Introduction
Primary bronchial lung cancer (referred to as lung
cancer) is the most common primary pulmonary malignant tumor,
despite progress in surgical techniques, chemotherapy and
radiotherapy. The 5-year survival rate of patients with lung cancer
remains very low. Jemal et al revealed that the 5-year
survival rate of patients with lung cancer was only 16% (1). It is currently thought that
abnormality of the multi-intracellular transmission pathway is
associated with lung cancer, and there is increasing interest in
the role of Notch signaling systems in tumorigenesis (2). Four notch genes encoding four types of
Notch receptors, Notch1–4, have been identified in mammals. At
least five types of ligand are capable of binding with these
receptors and altering their function. Previously, studies
concerned with the pathogenesis of lung cancer have identified a
differential expression of Notch components in different types of
lung cancer. It is hypothesized that Notch1 and 3 are associated
with non-small cell lung cancer (NSCLC) (3,4). The
majority of studies consider that Notch1 is involved in cancer
promotion (5,6) and, compared with Notch1, there are a
small number of studies regarding Notch3. Additionally, whether the
effects of Notch1 and 3 are similar remains controversial.
To further investigate the role of Notch signaling
in lung adenocarcinoma, on the basis of Notch1 research, cases of
surgical resection of small cell lung cancer (SCLC), lung squamous
cell carcinoma and adenocarcinoma for the past 2 years in our
hospital were reviewed. Immunohistochemistry and imaging analysis
were used to detect the expression of Notch3 in lung cancer and
negative tissue margins. Preliminary results indicated that the
differences in Notch3 expression in lung adenocarcinoma were the
most prominent and, accordingly, those specimens were collected
from adult thoracic (lung) cancer surgerical resections. Specimens
were later diagnosed as lung adenocarcinoma in post-operative
pathological sections or normal lung tissue samples between January
2011 and May 2012. The expression of Notch3 in lung cancer tissues
was detected by reverse transcription polymerase chain reaction
(RT-PCR) and western blot analysis to further explore the role of
Notch3 in adult lung adenocarcinoma.
Materials and methods
Study population and tissue samples
Fify-seven archived lung cancer wax blocks were
collected from thoracic surgical resections between January 2010
and December 2011 at the Jinshan Branch of the Sixth People’s
Hospital of Shanghai, Shanghai Jiao Tong University, Shanghai,
China. Of the 55 blocks, 20 cases were squamous cell carcinoma, 24
were adenocarcinoma and 13 were small cell carcinoma. The general
clinical data of these patients are shown in Table I. One cancerous tissue embedded in a
paraffin wax block was selected with the corresponding non-tumor
tissue of the wax block. Three groups of non-tumor tissue in the
wax block specimens were missing (partially absent). Additionally,
serial sections (4 μm) were cut for analysis.
 | Table I.General clinical data of 57 lung
cancer cases. |
Table I.
General clinical data of 57 lung
cancer cases.
| A (squamous cell
carcinoma) | A1 (negative
margin) | B
(adenocarcinoma) | B1 (negative
margin) | C (small cell
carcinoma) | C1 (negative
margin) |
|---|
| Case number | 20 | 18 | 24 | 21 | 13 | 11 |
| Mean age (years) | 68.9 | | 63.5 | | 64.9 | |
| Gender ratio
(male:female) | 17:3 | | 18:6 | | 10:3 | |
The specimens of adult lung cancer from thoracic
surgery, which were later diagnosed by post-operative pathological
section analysis as lung adenocarcinoma, were studied as the
experimental group, and the specimens of the corresponding
non-tumor tissues were studied as the control group. There were 13
lung adenocarcinoma patients, comprising 9 males and 4 females,
aged 40–72 years (mean, 59.5 years).
The study was conducted according to the principles
of the Declaration of Helsinki. Informed consent was obtained and
the Ethics Committee of Jinshan Branch of the Sixth People’s
Hospital, Shanghai Jiao Tong University approved the study. The
patient data, which are contained within this article, were
obtained by a hospital-based doctor at Jinshan Branch of the Sixth
People’s Hospital, Shanghai Jiao Tong University. Permission to use
these data in this report has been obtained from all the subjects
who participated in this study (7).
Hematoxylin and eosin (H&E) staining
and immunohistochemistry
Prepared paraffin sections from each group were
stained conventionally with H&E to determine whether there was
consistency with the pathological type determined in the original
study.
The aforementioned prepared paraffin sections were
studied according to standard procedures. The working
concentrations of Notch3 antibody (Cell Signaling Technology, Inc.,
Danvers, MA, USA) and glyceraldehyde 3-phosphate dehydrogenase
(GAPDH; Shanghai Kangcheng Biotechnology Co., Ltd., Shanghai,
China) were 1:200 and that of HRP-marked secondary antibody
(Shanghai Weiao Biotech Ltd., Shanghai, China) was 1:100.
Phosphate-buffered saline (PBS) was selected as the negative
control as opposed to the primary antibody.
Evaluation of Notch3 immunostaining
Cytoplasm and/or nuclei with brown particles were
declared as positive. The imaging analysis was performed by an
immunohistochemical slice set under an Olympus (Tokyo, Japan)
microscope (×200); the accurate position of the measured visual
field was determined and then three complete visual fields without
overlap were randomly selected for viewing. Image-Pro Plus 6.0
software was used to analyze the area, the integral optical density
(IOD), the number of positive cells and the total cell number in
each visual field. The mean IOD of each slice was set as the
measured value for the case.
Surgical specimens and HE staining
Fresh specimens obtained via surgical resection were
sent to the Department of Pathology at the Jinshan Branch of the
Sixth People’s Hospital of Shanghai, Shanghai Jiao Tong University,
Shanghai, China. Under the guidance and assistance of the
pathologist, the samples (two samples of lung cancer tissue and two
samples of normal control lung tissue) were immediately drawn,
placed into Eppendorf (EP) tubes (1.5 ml, sterilized and enzyme
inactivated) and stored at −80°C. Another sample from the same
position as the cancerous tissue and a normal control lung tissue
sample were drawn and fixed in 10% neutral formalin for >24 h
for H&E staining.
Semiquantitative RT-PCR
RNA extraction was carried out on the samples of
cancerous and normal control lung tissue with TRIzol reagent. The
RNA concentration and purity were detected with a
spectrophotometer. RT-PCR was performed with a Takara RT-PCR kit
according to the manufacturer’s instructions (Takara Bio., Inc.,
Shiga, Japan). The amplified products of Notch3 and GAPDH were 419
and 309 bp, respectively. The primer sequences were as follows:
Upstream, AATGCCAACTGAAGAGGATGAG and downstream,
AGTGTAAGGCTGATTTCCCAAG for Notch3; upstream, TCCCATCACCATCTTCCAG
and downstream, ATGAGTCCTTCCACGATACC for GAPDH. PCR was performed
in a 25-μl reaction mixture. The conditions for Notch3 and GAPDH
PCR were one cycle of denaturing at 94°C for 2 min, followed by 40
(Notch3) or 25 (GAPDH) cycles at 94°C for 30 sec, and at 60°C
(Notch3) or 55°C (GAPDH) for 30 sec, prior to a final extension at
72°C for 1 min.
Following agarose gel electrophoresis (80 V, 60
min), the electrophoresed PCR products were scanned by
densitometry. The relative value of the Notch3 band to the GAPDH
band was calculated in each sample.
Western blot analysis
The samples of cancerous tissue and normal control
lung tissue were lysed in a protein lysate. After the cellular
protein was extracted, equal amounts (40 μg) of protein were
resolved on precast gels (Gradipore Ltd., Frenchs Forest,
Australia) and transferred to polyvinylidene fluoride (PVDF)
membranes. The blots were probed with 1:200 Notch3 monoclonal
antibody (Cell Signaling Technology, Inc.), GAPDH monoclonal
antibody (Shanghai Kangcheng Biotechnology Co., Ltd.) and
hoseradish peroxidase (HRP)-marked secondary antibody (Shanghai
Weiao Biotech Ltd.). The bound antibodies were visualized with a
Photope-HRP Western Detection kit (Cell Signaling Technology,
Inc.). The relative value of the Notch3 band to the GAPDH band was
calculated in each sample with Quality-one software.
Statistical analysis
Statistical analyses were performed using the
Statistical Package for the Social Sciences (SPSS) 17.0 software
(SPSS Inc., Chicago, IL, USA). A one-way analysis of variance
(ANOVA) was used to test for significant differences in measured
variables between groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
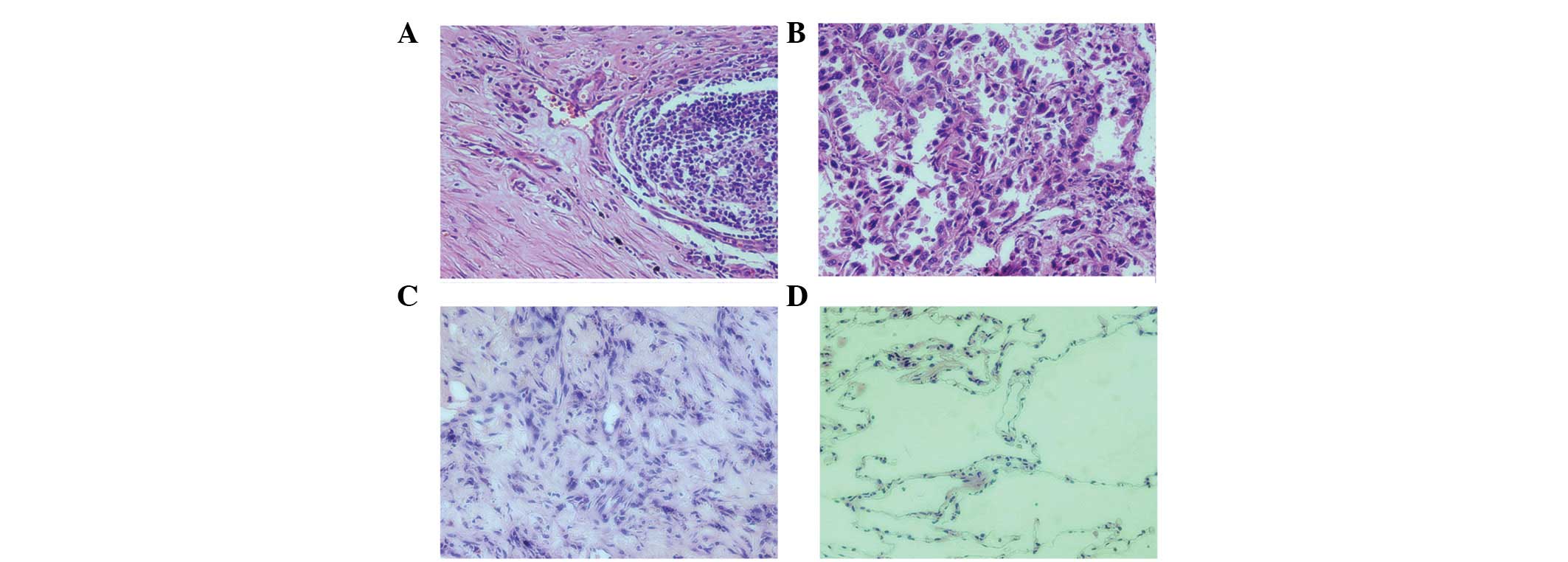
Pathomorphology of paraffin sections of
each group and immunohistochemical analysis for Notch3 protein
The selected specimens were confirmed to be
consistent with the lung cancer sub-types following observation
with H&E staining (Fig. 1).
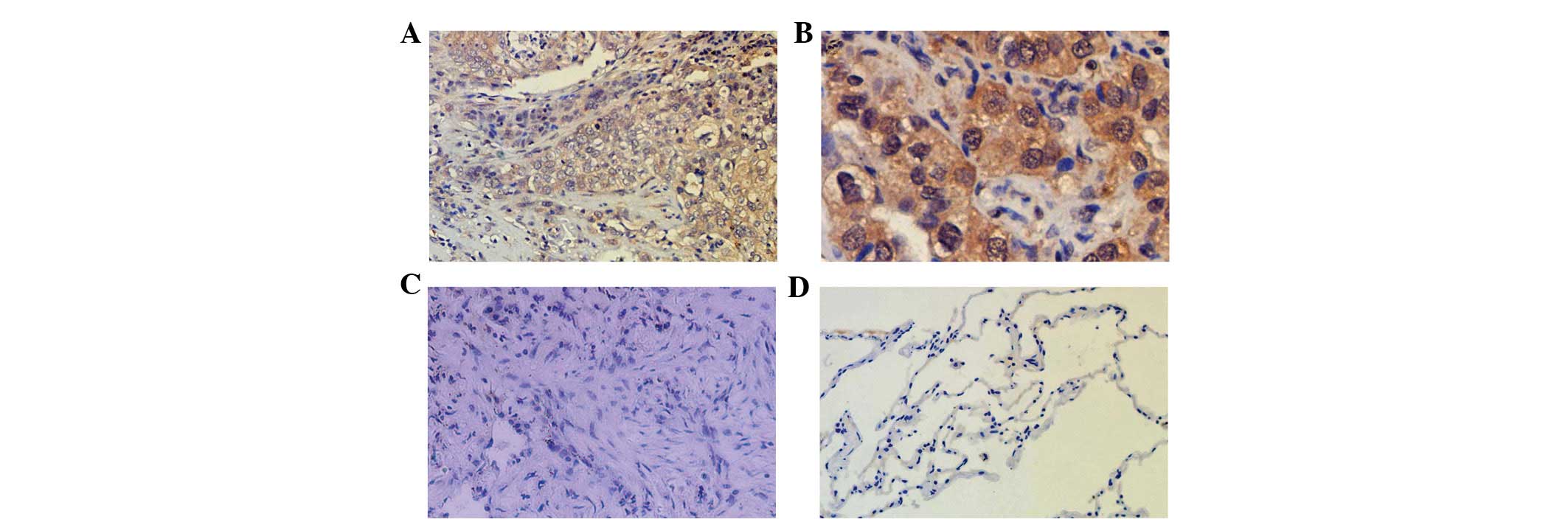
By immunohistochemisty, positive Notch3 expression
was observed in each group in the vascular smooth muscle of small
vessels, while weaker staining was observed in the alveolar
epithelial cells of non-tumor tissues. Positive Notch3 expression
was identified in the cytoplasm and nuclei in the squamous cell
carcinoma and adenocarcinoma groups, and the adenocarcinoma groups
demonstrated a stronger positive expression. However, the positive
staining was not clear in the SCLC group. Following the statistical
analysis of Notch3-positive cellular responses, the results
demonstrated that there were no significant differences in the
non-tumor tissue of the squamous cell carcinoma, adenocarcinoma and
small cell carcinoma groups (P>0.05). Notch3 expression in the
cancer tissue was stronger than that of the corresponding non-tumor
tissue in the squamous cell carcinoma and adenocarcinoma groups;
the difference was statistically significant (P<0.01). Notch3
expression in the cancer tissue was weaker than that of the
corresponding non-tumor tissue in the SCLC group; the difference
was statistically significant (P<0.01). The highest Notch3
expression was observed in the adenocarcinoma group compared with
the other two groups and the difference was statistically
significant (P<0.01; Fig. 2 and
Table II).
 | Table II.Notch3 expression in subtypes (mean ±
standard deviation). |
Table II.
Notch3 expression in subtypes (mean ±
standard deviation).
| Group | Number of cases | Integrated optical
density |
|---|
| Aa,d | 20 | 5920.56±928.60 |
| A1 | 18 | 1693.64±439.25 |
| Bb,c | 24 | 20074.32±1264.08 |
| B1 | 21 | 1740.38±360.32 |
| Cd | 13 | 265.09±78.31 |
| C1 | 11 | 1864.21±373.63 |
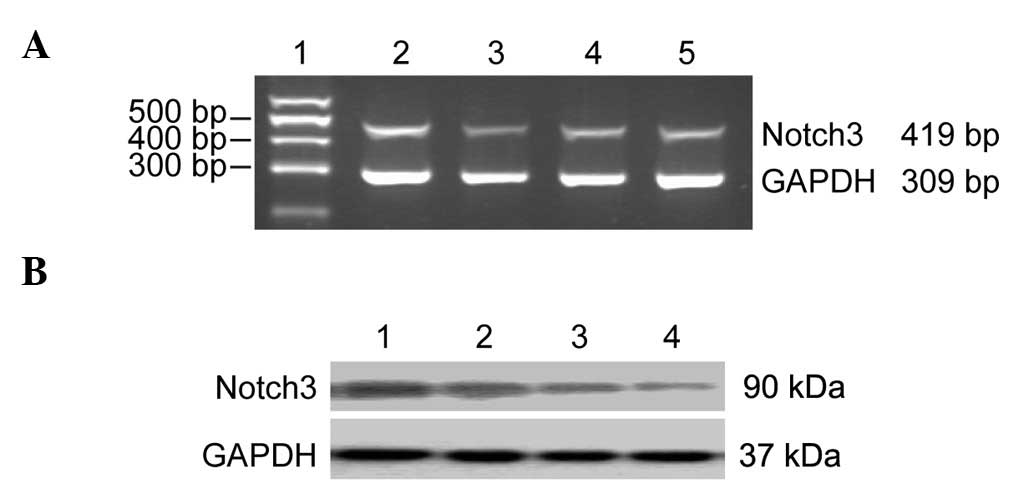
Semiquantitative RT-PCR analysis of
Notch3 mRNA
The observation of pathological sections with
H&E staining of selected lung tissue confirmed the specimens
were adenocarcinoma and paraneoplastic normal lung tissues, in
accordance with the findings of the Department of Pathology. There
were 13 adult lung adenocarcinomas and 13 paraneoplastic normal
lung tissue specimen cases. According to the RT-PCR results, the
expression of Notch3 mRNA in the lung adenocarcinoma group was
higher than that of the paraneoplastic normal lung tissues;
however, statistical analysis showed that the integrated optical
density (IOD) values were not significantly different between the 2
groups (P>0.05; Fig. 3 and
Table III).
 | Table III.Integral optical density values of
Notch3 mRNA in lung adenocarcinoma tissue (mean ± standard
deviation, n=13). |
Table III.
Integral optical density values of
Notch3 mRNA in lung adenocarcinoma tissue (mean ± standard
deviation, n=13).
| Group | Notch3 mRNA | P-value |
|---|
| Lung adenocarcinoma
group | 0.32±0.05 | |
| Non-tumor tissue
group | 0.28±0.06 | 0.05 |
Expression of Notch3 protein in lung
adenocarcinoma
According to the western blot analysis results, the
expression of Notch3 protein in the lung adenocarcinoma group was
higher than that of the paraneoplastic normal lung tissue, and the
statistical analysis demonstrated that the integrated optical
density (IOD) values were not significantly different (P<0.01;
Fig. 3 and Table IV).
 | Table IV.Integral optical density values of
Notch3 protein in lung adenocarcinoma tissue (mean ± standard
deviation, n=13). |
Table IV.
Integral optical density values of
Notch3 protein in lung adenocarcinoma tissue (mean ± standard
deviation, n=13).
| Group | Notch3 protein | P-value |
|---|
| Lung adenocarcinoma
group | 0.28±0.06 | |
| Paraneoplastic
group | 0.19±0.03 | <0.01 |
Discussion
Notch is a highly evolutionarily conserved single
transmembrane receptor protein that plays an important role in
deciding cell fate. It is a key regulatory factor in cell
differentiation, proliferation and apoptosis. Four types of Notch
genes have been found in mammals coding four Notch receptors
(Notch1–4). At least 5 types of ligand bind to Notch: Jagged1 and 2
and Delta1, 3 and 4. HES/HEY family members are genes that act
downstream of Notch, and have different gene distributions in
different tissues. One study regarding the integrative genomic
analyses of the HES/HEY family presumed that Notch-independent HES1
and 3 transcription occurred in undifferentiated ES cells, and that
Notch-dependent HES1 and 5, HEY1 and 2, and HEYL transcription
occurred in fetal, adult or cancer tissue (8). Notch signals are closely related to
certain regulatory factors in multiple tumorigenesis. The Notch
ligand, JAG1, which functions as a WNT-dependent Notch signaling
activator, is the key molecule maintaining homeostasis of stem and
progenitor cells (9). γ-secretase
cleaving Notch into the Notch intercellular domain (NICD) is the
key step in the conduction of Notch signals. One study
investigating tumor angiogenesis suggested that administration of
the γ-secretase inhibitor may be combined with disruption of eNOS
or interruption of VEGF signaling (10). This phenomenon may be explained by
the upstream regulation of Notch (11). Notch signals are located upstream of
signal transduction for other regulatory factors associated with
tumorigenesis, including NFAT, NF-κB, T-Bet and GATA3 (12). Therefore, the role of Notch in the
occurrence and development of certain tumors in lung cancer has
been confirmed. Abnormal expression of Notch1 was first identified
in human T lymphocytes in acute lymphoblastic leukemia (13). Subsequent studies have confirmed
that Notch subtypes are differentially expressed and have various
roles in different types of tumors (14–17).
There has been an increasing interest in Notch in
lung cancer research. Primary bronchogenic carcinoma of the lung
may be summarized as either SCLC or NSCLC, and NSCLC mainly
includes lung squamous cell carcinoma and adenocarcinoma. At
present, the role of Notch in lung carcinogenesis has not been
determined. The Notch signaling pathway either promotes or inhibits
lung cancer, and it is inferred that this may be related to a
number of factors, such as different types of tumor cells, subtypes
of Notch and ligands (4,18). Notch1 and 3 are considered to have a
relatively close correlation with lung cancer (3,4).
Notchl signal activation is considered to promote the growth of
tumor cells of NSCLC and inhibit the growth of SCLC (19,20).
Studies on Notch3, compared with Notch 1, remain small in number,
and whether Notch3 and Notch1 have a syntropic effect is unknown.
In our preliminary studies, the expression of Notch1 in lung
squamous cell carcinoma, adenocarcinoma and SCLC specimens was
detected by immunohistochemical methods. The experimental results
demonstrated a high Notch1 expression in lung adenocarcinoma and
squamous cell carcinoma, while low Notch1 expression was observed
in SCLC. Differences also exist in the two types of high expression
(21,22). The expression of Notch1 and 3 was
detected in the lung tissue of asthma model mice and they
demonstrated reverse changes (23).
To discover the role of Notch3 in lung cancer and
its correlation with Notch1, by reviewing the specimens of SCLC,
lung squamous cell carcinoma and adenocarcinoma removed during
surgery in the last two years, the expression of Notch3 in each
group of lung cancer tissue and its corresponding non-tumor tissue
was detected by an immunohistochemical method on the basis of the
study of Notch1. The results revealed that among the three groups
of specimens of lung cancer, the expression of Notch3 was the
highest in squamous cell carcinoma and adenocarcinoma, and was the
lowest in SCLC. The difference between the groups was statistically
significant. Among the three groups of lung cancer specimens, the
expression of Notch3 in lung adenocarcinoma was the highest.
Accordingly, we collected specimens of adult lung cancer during
thoracic surgery, which were later diagnosed as lung adenocarcinoma
and normal lung tissue, between January 2011 and May 2012 at the
Jinshan Branch of the Sixth People’s Hospital of Shanghai, Shanghai
Jiao Tong University, Shanghai, China. The expression of Notch3 in
the lung cancer tissue was detected by RT-PCR and western blot
analysis. The results demonstrated that the expression of Notch3
protein in the lung adenocarcinoma group was higher than that of
the normal paraneoplastic lung tissues. Additionally, the
expression of Notch3 mRNA in the lung adenocarcinoma group was
higher than that of the normal paraneoplastic lung tissues.
However, the IOD values showed no significant difference between
the two groups. The reasons for the inconsistency between the
levels of nucleic acid and protein expression, as analyzed, may be
associated with the small number of experimental specimens and less
precise experimental methods. This type of error may be corrected
by applying a real-time PCR method. It may be inferred from the
experimental results that Notch3 has a certain type of effect,
which varies in the different pathological types of lung cancer and
shows a syntropic effect with Notch1. However, whether there is an
additive effect requires further study. There are few studies on
other Notch subtypes in lung cancer. Clarification of the
correlation between Notch signaling and primary lung cancer
incidence has great significance.
Acknowledgements
This study was supported by the
Scientific Research Foundation of Science and Technology Commission
of Jinshan District of Shanghai (No. 2011-3-06) and the Scientific
Research Foundation of Shanghai Municipal Education Commission (No.
11YZ44), China.
References
|
1.
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar
|
|
2.
|
Tonon G, Modi S, Wu L, et al:
t(11;19)(q21;p13) translocation in mucoepidermoid carcinoma creates
a novel fusion product that disrupts a Notch signaling pathway. Nat
Genet. 33:208–213. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Collins BJ, Kleeberger W and Ball DW:
Notch in lung development and lung cancer. Semin Cancer Biol.
14:357–364. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Lin L, Mernaugh R, Yi F, Blum D, Carbone
DP and Dang TP: Targeting specific regions of the Notch3
ligand-binding domain induces apoptosis and inhibits tumor growth
in lung cancer. Cancer Res. 70:632–638. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Westhoff B, Colaluca IN, D’Ario G, et al:
Alterations of the Notch pathway in lung cancer. Proc Natl Acad Sci
USA. 106:22293–22298. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Baumgart A, Seidl S, Vlachou P, et al:
ADAM17 regulates epidermal growth factor receptor expression
through the activation of Notch1 in non-small cell lung cancer.
Cancer Res. 70:5368–5378. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Fan JY, Wang YF, Han B, Ji YR, Song HD and
Fan XQ: FOXL2 mutations in Chinese families with Blepharophimosis
syndrome (BPES). Transl Res. 157:48–52. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Katoh M: Integrative genomic analyses on
HES/HEY family: Notch-independent HES1, HES3 transcription in
undifferentiated ES cells, and Notch-dependent HES1, HES5, HEY1,
HEY2, HEYL transcription in fetal tissues, adult tissues, or
cancer. Int J Oncol. 31:461–466. 2007.
|
|
9.
|
Katoh M: Notch ligand, JAG1, is
evolutionarily conserved target of canonical WNT signaling pathway
in progenitor cells. Int J Mol Med. 17:681–685. 2006.PubMed/NCBI
|
|
10.
|
Zou YH, Cao YQ, Wang LX, Zhang YH, Yue ZJ
and Liu JM: γ-secretase inhibitor up-regulates vascular endothelial
growth factor receptor-2 and endothelial nitric oxide synthase. Exp
Ther Med. 2:725–729. 2011.
|
|
11.
|
Benedito R, Rocha SF, Woeste M, et al:
Notch-dependent VEGFR3 upregulation allows angiogenesis without
VEGF-VEGFR2 signalling. Nature. 484:110–114. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Ansel KM, Djuretic I, Tanasa B and Rao A:
Regulation of Th2 differentiation and Il4 locus accessibility. Annu
Rev Immunol. 24:607–656. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Ellisen LW, Bird J, West DC, et al: TAN-1,
the human homolog of the Drosophila notch gene, is broken by
chromosomal trans-locations in T lymphoblastic neoplasms. Cell.
66:649–661. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Purow BW, Haque RM, Noel MW, et al:
Expression of Notch-1 and its ligands, Delta-like-1 and Jagged-1,
is critical for glioma cell survival and proliferation. Cancer Res.
65:2353–2363. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Wu F, Stutzman A and Mo YY: Notch
signaling and its role in breast cancer. Front Biosci.
12:4370–4383. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Güngör C, Zander H, Effenberger KE, et al:
Notch signaling activated by replication stress-induced expression
of midkine drives epithelial-mesenchymal transition and
chemoresistance in pancreatic cancer. Cancer Res. 71:5009–5019.
2011.PubMed/NCBI
|
|
17.
|
Ye QF, Zhang YC, Peng XQ, Long Z, Ming YZ
and He LY: Silencing Notch-1 induces apoptosis and increases the
chemo-sensitivity of prostate cancer cells to docetaxel through
Bcl-2 and Bax. Oncol Lett. 3:879–884. 2012.PubMed/NCBI
|
|
18.
|
Ito T, Udaka N, Ikeda M, Yazawa T,
Kageyama R and Kitamura H: Significance of proneural basic
helix-loop-helix transcription factors in neuroendocrine
differentiation of fetal lung epithelial cells and lung carcinoma
cells. Histol Histopathol. 16:335–343. 2001.
|
|
19.
|
Eliasz S, Liang S, Chen Y, et al: Notch-1
stimulates survival of lung adenocarcinoma cells during hypoxia by
activating the IGF-1R pathway. Oncogene. 29:2488–2498. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Sriuranpong V, Borges MW, Ravi RK, et al:
Notch signaling induces cell cycle arrest in small cell lung cancer
cells. Cancer Res. 61:3200–3205. 2001.PubMed/NCBI
|
|
21.
|
Zhou M, Ma J and Guo X: The analysis of
Notch1 receptor protein expression in adult SCLC and NSCLDC. J Clin
Res (Chinese). 27:398–400. 2010.
|
|
22.
|
Fan Z, Zhou M and M: Express of Notch1
protein in adult adeno-carcinoma of lung and its significance. Int
J Respir (Chinese). 31:1625–1628. 2011.
|
|
23.
|
Guo XJ, Zhou M, Ren LP, Yang M, Huang SG
and Xu WG: Small interfering RNA-mediated knockdown of Notch1 in
lung. Chin Med J (Engl). 122:2647–2651. 2009.PubMed/NCBI
|