Introduction
Small cell lung cancer (SCLC) is a highly aggressive
and lethal type of cancer in humans. It constitutes approximately
15% of all cases of primary lung cancer (1). SCLC is sensitive to chemotherapy and
radiotherapy, but long-term survival is low and the majority of
patients eventually develop progressive disease. There is a high
rate of relapse even among patients who achieve a complete
response. High levels of expression of c-kit and its ligand, stem
cell factor (SCF), have been widely found in both SCLC tumors and
established cell cultures (2).
Imatinib mesylate (STI571) is an oral inhibitor of a
number of tyrosine kinases, which acts through occupation of the
highly conserved structure of tyrosine kinase at the ATP binding
site. With the success of imatinib mesylate in the treatment of
gastrointestinal stromal tumors expressing c-kit, its use in SCLC
serves as a novel molecular therapeutic approach. Autocrine or
paracrine activation of c-kit by its ligand has been postulated for
lung cancer, but this receptor can also be activated by mutations
of c-kit. The activity of imatinib mesylate is associated with
mutation of the c-kit gene exons 9 and 11 in gastrointestinal
stromal tumors (3,4). Certain studies have reported that
imatinib mesylate failed to demonstrate clinical activity in SCLC
(5–7). Boldrini et al (8) examined 60 SCLC samples to determine
mutations of the c-kit coding region. Expression of c-kit was
demonstrated in approximately 40% of SCLC samples. Their results
showed that two patients had mutations in exon 9 and three patients
had mutations in exon 11. The expression of c-kit and its
mutational status did not appear to be relevant to or have a
significant impact on survival (8).
The cause of this negative result with imatinib used in SCLC may be
due to the low incidence of c-kit exon 9 or 11 mutation.
Epidermal growth factor receptor (EGFR) tyrosine
kinase inhibitors have been widely used in non-small cell lung
cancer (NSCLC) (9–13), but have failed to treat relapsed
SCLC (14). The incidence of EGFR
mutation in NSCLC is higher in China than in the United States of
America and European countries (15,16).
There may be also differences in the incidence of c-kit gene
mutations between China and European countries. However, at present
no study examining imatinib mesylate treatment for SCLC in China is
available. To determine the expression and mutation of c-kit and
its correlation with prognosis of SCLC in China, we examined c-kit
expression and mutation status in 36 SCLC patients.
Patients and methods
Patient characteristics
A total of 36 SCLC surgical specimens were
retrospectively collected at the Zhejiang Cancer Hospital,
Hangzhou, China between 1998 and 2010. The median age of the
patients was 54 years and the range was 22–73 years. There were 4
female and 32 male patients. Samples were derived from the primary
tumor. The pathological type of all patients was conventional SCLC
based on the standard criteria defined by WHO classification. The
median pack-years of smoking history was 30. The cancer stage was
determined according to the seventh edition of TNM classification
for lung cancer: T1 8 cases, T2 14 cases, T3 10 cases and T4 4
cases; N0 12 cases, N1 10 cases, N2 14 cases and N3 0 cases
(Table I); IA 4 cases, IB 2 cases,
IIA 6 cases, IIB 5 cases, IIIA 17 cases and IIIB 2 cases. A total
of 23 patients (64%) were successfully followed-up. The median age
was 56 years and the range was 38–73 years. There was 1 female and
22 male patients. The cancer stage was determined according to the
seventh edition of TNM classification for lung cancer: T1 6 cases,
T2 7 cases, T3 7 cases and T4 3 cases; N0 10 cases, N1 5 cases, N2
8 cases and N3 0 cases (Table I);
IA 4 cases, IB 2 cases, IIA 3 cases, IIB 2 cases, IIIA 11 cases and
IIIB 1 case. The median pack-years of smoking history was 30
(Table I). A total of 82.6% of
patients received first-line chemotherapy after surgery; the most
common treatment was a combination regimen of etoposide and
cisplatin. The follow-up deadline was November 01, 2011. The
survival time was calculated from the date of histological
diagnosis. The use of tissue samples was approved by the Ethics
Review Committee of Zhejiang Cancer Hospital.
 | Table IBaseline characteristics of the study
population. |
Table I
Baseline characteristics of the study
population.
| Characteristic | No. of patients
(n=36) | Follow-up no. of
patients (n=23) |
|---|
| Age (years)
(median) | 54 | 56 |
| Gender
(female/male) | 4/32 | 1/22 |
| Smoking history
(median pack-years) | 30 | 30 |
| Stage |
| I | 6 | 6 |
| II | 11 | 5 |
| III | 19 | 12 |
| IV | 0 | 0 |
| T stage |
| T1 | 8 | 6 |
| T2 | 14 | 7 |
| T3 | 10 | 7 |
| T4 | 4 | 3 |
| N stage |
| N0 | 12 | 10 |
| N1 | 10 | 5 |
| N2 | 14 | 8 |
| N3 | 0 | 0 |
Immunohistochemistry
Slide sections (4 μm) of the specimens were heated
in a pressure cooker for 4 h at 56°C. Paraffin wax sections were
de-waxed and 0.01 mmol/l pH 9.0 TE buffer was added before slides
were placed in a water bath for 20 min at 95°C. After antigen
retrieval, the sections were rinsed under distilled water, and 3%
H2O2 was used to block endogenous-peroxidase
activity. Each section had 50 μl polyclonal rabbit anti-c-kit
antibody (1:400; Dako, Glostrup, Denmark) added for 60 min at 37°C,
and 50 μl Envision complex was added for 40 min at room
temperature. After each step the slides were rinsed 3 times with
0.01 mmol/l PBS (pH 7.4) for 5 min. The slides were colored by
0.04% DAB-0.03% H2O2 colored water, rinsed
under distilled water, and counterstained in hematoxylin solution
for 1 min. The slides were then dehydrated, made transparent and
covered with a coverslip. The sections incubated in PBS instead of
the primary antibody were used as negative controls, while the
known positive sections were used as positive controls. C-kit
expression of tumors were scored as negative (0) if <5% of cells
were positive, weak staining (+) if 5–25% of cells were positive,
moderate (++) if 26–50% of cells were positive and strong (+++) if
>50% of cells were positive (7).
DNA preparation
Genomic DNA was isolated and purified from
formalin-fixed paraffin-embedded tissues using the GT pure FFPE
Tissue DNA Extraction kit (Gene Tech, Shanghai, China).
C-kit pyrosequencing assay
For the amplification of the exon 9 and 11 fragments
of the c-kit gene isolated from the genomic DNA, polymerase chain
reaction (PCR) amplification primers were designed for
pyrosequencing: c-kit-9, forward: 5′-ATGGCACGGTTGAATGTAAGG-3′ and
reverse biotinylated primer: 5′-CAGAGCCTAAACATCCCCTT AAA-3′; and
c-kit-11, forward: 5′-AGGTGATCTATTTTTCCC TTTCTC-3′ and reverse
biotinylated primer: 5′-GGAAACT CCCATTTGTGATCAT-3′. Each PCR
reaction contained forward and reverse primers (each 4 pmol), 2 μl
template DNA solution and 2 unit hotstart Taq DNA Polymerase
(Takara, Shiga, Japan) in a 40 ml volume. PCR conditions consisted
of initial denaturing for 3 min at 95°C; annealing at 50 cycles of
15 sec at 95°C, 30 sec at 56°C, 30 sec at 72°C; and a final
extension of 5 min at 72°C. The PCR products were sequenced by the
Pyrosequencing PyroMark ID system (Qiagen, Hilden, Germany)
according to the manufacturer’s instructions. Using the two
pyrosequencing primers (5′-3′ orientation): c-kit-9,
CGATGTGGGCAAGACT; and c-kit-11, TCCCTTTCTCCC CAC, pyrosequencing
was performed using PyroMark Gold Q96 reagents (Qiagen) containing
an enzyme and substrate mixture, dATP-S, dCTP, dGTP and dTTP.
Statistical analysis
Data were analyzed using the statistical software
SPSS (version 15). Kaplan-Meier life-table curves were compared
using the log-rank test to estimate survival. To compare the
survival pattern of the different variables, logistic regression
analysis was applied. P<0.05 was considered to indicate a
statistically significant difference.
Results
The incidence of c-kit-positive expression was
83.3%, including 25.0% weak staining, 22.2% moderate staining and
36.1% strong staining (Table II).
The status of c-kit expression in the follow-up patients was
negative staining in 6 cases, weak in 6 cases, moderate in 5 cases
and strong staining in 6 cases (Fig.
1). The median survival time (MST) of the follow-up patients
was 20 months. There was no difference in overall survival (OS)
between the SCLC patients with c-kit negative expression and SCLC
patients with c-kit positive expression. There was also no
difference in OS between the SCLC patients with c-kit negative and
weak staining and SCLC patients with c-kit moderate and strong
staining. The OS of SCLC patients with c-kit strong staining was
shorter than those with c-kit not strong staining (P=0.030)
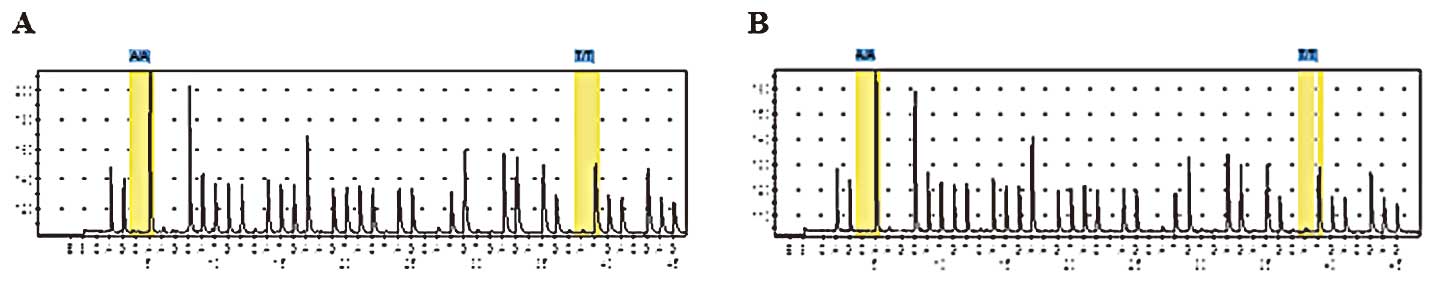
(Fig. 2). No mutation in c-kit
exons 9 and 11 was detected (Fig.
3).
 | Table IIClinical characteristics of patients
with c-kit-positive or c-kit-negative expression. |
Table II
Clinical characteristics of patients
with c-kit-positive or c-kit-negative expression.
| c-kit expression |
|---|
|
|
|---|
| Negative | + | ++ | +++ |
|---|
| No. of patients | 6 | 9 | 8 | 13 |
| Age (median) | 64 | 52 | 50 | 53 |
| Gender
(female/male) | 0/6 | 0/9 | 0/8 | 4/9 |
| History of smoking
(median pack-year) | 22 | 30 | 31 | 20 |
Discussion
The c-kit protein is a member of the type III
receptor tyrosine kinase family. A positive c-kit expression has
been observed in 37% of SCLC patients, and c-kit expression has
been demonstrated to be associated with decreased survival
(17). C-kit expression is of
particular clinical relevance for patients with advanced disease
and poor response to chemotherapy. C-kit serves as a new prognostic
factor for SCLC. Given the limited therapeutic options and
unfavorable prognosis of SCLC, clinical studies aimed at targeting
c-kit are necessary (17). In their
study, Blackhall et al demonstrated c-kit expression in 51%
of tumors, and concluded that c-kit expression is not predictive of
survival (18). A number of
different studies have classified c-kit-positive expression using a
variety of standards. The standard of c-kit positivity as
identified in a study by Micke et al (17) was determined when positive slides
demonstrated c-kit-positive cells including membrane staining in at
least 10% of all tumor cells. Slides demonstrating a weak
cytoplasmic signal without membrane staining were defined as
negative. The standard of c-kit positivity by Blackhall et
al (18) was determined when
c-kit expression was greater than 35%. The standard of c-kit
positivity by Boldrini et al (8) was determined when tumors with
immunoreactive cells had a threshold value above 1%. ‘Positive’ was
classified into three scales, as follows: weak staining as +,
moderate staining as ++ and strong staining as +++. These different
standards may lead to varying results of whether c-kit expression
may serve as a prognostic factor of SCLC. Therefore, it is
significant to incorporate the standard of c-kit positivity in all
studies. Our standard of c-kit positivity was in accordance with Dy
et al (7). In this standard,
c-kit positivity was classified as weak staining (+), moderate
staining (++) and strong staining (+++).
Combined SCLC has been reported to account for less
than 1–3.2% of all SCLC (19,20).
The surgical specimens reflect the clinicopathological status; a
high percentage of cases (28%) demonstrated that SCLC combined with
NSCLC in surgical specimens (21).
The specimens used in our study were obtained from surgical
resection, and pathological type of all our patients was
conventional SCLC. The specimens reported by Blackhall et al
were from SCLC biopsies (18). Our
study has shown that c-kit expression is high in SCLC, and strong
staining correlates with a worse survival prognosis.
Imatinib mesylate failed to demonstrate any clinical
activity despite patients being selected for c-kit-expressing SCLC,
and efficacy prediction of imatinib mesylate should not be based on
c-kit expression (8). Patients of
gastrointestinal stromal tumor with mutations in c-kit exons 9 and
11 are sensitive to imatinib mesylate. The incidence of EGFR
mutation of NSCLC in China is higher than in the United States of
America and European countries (15,16),
but no mutation of c-kit exons 9 or 11 was detected in our
study.
Compared to other genotyping and genetic detection
methods, pyrosequencing technology is unique. It delivers the ‘gold
standard’ of genetic analysis: the sequence itself. Other methods
only provide a ‘Yes/No’ signal. Unlike a fluorescent signal,
sequence information is intelligible. It is easy to communicate
these results in literature, and easy to transfer meaningful data
between research labs. Pyrosequencing assays are mutation-tolerant.
Unlike hybridisation-based assays, pyrosequencing analysis
generates a correct sequence regardless of the appearance of a new,
unexpected mutation. This is crucial to microbiological
applications: hybridisation-based assays can give false negatives
in the presence of a new mutation. With pyrosequencing analysis,
the sequence information is obtained, and the data is fully
quantitative; ideal for measuring the relative amounts of alleles.
This property allows the quantification of DNA methylation,
heterozygosity, ploidy levels, multi-copy genes, pooled DNA
samples, hematopoeitic chimerism and mixed genotypes in
heterogeneous samples (e.g., tumor and normal cells). In the study
by Boldrini et al, c-kit exons 9 and 11 were analyzed by
PCR-single-strand conformational polymorphism and automated
sequencing (8), while in our study
pyrosequencing technology was used to detect c-kit exons 9 and 11.
Boldrini et al (8)
demonstrated that the expression of c-kit and its mutational status
failed to appear relevant or to have a significant impact on
survival.
No mutation of c-kit exons 9 or 11 was detected in
our study. C-kit expression is high in SCLC, and strong staining
correlates to to worse survival prognosis. The incidence of EGFR
exons 19 and 21 mutation in SCLC is extremely low in China and
Japan (22–24), and gefitinib failed to demonstrate
benefit in relapsed SCLC patients (14). Given the limited therapeutic options
and unfavorable prognosis of SCLC, increased interest and studies
are required to develop targeted therapies to improve survival.
Acknowledgements
This study was funded by the Zhejiang Provincial
Natural Science Foundation of China (No.Y2110004), the Zhejiang
Province Medical Science Fund Project of China (No.2010KYA035), the
Zhejiang Province Traditional Medical Science Fund Project of China
(No.2010ZA006) and the Ningbo City Medical Science Fund Project of
China (No.2009A20).
Abbreviations:
|
SCLC
|
small cell lung cancer
|
|
EGFR
|
epidermal growth factor receptor
|
|
PCR
|
polymerase chain reaction
|
|
NSCLC
|
non-small cell lung cancer
|
|
MST
|
median survival time
|
|
OS
|
overall survival
|
References
|
1
|
Govindan R, Page N, Morgensztern D, et al:
Changing epidemiology of small-cell lung cancer in the United
States over the last 30 years: analysis of the surveillance,
epidemiologic, and end results database. J Clin Oncol.
24:4539–4544. 2006.PubMed/NCBI
|
|
2
|
Rygaard K, Nakamura T and Spang-Thomsen M:
Expression of the proto-oncogenes c-met and c-kit and their
ligands, hepatocyte growth factor/scatter factor and stem cell
factor, in SCLC cell lines and xenografts. Br J Cancer. 67:37–46.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cirocchi R, Farinella E, La Mura F, et al:
Efficacy of surgery and imatinib mesylate in the treatment of
advanced gastrointestinal stromal tumor: a systematic review.
Tumori. 96:392–399. 2010.PubMed/NCBI
|
|
4
|
Reichardt P: Optimal use of targeted
agents for advanced gastrointestinal stromal tumours. Oncology.
78:130–40. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Johnson BE, Fischer T, Fischer B, et al:
Phase II study of imatinib in patients with small cell lung cancer.
Clin Cancer Res. 9:5880–5887. 2003.PubMed/NCBI
|
|
6
|
Schneider BJ, Kalemkerian GP, Ramnath N,
et al: Phase II trial of imatinib maintenance therapy after
irinotecan and cisplatin in patients with c-Kit-positive,
extensive-stage small-cell lung cancer. Clin Lung Cancer.
11:223–227. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dy GK, Miller AA, Mandrekar SJ, et al: A
phase II trial of imatinib (ST1571) in patients with c-kit
expressing relapsed small-cell lung cancer: a CALGB and NCCTG
study. Ann Oncol. 16:1811–1816. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Boldrini L, Ursino S, Gisfredi S, et al:
Expression and mutational status of c-kit in small-cell lung
cancer: prognostic relevance. Clin Cancer Res. 10:4101–4108. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Douillard JY, Shepherd FA, Hirsh V, et al:
Molecular predictors of outcome with gefitinib and docetaxel in
previously treated non-small-cell lung cancer: data from the
randomized phase III INTEREST trial. J Clin Oncol. 28:744–752.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Maemondo M, Inoue A, Kobayashi K, et al:
Gefitinib or chemotherapy for non-small-cell lung cancer with
mutated EGFR. N Engl J Med. 362:2380–2388. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mok TS, Wu YL, Thongprasert S, et al:
Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N
Engl J Med. 361:947–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mitsudomi T, Morita S, Yatabe Y, et al:
Gefitinib versus cisplatin plus docetaxel in patients with
non-small-cell lung cancer harbouring mutations of the epidermal
growth factor receptor (WJTOG3405): an open label, randomised phase
3 trial. Lancet Oncol. 11:121–128. 2010. View Article : Google Scholar
|
|
13
|
Zhou C, Wu YL, Chen G, et al: Erlotinib
versus chemotherapy as first-line treatment for patients with
advanced EGFR mutation-positive non-small-cell lung cancer
(OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase
3 study. Lancet Oncol. 12:735–742. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Moore AM, Einhorn LH, Estes D, et al:
Gefitinib in patients with chemo-sensitive and chemo-refractory
relapsed small cell cancers: A Hoosier Oncology Group phase II
trial. Lung Cancer. 52:93–97. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shigematsu H, Lin L, Takahashi T, et al:
Clinical and biological features associated with epidermal growth
factor receptor gene mutations in lung cancers. J Natl Cancer Inst.
97:339–346. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu YL, Zhong WZ, Li LY, et al: Epidermal
growth factor receptor mutations and their correlation with
gefitinib therapy in patients with non-small cell lung cancer: a
meta-analysis based on updated individual patient data from six
medical centers in mainland China. J Thorac Oncol. 2:430–439. 2007.
View Article : Google Scholar
|
|
17
|
Micke P, Basrai M, Faldum A, et al:
Characterization of c-kit expression in small cell lung cancer:
prognostic and therapeutic implications. Clin Cancer Res.
9:188–194. 2003.PubMed/NCBI
|
|
18
|
Blackhall FH, Pintilie M, Michael M, et
al: Expression and prognostic significance of kit, protein kinase
B, and mitogen-activated protein kinase in patients with small cell
lung cancer. Clin Cancer Res. 9:2241–2247. 2003.PubMed/NCBI
|
|
19
|
Mangum MD, Greco FA, Hainsworth JD, et al:
Combined small-cell and non-small-cell lung cancer. J Clin Oncol.
7:607–612. 1989.PubMed/NCBI
|
|
20
|
Fraire AE, Johnson EH, Yesner R, et al:
Prognostic significance of histopathologic subtype and stage in
small cell lung cancer. Hum Pathol. 23:520–528. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nicholson SA, Beasley MB, Brambilla E, et
al: Small cell lung carcinoma (SCLC): a clinicopathologic study of
100 cases with surgical specimens. Am J Surg Pathol. 26:1184–1197.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lu HY, Sun WY, Chen B, et al: Epidermal
growth factor receptor mutations in small cell lung cancer patients
who received surgical resection in China. Neoplasma. 59:100–104.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tatematsu A, Shimizu J, Murakami Y, et al:
Epidermal growth factor receptor mutations in small cell lung
cancer. Clin Cancer Res. 14:6092–6096. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shiao TH, Chang YL, Yu CJ, et al:
Epidermal growth factor receptor mutations in small cell lung
cancer: a brief report. J Thorac Oncol. 5:195–198. 2011. View Article : Google Scholar : PubMed/NCBI
|