Introduction
Chronic myeloid leukemia (CML) is a
myeloproliferative disease that originates in abnormal pluripotent
bone marrow stem cells and is consistently associated with the
Philadelphia (Ph) chromosome which normally leads to BCR/ABL gene
fusion (1). The Ph chromosome
created as a result of t(9;22) (q34;q11) is observed in more than
90% of CML patients. The BCR-ABL fusion gene is formed by the
transposing of the 3′ portion of the ABL oncogene from 9q34 to the
5′ portion of the BCR gene on chromosome 22, and this fusion gene
encodes a constitutive active tyrosine kinase (2). Masked or variant Ph translocations
characterize 5–10% of CML cases. A masked Ph chromosome is found in
cases with a normal karyotype, as a result of a cryptic
rearrangement, or in patients with complex changes where the
typical t(9;22) (q34;q11) is not detectable by G-banding (3). These rearrangements are detected by
fluorescence in situ hybridization (FISH) (4). The variant Ph translocation is
cytogenetically classified as involving chromosomes 9 and 22, as
well as one or more other chromosomes (5,6).
Imatinib mesylate (Glivec, formerly STI571) was designed
specifically to inhibit the tyrosine kinase activity of the BCR/ABL
protein and other tyrosine kinases such as cABL, c-KIT and PDGF
(platelet-derived growth factor receptor). By binding to an active
site of the tyrosine kinase, Glivec switches off downstream
signaling, cells stop proliferating and apoptosis ensues (7). Various studies showed a high
efficiency of imatinib therapy to achieve a complete or major
cytogenetic response, i.e., a reduction to 0–34% Ph-positive cells.
This positive effect is achieved in cases with a simple t(9;22)
combined with complex translocations resulting in BCR/ABL gene
fusion, as well as in cases with cytogenetic clonal evolution
(8,9).
This study investigated a novel Ph
chromosome-positive CML case with a new complex rearrangement
formed by four chromosomes and new complex aberrations involving
four chromosomal breakpoints. Treatment with imatinib proved
successful. In this case, the high-resolution array-proven
multicolor banding (aMCB) technique was crucial in the detection of
genetic changes.
Materials and methods
Case report
A 43-year-old female was diagnosed as suffering from
CML in the chronic phase (CP) following a blood cell count that was
initiated in January 2004 due to a white blood cell count (WBC) of
8.0×109/l and fever. The patient was treated with
imatinib mesylate at a dose of 400 mg/day overall for 10 months.
During that period the patient showed no symptoms. However, in July
2006, the patient presented for the second time with a WBC of
4.8×109/l consisting of 61% neutrophils, 38% lymphocytes
and 1% immature cells. The platelet count was 375×109/l
and the hemoglobin level was 12.1 g/dl. The patient was treated
with imatinib mesylate at a dose of 400 mg/day overall for 30
months. A physical examination revealed no hepatomegaly or
splenomegaly, and a bone marrow trephine did not show any fibrosis.
The patient was lost during follow-up. In August 2009, she
succumbed to unknown causes.
Cytogenetic analysis
Chromosome analysis using GTG-banding was performed
according to standard procedures (10). A total of 20 metaphases derived from
the unstimulated bone marrow of the patient were analyzed.
Karyotypes were described according to the International System for
Human Cytogenetic Nomenclature (11).
Molecular cytogenetics
FISH using a LSI BCR/ABL dual-color dual fusion
translocation probe (Abbott Molecular/Vysis, USA), whole chromosome
painting (WCP) probe for chromosomes 12, 16 and 22 (MetaSystems,
Altlussheim, Germany) and an alpha satellite probe (CEP) for
chromosome 9 (Abbott Molecular/Vysis) were applied according to the
manufacturer’s instructions (12).
Array-proven multicolor banding probe (aMCB) sets based on
microdissection-derived region-specific libraries for chromosomes
9, 12, 16 and 22 were applied as previously described (13,14). A
total of 20 metaphase spreads were analyzed, using a fluorescence
microscope (AxioImager.Z1 mot, Zeiss) equipped with appropriate
filter sets to distinguish between a maximum of five fluorochromes
and the counterstain DAPI (4′,6-diamino-2-phenylindole). Image
capturing and processing were carried out using an ISIS imaging
system (MetaSystems) for the MCB evaluation.
Results
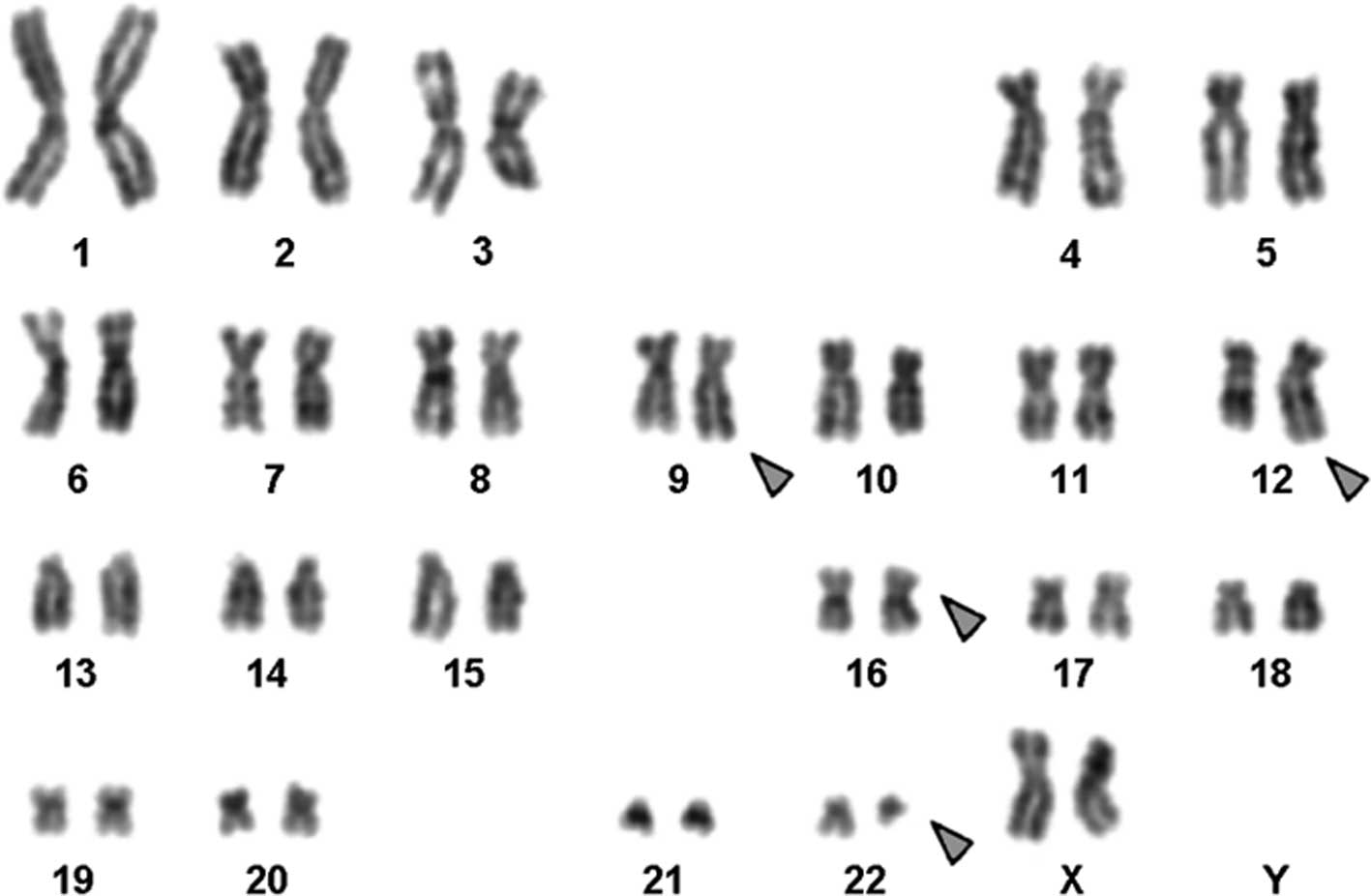
Karyotyping was performed following the initiation
of chemotherapy treatment, with various karyotypic changes. A
complex karyotype 46,XX,t(9;12;16;22)/46,XX was determined in the
GTG-banding (Fig. 1) and was
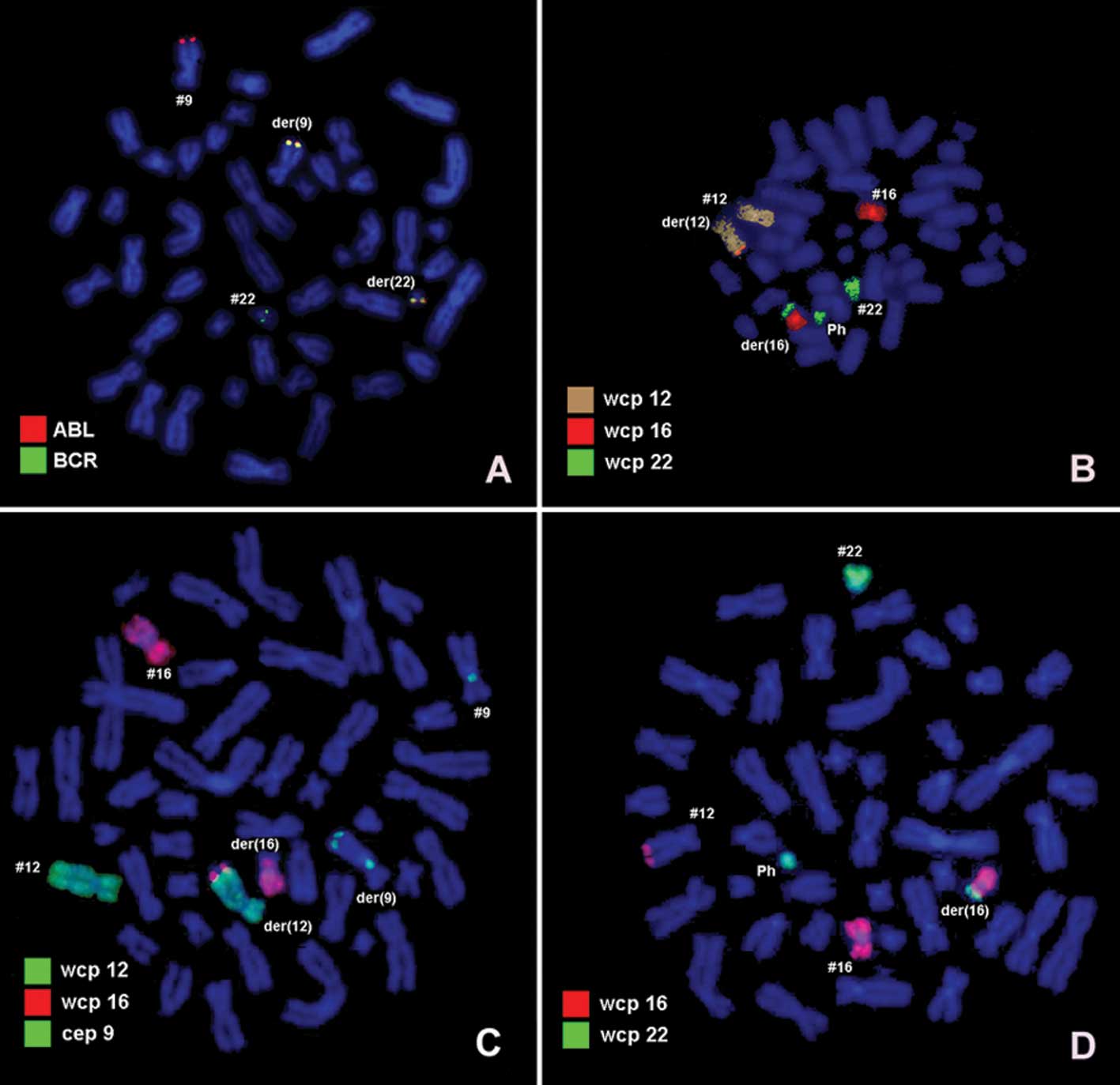
further specified by molecular cytogenetic studies (Figs. 2 and 3). A dual-color FISH using a probe
specific for BCR and ABL revealed a typical Ph chromosome with the
BCR/ABL fusion gene (Fig. 2A).
Dual-color FISH using WCP- and CEP-specific probes was applied to
evidence further rearrangements (Figs.
2B and D). Thus, the chromosomes 9, 12, 16 and 22 were found to
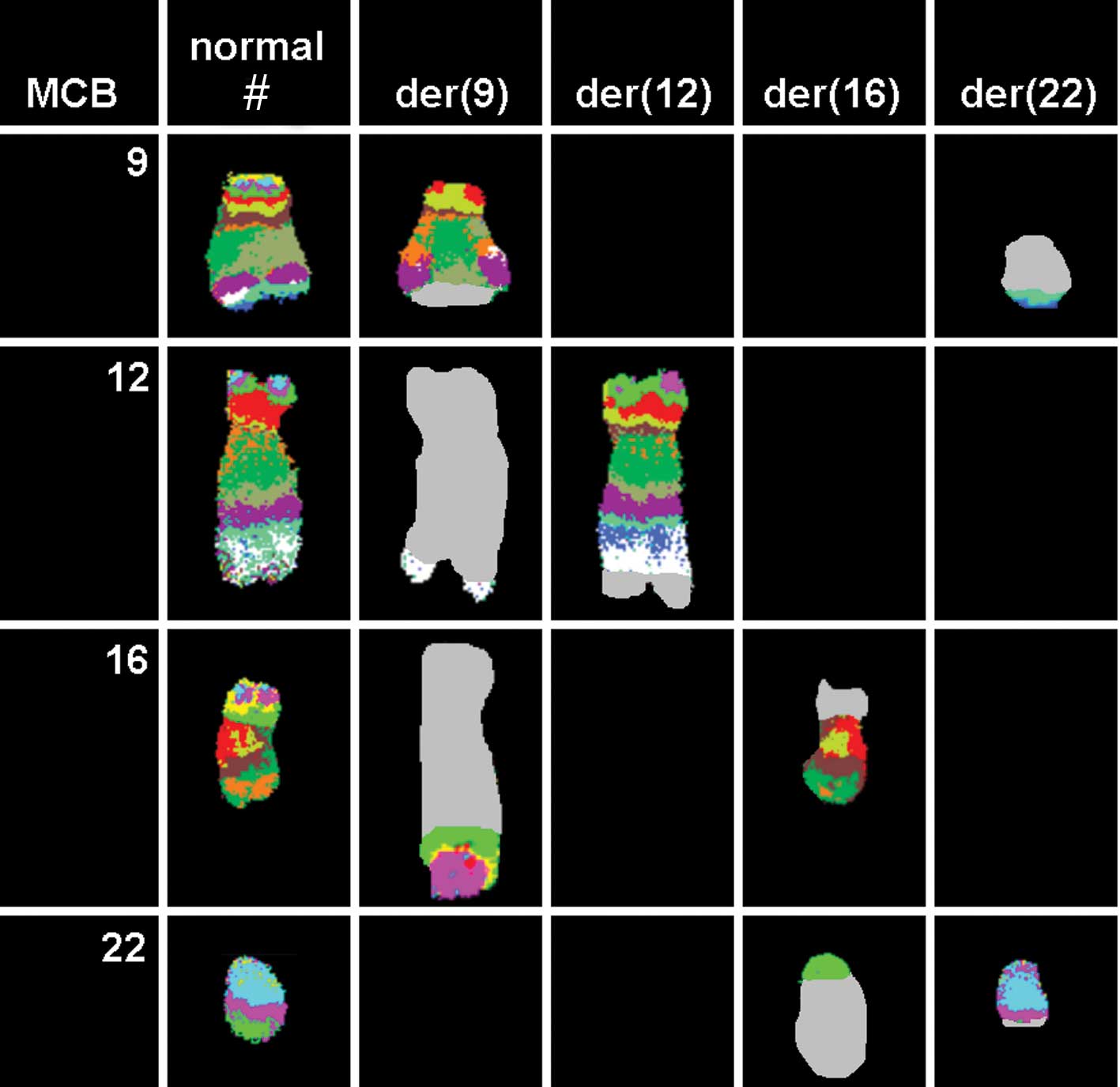
be involved in the karyotypic changes. aMCB using probes for the
corresponding chromosomes was performed as previously reported
(14). A complex translocation
among the four chromosomes was detected (Fig. 3), and the final karyotypes obtained
were: 46,XX,t(9;12;16;22)(q34;q24.2~24.31;p11.2;q11)[12]/
46,XX[8].
Discussion
We described a novel Philadelphia (Ph)
chromosome-positive CML case with the new complex variant
translocation t(9;12;16;22)(q34;q24.2–24.31;p11.2;q11). To the best
of our knowledge, this translocation has never previously been
observed in CML (15).
In 5–10% of Ph chromosome-positive CML cases,
complex translocations are noted in both chromosomes 9 and 22 and
other chromosomes, such as 12 and 16 (3). At present, it appears that variant
translocations are able to affect any chromosome. However, it has
been suggested that the distribution of the breakpoints is
non-random, with the chromosomal bands most susceptible to breakage
being 1p36, 3p21, 5q31, 6p21, 9q22, 10q22, 11q13, 12p13, 17p13,
17q21, 17q25, 19q13, 21q22, 22q12 and 22q13 (1).
Two potential mechanisms for variant translocation
formation have been suggested. The first is a single-event
rearrangement via the simultaneous breakage of a number of
chromosomes followed by mismatched joining (6). Nacheva et al proposed a
classical Ph translocation followed by further translocation
between chromosomes 9 and 22, in addition to a third chromosome
(16). The mechanism of the
formation of a variant Ph translocation may have prognostic
importance in that a two-event mechanism indicates clonal
evolution, whereas a variant translocation occurring via a single
genomic rearrangement may confer a similar prognosis to the
classical Ph translocation (17).
In conclusion, we report a novel case of a Ph
chromosome-positive CML in CP with a new complex variant Ph
translocation involving the four chromosomal aberrations of 9q34,
q24.2–24.31, 16p11.2 and 22q11. Notably, the reported patient had a
beneficial response to imatinib, although she succumbed to unknown
causes.
Acknowledgements
We thank Professor I. Othman, Director General of
Atomic Energy Commission of Syria (AECS), and Dr N. Mirali, Head of
Molecular Biology and Biotechnology Department, for their support.
This study was supported by the AECS, and in parts by the
Stefan-Morsch-Stiftung, Monika-Kutzner-Stiftung and the DAAD
(D/07/09624).
References
|
1
|
Johansson B, Fioretos T and Mitelman F:
Cytogenetic and molecular genetic evolution of chronic myeloid
leukemia. Acta Haematol. 107:76–94. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shtivelman E, Lifshitz B, Gale RP and
Canaani E: Fused transcript of abl and bcr genes in chronic
myelogenous leukemia. Nature. 315:550–554. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
La Starza R, Testoni N, Lafage-Pochitaloff
M, Ruggeri D, Ottaviani E, Perla G, Martelli MF, Marynen P and
Mecucci C: Complex variant Philadelphia translocations involving
the short arm of chromosome 6 in chronic myeloid leukemia.
Haematologica. 87:143–147. 2002.
|
|
4
|
Todorić-Zivanović B, Marisavljević D,
Surace C, Cemerikić V, Marković O, Krtolica K, Tatomirović Z,
Cikota B, Magić Z and Rocchi M: A Ph-negative chronic myeloid
leukemia with a complex BCR/ABL rearrangement and a
t(6;9)(p21;q34.1). Cancer Genet Cytogenet. 166:180–185.
2006.PubMed/NCBI
|
|
5
|
Naumann S and Decker HJ: Genesis of
variant Philadelphia chromosome translocations in chronic
myelocytic leukemia. Cancer Genet Cytogenet. 147:18–22. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fitzgerald PH and Morris CM: Complex
chromosomal translocations in the Philadelphia chromosome
leukemias. Serial translocations or a concerted genomic
rearrangement? Cancer Genet Cytogenet. 57:143–151. 1991. View Article : Google Scholar
|
|
7
|
Griffen J: The biology of signal
transduction The biology of signal transduction inhibition: basic
science to novel therapies. Semin Oncol. 28:3–8. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kantarjian H, Sawyers C, Hochhaus A,
Guilhot F, Schiffer C, Gambacorti-Passerini C, Niederwieser D,
Resta D, Capdeville R, Zoellner U, Talpaz M, Druker B, Goldman J,
O’Brien SG, Russell N, Fischer T, Ottmann O, Cony-Makhoul P, Facon
T, Stone R, Miller C, Tallman M, Brown R, Schuster M, Loughran T,
Gratwohl A, Mandelli F, Saglio G, Lazzarino M, Russo D, Baccarani M
and Morra E: International STI571 CML Study Group: Hematologic and
cytogenetic responses to imatinib mesylate in chronic myelogenous
leukemia. N Engl J Med. 346:645–652. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cortes JE, Talpaz M, Giles F, O’Brien S,
Rios MB, Shan J, Garcia-Manero G, Faderl S, Thomas DA, Wierda W,
Ferrajoli A, Jeha S and Kantarjian HM: Prognostic significance of
cytogenetic clonal evolution in patients with chronic myelogenous
leukemia on imatinib mesylate therapy. Blood. 101:3794–3800. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Claussen U, Michel S, Mühlig P, Westermann
M, Grummt UW, Kromeyer-Hauschild K and Liehr T: Demystifying
chromosome preparation and the implications for the concept of
chromosome condensation during mitosis. Cytogenet Genome Res.
98:136–146. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shaffer L, Slovak M and Cambell L; ISCN
(2009). An International System for Human Cytogenetic Nomenclature.
S. Karger; Basel: 2009
|
|
12
|
Al-Achkar W, Wafa A and Nweder MS: A
complex translocation t(5;9;22) in Philadelphia cells involving the
short arm of chromosome 5 in a case of chronic myelogenous
leukemia. J Exp Clin Cancer Res. 26:411–415. 2007.PubMed/NCBI
|
|
13
|
Weise A, Mrasek K, Fickelscher I, Claussen
U, Cheung SW, Cai WW, Liehr T and Kosyakova N: Molecular definition
of high-resolution multicolor banding probes: first within the
human DNA sequence anchored FISH banding probe set. J Histochem
Cytochem. 56:487–493. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liehr T, Heller A, Starke H, Rubtsov N,
Trifonov V, Mrasek K, Weise A, Kuechler A and Claussen U:
Microdissection based high resolution multicolor banding for all 24
human chromosomes. Int J Mol Med. 9:335–339. 2002.PubMed/NCBI
|
|
15
|
Mitelman F, Johansson B and Mertens F:
Mitelman Database of Chromosome Aberrations in Cancer. 2009,
http://cgap.nci.nih.gov/Chromosomes/Mitelman.
|
|
16
|
Nacheva E, Holloway T, Brown K, Bloxham D
and Green AR: Philadelphia-negative chronic myeloid leukaemia:
detection by FISH of BCR-ABL fusion gene localized either to
chromosome 9 or chromosome 22. Br J Haematol. 87:409–412. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Reid AG, Huntly BJP, Grace C, Green AR and
Nacheva EP: Survival implications of molecular heterogeneity in
variant Philadelphia-positive chronic myeloid leukaemia. Br J
Haematol. 121:419–427. 2003. View Article : Google Scholar : PubMed/NCBI
|