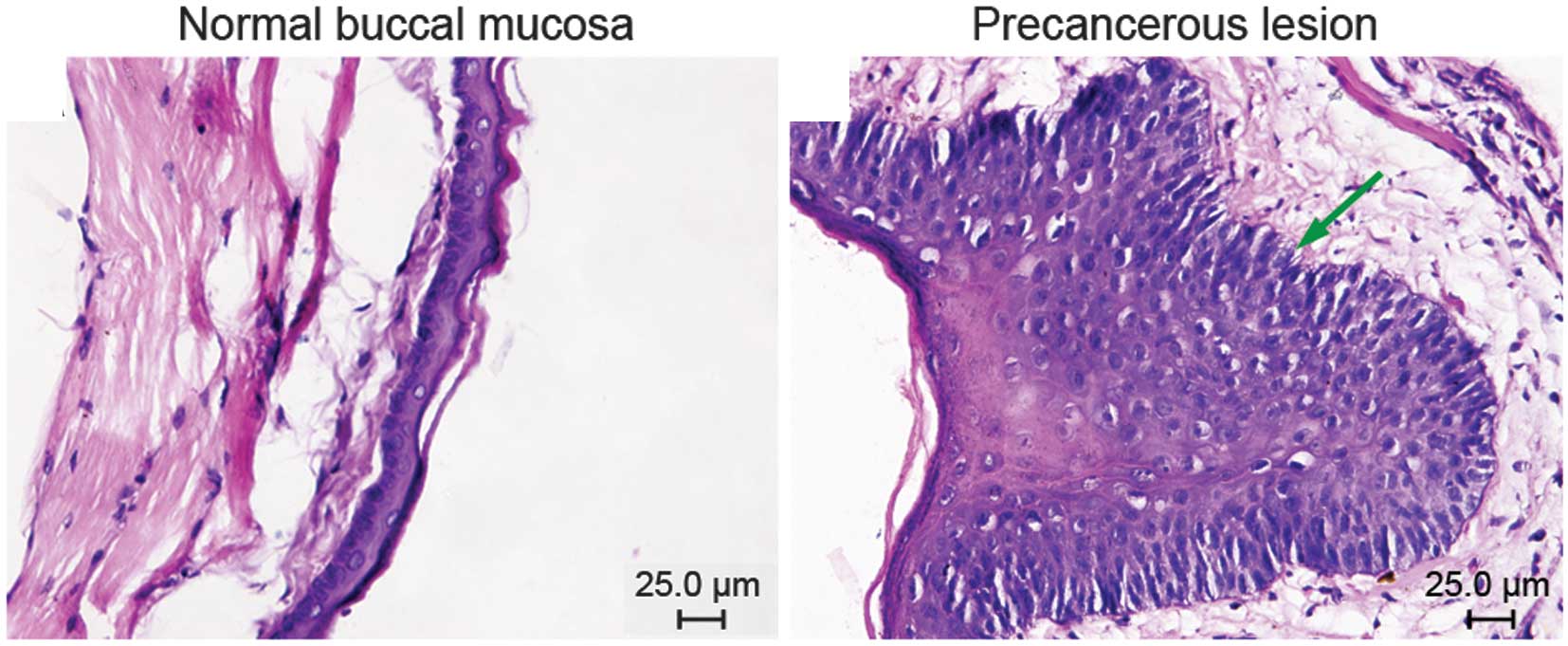
Conventional hematoxylin and eosin
staining of normal buccal mucosa and precancerous lesions
Group I comprised normal buccal mucosa. Cases in
group II were classified according to the WHO criteria for
precancerous lesions: 3 showed mild dysplasia and 7 moderate
dysplasia (Fig. 1).
Microarray hybridization results and
analysis of biological data Screen of differentially expressed
genes in normal buccal mucosa and precancerous lesions
A total of 1331 genes, including 1278 known, 53
unknown, 747 up-regulated and 584 down-regulated genes, were found
to be differentially expressed during the transformation of normal
buccal mucosa to precancerous lesions in golden hamsters.
Gene Ontology functional
classification
A total of 1331 differentially expressed genes were
divided into 8 major functional groups according to the GO
classification criteria (Table
II).
 | Table IIGene Ontology classification for
differentially expressed genes. |
Table II
Gene Ontology classification for
differentially expressed genes.
| Gene Ontology
functional classification | No. of up-regulated
genes | No. of
down-regulated genes | Total no. | Percentage |
|---|
| Genes related to
regulation of cell physiological processes | 239 | 187 | 426 | 30.78 |
| Genes related to
cell structure | 232 | 181 | 413 | 29.84 |
| Genes related to
molecular location | 87 | 68 | 155 | 11.20 |
| Genes related to
macromolecule metabolism | 61 | 48 | 109 | 7.88 |
| Genes related to
immune regulation | 58 | 46 | 104 | 7.51 |
| Genes related to
signal transduction | 38 | 30 | 68 | 4.91 |
| Genes related to
material transfer | 31 | 25 | 56 | 4.04 |
| Unkown genes | 36 | 17 | 53 | 3.83 |
Pathway analysis
Pathway analysis revealed a total of 14 gene
interaction pathways significantly associated with the 1278 known
differentially expressed genes (P<0.05). Only 21 genes in the
1278 differentially expressed genes enhanced the 14 pathways
(Table III).
 | Table IIIPathway analysis. |
Table III
Pathway analysis.
| Pathway | P-value | Gene symbol |
|---|
| Caspase-8 is formed
from pro-caspase-8 | 0.0366 | Up: 0
Down: Casp8 |
| Activation of
pro-caspase-8 | 0.0366 | Up: 0
Down: Casp8 |
| Switching of
origins to a post-replicative state | 0.0364 | Up:
Orc1L
Down: 0 |
| Association of
licensing factors with the pre-replicative complex | 0.0366 | Up:
Orc1L
Down: 0 |
| Signaling by G
protein-coupled receptor | 0.0224 | Up: CCL5;
CXCL12
Down: CCL20; P518/Qrfp |
| Class
A/1(rhodopsin-like receptors) | 0.0224 | Up: CCL5;
CXCL12
Down: CCL20; P518/Qrfp |
| Chemokine receptors
bind chemokines | 0.0106 | Up: CCL5;
CXCL12
Down: CCL20 |
| Formation of fibrin
clot | 0.0158 | Up: F5;
Serping1
Down: TFPI |
| Integrin cell
surface interactions | 0.0321 | Up: Vcam1;
Fn1
Down: 0 |
| Signaling in immune
system | 0.0447 | Up: Angpt2;
Lcp2; Vcam1
Down: Cxadr |
| Arachidonic acid
metabolism | 0.0120 | Up: 0
Down: Cyp2b13 |
| Metabolism of
xenobiotics by cytochrome P450 | 0.0400 | Up:0
Down: Cyp2b13 |
| IL5 | 0.0313 | Up: Lyn; IL5ra;
Hck; Btk
Down: 0 |
| IL6 | 0.0302 | Up: RGD1564385
(fes); Btk; Vav1; Lyn; Hck
Down: 0 |
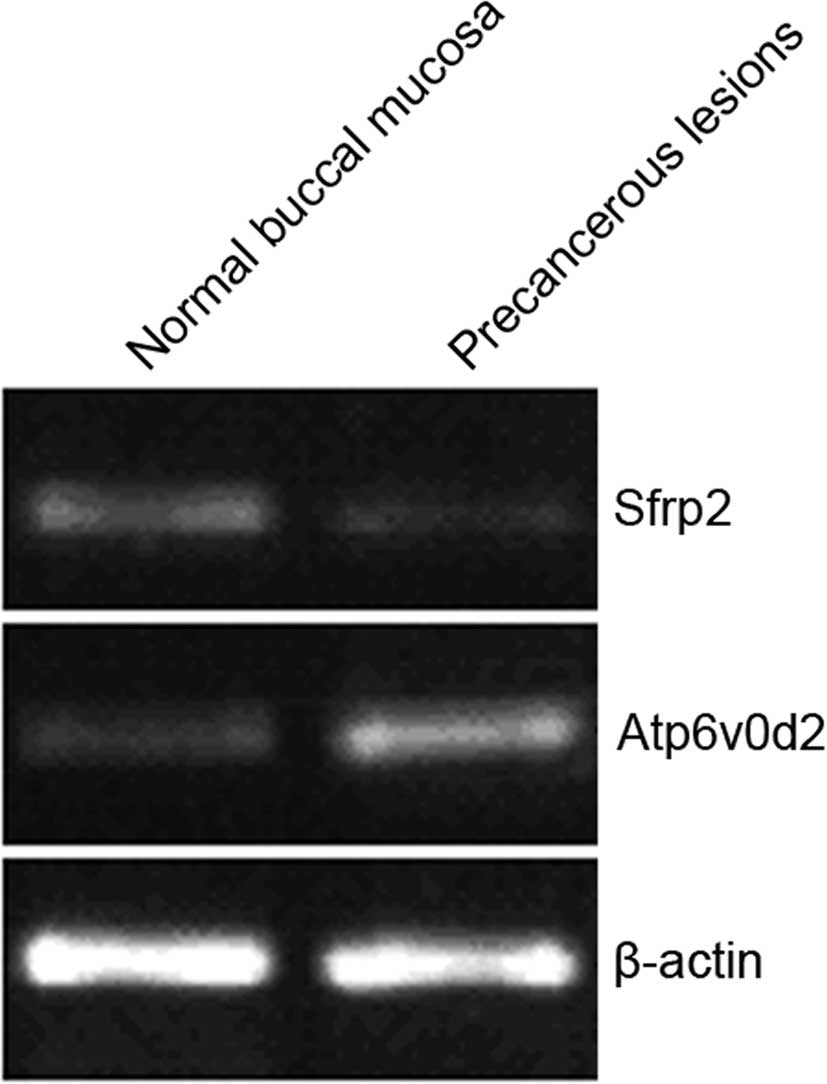
RT-PCR verification results
Atp6v0d2 up-regulation and Sfrp2 down-regulation
were verified using RT-PCR (Fig.
2). The optical density values of the PCR bands were analyzed
and compared with β-actin. Expression of Atp6v0d2 and Sfrp2 in
cheek pouch mucosa precancerous lesions was 6.21 and 0.231 times,
respectively, that of the normal cheek pouch mucosa, which was
consistent with the microarray results (Table IV).
 | Table IVComparison of RT-PCR results with
microarray results. |
Table IV
Comparison of RT-PCR results with
microarray results.
| Gene name | Gene mRNA/β-actin
mRNA (mean ± SD) | B/A ratio | Microarray
ratio |
|---|
|
| | |
|---|
| A | B | | |
|---|
| Atp6v0d2 | 0.1292±0.0023 | 0.8023±0.0143 | 6.21 | 6.70 |
| Sfrp2 | 0.4201±0.0034 | 0.09702±0.0007 | 0.231 | −4.65 |
Discussion
This study identified 1331 genes that were
differentially expressed during the transformation of normal buccal
mucosa to precancerous lesions in golden hamsters. Additionally,
the genes were found to act in 14 different pathways, suggesting
that the occurrence of precancerous oral buccal mucosa lesions in
golden hamsters is a complicated pathological process that involves
a number of genetic changes that affect numerous pathways. GO
analysis revealed that the differentially expressed genes can be
divided into 8 functional groups, in which 30.78% of the genes
identified are involved in the regulation of cell physiological
processes and 29.84% are related to cell structure. Together, these
two functional groups account for 60.62% of the 1331 genes
identified, indicating that these genes are crucial in the
formation of oral precancerous lesions.
Pathway analysis identified 14 gene interaction
pathways that were significantly correlated to the 1278 known
differentially expressed genes. Only 21 of the 1278 differentially
expressed genes were enhanced in the 14 pathways. Since pathways
express interactions between proteins, we suggest that the
following candidate genes are key in the development of
precancerous oral buccal mucosal lesions: Cyp2b13,
Orc1L, casp8, CCL5, CXCL12,
CCL20, Serping1, P518/Qrfp, F5,
TFPI, Vcam1, Fn1, Angpt2, Lcp2,
Cxadr, Lyn, Hck, Btk,
RGD1564385/fes, Vav1 and IL5ra.
In the 14 pathways, caspase-8 (casp8) is formed from
pro-casp8 following pro-casp8 activation in the apoptosis pathway.
The caspase family of proteases is a group of aspartate-specific
hydrolases containing cysteine. Following cascade activation of the
caspases, the Fas receptor is activated and induces apoptosis, for
which casp8 activation is the first step in cascade activation
(17,18). In this study, casp8 was
down-regulated, resulting in cell apoptosis.
The association of licensing factors with the
pre-replicative complex pathway was primarily correlated to the
regulation of DNA replication. DNA synthesis is required to induce
a post-replicative state in the cell. Origin recognition complex
(Orc) is a protein complex involved in the replication of
eukaryotic DNA that regulates cell proliferation and the cell cycle
(19). Orc1 is a key member of this
protein complex (20) and is a
crucial factor in the cell cycle progression from the G0 to G1/S
phase. In this study, Orc1L was up-regulated in two pathways and
promoted cell proliferation.
Chemokine receptors bind chemokines, and
rhodopsin-like receptor pathways are subordinate branches of the G
protein-coupled receptor (GPCR) signaling pathway. GPCRs are
members of the seven-pass transmembrane domain receptor
superfamily. GPCRs, combined with chemokines, activate PKC kinases
and Ras and Rho family members. Moreover, these receptors play a
key role in cell growth, adhesion and directional migration.
Previous studies showed that a high expression of chemokines CCL5
and CXCL12 promotes cell proliferation and cancer cell metastasis
(21,22). CCL5 and CXCL12 were up-regulated in
the three preceding pathways (signaling by GPCR, chemokine
receptors bind chemokines and rhodopsin-like receptor pathways) in
this study. CCL20 is known to be involved in the directional
migration of dendritic and T cells. CCL20 promotes the chemotaxis
of immature dendritic cells to tumor areas, activates T cells and
triggers an immune response, thus inhibiting tumor growth (23). Previous studies showed that CCL20 is
highly expressed in tumors (24).
However, in this study, CCL20 in precancerous lesions was expressed
at low levels in the three preceding pathways. This low expression
may have led to a decreased local immunity, thereby promoting
lesion development. P518/Qrfp is an endogenous GPCR ligand
and its involvement in tumor development has not previously been
reported.
Formation of the fibrin clot is a branch of the
hemostasis pathway (25). F5
encodes coagulation factor V and the Serping1 gene encodes
the C1 inhibitor. TPFI inhibits the extrinsic coagulation
process and activation of FIX by the FVIIa tissue factor complex.
In this pathway, F5 and Serping1 expression were up-regulated and
TFPI expression was down-regulated, suggesting that the main
characteristics of the vascular changes in precancerous lesions
enhanced coagulation and inhibited anticoagulation.
The integrin cell surface pathway primarily mediates
adhesion between cells and the extracellular matrix (ECM). Vcaml is
a member of the immunoglobulin superfamily and high expression
levels of Vcam1 enhance endothelial cell migration and promote
angiogenesis (26). High expression
levels of Fn, a macromolecular glycoprotein with a double-stranded
structure (27), increase cell-cell
and cell-ECM adhesion. In the integrin pathway, Vcam1 and Fn1 are
highly expressed, enhancing the adhesion between cells and the ECM,
thereby promoting angiogenesis.
We identified four abnormally expressed genes
(Angpt2, Lcp2, Vcam1 and Cxadr) that
were enhanced in signaling pathways involved in the immune system.
A high expression of Angpt2 and Vcam1 promotes angiogenesis
(26,28). Lcp2 plays a key role in T cell
activation; however, its role in tumor formation has not previously
been reported. Cxadr is a cell adhesion molecule and its decreased
expression weakens intercellular adhesion (29). During precancerous lesion formation,
we observed that Angpt2, Lcp2 and Vcam1 were
up-regulated, whereas Cxadr was down-regulated. The combined
effect of the three genes may promote angiogenesis.
The arachidonic acid (AA) and cytochrome P450
(CYP450)-dependent xenobiotic metabolic pathways are subordinate
branches of biological oxidation pathways. AA is an essential fatty
acid for the human body and previous studies have shown that
abnormal AA in the cyclooxygenase and lipoxygenase metabolic
pathways is associated with tumor occurrence and development
(30,31). CYP450 is a group of isoenzymes with
a similar structure and function encoded by a superfamily of genes.
CYP450 plays a key role in the metabolism of endogenous and
external carcinogens, chemical poisons and toxins. CYP450 induces
genetic mutations or inhibits gene expression (32). Cyp2b13 is a member of a second
CYP450 family and its expression in the two pathways was
down-regulated. This down-regulation may lead to the decreased
metabolism of exogenous toxins such as DMBA, and promote the
formation of precancerous lesions.
The IL5 and IL6 pathways are correlated to the
regulation of intracellular protein tyrosine kinase (PTK) activity.
PTKs are crucial for cell signal transduction and the regulation of
cell growth, proliferation and differentiation (33). Lyn, Btk, Hck and Fes are members of
the non-receptor-type PTK family. A high expression of Lyn, Btk,
Hck and Fes results in cell proliferation, anti-apoptosis and the
promotion of angiogenesis (34–36).
Up-regulation of Vav1 and IL5ra is correlated to tyrosine
phosphorylation and the activation of signal transduction.
Up-regulation of Lyn, IL5ra, Hck, Btk, RGD1564385 (fes) and Vav1 in
the pathway enhances tyrosine kinase activity, thereby promoting
cell proliferation and angiogenesis, and inhibiting apoptosis.
This study showed that precancerous buccal mucosal
lesions in hamsters not only involved the abnormal expression of a
number of genes, but that the identified differentially expressed
genes induce the occurrence of precancerous lesions through various
pathways. The overall mechanism involved in the formation of
hamster buccal mucosal precancerous lesions requires that the
chemical carcinogen DMBA act on the oral buccal mucosa. This
activity results in changes in gene expression including that of
Cyp2b13. Subsequently, DMBA metabolism (with cell and genetic
toxicity) is reduced through the metabolism of xenobiotics by the
CYP450 and AA metabolic pathways. The dual role of cell metabolism
disorders and DMBA toxicity likely results in changes in the
aforementioned genes and pathways, including activation of DNA
replication (Orc1L up-regulation) and inhibition of the cell
apoptosis pathway (casp8 down-regulation). The gene expression
changes induced cell proliferation, angiogenesis, apoptosis
inhibition, cell cycle regulation disorders and decreased local
immunity, resulting in the occurrence and development of
precancerous lesions. Therefore, we hypothesize that
Cyp2b13, Orc1L, casp8 and the remaining 21 genes are
candidates in the development of oral precancerous lesions.
Inhibitors that regulate these genes or pathways effectively may
improve treatment methods and chemoprophylaxis of precancerous
lesions.
Acknowledgements
The study was supported by the Medical Foundation of
Chongqing City Health Bureau, Chongqing, P.R. China (No.
03-2-073).
References
|
1
|
Kademani D, Bell RB, Schmidt BL,
Blanchaert R, Fernandes R, Lambert P and Tucker WM: Oral and
maxillofacial surgeons treating oral cancer: a preliminary report
from the American Association of Oral and Maxillofacial Surgeons
Task Force on Oral Cancer. J Oral Maxillofac Surg. 66:2151–2157.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bernier J and Cooper JS: Chemoradiation
after surgery for high-risk head and neck cancer patients: how
strong is the evidence? Oncologist. 10:215–224. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Carinci F, Lo Muzio L, Piattelli A, et al:
Genetic portrait of mild and severe lingual dysplasia. Oral Oncol.
41:365–374. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Choi P and Chen C: Genetic expression
profiles and biologic pathway alterations in head and neck squamous
cell carcinoma. Cancer. 104:1113–1128. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shiu MN and Chen TH: Impact of betel quid,
tobacco and alcohol on three-stage disease natural history of oral
leukoplakia and cancer: implication for prevention of oral cancer.
Eur J Cancer Prev. 13:39–45. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nagata M, Fujita H, Ida H, et al:
Identification of potential biomarkers of lymph node metastasis in
oral squamous cell carcinoma by cDNA microarray analysis. Int J
Cancer. 106:683–689. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kashiwazaki H, Hassan NM, Hamada J, et al:
Gene expression profile changes correlated with lymph node
metastasis in oral squamous cell carcinoma. Odontology. 96:38–43.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Arora S, Matta A, Shukla NK, Deo SV and
Ralhan R: Identification of differentially expressed genes in oral
squamous cell carcinoma. Mol Carcinog. 42:97–108. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Erdem NF, Carlson ER and Gerard DA:
Characterization of gene expression profiles of 3 different human
oral squamous cell carcinoma cell lines with different invasion and
metastatic capacities. J Oral Maxillofac Surg. 66:918–927. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mendez E, Cheng C, Farwell DG, et al:
Transcriptional expression profiles of oral squamous cell
carcinomas. Cancer. 95:1482–1494. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gottschlich S, Ambrosch P, Cordes C,
Gorogh T, Schreiber S and Hasler R: Gene expression profiling of
head and neck squamous cell carcinoma using cDNA microarrays. Int J
Oncol. 29:605–613. 2006.PubMed/NCBI
|
|
12
|
Estilo CL, O-charoenrat P, Talbot S, et
al: Oral tongue cancer gene expression profiling: identification of
novel potential prognosticators by oligonucleotide microarray
analysis. BMC Cancer. 9:112009. View Article : Google Scholar
|
|
13
|
Salley JJ: Experimental carcinogenesis in
the cheek pouch of the Syrian hamster. J Dent Res. 33:253–262.
1954. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Feng L and Wang Z: Chemopreventive effect
of celecoxib in oral precancers and cancers. Laryngoscope.
116:1842–1845. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gimenez-Conti IB and Slaga TJ: The hamster
cheek pouch carcinogenesis model. J Cell Biochem Suppl. 17F:83–90.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
World Health Organization. Report of a
meeting of investigators on the histological definition of
precancerous lesions. Geneva: World Health Organization; pp.
7311973
|
|
17
|
Stupack DG, Teitz T, Potter MD, et al:
Potentiation of neuroblastoma metastasis by loss of caspase-8.
Nature. 439:95–99. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
McKee AE and Thiele CJ: Targeting caspase
8 to reduce the formation of metastases in neuroblastoma. Expert
Opin Ther Targets. 10:703–708. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
DePamphilis ML: Cell cycle dependent
regulation of the origin recognition complex. Cell Cycle. 4:70–79.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tatsumi Y, Ohta S, Kimura H, Tsurimoto T
and Obuse C: The ORC1 cycle in human cells: I. cell cycle-regulated
oscillation of human ORC1. J Biol Chem. 278:41528–41534. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Burger JA and Kipps TJ: CXCR4: a key
receptor in the crosstalk between tumor cells and their
microenvironment. Blood. 107:1761–1767. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Vaday GG, Peehl DM, Kadam PA and Lawrence
DM: Expression of CCL5 (RANTES) and CCR5 in prostate cancer.
Prostate. 66:124–134. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fushimi T, Kojima A, Moore MA and Crystal
RG: Macrophage inflammatory protein 3alpha transgene attracts
dendritic cells to established murine tumors and suppresses tumor
growth. J Clin Invest. 105:1383–1393. 2000. View Article : Google Scholar
|
|
24
|
Rubie C, Frick VO, Wagner M, et al:
Chemokine expression in hepatocellular carcinoma versus colorectal
liver metastases. World J Gastroenterol. 12:6627–6633.
2006.PubMed/NCBI
|
|
25
|
Yu JL, May L, Lhotak V, et al: Oncogenic
events regulate tissue factor expression in colorectal cancer
cells: implications for tumor progression and angiogenesis. Blood.
105:1734–1741. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nakao S, Kuwano T, Ishibashi T, Kuwano M
and Ono M: Synergistic effect of TNF-alpha in soluble
VCAM-1-induced angiogenesis through alpha 4 integrins. J Immunol.
170:5704–5711. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gubala E, Wiench M, Oczko-Wojciechowska M,
et al: [Gene expression analysis by DNA microarray in papillary
thyroid cancer]. Endokrynol Pol. 56:752–757. 2005.
|
|
28
|
Koga K, Todaka T, Morioka M, et al:
Expression of angiopoietin- 2 in human glioma cells and its role
for angiogenesis. Cancer Res. 61:6248–6254. 2001.PubMed/NCBI
|
|
29
|
Zhang LL, He DL, Li X, Li L, Zhu GD, Zhang
D and Wang XY: Overexpression of coxsackie and adenovirus receptor
inhibit growth of human bladder cancer cell in vitro and in vivo.
Acta Pharmacol Sin. 28:895–900. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cao Y and Prescott SM: Many actions of
cyclooxygenase-2 in cellular dynamics and in cancer. J Cell
Physiol. 190:279–286. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Poff CD and Balazy M: Drugs that target
lipoxygenases and leukotrienes as emerging therapies for asthma and
cancer. Curr Drug Targets Inflamm Allergy. 3:19–33. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Brookman-Amissah N, Mackay AG and Swann
PF: Isolation and sequencing of the cDNA of a novel cytochrome P450
from rat oesophagus. Carcinogenesis. 22:155–160. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Konecny GE, Pegram MD, Venkatesan N, et
al: Activity of the dual kinase inhibitor lapatinib (GW572016)
against HER-2-overexpressing and trastuzumab-treated breast cancer
cells. Cancer Res. 66:1630–1639. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Suh HS, Kim MO and Lee SC: Inhibition of
granulocyte-macrophage colony-stimulating factor signaling and
microglial proliferation by anti-CD45RO: role of Hck tyrosine
kinase and phosphatidylinositol 3-kinase/Akt. J Immunol.
174:2712–2719. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Grosso S, Puissant A, Dufies M, et al:
Gene expression profiling of imatinib and PD166326-resistant CML
cell lines identifies Fyn as a gene associated with resistance to
BCR-ABL inhibitors. Mol Cancer Ther. 8:1924–1933. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Stephens LR, Anderson KE and Hawkins PT:
Src family kinases mediate receptor-stimulated, phosphoinositide
3-kinase-dependent, tyrosine phosphorylation of dual adaptor for
phosphotyrosine and 3-phosphoinositides-1 in endothelial and B cell
lines. J Biol Chem. 276:42767–42773. 2001. View Article : Google Scholar
|