Introduction
Endometrial cancer is one of the most common types
of female cancer and it seriously endangers the health of women.
Therefore, research on the pathogenesis and novel therapeutic
approaches to endometrial cancer is of great significance.
microRNAs (miRNAs) are a class of small non-coding RNAs of
approximately 22 nucleotides in length, which regulate gene
expression in animals and plants by targeting mRNAs at the
3’-untranslated regions (UTRs) for cleavage or translational
repression. The results of previous studies showed that over half
of the miRNAs may be aligned to genomic fragile sites or regions
associated with cancer (1),
indicating that miRNA may be significant in cancer pathogenesis,
development and metastasis. The aberrant expression of miRNA has
been reported in various tumors, including lung, colon, breast,
liver and gastric cancer (2–5),
indicating that there is a close correlation between miRNAs and
human malignancy.
The aberrant overexpression of miR-103 was first
identified in endometrial cancer by Boren et al (6) and Chung et al (7). Boren et al found that miR-103
may be associated with the development of endometrial cancer and
suggested tissue inhibitor of metalloproteinase 3 (TIMP-3) as a
potential target for miR-103. Later studies suggested that miR-103
was aberrantly overexpressed in various types of cancer. For
example, 10 miRNAs, including miR-103, were found to be upregulated
in bladder cancer (8). miR-103 maps
near the homeobox (HOX) genes, which are crucial for normal
development and oncogenesis. Yang et al (9) reported that the overexpression of
miR-103 may be involved in the tumorigenesis of erythroleukemia by
arresting erythroid maturation. Guo et al (10) found that the high expression of
has-miR-103/107 was correlated with poor survival by univariate and
multivariate analyses. More recently, the results of other studies
have indicated that miR-103 affects neuronal migration by
modulating CDK5R1 expression (11).
However, as yet, no studies have investigated the effects of
miR-103 in endometrial cancers. The mechanism by which miR-103
affects cancer development and progression is not fully understood.
Previous studies suggested that the attenuated expression of TIMP-3
exists in various types of cancer, including chorionic, prostatic
and esophageal carcinomas and melanotic cancer (12–15).
At present, no studies investigating TIMP-3 expression in
endometrial cancer have been published.
In this study, we aimed to evaluate the possible
involvement of miR-103 in endometrial carcinogenesis and to reveal
any correlation between miR-103 and TIMP-3. The results of this
study may aid the understanding of the effects of miR-103 in the
tumorigenesis and progression of endometrial cancer.
Materials and methods
Cell culture and reagents
The human endometrial cancer cell lines, HEC-1B
(estrogen receptor-negative; from our laboratory) and Ishikawa
(estrongen receptor-positive; donated by Professor Wei Lihui,
Medical College of Beijing University) were grown in α-MEM (Gibco,
Darmstadt, Germany) supplemented with 10% fetal bovine serum
(Gibco), 100 U/ml of penicillin and 100 μg/ml of streptomycin
(Gibco). The cells were incubated at 37°C in a humidified chamber
supplemented with 5% CO2. The anti-miR-103
oligonucleotide and control negative oligonucleotide (negative
control) were purchased from Ambion (Carlsbad, CA, USA). The TIMP-3
antibody was purchased from Abcam (Cambridge, MA, USA), GAPDH
antibody from Sigma (Steinheim, Germany) and goat anti-mouse IgG
from Bio-Rad (Hercules, CA, USA). The primer sequences used were:
miR-103 RT primer: GTCGTATCCAGTGCAGGGTCCGAGGTAT
TCGCACTGGATACGACTCAGCC; miR-103 forward: GAGCAGCATTGTACAG and
reverse: GTGCAGG GTCCGAGGT: U6 forward: CTCGCTTCGGCAGCACA and
reverse: AACGCTTCACGAATTTGCGT; TIMP-3 forward: AGTTACCCAGCCCTATGA
and reverse: GCA AAGGCTTAAACATCT; β-actin forward: CGTGGG
CCGCCCTAGGCACCA and reverse: TTGGCTTAGGG TTCAGGGGGG. The primers
were purchased from Shanghai Songon (Shanghai, China). Transwell
chambers (8 μm, 6.5 mm) were purchased from Millipore (Billerica,
MA, USA) and Matrigel was purchased from BD Biosciences (San Jose,
CA, USA).
Cell transfection
Suitable cells were seeded in six-well plates to
ensure that they would reach 30% confluence the following day. The
transfection of the anti-miR-103 or negative control
oligonucleotide was performed using the Lipofectamine 2000 reagent
(Invitrogen, Carlsbad, CA, USA) in antibiotic-free Opti-MEM
(Invitrogen) according to the manufacturer’s instructions, with a
final concentration of 15 nM in the Ishikawa cells and 30 nM in the
HEC-1B cells. Following 48 h of transfection, the cells were
harvested and processed for further analysis.
Quantitative reverse transcription
(qRT)-PCR analysis of mRNA and miRNA expression
Total RNA was isolated from the cells using TRIzol
reagent (Invitrogen) to obtain miRNA and mRNA. For the expression
analysis of miR-103 and TIMP-3, the stem-loop RT and real-time PCR
primers were designed as described above. Briefly, miRNAs and mRNA
were examined by SYBR real-time qRT-PCR (Takara, Dalian, China) on
a 7500 Real-Time PCR system (Applied Biosystems, Carlsbad, CA, USA)
according to the manufacturer’s instructions. The relative
expression was calculated using the ΔCT method and the expression
of miR-103 was normalized using the 2−ΔΔCT method
(Pfaffl, 2001) relative to U6-snRNA and TIMP-3-mRNA was normalized
to β-actin. The qRT-PCR assays were performed in duplicate and the
data were presented as the mean ± standard error of the mean
(SEM).
Western blot analysis
Cell lysates were prepared in lysis buffer
containing a protease inhibitor cocktail (Roche Diagnostics,
Mannheim, Germany) following a 48-h transfection with either
anti-miR-103 or the negative control oligonucleotide. The protein
concentrations of the total cell lysates were measured using the
Micro BCA protein assay kit (Pierce Biotechnology Inc., Rockford,
IL, USA) and 80 μg of total cell lysate per lane was separated by
10% SDS-PAGE, then transferred to a PVDF membrane (Millipore). The
membrane was then soaked in blocking solution (1X TBS, 10% non-fat
dried milk) for 2 h and incubated with anti-actin monoclonal
antibody at 4°C overnight. The following day, the membrane was
washed with TBST six times for 1 h, incubated with the secondary
antibody (goat anti-mouse IgG; 1:5,000; Santa Cruz Biotechnology,
Inc., Santa Cruz, CA, USA) for 2 h and washed with TBST six times
for 1 h. Finally, the enhanced chemiluminescence (ECL) system
western blotting detection reagents (Millipore) were used according
to the manufacturer’s instructions and exposed to X-ray film.
Luciferase reporter assay
The 3’-UTR segments of TIMP-3 mRNA containing the
miR-103 binding sites were amplified by PCR from human genomic DNA
and inserted into the Xbal site of the pGL3 vector (Promega
Corporation, Madison, WI, USA), designated pGL3-TIMP-3-wt.
pGL3-PTEN-mut, which had mutations in the predicted miR-103 binding
sites, was constructed as a template. The wild-type or mutant
reporter (300 ng) was co-transfected into Ishikawa and HEC1B cells
in 12-well plates with a Renilla plasmid (10 ng) using
Lipofectamine 2000 (Invitrogen). At 6 h after transfection, the
cells were transfected with anti-miR-103 or the negative control
oligonucleotide. The reporter assay was performed 42 h
post-transfection using the Dual luciferase assay system (Promega)
and the firefly luciferase activity of each sample was normalized
to the Renilla luciferase activity.
Cell invasion assays
Transwell invasion experiments were performed in
24-well Matrigel-coated invasion chambers. Approximately
2×104 cells in 0.2 ml serum-free α-MEM were added into
each filter of the upper chamber and 0.6 ml of complete α-MEM was
added to the lower chambers. Following culture for 24 h, the cells
on the upper side of the filters were removed with cotton-tipped
swabs and the cells that had invaded into the lower side of the
filter were fixed in methanol for 15 min, stained with 0.05%
crystal violet in PBS for 15 min and microscopically observed and
counted in 5 fields of view at a magnification of ×200. The
invasive activity of the cancer cells was expressed as the mean
number of cells that invaded to the lower side of the filter and
the results were shown as the mean ± SD of the number of cells per
field of view. The experiments were repeated at least three
times.
Cell proliferation assays
Cell proliferation was assessed using an MTT assay.
Cells (5.0×103 cells/well) were seeded on 96-well
plates. Following the transfection of the Ishikawa cells by
anti-miR-103 for varying time periods (20, 44, 68 and 92 h), 50 μl
of MTT (5 mg/ml; Sigma, Chicago, IL, USA) was added to each well
and the cells were incubated for a further 4 h at 37°C. The plates
were then agitated for 30 sec and the optical density (OD) was
measured at 490 nm on an enzyme immunoassay analyzer (Bio-Rad,
Tokyo, Japan).
Evaluation of apoptosis
Apoptosis was detected by a flow cytometric analysis
of Annexin V and PE staining. The Annexin V-FITC versus PE assay
was performed following the manufacturer’s instructions. Briefly,
adherent cells were harvested and suspended in the binding buffer
(1×106 cells/ml). The cells were then incubated with
Annexin V-FITC and PE for 15 min at room temperature in the dark
and immediately analyzed by flow cytometry (BD Biosciences).
Statistical analysis
Statistical analysis was performed using the SPSS
13.0 software (SPSS, Inc., Chicago, IL, USA). The values are
presented as the mean ± SEM. The differences/correlations between
the groups were calculated using the Student’s t-test and analysis
of variance. P<0.05 was considered to indicate a statistically
significant result.
Results
miR-103 directly targets TIMP-3
3’-UTRs
The mature sequence of miR-103
(5’-AGCAGCAUUGUACAGGGCUAUGA-3’) was retrieved from the miRBase
Sequence Database, release 14 (http://microrna.sanger.ac.uk/sequences/). A
bioinformatics search (http://diana.pcbi.upenn.edu/cgi-bin/TargetCombo.cgi),
revealed that an evolutionarily conserved target sequence for
miR-103 is located in the 3’-UTR of TIMP-3 (Fig. 1C). This suggests that it is a target
for miR-103. To validate whether TIMP-3 is a direct target of
miR-103, we performed a luciferase reporter assay. The decreased
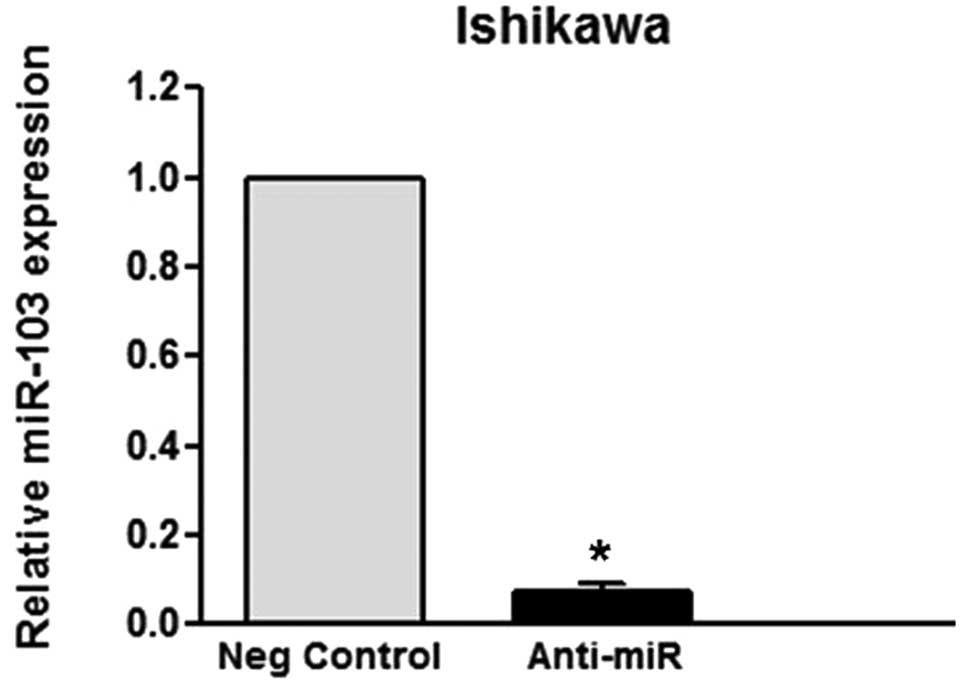
level of expression of miR-103 upon transfection, confirmed by
qRT-PCR (Fig. 1A and B),
significantly affected the luciferase expression, measured as
relative luciferase activity (Fig. 1D
and E). The results revealed a statistically significant
upregulation of luciferase activity in Ishikawa and HEC-1B cells
when these cells were transfected with pGL3-TIMP-3-wt together with
the miR-103 inhibitor, but not with any other combination. These
results confirm the bioinformatic predictions, indicating that the
TIMP-3 3’-UTR is a direct target of miR-103.
Inhibition of miR-103 upregulates TIMP-3
protein expression in endometrial cancer cells
If miR-103 targets the TIMP-3 3’-UTR, the TIMP-3
protein expression levels should be inversely correlated with
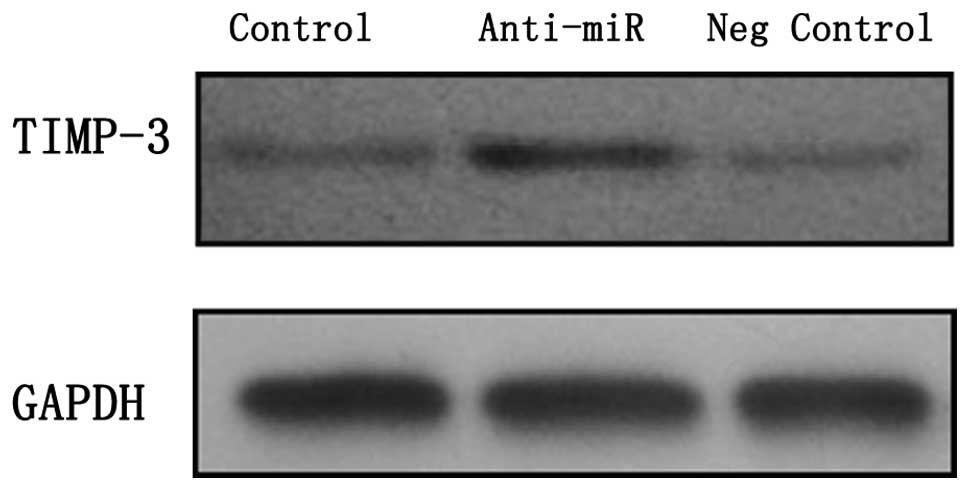
miR-103 expression levels. A western blot analysis of TIMP-3
expression was performed in two endometrial cancer cell lines, in
which the knockdown of miR-103 expression following transfection
was confirmed by qRT-PCR. As expected, the TIMP-3 protein levels
were significantly increased in the two cell lines which were
transfected with anti-miR-103, compared with those transfected with
the negative control oligonucleotide and the untreated endometrial
cancer cells (Fig. 2). These
results are consistent with the hypothesis that miR-103 is able to
target TIMP-3 and thereby influence the amount of TIMP-3 protein in
the cells. We also measured the TIMP-3 mRNA expression by qRT-PCR,
but the difference was not statistically significant (data not
shown).
Inhibition of miR-103 reduces the growth
and invasion of endometrial cancer cells
The impact of the modulation of miR-103 expression
on cell growth and viability following transfection was tested
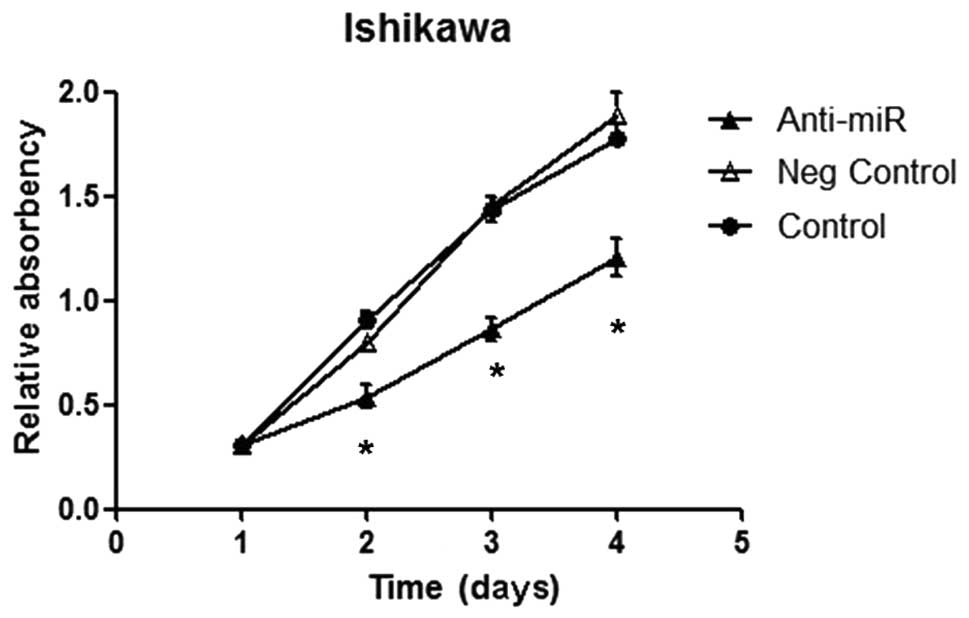
using the MTT assay. No significant difference was observed in the
light absorption values between the untreated and negative control
groups (Fig. 3A). The cells that
were transfected with anti-miR-103 showed slow proliferation and
the inhibition rates at 48, 72 and 96 h following transfection were
20.70, 40.63 and 35.92%, respectively, indicating that cell
proliferation was suppressed by the transfection of anti-miR-103
(Fig. 3A). To evaluate the effect
of miR-103 downregulation on the invasion of endometrial cancer
cells, a cell invasion assay was performed. Cells were grown in the
upper chamber and assessed for invasion through the Matrigel toward
a chemoattractant (10% serum) in the lower chamber. The number of
invasive HEC-1B and Ishikawa cells transfected with anti-miR-103
were significantly reduced relative to those with control
oligonucleotide (Fig. 3A and B).
miR-103 silencing resulted in a 42.86% decrease in the number of
invasive Ishikawa cells (Fig. 3C;
P<0.05). Similarly, the silencing of miR-103 in HEC-1B cells
significantly inhibited cell invasion, with a 45.45% decrease in
the number of invasive cells (Fig.
3C; P<0.05). The Annexin V-FITC versus PE assay showed that
anti-miR-103 had no effect on the apoptosis of the Ishikawa and
HEC-1B cells. Taken together, these data suggest that miR-103 is
essential for tumor cell growth and invasion in vitro.
Discussion
This is the first study to show that the tumor
suppressor TIMP-3 is negatively regulated by miR-103 at the
post-transcriptional level, via a specific target site within the
3’-UTR. This is also the first study to demonstrate that miR-103
induces growth and invasion in endometrial cancer cells.
miRNAs induce the degradation or translational
repression of target mRNAs and are thereby involved in the
initiation and progression of human cancer (16), although the underlying mechanism is
largely unknown.
To investigate the correlation between TIMP-3 and
miR-103, we first performed a bioinformatics search. It revealed a
conserved target site for miR-103 within the TIMP-3 3’-UTR. A
miR-103 inhibitor was then transfected into the endometrial cancer
cells. As expected, the epigenetic inhibition of the expression of
miR-103 resulted in the upregulation of the TIMP-3 protein. To
analyse whether the predicted miR-103 target site in the 3’-UTR of
TIMP-3 was responsible for its regulation, we performed a
luciferase reporter assay, the results of which were in accordance
with those of the western blot analysis. These data suggest that
miR-103 is important in the regulation of TIMP-3. Additionally, the
aberrant expression of TIMP-3 in endometrial cancer may be caused
by the aberrant expression of miR-103. Therefore, we conclude that
miR-103 directly targets TIMP-3 3’-UTRs.
The roles of TIMP-3 in cancer progression have been
highlighted in studies that have reported that the adenoviral-
mediated overexpression of TIMP-3 reduced the invasion of HeLa and
HT1080 cells (17) and the
vector-mediated expression of TIMP-3 in cancer cells reduced
metastasis (18). In breast cancer,
the overexpression of miR-21 was found to promote cell invasion via
the regulation of TIMP-3 (19).
TIMP-3 also induced cells to undergo apoptosis (20). Moreover, TIMP-3 was found to inhibit
tumor growth in lung cancer cells in vivo (21).
In the present study, the results of the MTT assay
revealed that the growth of cells transfected with anti-miR-103 was
inhibited compared with the untreated and negative control groups.
This finding suggests that the upregulation of TIMP-3 reduces cell
proliferation, which is in agreement with the study by Finan et
al (21), while the impact of
modulated TIMP-3 on endometrial cancer in vivo requires
further study. Notably, the results of the Annexin V-FITC versus PE
assay showed that anti-miR-103 had no effect on the apoptosis of
the Ishikawa and HEC-1B cells. One possible reason for this is that
the biological effects of TIMP-3 are further defined by the
specific genetic and epigenetic background of each cell line.
Metastasis is a significant cause of cancer-related mortality.
Therefore, identifying the role of miR-103 in invasion and
metastasis has direct clinical implications. The results of the
transwell invasion experiment showed that the transfection of
anti-miR-103 reduced the invasion of endometrial cancer cells.
In conclusion, the results of the present study
suggest that the tumor suppressor TIMP-3 is negatively regulated at
the post-transcriptional level by miR-103 via a specific target
motif at the TIMP-3 3’-UTR. Furthermore, miR-103 induces invasion
and proliferation in endometrial cancer cells. Additional
mechanisms and targets of miR-103 besides TIMP-3 are likely to
contribute to miR-103-induced tumor cell proliferation and
invasion. Although the aim of this study was to gain a better
understanding of the function of miR-103 in endometrial cancer,
future in vivo studies are required to address the
therapeutic potential of miR-103.
Acknowledgements
The study was funded by general project H200942,
supported by the Department of Public Health of Jiangsu
Province.
References
|
1
|
Calin GA, Sevignani C, Dumitru CD, Hyslop
T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M
and Croce CM: Human microRNA genes are frequently located at
fragile sites and genomic regions involved in cancers. Proc Natl
Acad Sci USA. 101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yanaihara N, Caplen N, Bowman E, Seike M,
Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et
al: Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xi Y, Shalgi R, Fodstad O, Pilpel Y and Ju
J: Differentially regulated micro-RNAs and actively translated
messenger RNA transcripts by tumor suppressor p53 in colon cancer.
Clin Cancer Res. 12:2014–2024. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wickramasinghe NS, Manavalan TT, Dougherty
SM, Riggs KA, Li Y and Klinge CM: Estradiol downregulates miR-21
expression and increases miR-21 target gene expression in MCF-7
breast cancer cells. Nucleic Acids Res. 37:2584–2595. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Boren T, Xiong Y, Hakam A, Wenham R, Apte
S, Wei Z, Kamath S, Chen DT, Dressman H and Lancaster JM: MicroRNAs
and their target messenger RNAs associated with endometrial
carcinogenesis. Gynecol Oncol. 110:206–215. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chung TK, Cheung TH, Huen NY, Wong KW, Lo
KW, Yim SF, Siu NS, Wong YM, Tsang PT, Pang MW, et al: Dysregulated
microRNAs and their predicted targets associated with endometrioid
endometrial adenocarcinoma in Hong Kong women. Int J Cancer.
124:1358–1365. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gottardo F, Liu CG, Ferracin M, Calin GA,
Fassan M, Bassi P, Sevignani C, Byrne D, Negrini M, Pagano F, et
al: Micro-RNA profiling in kidney and bladder cancers. Urol Oncol.
25:387–392. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang GH, Wang F, Yu J, Wang XS, Yuan JY
and Zhang JW: MicroRNAs are involved in erythroid differentiation
control. J Cell Biochem. 107:548–556. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guo Y, Chen Z, Zhang L, Zhou F, Shi S,
Feng X, Li B, Meng X, Ma X, Luo M, et al: Distinctive microRNA
profiles relating to patient survival in esophageal squamous cell
carcinoma. Cancer Res. 68:26–33. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Moncini S, Salvi A, Zuccotti P, Viero G,
Quattrone A, Barlati S, De Petro G, Venturin M and Riva P: The role
of miR-103 and miR-107 in regulation of CDK5R1 expression and in
cellular migration. PLoS One. 6:e200382011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Feng H, Cheung AN and Xue WC:
Down-regulation and promoter methylation of tissue inhibitor of
metalloproteinase 3 in choriocarcinoma. Gynecol Oncol. 94:375–382.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yegnasubramanian S, Kowalski J, Gonzalgo
ML, Zahurak M, Piantadosi S, Walsh PC, Bova GS, De Marzo AM, Isaacs
WB and Nelson WG: Hypermethylation of CpG islands in primary and
metastatic human prostate cancer. Cancer Res. 64:1975–1986. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Darnton SJ, Hardie LJ and Muc RS: Tissue
inhibitor of metalloproteinase-3 (TIMP-3) gene is methylated in the
development of esophageal adenocarcinoma: loss of expression
correlates with poor prognosis. Int J Cancer. 115:351–358. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bachman KE, Herman JG, Corn PG, Merlo A,
Costello JF, Cavenee WK, Baylin SB and Graff JR:
Methylation-associated silencing of the tissue inhibitor of
metalloproteinase-3 gene suggest a suppressor role in kidney,
brain, and other human cancers. Cancer Res. 59:798–802.
1999.PubMed/NCBI
|
|
16
|
Kumar MS, Lu J, Mercer KL, Golub TR and
Jacks T: Impaired microRNA processing enhances cellular
transformation and tumorigenesis. Nat Genet. 39:673–677. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Baker AH, George SJ, Zaltsman AB, Murphy G
and Newby AC: Inhibition of invasion and induction of apoptotic
cell death of cancer cell lines by overexpression of TIMP-3. Br J
Cancer. 79:1347–1355. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lou Y, Yang X, Wang F, Cui Z and Huang Y:
MicroRNA-21 promotes the cell proliferation, invasion and migration
abilities in ovarian epithelial carcinomas through inhibiting the
expression of PTEN protein. Int J Mol Med. 26:819–27.
2010.PubMed/NCBI
|
|
19
|
Song B, Wang C, Liu J, Wang X, Lv L, Wei
L, Xie L, Zheng Y and Song X: MicroRNA-21 regulates breast cancer
invasion partly by targeting tissue inhibitor of metalloproteinase
3 expression. J Exp Clin Cancer Res. 29:292010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bian J, Wang Y, Smith MR, Kim H, Jacobs C,
Jackman J, Kung HF, Colburn NH and Sun Y: Suppression of in vivo
tumor growth and induction of suspension cell death by tissue
inhibitor of metalloproteinases (TIMP)-3. Carcinogenesis.
17:1805–1811. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Finan KM, Hodge G, Reynolds AM, Hodge S,
Holmes MD, Baker AH and Reynolds PN: In vitro susceptibility to the
proapoptotic effects of TIMP-3 gene delivery translates to greater
in vivo efficacy versus gene delivery for TIMPs-1 or -2. Lung
Cancer. 53:273–284. 2006. View Article : Google Scholar : PubMed/NCBI
|