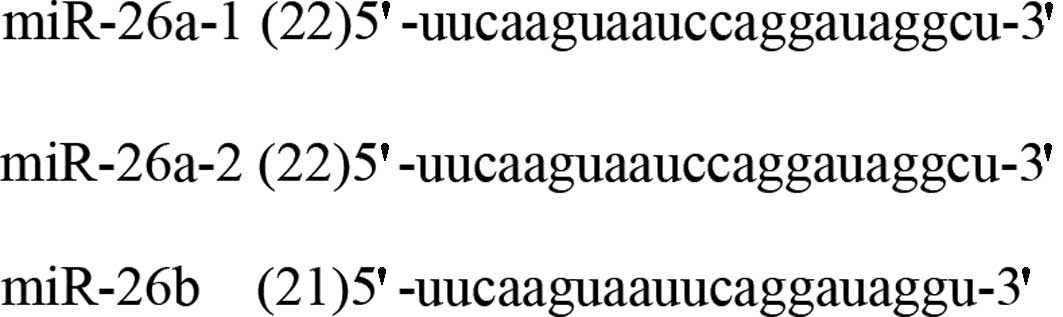1. Processing and general functions of
miRNAs
MicroRNAs (miRNAs) are a class of endogenously small
(approximately 22-nucleotide) non-coding RNAs present extensively
in eukaryotes, which were first discovered in a 1994 investigation
into eelworms. Pri-miRNA (approximately 1000 bp transcription
mostly through RNA polymerase II and nearly 20% of the remaining
through RNA polymerase III) forms a distinctive ‘hairpin’ stem-loop
secondary structure and enters the miRNA processing complex.
Through a series of processing by Drosha (intranuclear), Exp-5 and
Dicer (intracytoplasm), the precursor miRNA (pre-miRNA;
approximately 60–70 nt, 5′ end phosphorylated 3′ end of the
suspended 2 nt sudden) (1) is
gradually processed into the miRNA* duplex, and then forms the
single-stranded structure of mature miRNA and a miRNA* by helicase.
The miRNA* is rapidly degraded. Simultaneously, the mature miRNA is
involved in the formation of RNA-induced silencing complex (RISC)
sand target mRNA through the 3′ untranslated region (UTR) complete
complemenary (plants) or imperfect complementary (animals), which
mediates target mRNA degradation or translation inhibition, thereby
regulating almost 33% of the protein coding gene (2–5).
The regulation is based on the imperfect complement
binding between the seed region of miRNA and 3′-UTR of target mRNA.
miRNAs then alter the expression of target genes at transcriptional
and post-transcriptional levels, including genes encoding
transcription factors and RNA regulatory proteins. The subsequent
effects of this process may alter the levels of other mRNA (or
protein interaction). miRNA may therefore affect the expression of
multiple genes and play a role in a variety of biological processes
through transcriptional and post-transcriptional regulation
(1). miRNA may change mRNA
stability by binding with the 3′-UTR of target mRNA (1). Consequently, researchers have paid
substantial attention to miRNA and numerous reports are available
on the biogenetic basic function of miRNA and its significant role
in disease and normal tissue physiological processes (6–12).
Tumor miRNA expression profiles have shown that the tumor has
different relatively specific miRNA expression profiles, and that
the same miRNA may differ between tumor types (13). miRNAs act as oncogenes or tumor
suppressor genes involved in tumorigenesis (14). Calin et al reported that
52.5% of miRNA genes located in fragile sites are associated with
cancer (15). Different miRNA
expression profiles exist in different stages of normal tissue
differentiation and development processes.
A study regarding single-nucleotide polymorphisms
(SNPs) of miRNA in the human genome revealed that miRNA had a lower
SNP density compared to the genome average, with only 24 SNPs
located in the 325 miRNAs studied. Findings of this study also
showed 2 miRNA regions, hsa-mir-26a-2 in the CTDSP2 gene and
hsa-mir-128-1 in the R3HDM1 gene, among a Spanish population
(16). Concomitantly, Diederichs
and Haber reported that the expression levels of miRNAs were
globally reduced in cancer compared with matched normal tissues. A
panel of 91 cancer-derived cell lines was analyzed for sequence
variations in 15 miRNAs involved in tumorigenesis by virtue of
their known target transcripts or their localization to sites of
frequent chromosomal instability. One of these miRNAs is miR-26
(17).
miR-26 is a functional miRNA that has merited
previous investigation. Various microarray expression profiles
showed that miR-26 expression is disordered in a number of human
tumors (18,19). The expression of miR-26 is altered
during normal tissue growth and development processes such as
myogenesis. Consequently, this review summarizes previous
investigations into the expression of miR-26 in different types of
diseases and different stages of growth and development.
2. Structure and functions of miR-26
MiR-26a-1, miR-26a-2 and miR-26b are the only 3
subtypes of the hsa-miR-26 family, and are located in chromosomes
3, 12 and 2, respectively. The mature miRNA of miR-26a-1 and
miR-26a-2 possesses the same sequence, with the exception of 2
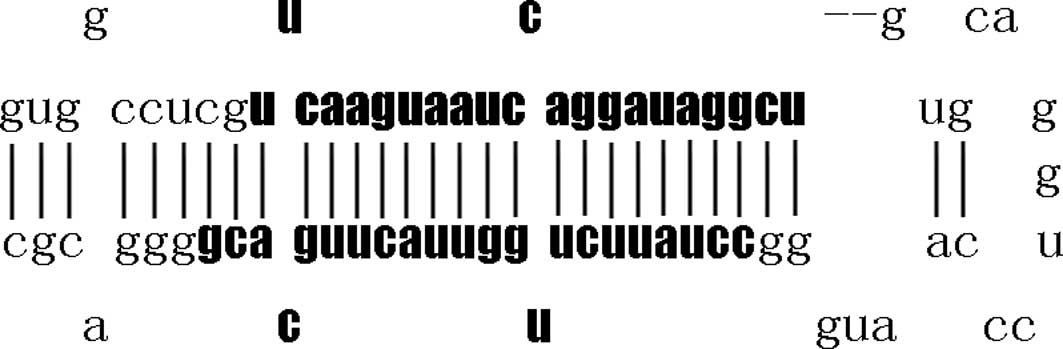
different nucleotides in mature miR-26b (Fig. 1). Pre-miR-26 with stem-loop
structure (Fig. 2) processed into
mature miR-26 by a series of enzymes of intranuclear and
intracytoplasm. The mature miR-26 was 21–22 nucleotides in length,
with a seed region of approximately 6–7 nucleotides. The sequence
of miR-26 seed region, an important region for binding to target
mRNA, is highly consistent in different genera members. Numerous
tumors and normal tissues exhibit different expression of miR-26
during growth, development and tumorigenesis and miR-26 may
participate in various biological processes through imperfect
sequence complementarity binding between seed region and 3′UTR of
target mRNA. miR-26 may repress the target gene translation and
decrease expression levels of target gene-coding protein. miRNA has
numerous significant target genes in regulatory networks for gene
therapy, which renders it important in tumorigenesis and tumor
therapy. It has been observed that expression of miR-26 is
disordered in many tumors and that it has specific functions in
different tumors.
3. miR-26 and tumors: down- and
up-regulation
Increasingly, studies have shown that miRNAs are
involved in tumorigenesis and act as oncogenic or tumor-suppressive
genes in various tumor types. miRNAs are therefore divided into two
types: oncogenic miRNA (oncomir) such as miR-17–92 cluster, and
tumor-suppressive miRNA such as miR-34. As stated above, the
expression of miR-26 is disordered in a number of tumors, but its
functions remain unknown.
Down-regulation
MiR-26 is down-regulated in various tumor types and
may exhibit tumor-suppressive activity during tumorigenesis in
these tumors. Wang et al reported that the expression of
miR-26 is down-regulated in groups T1 (differentiated grade 1–2)
and T2 (differentiated grade 1–3) bladder cancer. miR-26 is the
most significant of the 4 down-regulated miRNAs in the T2 group
bladder tumors. The decrease in expression of 4 miRNAs is common
for all bladder tumors regardless of cancer stage or tumor
differentiation. miR-26 may therefore be a significant marker in
bladder cancer (20). Maillot et
al observed that E2-repressed miR-26a and miR-181a regulated
numerous genes associated with cell growth and proliferation
through estrogen receptors and transcription factors. Additionally,
miRNA expression was regulated in breast cancer in women who had
received anti-estrogen neoadjuvant therapy. This regulation
indicated that miRNA was correlated to anti-estrogen resistance of
breast cancer (21). Zhang et
al noted that miR-26a was down-regulated in breast cancer
specimens and cell lines, and that it initiated apoptosis through
endogenous and exogenous pathways activated by caspase-8 and 9 as
well as through binding to the 3′-UTR of MTDH and EZH2 directly.
MiR-26 impairs the in vitro colony-forming and in
vivo tumor-loading abilities of MCF7 cells (22). Moreover, Yu et al found that
the expression of miR-26 in oral squamous cell carcinoma in Syrian
hamsters was decreased (23).
Visone et al reported that miR-26a was significantly
decreased in anaplastic carcinomas (ATC) in comparison to normal
thyroid tissue. The overexpression of miR-26 in 2 human ATC-derived
cell lines significantly decreased thyroid carcinogenesis,
suggesting a crucial role for miR-26a down-regulation in thyroid
carcinogenesis. miR-26a may exhibit a tumor-suppressive activity
since cell-growth inhibition was achieved (24). In their study, Ciarapica et
al compared a group of 5 rhabdomyosarcoma (RMS1–5) with two
muscle tissues as controls (MT1 and MT2). These authors observed
that miR-26a exhibited a significant negative fold-change in all
RMS1–5 compared with MT1 and MT2, indicating that its
down-regulation has a potential role in rhabdomyosarcoma. They also
confirmed that EZH2, a validated target gene of miR-26a, was
up-regulated in rhabdomyosarcoma, indicating that a
miR-26a-dependent regulation of EZH2 may be active in
rhabdomyosarcoma cells (25).
Myc is a significant oncogene that is always mutated
or amplified in various types of human cancer. Myc is associated
with cell growth and proliferation and is crucial to tumorigenesis
and progression. Following the study of cell lines and murine
lymphoma models (26), a study on
miRNA expression profiles in Myc-driven tumorigenesis reported that
the expression of miR-26 decreased in Burkitt lymphoma (BL; an
aggressive variant of non-Hodgkin’s B-cell lymphoma).
Overexpression of miR-26a in human BL-derived cell lines using the
episomal expression system produces an increased percentage of
cells in the G1 phase and fewer cells in the S/G2 phase within the
miR-26a-expressing cell lines as compared to the empty vector
controls 72 h after transfection, indicating that miR-26 arrested
cell cycle progression. miR-26a was consistently repressed by Myc
in multiple tumors, indicating that this miRNA may have a strong
tumor-suppressor function in Myc-induced lymphomas. These authors
attempted to elucidate the effector pathway for miR-26a. The focus
was on potential targets, nominated in at least 2 different
databases, which may have been involved in the observed G1 arrest
by miR-26a overexpression. The results showed that a significant
degression of EZH2 (Enhancer of Zeste Homolog 2) in the gene
expression profile was induced by miR-26a over-expression, in both
human BL-derived and murine lymphoma cell lines. Myc may thus
contribute to the up-regulation of EZH2 via the down-regulation of
its targeting miRNA. The suppression of the miR-26a-mediated
attenuation of EZH2 expression by Myc was shown to play a critical
role in lymphomagenesis. A positive feedback loop comprising Myc
and EZH2 was involved in the formation of the malignant lymphoma
phenotype (26). Kota et al
reported that the expression of miR-26 was down-regulated in
hepatocellular carcinoma (HCC) cells and that overexpression of
miR-26a in liver cancer cells in vitro induced an increase
in cells of G1 stage as well as a decrease in cells of the S stage,
indicating that miR-26a induced a G1 arrest. Systemic
administration of this miRNA to a mouse model of HCC using
adeno-associated virus resulted in the inhibition of cancer cell
growth and proliferation, and increased tumor-specific apoptosis.
This process indicated that miR-26a was a tumor-suppressor miRNA
(27). A study is available
pertaining to miRNA expression, survival and response to interferon
in 455 patients with HCC who had undergone radical tumor resection.
Expression of miR-26a and miR-26b was found to be higher in female
than in male individuals in the non-tumor liver tissue of
hepatocellular carcinoma patients, indicating that a higher
expression of miR-26 may explain the lower morbidity. Expression of
miR-26 was down-regulated in tumors compared with paired
non-cancerous tissues, indicating that the sexpression of miR-26
was associated with HCC. Moreover, tumors with a reduced miR-26
expression exhibited a distinct transcriptomic pattern and
activated the signaling pathways between nuclear factor κB and
interleukin-6, which may play a role in tumor development according
to gene networks information. Patients with a lower miR-26
expression in HCC had a shorter survival but a more favorable
response to interferon therapy than those with a higher miR-26
expression in HCC, indicating that miR-26 was associated with
post-operative survival (28).
The studies of miR-26 expression profiles in tumors
raise 3 significant points. First, miR-26 expression decreases in
bladder tumor, breast cancer, oral squamous cell carcinoma,
anaplastic carcinomas, Burkitt lymphoma HCC and rhabdomyosarcoma,
and it may be a suppressor miRNA in those tumors. Second, Myc is a
significant oncogene associated with tumorigenesis and is always
mutated or amplified in various types of human cancer. miR-26a is
one of the miRNAs consistently repressed by Myc in multiple tumors
(26). Third, the 3′-UTR of EZH2, a
crucial subunit of Polycomb repressive complex 2, has binding sites
with a seed region of miR-26. EZH2 is an important transcription
regulation factor in tumorigenesis, and since it regulates the
global level of gene expression it may be one of the downstream
target genes of miR-26 (26,27).
The expression of EZH2 is up-regulated in numerous tumor types and
miR-26 may exhibit functions of tumor suppression via
down-regulation of the translation of EZH2.
Up-regulation
miR-26 expression reportedly decreased in various
tumor types, where it functioned as tumor-suppressor miRNA.
However, various recent studies revealed that the expression of
miR-26 was up-regulated in tumors such as glioma (29,30).
Huse et al reported that miR-26a was overexpressed in
high-grade glioma (the most prevalent diagnostic category of
primary brain tumor in the adult population) and directly targeted
PTEN. MiR-26a was frequently amplified at the DNA level in a subset
of human high-grade gliomas and its over-expression was strongly
associated with monoallelic PTEN loss. Overexpression of miR-26a in
a murine glioma model using the RCAS/tv-a system revealed that
miR-26a repressed the endogenous PTEN protein effectively by
binding to 3 potential binding sites in the PTEN 3′-UTR in a
relevant glioma model system, promoting tumorigenesis. miR-26 may
therefore be an oncomir in glioma. Notably, the study by Huse et
al indicated that miR-26a over-expression in LN-18 cells also
decreased the expression of EZH2 and SMAD1, indicating that the
transcripts of the two proteins were effectively targeted by miRNA
during gliomagenesis (29). This
review suggests that miR-26 regulates numerous target genes
simultaneously and that its role is completely different in certain
tumors. In their study on an oncomir/oncogene cluster regulating
glioblastoma survivorship, Kim et al noted that miR-26a
regulated PTEN expression and AKT activation and inhibited RB1 and
MAP3K2/MEKK2 expression and JNK-dependent apoptosis in glioblastoma
multiforme (GBM). PTEN was therefore considered to be one of the
downstream target genes of miR-26a in GBM. Overexpression of
miR-26a increased GBM cell growth compared to the control cells,
consistent with the characteristics of AKT activation and miR-26
overexpression by the miR-26 mimic or lentiviral in U87 GBM cells,
lack of functional PTEN, decreased RB1 expression and increased DNA
synthesis. The MAP3K2 gene encodes MEKK2, which is involved in JNK
and ERK5 activation, and JNK activation can promote apoptosis in
GBM cells. miR-26a therefore decreased JNK-dependent apoptosis by
inhibiting MAP3K2/MEKK2 expression in GBM cells, whereas the
miR-26a inhibitor increased this expression. PTEN was therefore not
the only target gene of miR-26a in GBM (30).
The above-mentioned studies regarding miR-26
expression in glioma raise 3 significant points. First, miR-26a
expression increases in GBM and promotes tumor cell growth and
proliferation function as an oncomir in GBM. Second, PTEN may be
one of the downstream target genes of miR-26a in GBM, due to the 3
potential binding sites with miR-26a in its 3′-UTR. miR-26a
regulates PTEN expression and AKT activation and inhibits RB1 and
MAP3K2/MEKK2 expression and JNK-dependent apoptosis in GBM. Third,
miR-26 regulates the expression of a set of target genes and the
role of these genes may be completely different in specific tumors.
The involvement of miR-26 in these tumors therefore depends on
those target genes and their corresponding pathways.
4. miR-26 and non-tumor disease
miR-26 expression is not only disordered in
tumorigenesis but also alterable in non-tumor diseases. Primary
billiary cirrhosis (PBC) caused by chronic cholestasis is often
accompanied by autoimmune diseases such as rheumatoid arthritis and
scleroderma. Padgett et al observed that a total of 35
independent miRNAs in the miRNA expression profile are disordered
and that miR-26a is one of the down-regulated miRNAs. The predicted
targets of these alternative miRNAs are known to affect cell
proliferation, apoptosis, inflammation, oxidative stress and
metabolism associated with the development of PBC (31). The roles that miR-26 plays in
non-tumor diseases have yet to be clarified, and further studies
are required.
5. Relationship between miR-26 and normal
tissue growth and development
miRNAs play crucial roles in numerous biological
processes via their target genes. It is known that miR-26 plays a
significant role in the growth, development and cell
differentiation of different tissues. Murine fetal hepatoblast
cells can be induced to differentiate between the hepatocyte and
cholangiocyte, and the expression of miRNAs is altered during the
differentiation processes. MiR-23b cluster miRNAs including miR-26a
have a gradient of effects on cell fate choice in the fetal mouse
liver via transforming growth factor-β (TGFβ)/bone morphogenetic
protein signal pathway. Low levels of the miR-23b miRNAs are
required in cholangiocytes to allow TGFβ signaling and bile duct
formation (32). In osteogenesis
several miRNAs (for example miR-26a) regulate osteoblast cell
growth and differentiation in human adipose tissue-derived stem
cells (33). The function of miR-26
in myogenesis is clearest in studies of miRNA in normal tissue
growth and development. Wong and Tellam (34) investigated the miRNA expression
profile of myogenesis. These authors identified 6 miRNAs with
2-fold or greater significant expression alteration in myotubes.
These miRNAs were divided into 3 groups according to the level of
expression. MiR-26a, an up-regulated miRNA in Cluster II, was
up-regulated more gradually during the course of myogenesis.
Overexpression of miR-26a in murine myogenic C2C12 cells induced
creatine kinase activity, an enzyme that markedly increased during
myogenesis. myoD and myogenin mRNA expression levels were also
up-regulated, and EZH2 was identified as a potential target of
miR-26a. Overexpression of miR-26a decreased EZH2 mRNA expression
and suppressed the activity of a luciferase reporter construct
fused with the 3′UTR of EZH2. The up-regulated expression of
miR-26a was required during terminal differentiation, thus the
negative regulator of myogenesis EZH2 is rapidly and efficiently
silenced, thereby promoting myogenesis and terminal differentiation
(34). miR-26 was found to play a
role in normal tissue growth and development and to have an impact
upon cell proliferation and differentiation; however, the mechanism
remains to be clarified.
6. Conclusions
miR-26, a functional miRNA, has merited
investigation, and has been found to possess different roles in
different tumors. The development of microarray techniques has led
to significant progress in the investigation of the expression of
miR-26 in numerous tumor types and certain normal tissue growth and
development. However, the target genes and molecule mechanisms
remain to be elucidated. The expression of miR-26 has been found to
decrease in bladder and breast cancer, oral squamous cell
carcinoma, anaplastic carcinomas, Burkitt lymphoma, HCC and
rhabdomyosarcoma, and is considered to be a suppressor miRNA in
those tumors. Expression of miR-26 increased in GBM, where it
promoted tumor cell growth and proliferation as an oncomir. MiR-26
is instrumental to normal tissue growth and development by
impacting on cell proliferation and differentiation. Various
studies also showed that EZH2, PTEN, SMAD1 and MTDH are potential
downstream target genes of miR-26. In conclusion, the molecular
mechanisms of miR-26 and the target genes in different tumors
remain unclear and should be investigated.
References
|
1
|
Liu X, Chen Z, Yu J and Zhou X: MicroRNA
profiling and head and neck cancer. Comp Funct Genomics.
837514:1–11. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lewis BP, Shih I, Jones-Rhoades MW, Bartel
DP and Burge CB: Prediction of mammalian microRNA targets. Cell.
115:787–798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xiaohui X, Jun L, Kulbokas EJ, et al:
Systematic discovery of regulatory motifs in human promoters and 3′
UTRs by comparison of several mammals. Nature. 434:338–345.
2005.
|
|
5
|
Williams AE: Functional aspects of animal
microRNAs. Cell Mol Life Sci. 65:545–562. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Stefani G and Slack FJ: Small non-coding
RNAs in animal development. Nat Rev Mol Cell Biol. 9:219–230. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bushati N and Cohen SM: microRNA
functions. Annu Rev Cell Dev Biol. 23:175–205. 2007. View Article : Google Scholar
|
|
10
|
Chang TC and Mendell TJ: microRNAs in
vertebrate physiology and human disease. Annu Rev Genomics Hum
Genet. 8:215–239. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kloosterman WP and Plasterk RHA: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tong AW and Nemunaitis J: Modulation of
miRNA activity in human cancer: a new paradigm for cancer gene
therapy? Cancer Gene Ther. 15:341–355. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen CZ: MicroRNAs as oncogenes and tumor
suppressors. N Engl J Med. 353:1768–1771. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Calin GA, Sevignani C, Dumitru CD, et al:
Human microRNA genes are frequently located at fragile sites and
genomic regions involved in cancers. Proc Natl Acad of Sci U S A.
101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Muinos-Gimeno M, Montfort M, Bayes M,
Estivill X and Espinosa-Parrilla Y: Design and evaluation of
single-nucleotide polymorphisms in microRNA genomic regions for
association studies in human disease. Eur J Hum Genet. 18:218–226.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Diederichs S and Haber DA: Sequence
variations of microRNAs in human cancer: alterations in predicted
secondary structure do not affect processing. Cancer Res.
66:6097–6104. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Iorio Marilena V, Ferracin M, Liu CG, et
al: MicroRNA gene expression deregulation in human breast cancer.
Cancer Res. 65:7065–7070. 2005.PubMed/NCBI
|
|
20
|
Wang G, Zhang H, He H, et al:
Up-regulation of microRNA in bladder tumor tissue is not common.
Int Urol Nephrol. 42:95–102. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Maillot G, Lacroix-Triki M, Pierredon S,
et al: Widespread estrogen-dependent repression of microRNAs in
breast tumor cell growth. Cancer Res. 69:8332–8340. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang B, Liu XX, He JR, et al:
Pathologically decreased miR-26a antagonizes apoptosis and
facilitates carcinogenesis by targeting MTDH and EZH2 in breast
cancer. Carcinogenesis. 32:2–9. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yu T, Wang XY, Gong RG, et al: The
expression profile of microRNAs in a model of
7,12-dimethyl-benz[a]anthrance- induced oral carcinogenesis in
Syrian hamster. J Exp Clin Cancer Res. 28:64–73. 2009.PubMed/NCBI
|
|
24
|
Visone R, Pallante P, Vecchione A, et al:
Specific microRNA are downregulated in human thyroid anaplastic
carcinomas. Oncogene. 26:7590–7595. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ciarapica R, Russo G, Verginelli F,
Donfrancesco A, Rota R and Giordano A: Deregulated expression of
miR-26a and EZH2 in rhabdomyosarcoma. Cell Cycle. 8:172–175. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sandrine S, Lars B and Thomas W:
Repressing the repressor: a new mode of MYC action in
lymphomagenesis. Cell Cycle. 8:556–559. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kota J, Chivukula RR, O’Donnell KA, et al:
Therapeutic microRNA delivery suppresses tumorigenesis in a murine
liver cancer model. Cell. 137:1005–1017. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ji J, Shi J, Budhu A, et al: MicroRNA
expression, survival, and response to interferon in liver cancer. N
Engl J Med. 361:1437–1447. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huse JT, Brennan C, Hambardzumyan D, et
al: The PTEN-regulating microRNA miR-26a is amplified in high-grade
glioma and facilitates gliomagenesis in vivo. Genes and Dev.
23:1327–1337. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kim H, Huang W, Jiang X, Pennicooke B,
Park PJ and Johnson MD: Integrative genome analysis reveals an
oncomir/oncogene cluster regulating glioblastoma survivorship. Proc
Natl Acad of Sci USA. 107:2183–2188. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Padgett KA, Lan RY, Leung PC, et al:
Primary billiary cirrhosis is associated with altered hepatic
microRNA expression. J Autoimmun. 32:246–253. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rogler CE, LeVoci L, Ader T, Massimi A,
Tchaikovskaya T, Norel R and Rogler LE: MicroRNA-23b cluster
microRNA regulate transforming growth factor-beta/bone
morphogenetic protein signaling and liver stem cell differentiation
by targeting Smads. Hepatology. 50:575–584. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Itoh T, Nozawa Y and Akao Y: MicroRNA-141
and -200a are involved in bone morphogenetic protein-2-induced
mouse pre-osteoblast differentiation by targeting distal-less
homeobox 5. J Biol Chem. 284:19272–19279. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wong CF and Tellam RL: MicroRNA-26a
targets the histone methyltransferase enhancer of Zeste homolog 2
during myogenesis. J Biol Chem. 283:9836–9843. 2008. View Article : Google Scholar : PubMed/NCBI
|