Introduction
Breast cancer is the most frequently diagnosed
cancer and is the leading cause of cancer-related mortality in
females (1). Generally, the
incidence and mortality of breast cancer is subject to
international variations, which are largely attributed to the
differences in reproductive and hormonal factors and the
availability of early detection services (1,2).
Currently, one of the major obstacles of breast cancer management
is the lack of sensitive and specific biomarkers that facilitate
the early detection and allow the monitoring of cancer progression
and treatment response (3,4). Currently, only two markers are
well-established and routinely used in the evaluation of breast
cancer: the estrogen receptor (ER), which predicts the response to
endocrine therapy, and the epidermal growth factor receptor-2
(HER-2/neu), which predicts the response to Trastuzumab (5). However, the variant expression and the
low sensitivities of the detection of ER and HER-2/neu restrict
their use as prognostic predictors. Great efforts have been made to
find new putative markers in the management of breast cancer.
MicroRNAs (miRNAs) are small, single-stranded,
non-coding RNAs of 19–24 nucleotides in length (6) with highly conserved sequences in the
context of their natural evolvement. miRNAs simultaneously regulate
the expression of multiple genes at the post-transcriptional level.
miRNAs inhibit mRNA translation and promote degradation via the
base pairing of semi-complementary sequences within the
3′-untranslated region (3-UTR) of target mRNAs (6,7). Over
previous years, miRNAs have been demonstrated to be involved in the
regulation of a wide spectrum of biological processes, including
cellular proliferation, apoptosis, development, differentiation and
metabolism (8,9). A variety of miRNAs have been
identified with altered expression in various types of human cancer
(9,10). Bioinformatic analysis has indicated
that each miRNA is capable of controlling a number of target genes,
underscoring the potential effect of miRNAs on almost every genetic
pathway (7,10). An ‘miRNA signature’ reflecting the
common characteristics of miRNA dysregulation in human tumors has
emerged from miRNA microarray analyses of more than 1,000 types of
primary tumors. miRNA profiling has shown that almost every type of
cancer exhibits a significantly different miRNA profile than its
normal counterpart tissue, suggesting the importance of
dysregulated miRNAs in human tumors (6).
In breast cancer, dysregulated miRNA may have the
potential to be used as a prognostic tool (11,12).
Among a wide spectrum of miRNAs, miR-155 is markedly overexpressed
in breast cancer tissues and is one of the most potent miRNA
suppressors of apoptosis in breast cancer cells (13). miR-155 may decrease the expression
of tumor protein p53-induced nuclear protein 1 (TP53INP1) and thus
attenuate TP53INP1-induced cell cycle arrest and apoptosis
(14). miR-155 also contributes to
oncogenicity in several other types of cancer (15,16).
miR-31 has been identified as an inhibitor of
multiple steps of the invasion-metastasis cascade in breast cancer
(17,18). More than 200 mRNAs are predicted to
be regulated by miR-31. Gene ontology has revealed that these
targets include a disproportionately large number of genes encoding
proteins involved in motility-related processes, including cell
adhesion, cytoskeletal remodeling and cell polarity (18,19).
It has been previously reported that miR-31 plasma levels may be
used as a diagnostic marker for oral squamous cell carcinoma
(20). However, the significance of
miR-31 as a prognostic predictor for breast cancer has not yet been
evaluated. The correlation between miR-31 and any other
clinicopathological parameters of breast cancer remains
obscure.
A noteworthy property of miRNA is its evident
stability (9,21,22).
miRNA is well-preserved in tissue samples, even several years after
formalin-fixation and paraffin embedment and is efficiently
extracted from and quantified in such specimens. Cancer-specific
miRNAs in the circulation are an emerging and exciting field in
cancer research (23). Their unique
stability and resilience make it possible for miRNAs to be present
in the circulation of cancer patients, therefore circulating miRNAs
have been explored as novel biomarkers for various types of adult
cancer (24). If this strategy
holds true for breast cancer, it would bring us closer to the
development of novel, sensitive and noninvasive biomarkers for this
common disease.
The current study determined the expression of
miR-155 and miR-31 in breast cancer tissues and their matched
adjacent normal tissues from 67 patients with stage I–III breast
cancer. The plasma levels of miR-135 and miR-31 in these patients,
compared with those in 70 age- and gender-matched healthy adults as
controls, were analyzed. This study further sought to investigate
whether the tissue expression and plasma levels of miR-135 and
miR-31 correlated with the clinicopathological characteristics of
these breast cancer patients.
Materials and methods
Patients and tissue samples
Anonymous cancerous and paired noncancerous tissues
from patients with pathologically confirmed breast cancer were
acquired from the Tissue Bank at the Cancer Hospital of Zhengzhou
University (Zhengzhou, China). These patients attended the Cancer
Hospital betweem September 2009 and September 2010. The following
inclusion criteria were employed: i) the patient had received no
radiotherapy or chemotherapy prior to surgery; ii) the patient had
pathological stage I–III breast cancer according to the American
Joint Committee on Cancer (AJCC) system. All samples were obtained
from patients who had undergone radical resections and samples from
normal mammary gland tissue were harvested at a distance of ≥5 cm
from the incisional margin as controls. All samples were
well-preserved in liquid nitrogen until total RNA of the samples
was extracted. In addition, blood samples were collected from all
67 patients prior to surgery. A total of 70 healthy adults served
as controls for plasma analysis. Whole blood (5 ml) was collected
from each subject in an anticoagulant EDTA tube and then
centrifuged at 3000 rpm at 4°C for 5 min. Following centrifugation,
the supernatant was harvested and stored at −80°C until required.
Acquisition of the tissue specimens was approved by the local
Institutional Review Board and all subjects provided written
informed consent for donating the blood samples.
Total RNA extraction and quantitative
real-time-PCR (qRT-PCR) analysis
Total cellular or plasma RNA, including miRNA, was
isolated from tissues or plasma samples using the Recover All™
Total Nucleic Acid Isolation kit (Invitrogen, Beijing, China)
according to the manufacturer’s instructions. Complementary DNA
(cDNA) was reverse transcribed from total cellular RNA using
specific miRNA primers from PrimeScript™ RT reagent kit and
TaqMan® MicroRNA Reverse Transcription kit
(Takara Bio, Inc., Shiga, Japan). The reverse transcriptase
reactions were run in a 20-μl volume containing 20 ng RNA at
16°C for 30 min, 42°C for 30 min and 85°C for 5 min. The products
were then kept at 4°C.
The expression levels of miRNAs were quantified
using a two-step RT-PCR method. The PCR primer pairs for miR-31 and
miR-155 were obtained from Invitrogen (Carlsbad, CA, USA) with the
following sequence: miR-31, 5′-AGGCAAGAT GCTGGCATAGCT-3′ (sense)
and miR-155, 5′-TTAATGCTA ATCGTGATAGGGGT-3′ (sense). For tissue
samples, RUNU6B (sense: 5′-GCGAGCACAGAATTAATACGAC-3′) was
used as a control to normalize the expression level of miRNAs by
correcting the differences of the cDNA template amount for the
quantitative PCR. For plasma samples, 18S ribosomal RNA (rRNA;
sense: 5′-GGATGAGCCTACAAC TGGCTT-3′) was used as a control to
normalize the expression level of target miRNAs. qRT-PCR was
performed using the SYBR® Premix Ex Taq™ PCR kit
(Takara Bio, Inc.). RT-PCR was run at 95°C for 10 min followed by
50 cycles of 15 sec at 95°C and 1 min at 60°C. All reactions were
run in triplicate using the ABI 7900 HT Fast Real-Time PCR System
with SDS software (version 2.3; Applied Biosystems China, Beijing,
China) and the average threshold cycle number (Ct) of each miRNA
was analyzed with the RQ Manager software (Version 1.2, Applied
Biosystems China).
The 2−ΔCt method
was used for analysis. The ΔCt value is the difference between the
Ct value of the specific miRNA and the Ct value of control, ΔCt =
Ct(miRNA)–Ct(RUNU6B/18S rRNA). The value of
ΔCt represents the miRNA expression level or content of each
sample. The value of 2−ΔCt
represents the relative quotient (RQ) of the expression of the
target gene to that of the control gene. The value of miRNA
expression is shown as mean ± SD. In the present study, the value
of RQ represents the ratio of miRNA expression in target samples to
that in control samples. An RQ<1 indicated that the expression
levels of miRNA in the target samples were lower than those in the
control samples. Conversely, an RQ>1 indicated higher miRNA
expression in target samples.
Immunohistochemistry
Frozen primary tumor samples were paraffin-embedded
and sliced into 4-μm thick sections. The tissue sections
were immunohistochemically stained by the Envision method using
primary monoclonal antibodies against the ER, progesterone receptor
(PR) and HER-2 (Novocastra™, Newcastle upon Tyne, UK). Positively
archived tissue sections were used as positive controls for each
antibody. In negative controls, the primary antibodies were
substituted by phosphate-buffered saline. H-scores were derived by
manual assessment of the intensity (scored 0–3) and the percentage
(%) of ER- and PR-positive tumor cells in 10 high-powered fields of
the same section. Sections were scored by two experienced
investigators who were blinded to the clinicopathological and
molecular variables of the patients. The two measurements were
multiplied to give an H-score varying from 0 to 300. In all areas,
only malignant cells with scores of ≥50 were considered positive
for either receptor. The DAKO scoring system was used for the
examination of the HER-2 status. A score of 0 or 1+ was considered
negative and a score of 3+ was considered positive for Her2. A
score of 2+ was identified by the FISH (fluorescence in situ
hybridization) test, if FISH+ was considered positive, FISH- was
considered negative.
Statistical analysis
The data are expressed as mean ± SD and were
analyzed with the SPSS software (Version 17.0; SPSS Inc., Chicago,
IL, USA). The paired sample t test was applied to compare the
amount of miRNA. Pearson’s and Spearman’s rank tests were applied
to analyze the correlation of values of the miRNA. The Mann-Whitney
U test and Kruskal-Wallis test were used to evaluate the
correlation between miRNA expression and the clinicopathological
parameters. P<0.05 was considered to indicate statistically
significant differences.
Results
Clinicopathological characteristics
A total of 67 patients met the inclusion criteria
and were selected for this study. The clinicopathological
characteristics of the breast cancer patients are shown in Table I. Their median age was 67 years
(range, 32–74) and 53.7% (36/67) were >52 years of age. The
majority of the patients had grade II (89.6%) and TNM stage II
(68.7%) disease and tumor size 2–5 cm (86.6%). The expression
levels of ER and PR were 77.6 and 83%, respectively. The majority
of the patients were HER-2 negative (73.1%).
 | Table IExpression of miR-155 and miR-31 in
the tissues of 67 breast cancer patients. |
Table I
Expression of miR-155 and miR-31 in
the tissues of 67 breast cancer patients.
| Clinicopathological
characteristics | RNA samples n
(%) | miR-155 (mean ±
SD) | miR-31 (mean ±
SD) |
|---|
| Age (years) | | | |
| ≥52 | 36 (53.7) | 0.1510±0.0035 |
0.02737±0.02097 |
| <52 | 31 (46.3) | 0.0076±0.0013 | 0.0032±0.0004 |
| Differentiation
grade | | | |
| I | 0 (0.0) | | |
| II | 60 (89.6) | 0.01167±0.0023 |
0.01799±0.01304 |
| III | 7 (10.4) | 0.0111±0.0024 | 0.0053±0.00174 |
| TNM stage | | | |
| I | 3 (4.5) | 0.0071±0.005 |
0.00281±0.00159 |
| II | 46 (68.7) | 0.01315±0.0029 |
0.02187±0.01643 |
| III | 18 (26.9) | 0.0084±0.00143 |
0.00383±0.00078 |
| Lymph node
metastasis | | | |
| 0 | 31 (46.3) | 0.01285±0.004 | 0.029±0.0244 |
| ≤3 | 19 (28.4) | 0.0122±0.0023 | 0.0063±0.0019 |
| >3 | 17 (25.3) | 0.0087±0.0015 | 0.0039±0.0008 |
| Tumor size
(cm) | | | |
| ≤2 | 3 (4.5) | 0.0067±0.0008 | 0.00312±0.0002 |
| 2–5 | 58 (86.6) | 0.01541±0.005 | 0.0221±0.0044 |
| >5 | 6 (8.9) | 0.0071±0.0004 | 0.0041±0.0007 |
| ER | | | |
| − | 27 (40.3) | 0.01852±0.0045 | 0.00487±0.0009 |
| + | 25 (37.3) | 0.00586±0.0010 |
0.00482±0.00141 |
| ++ | 10 (14.9) | 0.00786±0.0029 | 0.08035±0.0755 |
| +++ | 5 (7.5) | 0.01057±0.0053 |
0.00565±0.00323 |
| PR | | | |
| − | 34 (50.7) | 0.0163±0.00364 | 0.00413±0.0007 |
| + | 23 (32.3) | 0.0060±0.0010 | 0.00516±0.0015 |
| ++ | 5 (8.5) | 0.0064±0.0053 | 0.1592±0.1502 |
| +++ | 5 (8.5) | 0.1057±0.0053 | 0.00565±0.0032 |
| HER-2 | | | |
| Negative | 49(73.1) | 0.01098±0.0026 | 0.0200±0.01540 |
| Positive | 18(26.9) | 0.01335±0.0027 | 0.00575±0.0014 |
miR-155 and miR-31 were overexpressed in
breast cancer patients compared with healthy tissue and plasma
samples
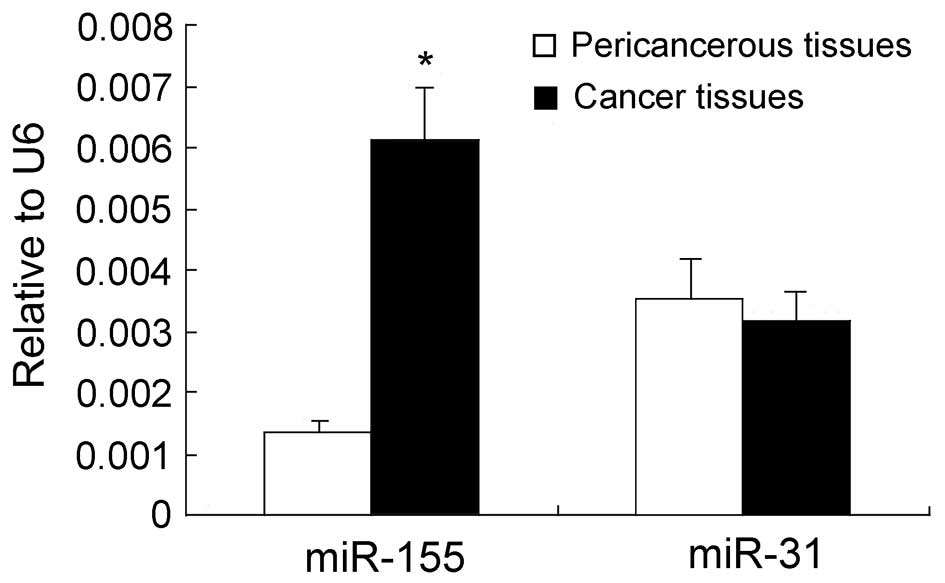
The expression levels of miR-155 and miR-31 in
breast cancer tissues were examined and matched to the adjacent
normal tissues by qRT-PCR assays. The results revealed that the
expression levels of miR-155 in breast cancer tissues were ∼6-fold
higher than those in the adjacent noncancerous tissues (P<0.05;
Fig. 1). Conversely, no
statistically significant difference was observed in the expression
levels of miR-31 between breast cancer and the adjacent
noncancerous tissues (P>0.05; Fig.
1). Notably, in patients who were >52 years old, the
expression levels of miR-155 and miR-31 were 20 and 8.6 times
higher than those in patients whose age was <52 years old
(Table I). We further investigated
whether there were any changes in the expression of miR-155 and
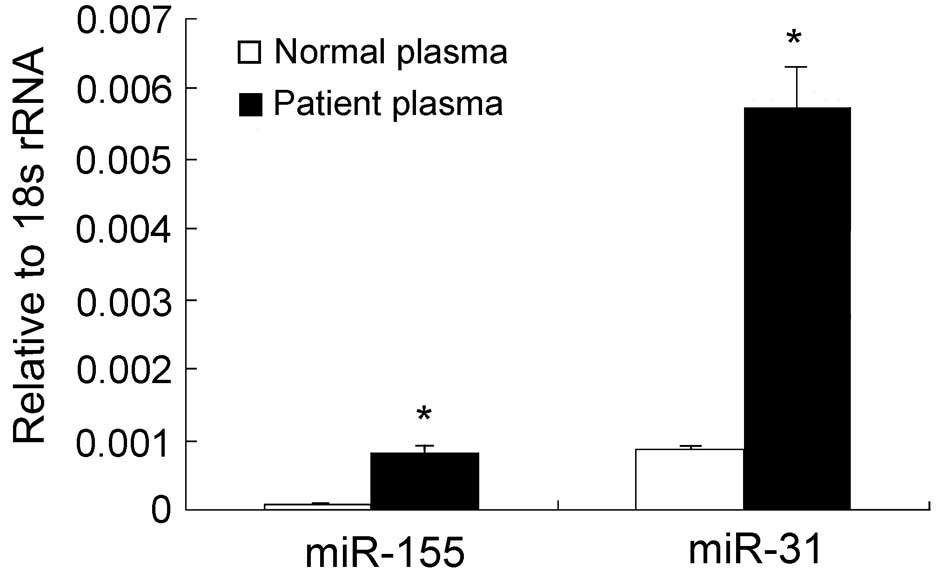
miR-31 in the plasma of breast cancer patients. The RT-PCR assays
revealed that the plasma levels of miR-155 in breast cancer
patients were ∼6-fold higher than those of the healthy subjects
(P<0.05; Fig. 2). Although no
difference in the miR-31 levels was detectable between cancerous
and noncancerous tissues, a significant difference was observed in
the plasma levels of miR-31 between breast cancer patients and
healthy controls (Table II).
Furthermore, the expression levels of miR-155 in the cancerous
tissues revealed a significant correlation with its expression
levels in the plasma (r=0.820; P=0.000), but no correlation was
noted between miR-31 expression in the cancerous tissues and plasma
samples of breast cancer patients.
 | Table IIExpression of miR-155 and miR-31 in
the plasma of 67 breast cancer patients. |
Table II
Expression of miR-155 and miR-31 in
the plasma of 67 breast cancer patients.
| Clinicopathological
characteristics | RNA samples n
(%) | miR-155 (mean ±
SD) | miR-31 (mean ±
SD) |
|---|
| TNM stage | | | |
| I | 3 (4.5) |
0.00024±0.00013 |
0.00112±0.00068 |
| II | 46 (68.7) | 0.0002±0.00005 |
0.00146±0.00040 |
| III | 18 (26.9) |
0.00016±0.00006 | 0.0011±0.00039 |
| ER | | | |
| − | 27 (40.3) |
0.00020±0.00006 |
0.00136±0.00037 |
| + | 25 (37.3) |
0.00010±0.00004 | 0.00078±0.0002 |
| ++ | 10 (14.9) |
0.00013±0.00004 |
0.00072±0.00022 |
| +++ | 5 (7.5) |
0.00045±0.00017 |
0.00340±0.00151 |
| PR | | | |
| − | 34 (50.7) |
0.00021±0.00006 | 0.00121±0.0003 |
| + | 23 (32.3) |
0.00016±0.00006 | 0.00100±0.0003 |
| ++ | 5 (8.5) |
0.00092±0.00002 | 0.00054±0.0001 |
| +++ | 5 (8.5) |
0.00049±0.00023 | 0.0043±0.0020 |
| HER-2 | | | |
| Negative | 49 (73.1) |
0.00019±0.00004 |
0.00130±0.00031 |
| Positive | 18 (26.9) |
0.00036±0.00016 |
0.00257±0.00124 |
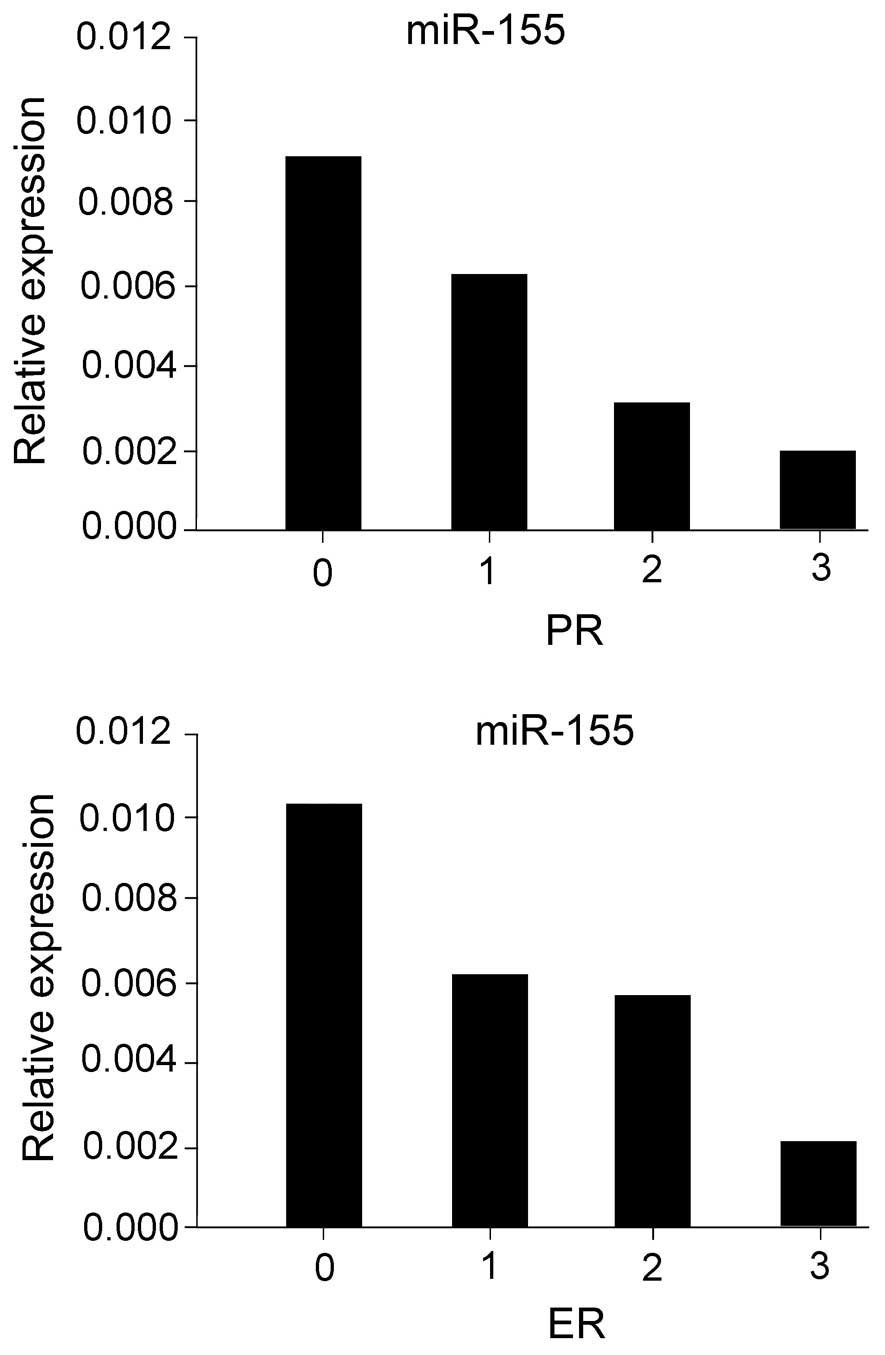
miR-155 expression is inversely
correlated with the ER and PR status of breast cancer patients
The key roles of ER, PR and HER-2, which promote
tumorigenesis, proliferation and distant metastasis, are
well-established in the endocrine and target therapeutic regimens
of breast cancer. Therefore, we investigated the correlation
between miR-155 and miR-31 expression in breast
cancer tissues and ER/PR/HER-2 status. An inverse correlation was
observed between the expression levels of ER/PR and those of
miR-155 in breast cancer patients (ER, r=−0.353, P=0.003;
PR, r=−0.357, P=0.003) (Fig. 3).
However, no correlation was observed between the expression levels
of ER, PR and miR-31 (ER, r=−0.353, P=0.003; PR, r=−0.357,
P=0.003). The correlation between miR-155 and miR-31 levels in the
plasma and ER/PR/HER-2 status in breast cancer patients was further
investigated. No correlation was observed between the expression
levels of ER, PR and the plasma levels of miR-155 (ER,
r=−0.353, P=0.003; PR, r=−0.357, P=0.003) as well as miR-31
(ER, r=−0.353, P=0.003; PR, r=−0.357, P=0.003). The correlation
between HER-2, an independent predictor of breast cancer prognosis,
miR-155 and miR-31 expression in breast cancer tissues and plasma
levels was also examined. It was demonstrated that HER-2 status was
not correlated with miR-155 (r=−0.353, P=0.003) or miR-31
expression (r=−0.353, P=0.003) in breast cancer tissues. Similarly,
HER-2 was not correlated with plasma miR-155 (r=−0.353, P=0.003)
and plasma miR-31 (r=−0.353, P=0.003).
The correlations of miR-155 and miR-31 with patient
clinicopathological characteristics, including TNM stage, lymph
node metastasis and tumor size were also analyzed. The expression
of miR-155 and miR-31 in TNM stage II tissues were higher than
those in other TNM stages (I and III) and miR-155 was highly
expressed in the groups of 0 or ≤3 lymph node metastases. High
miR-31 expression was identified in non-lymph node metastasic
tissues. In patients with tumor sizes of 2–5 cm, miR-155 and miR-31
were more abundant than in other sizes (Table I). The analysis of a correlation
between the other clinicopathological characteristics examined,
miR-155 and miR-31 expression in tissues and in plasma did not
reveal any significant difference.
Discussion
miR-155 is overexpressed in various types of solid
tumors, including breast cancer (25), pancreatic ductal adenocarcinoma
(26) and lung cancer (27). miR-155 is considered to be a
biological oncomir marker for poor prognosis. The significance
analysis of microarrays (SAM) and prediction analysis of
microarrays (PAM) from six types of solid tumors (lung, breast,
colon, gastric, prostate and endocrine pancreatic tumors) revealed
miR-21 and miR-155 to be at the top of the oncomir list (28). The dysregulation of miR-155
expression is involved in numerous pathological processes and
oncogenic signaling (15,16,29).
It was revealed that the miR-155 expression levels in breast cancer
tissues were significantly elevated compared with those in paired
normal tissues. miRNAs in the circulation have been shown to be
stable (12,30) and are readily accessible. Sampling
of miRNA in the blood is minimally invasive. Thus, the detection of
miRNA in patient plasma has numerous advantages for possible clinic
application. The plasma levels of miR-155 were significantly
increased in breast cancer patients compared with those in healthy
adults. These results are consistent with the observations in
tissue samples. Furthermore, miR-155 expression was inversely
correlated with ER and PR expression, regardless of HER-2 status.
These findings suggest that miR-155 expression is dysregulated in
breast cancer and is involved in the oncogenesis of breast
tumors.
miR-31, which is encoded by a single genomic locus
and is expressed in a variety of human tissues (19,31),
is one of the pleiotropically cancer-relevant miRNAs. In a previous
study, Valastyan et al identified miR-31 as a regulator of
metastatic progression in human breast cancer via a PCR-based
screening (17). The authors
demonstrated an inverse correlation between miR-31 expression and
the invasive capability in 15 different breast epithelial cell
lines. Additionally, miR-31 levels in primary human breast tumors
were revealed to be inversely associated with the propensity of
clinically detectable distant metastases (17). In contrast to the existing clinical
biomarkers for breast cancer, miR-31 expression was not correlated
with disease relapse. In the current study, miR-31 expression was
significantly elevated in the plasma samples of patients with
breast cancer compared with that in healthy adults, whereas no
significant difference was revealed between cancerous and
noncancerous tissues. The observation that the level of miR-31
expression in tissue samples was different from the level of plasma
samples reflects the complexity of the miRNA machinery regulation.
One possible explanation is that numerous miRNAs are released into
the circulation of the patients in a selective manner. The results
of the current study did not demonstrate a close correlation
between miR-31 and any clinicopathological characteristics. Unlike
miR-31, miR-155 expression in tissue samples was consistent with
that in plasma samples, indicating that the miR-155 level is a
direct reflection of miR-155 expression in cancerous tissues. These
results offer an experimental basis for the use of miRNAs as
therapeutic targets. The development of modified miRNAs with longer
in vivo half-lives and greater efficiency is the first step
towards translating these fundamental research advances into
medical practice. Future in vivo experiments of miRNA
transgenics and knockouts are likely to offer more valuable
information concerning the safety and efficacy of miRNA based
therapy.
References
|
1.
|
A JemalF BrayMM CenterJ FerlayE WardD
FormanGlobal cancer statisticsCA Cancer J
Clin616990201110.3322/caac.20107
|
|
2.
|
MV IorioM FerracinCG LiuMicroRNA gene
expression deregulation in human breast cancerCancer
Res6570657070200510.1158/0008-5472.CAN-05-178316103053
|
|
3.
|
H MouridsenA Giobbie-HurderA GoldhirschBIG
1-98 Collaborative Group: Letrozole therapy alone or in sequence
with tamoxifen in women with breast cancerN Engl J
Med361766776200910.1056/NEJMoa081081819692688
|
|
4.
|
JF ForbesJ CuzickA BuzdarA HowellJS
TobiasM BaumArimidex, Tamoxifen, Alone or in Combination
GroupEffect of anastrozole and tamoxifen as adjuvant treatment for
early-stage breast cancer: 100-month analysis of the ATAC
trialLancet Oncol94553200810.1016/S1470-2045(07)70385-618083636
|
|
5.
|
Early Breast Cancer Trialist’s
Collaborative GroupEffects of chemotherapy and hormonal therapy for
early breast cancer on recurrence and 15-year survival: an overview
of the randomised trialsLancet36516871717200515894097
|
|
6.
|
GA CalinCM CroceMicroRNA signatures in
human cancersNat Rev Cancer6857866200610.1038/nrc199717060945
|
|
7.
|
GA CalinCM CroceMicroRNA-cancer
connection: the beginning of a new taleCancer
Res6673907394200610.1158/0008-5472.CAN-06-080016885332
|
|
8.
|
K JeyaseelanWB HerathA ArmugamMicroRNAs as
therapeutic targets in human diseasesExpert Opin Ther
Targets1111191129200710.1517/14728222.11.8.111917665982
|
|
9.
|
CE Stahlhut EspinosaFJ SlackThe role of
microRNAs in cancerYale J Biol Med791311402006
|
|
10.
|
A Esquela-KerscherFJ SlackOncomirs -
microRNAs with a role in cancerNat Rev
Cancer6259269200610.1038/nrc1840
|
|
11.
|
SF TavazoieC AlarcónT OskarssonEndogenous
human microRNAs that suppress breast cancer
metastasisNature451147152200810.1038/nature0648718185580
|
|
12.
|
W ZhuW QinU AtasoyER SauterCirculating
microRNAs in breast cancer and healthy subjectsBMC Res
Notes289200910.1186/1756-0500-2-8919454029
|
|
13.
|
I FaraoniFR AntonettiJ CardoneE
BonmassarmiR-155 gene: a typical multifunctional microRNABiochim
Biophys Acta1792497505200910.1016/j.bbadis.2009.02.01319268705
|
|
14.
|
RM O’ConnellAA ChaudhuriDS RaoD
BaltimoreInositol phosphatase SHIP1 is a primary target of
miR-155Proc Natl Acad Sci USA10671137118200919359473
|
|
15.
|
S JiangHW ZhangMH LuMicroRNA-155 functions
as an OncomiR in breast cancer by targeting the suppressor of
cytokine signaling 1 geneCancer
Res7031193127201010.1158/0008-5472.CAN-09-425020354188
|
|
16.
|
E TiliJJ MichailleD WernickeMutator
activity induced by microRNA-155 (miR-155) links inflammation and
cancerProc Natl Acad Sci
USA10849084913201110.1073/pnas.110179510821383199
|
|
17.
|
S ValastyanF ReinhardtN BenaichA
pleiotropically acting microRNA, miR-31, inhibits breast cancer
metastasisCell13710321046200910.1016/j.cell.2009.03.04719524507
|
|
18.
|
S ValastyanA ChangN BenaichF ReinhardtRA
WeinbergActivation of miR-31 function in already-established
metastases elicits metastatic regressionGenes
Dev25646659201110.1101/gad.200421121406558
|
|
19.
|
S ValastyanRA WeinbergmiR-31: a crucial
overseer of tumor metastasis and other emerging rolesCell
Cycle921242129201010.4161/cc.9.11.1184320505365
|
|
20.
|
CJ LiuSY KaoHF TuMM TsaiKW ChangSC
LinIncrease of microRNA miR-31 level in plasma could be a potential
marker of oral cancerOral
Dis16360364201010.1111/j.1601-0825.2009.01646.x20233326
|
|
21.
|
RA SiniB TrinkA NissanThe role of microRNA
in tumorigenesis: key players or innocent bystandersJ Surg
Oncol99135136200910.1002/jso.2121319072979
|
|
22.
|
NB TsuiEK NgYM LoStability of endogenous
and added RNA in blood specimens, serum, and plasmaClin
Chem4816471653200212324479
|
|
23.
|
PS MitchellRK ParkinEM KrohCirculating
microRNAs as stable blood-based markers for cancer detectionProc
Natl Acad Sci
USA1051051310518200810.1073/pnas.080454910518663219
|
|
24.
|
H ZhaoJ ShenL MedicoD WangCB AmbrosoneS
LiuA pilot study of circulating miRNAs as potential biomarkers of
early stage breast cancerPLoS
One5e13735201010.1371/journal.pone.001373521060830
|
|
25.
|
J ZhuXQ HuGL GuoExpression and its
clinical significance of miR-155 in human primary breast
cancerZhonghua Wai Ke Za Zhi482052082010(In Chinese)
|
|
26.
|
N HabbeJB KoorstraJT MendellMicroRNA
miR-155 is a biomarker of early pancreatic neoplasiaCancer Biol
Ther8340346200910.4161/cbt.8.4.733819106647
|
|
27.
|
T DonnemK EkloT BergPrognostic impact of
MiR-155 in non-small cell lung cancer evaluated by in situ
hybridizationJ Transl Med96201110.1186/1479-5876-9-621219656
|
|
28.
|
S VoliniaGA CalinCG LiuA microRNA
expression signature of human solid tumors defines cancer gene
targetsProc Natl Acad Sci
USA10322572261200610.1073/pnas.051056510316461460
|
|
29.
|
E TiliCM CroceJJ MichaillemiR-155: on the
crosstalk between inflammation and cancerInt Rev
Immunol28264284200910.1080/0883018090309379619811312
|
|
30.
|
HM HeneghanN MillerAJ LoweryKJ SweeneyJ
NewellMJ KerinCirculating microRNAs as novel minimally invasive
biomarkers for breast cancerAnn
Surg251499505201010.1097/SLA.0b013e3181cc939f20134314
|
|
31.
|
CH StueltenDS SalomonmiR-31 in cancer:
location mattersCell
Cycle946084609201010.4161/cc.9.23.1392821260945
|