Introduction
Genetic linkage analysis has been highly successful
in mapping the genes responsible for Mendelian diseases. During the
past decade, attempts have been made to extend this approach to
multifactorial disorders and other health-related traits. However,
since complex diseases, including colorectal cancer, are
characterized by the modest contributions of each susceptibility
gene, identification of strong and replicable linkages has proven
to be difficult (1,2). In this regard, the results of our
linkage studies on human leukocyte antigen (HLA) allele
polymorphisms and cancers have also been discouraging owing to
inconsistent findings and weak linkage signals; however, these
studies have indicated that genetic factors may affect the early
onset of cancer. Therefore, we developed a new study design based
on the age of onset of cancer.
Cancer is typically a complex disorder to which,
until now, an unknown number of genes contribute by interacting
with each other and the environment (3,4). The
identities of these genes have remained elusive in spite of the
rapid pace of development of molecular technology and the increase
in genome sequence information. This scenario may be partly
attributed to by the absence of methods for high-efficacy analysis
of genetic information. Single nucleotide polymorphisms (SNPs) are
currently popular allelic markers (5,6);
however, their value is largely qualitative. They may not be
satisfactory indicators of complex diseases owing to their low
quantitative efficacy. Our new study design uses the age of onset
of cancer as an indicative factor that may provide a quantitative
rather than a qualitative estimate (by assessing allele frequency
at the locus). The efficacy of analysis may be improved since a
quantitative measure is more statistically robust than a
qualitative one.
The HLA complex genes are located on the short arm
of chromosome 6 and are the most polymorphic loci within the human
genome. The primary function of HLA is to stimulate the immune
system to identify infectious pathogens and eliminate them. The
status of HLA alleles is important in immune responses and
immunological tolerance (7).
Specific antigens of the HLA system are associated with numerous
solid tumors (8–10). Specifically, persistent pro-tumor
immune responses, now generally accepted to potentiate primary
tumor development, are being recognized as mediators of cancer
metastasis (11–13). The aim of the present study was to
determine whether polymorphisms in the HLA-DQB1 alleles are
associated with colorectal cancer and to identify an efficient
research method for the identification of cancer-related genes.
Materials and methods
Subjects
A total of 100 Chinese patients (200 chromosomes),
52 males and 48 females with a mean age of 61.77±11.42 years, who
wished to participate in the study, were included. Patients with
colorectal cancer were evidenced by surgical intervention and their
ages at the time of first surgery were recorded. All patients gave
written informed consent, and the study was approved by the
Institutional Ethics Committee of Dalian Medical University.
HLA-DQB1 allele typing
Genomic DNA was extracted from white blood cells
using standard techniques for HLA typing. The samples were typed
using an HLA-DQB1 ‘low resolution’ polymerase chain reaction
using a sequence-specific primers (PCR-SSP) typing kit (Pel-Freez
Clinical Systems, Brown Deer, WI, USA) including allele-specific
primers for DQB1*02, DQB1*03, DQB1*04, DQB1*05 and DQB1*06. The
commercial tests were run according to the manufacturer’s
instructions. Products were separated by electrophoresis in 2%
agarose gel and visualized by ethidium bromide staining and
ultraviolet (UV) transillumination. Automated gel reading was
performed using Pel-Freez software.
Experimental design and statistical
analysis
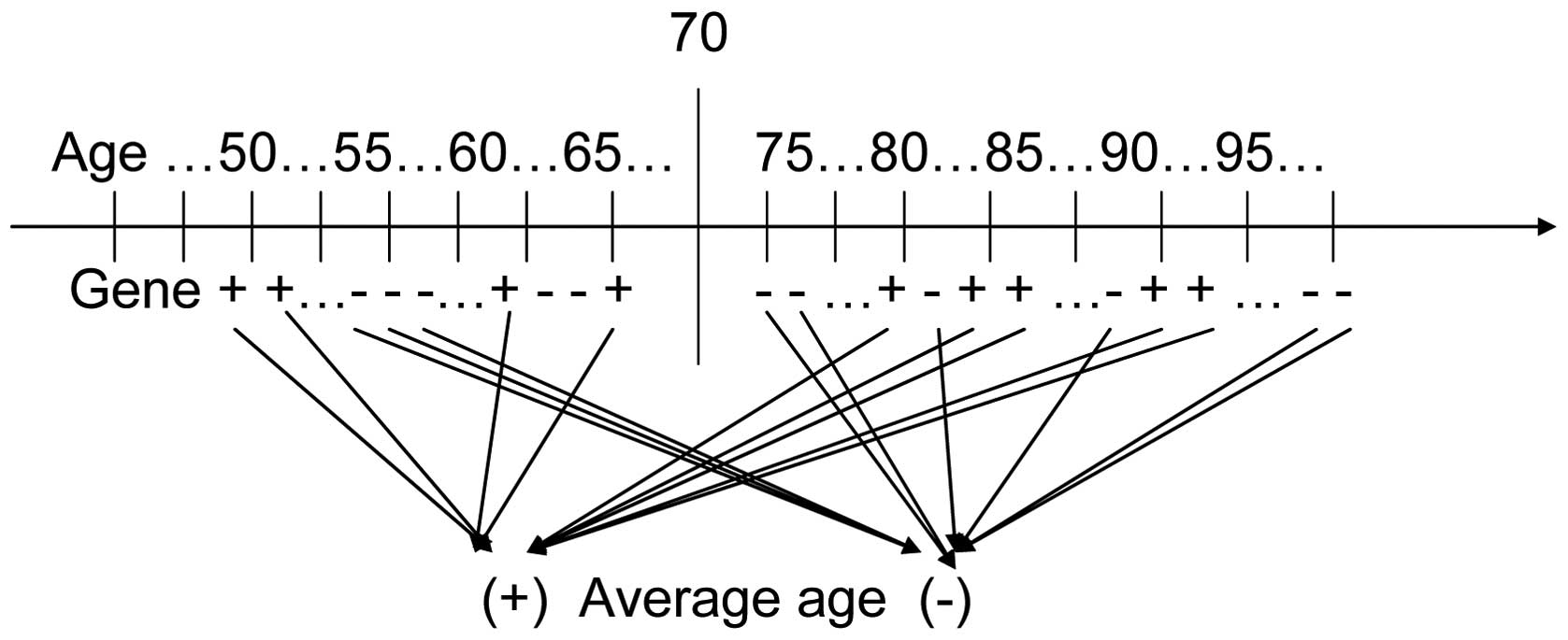
The basic concept of the new analysis method is to
consider the age of onset as a pathogenic effect of a gene. By
comparing the average ages of subjects carrying a gene and those
not carrying the gene, the association between the gene and the
disease may be ascertained. Fig. 1
shows the experimental design.
In the present study, the age of the subject was
considered to be the dependent variable, and the gene carriers
(subjects carrying or not carrying the gene) were considered to be
the grouping variable. A Student’s t-test was used to assess the
difference between the mean ages of the subjects that carry the
genes and those that do not carry the genes. P<0.05 was
considered to indicate a statistically significant difference
(two-tailed test), and a Bonferroni correction was applied. If the
average age of the subjects who carried a certain gene was
significantly lower or higher than that of the subjects who did not
carry the gene, then the gene was regarded as a disease-related
gene (susceptibility or resistance gene). Analyses were performed
using the SPSS 13.0 statistical software package.
Results
A total of 100 Chinese patients were included in
this study, and the age at onset of colorectal cancer was observed.
The patient samples were typed using a HLA-DQB1 PCR-SSP typing kit.
The average ages of the patients with and without the alleles were
calculated. Results showed that the mean age of subjects with the
HLA-DQB1*02 allele was less than that of subjects without this
allele (p<0.05), as shown in Table
I. No significant differences were observed between the mean
ages of the subjects with and without other HLA-DQB1 alleles.
 | Table IAverage age differences between
subjects with and without certain HLA-DQB1 alleles. |
Table I
Average age differences between
subjects with and without certain HLA-DQB1 alleles.
| DQB1* | Subjects with the
allele | Subjects without the
allele | p-value |
|---|
|
|
| |
|---|
| n | Average age | n | Average age | |
|---|
| 02 | 43 | 57.49±12.71 | 157 | 62.94±10.79 | 0.005 |
| 03 | 73 | 63.03±10.10 | 127 | 61.05±12.09 | 0.239 |
| 04 | 12 | 61.42±8.50 | 188 | 61.79±11.60 | 0.912 |
| 05 | 16 | 64.63±14.63 | 184 | 61.52±11.11 | 0.298 |
| 06 | 56 | 62.68±11.06 | 144 | 61.42±11.57 | 0.484 |
Discussion
Studies regarding to the pathogenic effects of a
gene on organisms should include factors such as the differences
between the frequency distributions of the gene in the disease and
the control groups (the stronger the effect of a gene, the greater
the differences in frequency distribution) and early or late age of
disease onset in gene carriers (the stronger the effects, the
earlier the onset). However, the traditional methods for
identifying disease-related genes have solely been based on
differences between the gene frequencies in the disease group and
the control group. Owing to enumeration data, the frequency
analysis requires large sample sizes and has low efficacy. Our new
analysis method is a quantitative one, and its statistical
efficiency is much higher than that of the traditional frequency
analysis. Therefore, unlike the methods used in other studies, this
method may be used to bypass the requirement for large sample
sizes, thus improving analytic efficacy.
In this investigation, we adopted a reliable assay
for HLA-DQB1 genotypes; the procedures were performed in
conditions that were in accordance with international standards.
Our results indicated that the mean age of subjects with the
HLA-DQB1*03 allele was less than that of subjects without the
allele (p<0.05), which implied that this allele was associated
with colorectal cancer susceptibility. These results suggest that
persistent pro-tumor immune responses may change with the HLA
allelic polymorphism and that the new analysis method is an
efficient and reliable approach for the identification of complex
disease-causing genes.
We expect that the increasing integration of
genetics, epidemiology and clinical trials through the sharing of
valuable data among laboratories may lead to new study methods that
may identify colorectal cancer-related genes and also clarify their
interactions with other risk factors.
Acknowledgements
The study was supported by grant LR201014 from the
Program for Liaoning Excellent Talents in the University of
China.
References
|
1
|
Sham P: Shifting paradigms in gene-mapping
methodology for complex traits. Pharmacogenomics. 2:195–202. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bracken MB: Genomic epidemiology of
complex disease: the need for an electronic evidence-based approach
to research synthesis. Am J Epidemiol. 162:297–301. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Velculescu VE: Defining the blueprint of
the cancer genome. Carcinogenesis. 29:1087–1091. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Christiani DC, Mehta AJ and Yu CL: Genetic
susceptibility to occupational exposures. Occup Environ Med.
65:430–436. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Brohede J, Dunne R, McKay JD and Hannan
GN: PPC: an algorithm for accurate estimation of SNP allele
frequencies in small equimolar pools of DNA using data from high
density microarrays. Nucleic Acids Res. 33:E1422005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nguyen TH, Liu C, Gershon ES and McMahon
FJ: Frequency Finder: a multi-source web application for collection
of public allele frequencies of SNP markers. Bioinformatics.
20:439–443. 2004.PubMed/NCBI
|
|
7
|
Fischer GF and Mayr WR: Molecular genetics
of the HLA complex. Wien Klin Wochenschr. 113:814–824.
2001.PubMed/NCBI
|
|
8
|
Schirle M: Identification of
tumor-associated HLA-ligands in the post-genomic era. J Hematother
Stem Cell Res. 11:873–881. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Seliger B, Abken H and Ferrone S: HLA-G
and MIC expression in tumors and their role in anti-tumor immunity.
Trends Immunol. 24:82–87. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cabrera T, Lopez-Nevot MA, Gaforio JJ,
Ruiz-Cabello F and Garrido F: Analysis of HLA expression in human
tumor tissues. Cancer Immunol Immunother. 52:1–9. 2003.PubMed/NCBI
|
|
11
|
Sun Y, Song M, Jäger E, Schwer C,
Stevanovic S, Flindt S, Karbach J, Nguyen XD, Schadendorf D and
Cichutek K: Human CD4+ T lymphocytes recognize a
vascular endothelial growth factor receptor-2-derived epitope in
association with HLA-DR. Clin Cancer Res. 14:4306–4315. 2008.
|
|
12
|
Mangalam A, Luckey D, Basal E, Behrens M,
Rodriguez M and David C: HLA-DQ6 (DQB1*0601)-restricted T cells
protect against experimental autoimmune encephalomyelitis in
HLA-DR3.DQ6 double-transgenic mice by generating anti-inflammatory
IFN-γ. J Immunol. 180:7747–7756. 2008.
|
|
13
|
Kim S, Sunwoo JB, Yang L, Choi T, Song YJ,
French AR, Vlahiotis A, Piccirillo JF, Cella M, Colonna M,
Mohanakumar T, Hsu KC, Dupont B and Yokoyama WM: HLA alleles
determine differences in human natural killer cell responsiveness
and potency. Proc Natl Acad Sci USA. 105:3053–3058. 2008.
View Article : Google Scholar : PubMed/NCBI
|