Introduction
Lung cancer is one of the most common types of
cancer and it remains the leading cause of cancer mortality in
males and females worldwide. There are two major types of lung
cancer: small cell lung cancer and non-small cell lung cancer
(NSCLC) (1,2). NSCLC, which is the most common type of
lung cancer, has a low 5-year survival rate (<15%), a poor
prognosis and a high rate of relapse (3). At present, various chemotherapies,
including combination therapies and targeted therapies, have been
utilized to treat NSCLC; however, the efficacy of these therapies
is insufficient. Therefore, the development of other therapies
against NSCLC progression is required for improving the efficacy of
cancer treatment.
As a novel therapeutic strategy, immunomodulatory
drugs have been utilized to treat NSCLC. The immunomodulatory drug
lenalidomide was originally approved to treat myelodysplastic
syndromes and multiple myeloma (4,5).
However, lenalidomide has been investigated for treating other
types of cancer, including NSCLC, malignant melanoma and prostate
cancer (6–11). Lenalidomide has been reported to
alter the production of cytokines and growth factors, which in turn
enhances the immune response against tumor cells and inhibits tumor
angiogenesis (12,13). Moreover, it has been reported that
lenalidomide is a potent co-stimulator of T-cell activation,
leading to increased T-cell cytokine production and activation of
CD8+ T cells and NK cells (14,15).
Furthermore, lenalidomide has direct antiproliferative effects on
tumor cells in the absence of immune effector cells (16). Lenalidomide causes
concentration-dependent cell cycle arrest in G0-G1 phase by
upregulating the CDK inhibitor p21 waf-1, a key cell cycle
regulator that modulates the activity of CDKs, and down-regulating
the activities of the prosurvival kinases ERK1/2 and Akt (17,18).
Lenalidomide inhibits the translation of C/EBPβ by downregulating
eIF4E, and IRF4 downregulation has been reported to be a critical
factor controlling multiple myeloma survival and as a prognostic
marker in patients with multiple myeloma associated with poor
survival (19,20). In NSCLC, objective responses have
been observed with lenalidomide-based therapy, suggesting that
lenalidomide is a potent drug for NSCLC treatment. Despite the
clinical effects of lenalidomide, studies of the precise mechanisms
of its action in NSCLC have not yet been performed. Studies of the
mechanisms of the anti-cancer properties of lenalidomide are
critical for improving the efficacy of this drug against NSCLC. In
the present study, we investigated the antiproliferative activity
of lenalidomide against NSCLC cell lines and the gene expression
profile changes in lenalidomide-treated NSCLC cells.
Materials and methods
Cell culture and lenalidomide
treatment
The human NSCLC cell lines Lu-99, H1299, A549, EBC1
and H460 were purchased from the ATCC (Manassas, VA, USA) and
cultured in RPMI-1640 medium containing 10% fetal bovine serum and
antibiotics at 37°C in a humidified chamber containing 5%
CO2. Cells were seeded into 60-mm culture dishes
(2x105 cells per dish) with various concentrations of
lenalidomide (Celgene, Summit, NJ, USA) and incubated for various
times.
RNA preparation and cDNA synthesis
Following lenalidomide treatment, total RNA was
extracted from cells using TRIzol reagent (Invitrogen, Carlsbad,
CA, USA) according to the manufacturer’s instructions. For the
microarray studies, both the quality and concentration of the RNA
samples were determined using an Agilent 2100 Bioanalyzer (Agilent
Technologies, Santa Clara, CA, USA) and a MaestroNano
Spectrophotometer (Maestrogen, Las Vegas, NV, USA). The recommended
RNA quality parameters for microarray analysis are as follows: UV
spectroscopy A260/A280 ratio of 1.8–2.0 and an A260/A230 ratio
>1.8; an 18S/28S rRNA ratio of 1.8–2.1; and an RNA integrity
number >8.0. To synthesize cDNA, 1 μg RNA was incubated
with oligo dT primers at 94°C for 10 min and reverse transcribed
with reverse transcriptase (Enzynomics, Seoul, Korea) at 37°C for 1
h.
DNA microarray analysis
DNA microarray analysis was performed using a
HumanHT-12 v4.0 Expression Beadchip kit (Illumina, San Diego, CA,
USA) according to the manufacturer’s instructions. The derived data
were analyzed using Genespring GX 11 (Agilent Technologies). The
raw data were filtered using the flag test and t-test, therefore
the miRNAs showing detectable expression levels (flag value =
present) were selected, and subjected to fold-change analysis.
Significant genes were determined using the fluorescence ratio
between the control and lenalidomide-treated samples, and genes
displaying a >2-fold increase or decrease were selected for
analysis.
Polymerase chain reaction (PCR)
analysis
The expression levels of BH3-interacting domain
death agonist (BID), v-fos FBJ murine osteosarcoma viral oncogene
homolog (FOS) and NK2 homeobox1 (NKX2-1) mRNA were determined by
PCR with their specific primers as follows: GADPH, (F)
5′-TTGCCATCAATGACCCCTTCA-3′ and (R) 5′-CGC CCCACTTGATTTTGGA-3′;
BID, (F) 5′-ATGGAC TGTGAGGTCAACAACGG-3′ and (R) 5′-CACGTA
GGTGCGTAGGTTCTGGTTA-3′; Fos, (F) 5′-CCAACT TCAT TCCCACG GTCAC-3′
and (R) 5′-TG GCA A TCTCGGTCTGCAAA-3′; NKX2-1, (F) 5′-ATGTCG
ATGAGTCCAAAGCA-3′ and (R) 5′-ACCG TATAGCA AGGTGGAGCA-3′.
Cell viability assay
Cell proliferation was determined using the WST-1
assay (EZ-Cytox Cell Viability Assay kit, ITSBIO, Seoul, Korea)
according to the manufacturer’s instructions. In brief, cells were
incubated in 96-well plates and treated with DMSO or various
concentrations of lenalidomide in RPMI-1640 medium containing 10%
FBS for 2 or 3 days. After incubation, the kit solution was added
to the cultured cells, which were incubated at 37°C for 2 h. Cell
viability was measured using an iMark microplate reader (Bio-Rad,
Hercules, CA, USA) at 450 nm using a 620-nm reference filter.
Statistical analysis
Statistical analysis was performed using the
χ2 test or Fisher’s exact test and Spearman rank
correlation coefficient analysis. P<0.05 was considered to
indicate a statistically significant result.
Results and Discussion
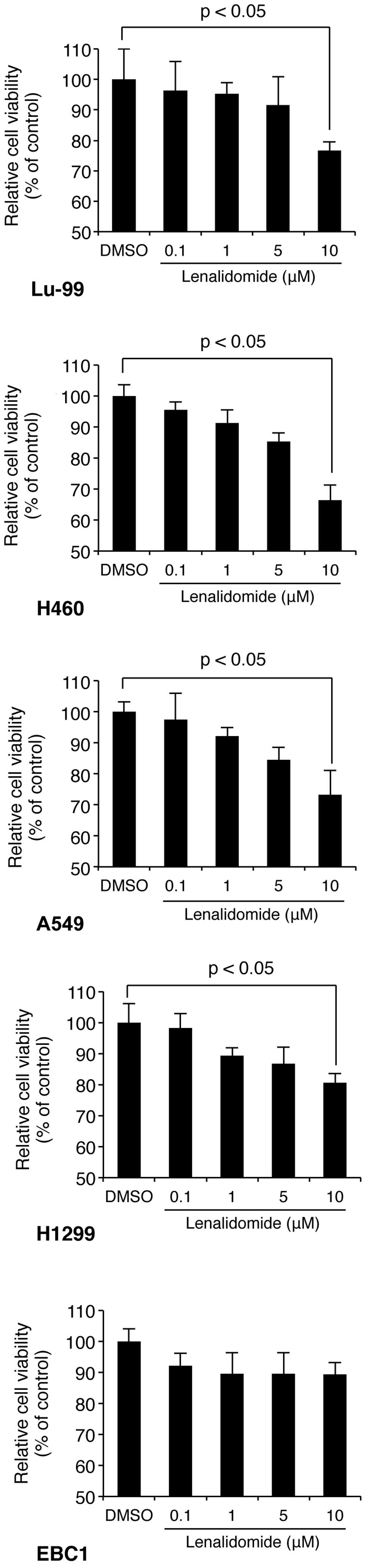
We first examined whether the immunomodulatory drug
lenalidomide exhibited direct antitumor activity in NSCLC cell
lines. To this end, Lu-99, H1299, H460, EBC1 and A549 cells were
exposed to a series of increasing concentrations of lenalidomide
for 72 h and the proliferation of the cells was measured by
analyzing the activity of mitochondrial dehydrogenases using the
WST-1 assay (described in Materials and methods). The assays
revealed that lenalidomide significantly inhibited the
proliferation of NSCLC cells (Lu-99, H1299, H460 and A549) in a
concentration-dependent manner (Fig.
1). In particular, H460 cells had the highest sensitivity for
lenalidomide, and therefore, we used this cell line to perform the
subsequent experiments.
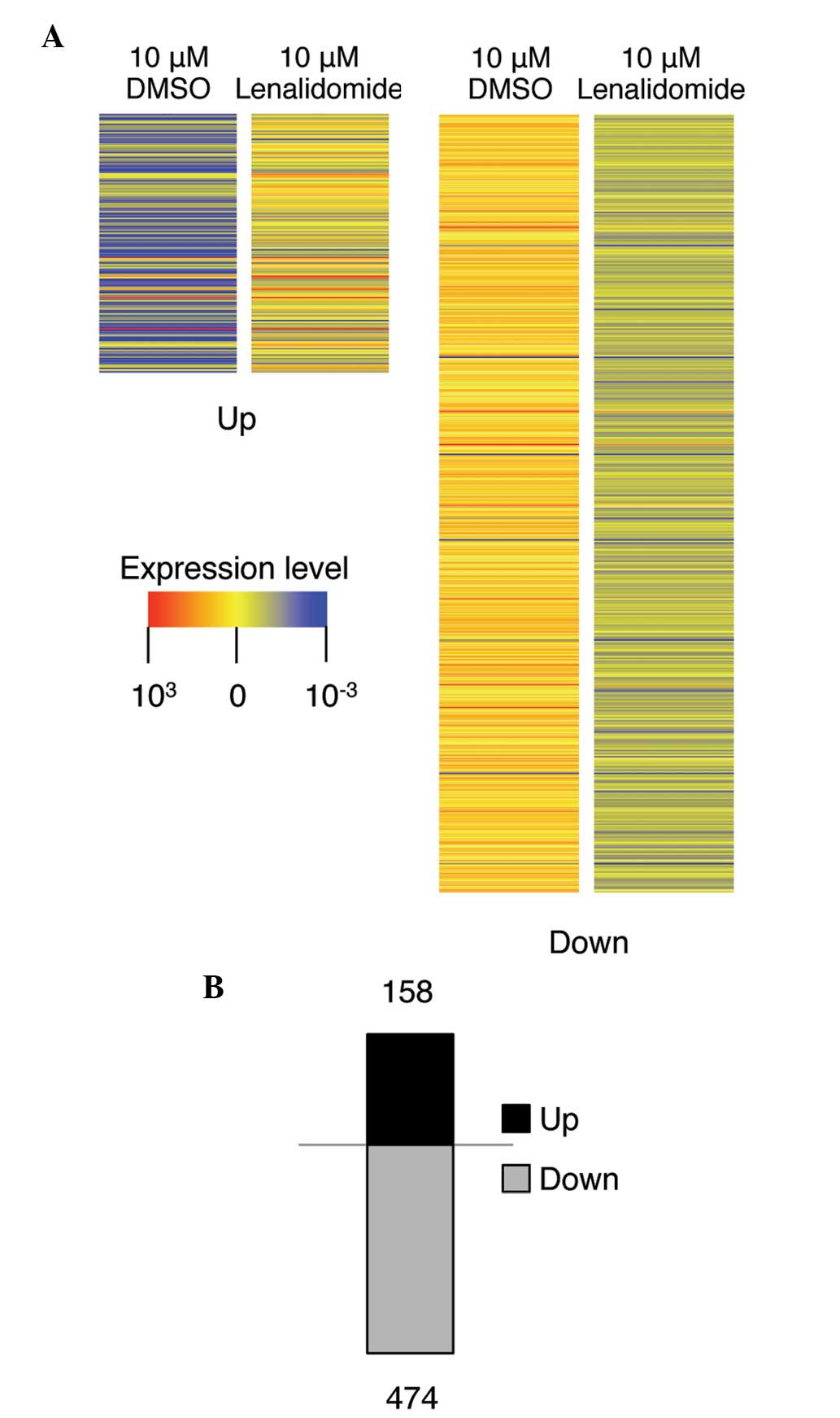
We next performed gene expression profiling analysis
to gain insight into the mode of action of lenalidomide and to
identify the molecular targets of this drug. H460 cells were
treated with the control DMSO or 10 μM lenalidomide for 72 h
and RNA samples were purified. The quality of each sample was
confirmed using Agilent software. We then compared the gene
expression profiles of the lenalidomide- and DMSO-treated samples
(see Materials and methods). As shown in Fig. 2A, lenalidomide regulated the
transcription of genes, and the genes displaying >1.5-fold
changes in transcription compared with that in control cells were
selected and identified by color contrast. Notably, 3-fold more
mRNAs were downregulated (474 mRNAs) than upregulated (158 mRNAs)
by lenalidomide treatment in H460 cells, indicating that
lenalidomide has an inhibitory effect on gene transcription. To
further analyze the transcriptional effect of lenalidomide, the 50
genes most strongly affected by lenalidomide treatment were
selected, as shown in Table I.
Furthermore, using the bioinformatics tool of Gene Set Enrichment
Analysis (http://www.broadinstitute.org/gsea/index.jsp), the
genes with >1.5-fold changes in expression were categorized into
different functional categories, such as homology or biochemical
activity, using the Gene Ontology project for gene sharing
(Table II). Specifically,
transcription factors, tumor suppressors and oncogenes were
relatively more abundant than other biological function-related
genes in Table II.
 | Table I.Top 50 genes displaying >1.5-fold
changes in expression following lenalidomide treatment. |
Table I.
Top 50 genes displaying >1.5-fold
changes in expression following lenalidomide treatment.
Upregulated genes
| Downregulated genes
|
|---|
| Gene name | FC | Gene name | FC |
|---|
| C14ORF153 | 2.92 | ZNF121 | 2.91 |
| GINS4 | 2.53 | NMI | 2.01 |
| XRCC2 | 2.08 | OR5T1 | 1.98 |
| SNN | 2.01 | ALOX15 | 1.87 |
| GRIPAP1 | 1.93 | KCNA4 | 1.84 |
| FKBP14 | 1.92 | ARR3 | 1.78 |
| BPNT1 | 1.87 | OR11A1 | 1.78 |
| RBBP9 | 1.85 | HFM1 | 1.75 |
| BLZF1 | 1.85 | NKX2-1 | 1.75 |
| HMG1L1 | 1.81 | LOXL1 | 1.74 |
| QRFPR | 1.81 | WDR21C | 1.74 |
| LMOD3 | 1.78 | PENK | 1.73 |
| ZNF483 | 1.77 | CCDC121 | 1.73 |
| DCDC1 | 1.77 | NOS2A | 1.71 |
| SHROOM4 | 1.77 | LONRF3 | 1.70 |
| DUSP19 | 1.76 | SS18 | 1.70 |
| ZNF549 | 1.75 | AIM1L | 1.69 |
| CHRNAS | 1.75 | EFCBP1 | 1.69 |
| DEM1 | 1.75 | KLC3 | 1.68 |
| USP49 | 1.74 | CSAG3B | 1.68 |
| PDP2 | 1.74 | NUCKS1 | 1.67 |
| ZMAT3 | 1.74 | MORF4L1 | 1.67 |
| MYO3B | 1.74 | LY6G5C | 1.67 |
| ZNF69 | 1.73 | ESRRG | 1.66 |
| OVOS2 | 1.73 | LYPD4 | 1.66 |
| LRRFIP1 | 1.72 | LRRN3 | 1.66 |
| TTTY22 | 1.72 | KIF5A | 1.66 |
| TRIM13 | 1.72 | FBXO47 | 1.66 |
| GNB4 | 1.72 | PRDM13 | 1.65 |
| CCBE1 | 1.72 | SYT15 | 1.65 |
| ZNF14 | 1.71 | UPK3B | 1.65 |
| FOS | 1.71 | CPM | 1.65 |
| IL17RD | 1.70 | BZW1 | 1.65 |
| MAGT1 | 1.70 | ASB4 | 1.64 |
| TDP1 | 1.67 | MEST | 1.64 |
| EIB2B | 1.67 | LGR6 | 1.63 |
| XCL1 | 1.67 | GEM | 1.63 |
| CDKN2A | 1.67 | MICALL2 | 1.62 |
| BID | 1.66 | PCDHGA1 | 1.62 |
| DDX51 | 1.65 | PCLO | 1.62 |
| ARL16 | 1.63 | SYPL2 | 1.62 |
| AGER | 1.63 | NR1I2 | 1.61 |
| NLRP8 | 1.63 | EPHA10 | 1.61 |
| FUT6 | 1.61 | DGKB | 1.61 |
| NSBP1 | 1.61 | GRHL1 | 1.61 |
| CREB1 | 1.61 | SLC6A5 | 1.61 |
| CDH24 | 1.60 | MCM9 | 1.61 |
| DMC1 | 1.56 | RGN | 1.61 |
| PHAX | 1.53 | CDC25B | 1.61 |
| MTSS1L | 1.51 | STATH | 1.60 |
 | Table II.Genes sharing a common feature such
as homology or biochemical activity. |
Table II.
Genes sharing a common feature such
as homology or biochemical activity.
| Gene feature | Cytokines and
growth factors | Transcription
factors | Homeodomain
proteins | Cell
differentiation markers | Protein
kinases | Translocated cancer
genes | Oncogenes | Tumor
suppressors |
|---|
| Tumor
suppressor | 1 (1/0) | 1 (0/1) | 0 | 1 | 0 | 0 | 0 | 3 (1/2) |
| Oncogenes | 0 | 3 (1/2) | 0 | 0 | 0 | 7 (2/5) | 7 (2/5) | |
| Translocated cancer
genes | 0 | 3 (1/2) | 0 | 0 | 0 | 7 (2/5) | | |
| Protein
kinases | 0 | 0 | 0 | 0 | 7 (1/6) | | | |
| Cell
differentiation markers | 0 | 0 | 0 | 2 (0/2) | | | | |
| Homeodomain
proteins | 0 | 1 (0/1) | 3 (1/2) | | | | | |
| Transcription
factors | 0 | 24 (8/16) | | | | | | |
| Cytokines and
growth factors | 6 (2/4) | | | | | | | |
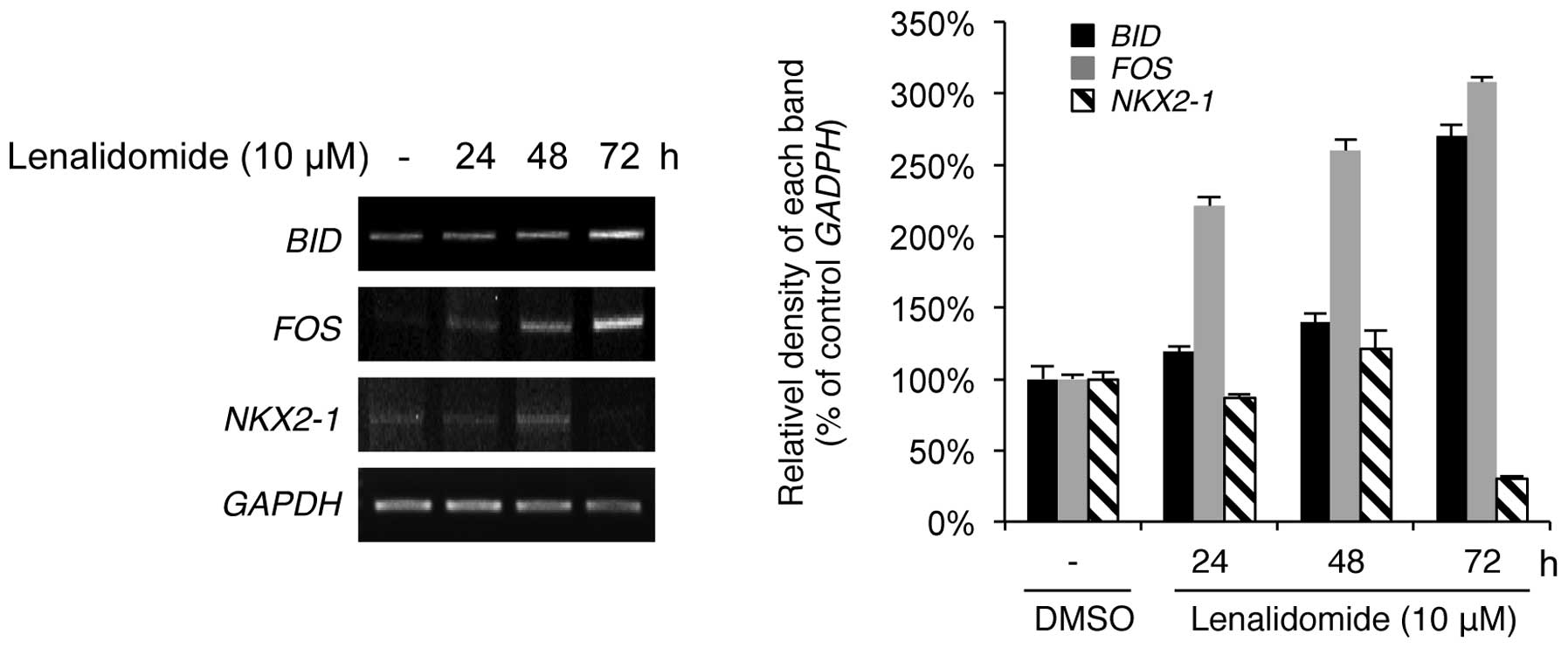
To verify the microarray data, we performed
reverse-transcription PCR (RT-PCR) analysis of several genes
selected on the basis of their roles in apoptosis, cell cycle
regulation, transcription and oncogenesis in NSCLC cells. These
genes were BID, FOS and NKX2-1. BID is a member of the proapoptotic
BCL2 family and is a key component of death receptor-mediated
caspase activation (21). FOS is a
transcription factor that binds to Jun and creates the
transcription factor activator protein 1, which regulates cell
proliferation, differentiation and apoptosis (22). Although the experimental condition
of the microarray was based on the single time point (72 h) of
lenalidomide treatment, as shown in Fig. 1, RT-PCR was performed to clarify the
results of the expression levels of BID, FOS and NKX2-1 by treating
cells with lenalidomide for 24, 48 and 72 h. As shown in Fig. 3, the results revealed good
concordance in gene expression between the microarray and RT-PCR
data. BID and FOS, which were upregulated by lenalidomide in the
array data, were markedly upregulated in a time-dependent manner;
conversely, NKX2-1 was downregulated by lenalidomide in the array
and RT-PCR data. Thus, lenalidomide-induced antiproliferative
effects may be mediated by regulating the expression of genes
associated with apoptosis, cell survival and transcription
factors.
In NSCLC, the NKX2-1 (also known as TTF1) gene is an
immunohistochemical marker for predicting the adenocarcinoma
subtype (23,24). The NKX2-1 gene is amplified in
10–15% of lung adenocarcinomas, and results of in vitro
studies further support the hypothesis that NKX2-1 acts as a
lineage-specific oncogene (24,25).
However, a tumor suppressor role has been observed in the same type
of cancer for NKX2-1 (26). The
expression of exogenous NKX2-1 limited tumor progression, resulting
in fewer tumors with an advanced histopathological grade.
Therefore, NKX2-1 has both oncogenic and tumor-suppressive
functions in lung cancer, suggesting that NKX2-1 downregulation
following lenalidomide treatment in NSCLC cells is a meaningful
result that requires further investigation.
These findings further the knowledge of the
signaling pathways targeted by lenalidomide and suggest that
lenalidomide causes changes in the gene expression profile of NSCLC
cells. In addition, these genes may be prospective target molecules
for the mechanism involved in the lenalidomide-induced
anti-proliferative effect in NSCLC cells.
Acknowledgements
The authors are grateful to all other
members of our research group for support and advice regarding this
study. This study was supported by a grant from the Ministry of
Education, Science, and Technology (grant 20110028646) of the
Republic of Korea.
References
|
1.
|
Ferlay J, Shin HR, Bray F, et al: GLOBOCAN
2008, Cancer incidence and mortality worldwide. IARC CancerBase No.
10. Lyon, France: International Agency for Research on Cancer;
2010, http://globocan.iarc.fr.
|
|
2.
|
American Cancer Society: 2010 Cancer facts
and figures. Atlanta, GA: American Cancer Society; 2010
|
|
3.
|
Herbst RS, Heymach JV and Lippman SM: Lung
cancer. N Engl J Med. 359:1367–1380. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
List A, Kurtin S, Roe DJ, et al: Efficacy
of lenalidomide in myelodysplastic syndromes. N Engl J Med.
352:549–557. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Hideshima T, Richardson PG and Anderson
KC: Current therapeutic uses of lenalidomide in multiple myeloma.
Expert Opin Investig Drugs. 15:171–179. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Eisen T, Trefzer U, Hamilton A, et al:
Results of a multicenter, randomized, double-blind phase 2/3 study
of lenalidomide in the treatment of pretreated relapsed or
refractory metastatic malignant melanoma. Cancer. 116:146–154.
2010.
|
|
7.
|
Kalmadi S, Davis M, Dowlati A, et al:
Phase I trial of three-weekly docetaxel, carboplatin and oral
lenalidomide (Revlimid) in patients with advanced solid tumors.
Invest New Drugs. 25:211–216. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Miller AA, Case D, Harmon M, et al: Phase
I study of lenalidomide in solid tumors. J Thorac Oncol. 2:445–449.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Dahut WL, Aragon-Ching JB, Woo S, et al:
Phase I study of oral lenalidomide in patients with refractory
metastatic cancer. J Clin Pharmacol. 49:650–660. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Bartlett JB, Tozer A, Stirling D and
Zeldis JB: Recent clinical studies of the immunomodulatory drug
(IMiD) lenalidomide. Br J Cancer. 93:613–619. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Tohnya TM, Ng SS, Dahut WL, et al: A phase
I study of oral CC-5013 (lenalidomide, Revlimid), a thalidomide
derivative, in patients with refractory metastatic cancer. Clin
Prostate Cancer. 2:241–243. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Gupta D, Treon SP, Shima Y, et al:
Adherence of multiple myeloma cells to bone marrow stromal cells
up-regulates vascular endothelial growth factor secretion:
therapeutic applications. Leukemia. 15:1950–1961. 2001. View Article : Google Scholar
|
|
13.
|
Teo SK: Properties of thalidomide and its
analogues: implications for anticancer therapy. AAPS J. 7:E14–E19.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Corral LG, Haslett PA, Muller GW, et al:
Differential cytokine modulation and T cell activation by two
distinct classes of thalidomide analogues that are potent
inhibitors of TNF-alpha. J Immunol. 163:380–386. 1999.PubMed/NCBI
|
|
15.
|
Dredge K, Marriott JB, Todryk SM, et al:
Protective antitumor immunity induced by a costimulatory
thalidomide analog in conjunction with whole tumor cell vaccination
is mediated by increased Th1-type immunity. J Immunol.
168:4914–4919. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Bartlett JB, Dredge K and Dalgleish AG:
The evolution of thalidomide and its IMiD derivatives as anticancer
agents. Nat Rev Cancer. 4:314–322. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Verhelle D, Corral LG, Wong K, et al:
Lenalidomide and CC-4047 inhibit the proliferation of malignant B
cells while expanding normal CD34+ progenitor cells. Cancer Res.
67:746–755. 2007.PubMed/NCBI
|
|
18.
|
Kotla V, Goel S, Nischal S, et al:
Mechanism of action of lenalidomide in hematological malignancies.
J Hematol Oncol. 2:362009. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Li S, Pal R, Monaghan SA, et al: IMiD
immunomodulatory compounds block C/EBP{beta} translation through
eIF4E down-regulation resulting in inhibition of MM. Blood.
117:5157–5165. 2011.PubMed/NCBI
|
|
20.
|
Lopez-Girona A, Heintel D, Zhang LH, et
al: Lenalidomide down-regulates the cell survival factor,
interferon regulatory factor-4, providing a potential mechanistic
link for predicting response. Br J Haematol. 154:325–336. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Puthalakath H and Strasser A: Keeping
killers on a tight leash: transcriptional and post-translational
control of the pro-apoptotic activity of BH3-only proteins. Cell
Death Differ. 9:505–512. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Durchdewald M, Angel P and Hess J: The
transcription factor Fos: a Janus-type regulator in health and
disease. Histol Histopathol. 24:1451–1461. 2009.PubMed/NCBI
|
|
23.
|
Chhieng DC, Cangiarella JF, Zakowski MF,
Goswami S, Cohen JM and Yee HT: Use of thyroid transcription factor
1, PE-10, and cytokeratins 7 and 20 in discriminating between
primary lung carcinomas and metastatic lesions in fine-needle
aspiration biopsy specimens. Cancer. 93:330–336. 2001. View Article : Google Scholar
|
|
24.
|
Tanaka H, Yanagisawa K, Shinjo K, et al:
Lineage-specific dependency of lung adenocarcinomas on the lung
development regulator TTF-1. Cancer Res. 67:6007–6011. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Kwei KA, Kim YH, Girard L, et al: Genomic
profiling identifies TITF1 as a lineage-specific oncogene amplified
in lung cancer. Oncogene. 27:3635–3640. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Winslow MM, Dayton TL, Verhaak RG, et al:
Suppression of lung adenocarcinoma progression by Nkx2-1. Nature.
473:101–104. 2011. View Article : Google Scholar : PubMed/NCBI
|