Introduction
Although it accounts for less than 0.5% of all types
of cancer, osteosarcoma (OS) is the most frequent primary
malignancy of the bone and occurs mainly in adolescents and young
adults (1). The initiation of
combinational chemotherapy with aggressive surgical resection has
markedly improved the prognosis of OS patients during the last few
decades (2). However, the current
neoadjuvant chemotherapy outcome for OS remains unsatisfactory in
the presence of metastases (3–5).
Despite the various efforts of basic research and clinical
practice, the molecular genetic mechanisms and the biology involved
in OS remain poorly understood. A greater understanding of OS is
essential for developing novel approaches to increase survival
rates (3).
As a large family of naturally occurring small
non-coding RNAs, microRNAs (miRNAs or miRs) employ a
post-transcriptional gene regulation mechanism that is involved in
numerous cellular processes, playing a role in development
regulation, differentiation, cell proliferation, differentiation,
apoptosis, cell cycle and tumorigenesis (6). Previous studies have shown that miRNAs
may play complex regulatory roles by binding to the 3′ untranslated
region of mRNAs; a single miRNA affects the expression of hundreds
of protein-coding target genes, while a protein-coding target gene
is regulated by a variety of miRNAs (7).
There is growing evidence that the aberrant
expression of specific miRNAs is correlated with various human
tumors, including breast cancer, hepatocellular carcinoma, leukemia
and colon cancer (8,9). It has been reported that miRNAs
regulate cancer cell apoptosis, cell cycle arrest, migration and
invasion. The alteration of specific miRNAs may lead to various
responses to the chemotherapy and several miRNAs have been
demonstrated to participate in the development of tumor metastasis
(10).
The current study was designed to investigate the
differential expression profiles of miRs between an OS and
osteoblast cell line. miRNA expression levels were determined using
bead-based array performing oligonucleotide capture probes specific
for miRNAs, which is feasible and attractive for its high speed and
heightened accuracy. In this study, the differential miRNAs were
explored through screening 1,146 mature miRNAs between the MG-63
and hFOB1.19 (HOB) cell lines and the expression of selected miRNAs
was confirmed using real-time quantitative PCR (RT- qPCR) in these
two cell lines. The tumor function-associated targeted mRNAs of
selected miRNAs by bioinformatics and previous literature were also
investigated. These findings provide insights into the role of
miRNAs in OS.
Materials and methods
Cell lines and reagents
Human OS MG-63 and osteoblast HOB cell lines were
obtained from the Type Culture Collection of Chinese Academy of
Sciences (Shanghai, China). MG-63 cells were cultured in MEM/EBSS
(Hyclone, Logan, UT, USA) supplemented with 10% heat-inactivated
fetal bovine serum (FBS, Hyclone), 50 U/ml penicillin and 50 mg/ml
streptomycin in a humidified incubator with 5% CO2 at
37°C. The HOB cells were maintained in the same conditions, except
that DMEM/F12 (v/v: 1:1, Hyclone) supplemented with 10% FBS and 0.3
mg/ml G418 (Sigma, St. Louis, MO, USA) was used.
RNA extraction
Total RNA was extracted from each cell line using an
miRNeasy Mini kit (Qiagen, Hilden, Germany) according to the
manufacturer’s instructions. This effectively recovered mRNA and
miRNA. RNA concentration and quality were measured using the
spectrophotometer (ND-2000, NanoDrop, Wilmington, DE, USA).
miRNA expression profiling by Illumina
miRNA microassay
The Illumina® TotalPrep™ RNA
amplification kit (Ambion, Austin, TX, USA) was used in cDNA
synthesis and purification with 200 ng total RNA from each treated
cell, followed by hybridizing on Human MicroRNA Expression
profiling v2 panels (Illumina, San Diego, CA, USA) according to the
manufacturer’s instructions (Illumina MicroRNA Expression Profiling
Assay Guide) and the method described previously (11). The Human v2 MicroRNA Expression
Profiling kit contains 1,146 assays for detecting >97% of the
miRNAs described in the miRBase database, plus additional novel
content derived using Illumina sequencing technology.
Array data processing and analysis were performed
with Illumina BeadStudio software (www.illumina.com).
The microRNA expression array was scanned and extracted using
BeadScan, with the data corrected by background subtraction in the
GenomeStudio module. The array intensity data were imported into
BeadStudio v3.2 (Illumina), a software package that permits
visualization and normalization of the data. The ‘Average’
normalization method was used for all analyses reported, with the
exception of assay reproducibility, given the number of replicates.
The normalized intensities and detection P-values were exported and
further analyzed using the R environment (version 2.6), in
combination with Bio-conductor packages (12).
miRNAs were considered significantly differentially
expressed if the P-values were <0.05 and the fold change ratio
(FCR) was >2.
RT-qPCR of specific miRNAs
Validation of differential gene expression was
performed for selected miRNAs, including miR-181a, miR-148a,
miR-99a, miR-195, miR-9, miR-335, miR-143, miR-145 and miR-539.
These miRNAs were amplified using the Bulge-Loop™ miRNA qRT-PCR
Primer Set (Ribobio, Guangzhou, China) (13). The thermal profile for the RT-qPCR
was at 95°C for 1 min, followed by 40 cycles of 95°C for 10 sec,
60°C for 20 sec and 72°C for 5 sec on a Bio-Rad CFX96 RT-qPCR
system (Bio-Rad, Hercules, CA, USA). All qPCR reactions, including
no-template controls, were performed in triplicate. Expression
levels of each miRNA were evaluated using a comparative threshold
cycle (Ct) method normalized to that of U6. The fold changes of
each miRNA were calculated from the expression levels in the MG-63
and HOB cell lines.
Bioinformatics analysis
The TarBase 6.0 database (http://diana.cslab.ece.ntua.gr/) was used to
investigate the validated target genes in cancer research (14). Moreover, following the collection of
all the validated genes, the function of these genes was analyzed
by previous literature and bioinformatics research. The chromosome
location of these miRNAs and their target genes was investigated to
exclude the potential bias of sex chromosomes and illustrate the
complex and comprehensive mechanisms in miRNA regulation of the
target gene expression.
Results
miRNA expression in the MG-63 and HOB
cell lines
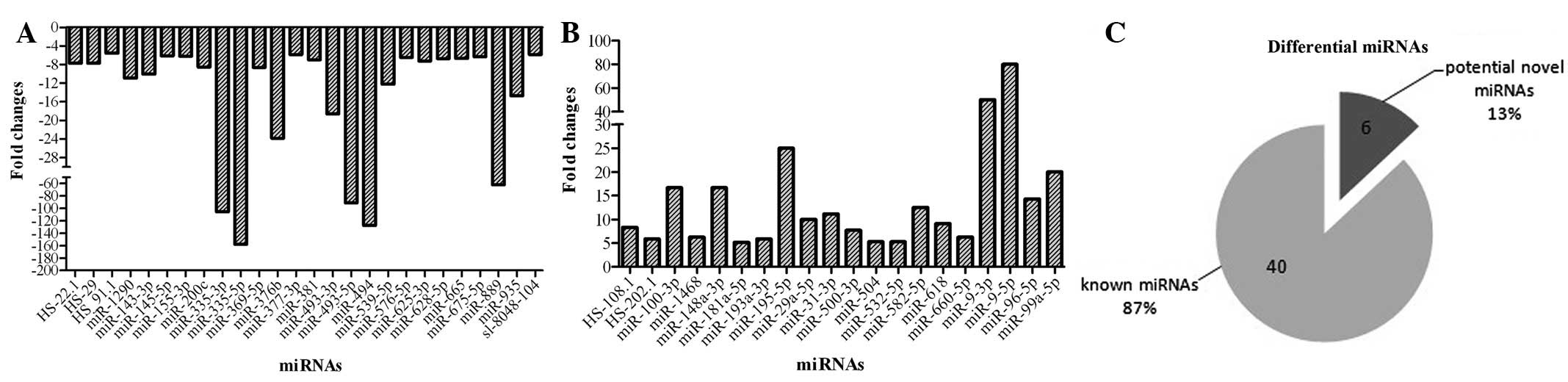
The fold changes (MG-63/HOB) were auto-analyzed by
software (BeadStudio v3.2, Illumina). Of the 1,146 miRNAs detected
in the microarray, 159 miRNAs were shown to be decreased and 109
miRNAs as increased. The various miRNAs were selected for further
analysis as follows: i) the minimum value should be >100 in the
two cell lines to eliminate the background value; ii) the fold
changes should be >5 for improved accuracy. As Fig. 1 shows, 46 miRNAs were selected as
differentially expressed between the MG-63 and HOB cell lines, of
which 26 were underexpressed and 20 were overexpressed. The fold
change was mainly <10, while several miRNAs in MG-63 cells were
markedly changed compared with the HOB cell line, including
miR-335, miR-493, miR-494, miR-195 and miR-9.
Validation of miRNAs in the MG-63 and HOB
cell lines
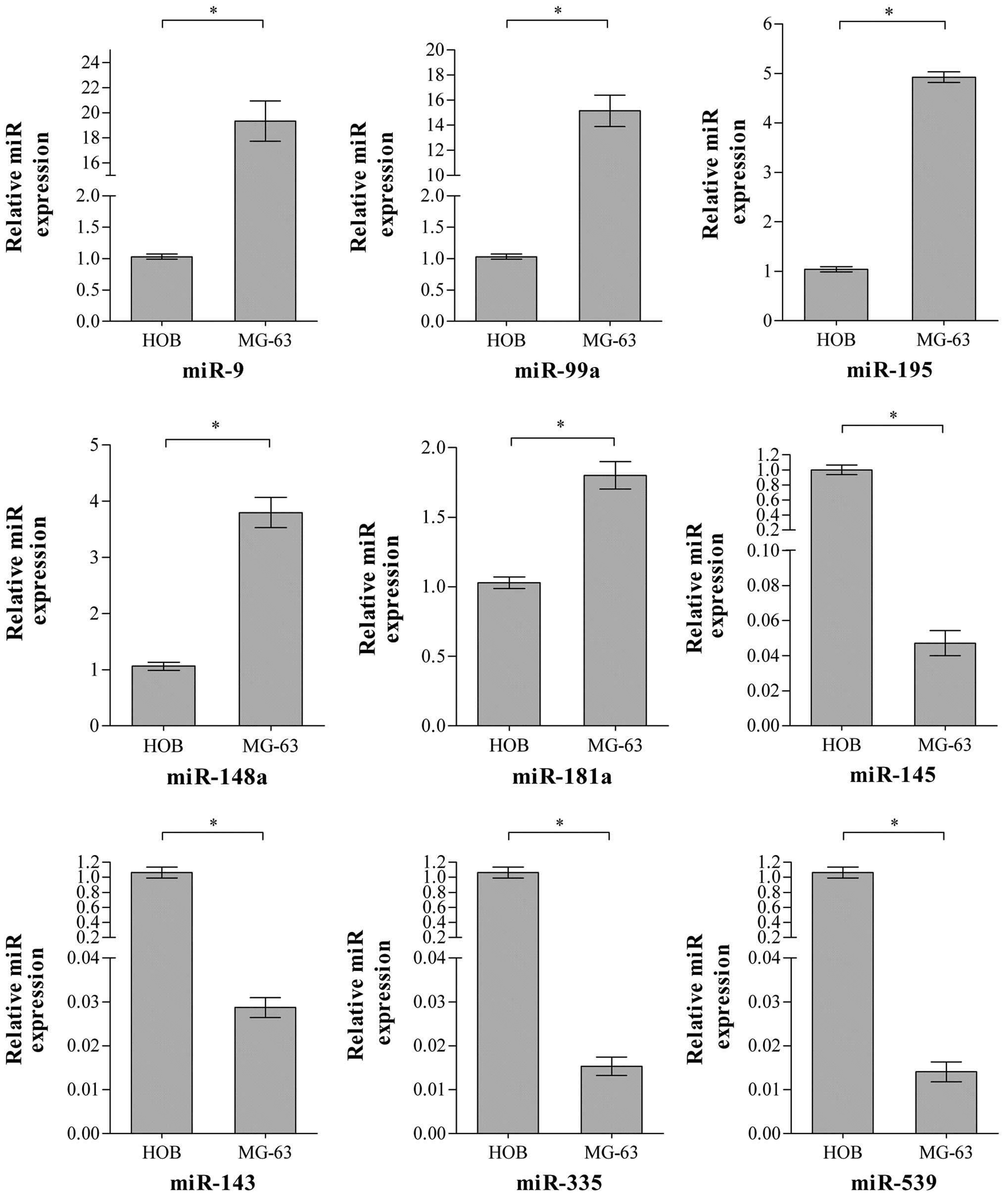
The RT-qPCR was employed to validate the
differential expression of selected miRNAs. As Fig. 2 shows, it was revealed that 9
specific miRNAs were differentially expressed between the OS MG-63
and HOB cell lines and the differences are consistent with the
microarray results shown. Therefore, it was demonstrated that these
are the differentially expressed miRNAs in OS MG-63 compared with
the HOB cell line.
Bioinformatics research on these
differential miRNAs and their target genes
Tables I and
II show the target genes involved
in the biological behavior of cancer and validated by previous
literature. The functions of these target genes are complex as they
are correlated with various cellular processes, including cell
proliferation, differentiation, cell cycle, apoptosis, signaling,
migration and invasion. Moreover, miRNAs may regulate the
expression and function of their target genes although they are
located on various chromosomes.
 | Table IValidated target genes of the miRNAs
in the increased group. |
Table I
Validated target genes of the miRNAs
in the increased group.
| miRNA | Location | Validated targets
(ref.) | Location | Functions in
cancer |
|---|
| miR-195 | 17p13.1 | CDK6 (15) | 7q21-q22 | Cell cycle and
arrest |
| | E2F3 (15) | 6p22 | Cell cycle and
arrest |
| | CCND1 (15) | 11q13 | Cell cycle and
arrest |
| | VEGFA (16) | 6p12 | Angiogenesis and
metastasis regulation |
| | Bcl-2 (17) | 18q21.3 | Apoptosis
regulation |
| | SKI (18) | 1q22-q24 | Proto-oncogene |
| | BCL2L11 (18) | 2q13 | Apoptosis
regulation |
| | CDK4 (19) | 12q14 | Cell cycle and
arrest |
| miR-99a | 21q21.1 | MTOR (20) | 1p36.2 | Response to
anti-cancer drugs |
| | FGFR3 (20) | 4p16.3 | Mitogenesis and
differentiation |
| | SKI (21) | 1q22-q24 | Proto-oncogene |
| | IGF1 (22) | 12q23.2 | Anti-apoptosis |
| miR-9 | 1q22 | NF-KB1 (23) | 4q24 | Transcription
regulation |
| | CDH1 (24) | 16q22.1 | Metastasis
regulation |
| | VEGFA (24) | 6p12 | Angiogenesis and
metastasis regulation |
| | VIM (24) | 10p13 | Cell attachment,
migration and signaling |
| | MMP13 (24) | 11q22.3 | Invasion and
metastasis regulation |
| | BIK (25) | 22q13.31 | Apoptosis
regulation |
| miR-148a | 7p15.2 | CDC25B (26) | 20p13 | Cell cycle and
arrest |
| | PTPN4 (21) | 2q14.2 | Cell growth,
differentiation, mitotic cycle and oncogenic transformation |
| | CDK19 (21) | 6q21 | Cell cycle and
arrest |
| | ROCK1 (27) | 18q11.1 | Invasion and
metastasis |
| miR-181a | 1q32.1 | CDKN1B (28) | 12p13.1-p12 | Cell cycle and
arrest |
| | Bcl-2 (29) | 18q21.3 | Apoptosis
regulation |
| | Hras (30) | 11p15.5 | Signal
transduction |
| | CDX2 (31) | 13q12.3 | Cell growth and
differentiation |
| | S100A1 (31) | 1q21 | Cell cycle and
differentiation |
| | KLF6 (32) | 10p15 | Tumor
suppressor |
 | Table IIValidated target genes of the miRNAs
in the decreased group. |
Table II
Validated target genes of the miRNAs
in the decreased group.
| miRNA | Location | Validated targets
(ref.) | Location | Functions in
cancer |
|---|
| miR-145 | 5q32 | FSCN1 (33) | 7p22 | Cell migration,
motility, adhesion and cellular interactions |
| | MMP1 (34) | 11q22.3 | Invasion and
metastasis regulation |
| | MMP12 (34) | 11q22.3 | Invasion and
metastasis regulation |
| | MMP14 (34) | 14q11-q12 | Invasion and
metastasis regulation |
| | TP53 (35) | 17p13.1 | Proliferation and
apoptosis regulation |
| miR-143 | 5q32 | MAPK7 (36) | 17p11.2 | Proliferation,
differentiation, transcription regulation |
| | MMP13 (37) | 11q22.3 | Invasion and
metastasis regulation |
| | Bcl-2 (38) | 18q21.3 | Apoptosis
regulation |
| | Hras (39) | 11p15.5 | Signal
transduction |
| | TP53 (35) | 17p13.1 | Proliferation and
apoptosis regulation |
| miR-335 | 7q32.2 | SP1 (40) | 12q13.1 | Cell proliferation,
differentiation, apoptosis regulation |
| | IGF1R (40) | 15q26.3 | Anti-apoptosis |
| | BRCA1 (40) | 17q21 | Tumor
suppressor |
| | BIK (41) | 22q13.31 | Apoptosis
regulation |
| | SMAD3 (41) | 15q22.33 | Carcinogenesis |
| | SMAD9 (41) | 2q26 | Carcinogenesis |
| | PML (41) | 15q22 | Tumor
suppressor |
| miR-539 | 14q32.31 | MITF (42) | 3p14.2-p14.1 | Cell
proliferation |
Discussion
The significance of miRNAs in the regulation of
cellular processes has been increasingly noted (43). Up- and/or downregulation of miRNA
expression in cancer suggests that miRNAs function as classical
tumor suppressor genes or oncogenes (6–8). The
expression fold changes of several miRNAs may aid in tumor
stratification and clinical outcome prognosis (44,45).
Previous research shows that specific miRNA expression may be
correlated with cancer recurrence. Therefore, the distinct
difference between normal and abnormal cells may be correlated with
the early diagnosis and treatment of the primary cancer or its
recurrence.
Although there are several studies concerning
specific miRNAs as the key biomarkers for the diagnosis, treatment
and evaluation of the chemoresponse or chemoprevention in cancer
therapy (46,47), little is known about miRNA profiling
and its signature in OS. The explosion of microarray technology has
led to its wide application in miRNA expression analysis. The miRNA
microarray was employed to detect the miRNA profiling in the OS and
HOB cell lines, respectively, and the real-time PCR was employed to
validate the miRNA of interest or marked differences between these
two cell lines. In the current study, 1,146 miRNAs were detected by
the micro-array, revealing 159 miRNAs as being part of the
decreased group and 109 miRNAs as the increased group. Following
further analysis of the miRNA microarray result, 46 miRNAs were
selected as the differentially expressed miRNAs between the MG-63
and HOB cell lines.
Furthermore, based on previous research and the
potential biological targets predicted by the various databases,
including Targetscan and PicTar, 9 miRNAs were selected to validate
their expression and demonstrate the difference between the two
cell lines. The stem-loop RT-PCR method described in the present
study is designed to detect and analyze mature miRNAs in a fast,
specific, accurate and reliable manner (48). Therefore, RT-qPCR was employed to
validate the expression of specific miRNAs of interest. Fig. 2 shows the expression of the selected
9 miRNAs quantified in the MG-63 and HOB cell lines. The
differences of these miRNAs between the two cell lines are
consistent with the microarray results shown. Therefore, these 9
miRNAs are accepted as the differentially expressed miRNAs between
the MG-63 and HOB cell lines.
It is well known that the miRNAs involved in complex
cancer-related cellular processes by regulating the various target
mRNA expression and miRNAs are thought to be components of vast
regulatory networks (49,50). Previous research has confirmed that
multiple miRNAs target the same gene, suggesting that the
correlation between miRNAs and target genes are complex and
interactive (51). Therefore,
analysis of the target genes of these differential miRNAs may
reveal their functions as oncogenes or anti-oncogenes. Furthermore,
this analysis is likely to aid the fuller understanding of the
biological function of specific miRNAs through analysis of their
target genes and vice versa (52–54).
The validated target genes of these 9 miRNAs were
obtained from bioinformatics research. Tables I and II show the validated target genes which
have been demonstrated by previous research to be involved in
various cellular process in cancer biology, including
proliferation, differentiation, cell cycle, apoptosis, signaling,
migration and invasion. These target genes are located on different
chromosomes, suggesting that miRNAs may regulate the expression and
function of mRNA although they are located on various chromosomes.
These 9 miRNAs may play a significant role in the biological
behavior of cancer, although they alter the target gene expression
in different directions (7).
A total of 9 miRNAs have been reported that may act
as biomarkers in the diagnosis, treatment and prediction of the
prognosis of cancer. Previous evidence has demonstrated that
miR-143 and miR-145, that belong to the same miRNA cluster,
regulate the expression and function of various target genes. It is
well known that the underexpression of the miR-143/145 cluster, the
expression of which was decreased in OS in the current study, are
strongly associated with carcinogenesis in various tumor types,
suggesting that they may act as significant tumor suppressors
(37). As Tables I and II demonstrate, tumor protein p53(TP53),
fascin homolog 1(FSCN1), several matrix metallopeptidases (MMPs)
and mitogen-activated protein kinases (MAPKs), which are regulated
by the miR-143/145 cluster, may be involved in cancer cell
proliferation, differentiation, gene transcription, apoptosis,
migration and invasion (55). The
similar effect of miR-143 and/or miR-145 has been demonstrated in
OS cell lines by previous research (56). Therefore, we may conclude that the
aberrant expression levels of the miR-143/145 clusters are
correlated with the carcinogenesis and development of OS compared
with the HOB cell line.
miR-9 is another of the widely-researched miRNAs in
cancer biology (23–25). It has been shown that miR-9 is
over-expressed in various tumor types, particularly in tumors with
micro-metastasis. The validated target genes (NF-KB1, CDH1, VEGFA,
VIM, MMP13 and BIK) shown in Table
I suggest that miR-9 may act as a significant regulatory miRNA,
which was also increased in OS in the current study compared with
the HOB cell line.
A previous study demonstrated that 6 other miRNAs
that were validated in the current study are also involved in the
biology of cancer. Of these, miR-99a may play a significant role in
cell growth and correlates with the prognosis of patients with
specific tumors (57,58). The aberrant expression of miR-195 in
certain types of cancer may be an effective biomarker in diagnosis
(59,60). The differential expression of
miR-148a has been reported to be a potential marker for colorectal
cancer screening and prognosis (61). miR-181a, which has been recently
demonstrated to be overexpressed miRNA in OS tissue, is correlated
with cancer development, apoptosis evasion and cell proliferation
(62). With regard to the reduced
expression group, miR-335, which is also underexpressed in certain
types of cancer, has also been demonstrated to be significant as a
biomarker of metastatic tumor and maintain differentiation
(63–65). Recently, it has been demonstrated
that miR-539 may inhibit cell proliferation through suppressing the
MITF expression (42). However, the
functional study of the 6 miRNAs in the OS cell line are limited
and ongoing research concerning their function may illustrate the
further mechanism of these miRNAs in OS oncogenesis, development
and metastasis.
In conclusion, the aberrant expression levels of
specific miRNAs, including miR-9, miR-99a, miR-195, miR-148a,
miR-181a, miR-143, miR-145, miR-335 and miR-539, may act as
potential biomarkers in the diagnosis, treatment and prognosis
prediction of OS. Further research on the function of their target
genes may provide new insights into the biology and treatment of
OS.
Acknowledgements
This study was supported by the
Natural Science Foundation of China (No.30772185), the Fundamental
Research Funds for the Central Universities (201130302020010).
References
|
1.
|
A JemalR SiegelE WardT MurrayJ XuMJ
ThunCancer statistics, 2007CA Cancer J
Clin574366200710.3322/canjclin.57.1.43
|
|
2.
|
PJ MesserschmittAN RettewRE BrookoverRM
GarciaPJ GettyEM GreenfieldSpecific tyrosine kinase inhibitors
regulate human osteosarcoma cells in vitroClin Orthop Relat
Res46621682175200810.1007/s11999-008-0338-918607665
|
|
3.
|
G BacciA LonghiM VersariM MercuriA
BriccoliP PicciPrognostic factors for osteosarcoma of the extremity
treated with neoadjuvant chemotherapy: 15-year experience in 789
patients treated at a single
institutionCancer10611541161200616421923
|
|
4.
|
A LonghiC ErraniM De PaolisM MercuriG
BacciPrimary bone osteosarcoma in the pediatric age: state of the
artCancer Treat
Rev32423436200610.1016/j.ctrv.2006.05.00516860938
|
|
5.
|
Y ZhangRX WeiXB ZhuL CaiW JinH
HuTanshinone IIA induces apoptosis and inhibits the proliferation,
migration and invasion of the osteosarcoma MG-63 cell line in
vitroAnticancer
Drugs23212219201210.1097/CAD.0b013e32834e559222126901
|
|
6.
|
E KobayashiFJ HornicekZ DuanMicroRNA
involvement in
osteosarcomaSarcoma2012359739201210.1155/2012/35973922550419
|
|
7.
|
RR LullaFF CostaJM BischofIdentification
of differentially expressed microRNAs in
osteosarcomaSarcoma2011732690201110.1155/2011/73269021789031
|
|
8.
|
A LujambioSW LoweThe microcosmos of
cancerNature482347355201210.1038/nature1088822337054
|
|
9.
|
V RottiersAM NäärMicroRNAs in metabolism
and metabolic disordersNat Rev Mol Cell
Biol13239250201210.1038/nrm331322436747
|
|
10.
|
W ZhangME DolanThe emerging role of
microRNAs in drug responsesCurr Opin Mol
Ther12695702201021154161
|
|
11.
|
L WangAL ObergYW AsmannGenome-wide
transcriptional profiling reveals microRNA-correlated genes and
biological processes in human lymphoblastoid cell linesPLoS
One11e5878200910.1371/journal.pone.000587819517021
|
|
12.
|
H ZhaoJ ShenL MedicoD WangCB AmbrosoneS
LiuA pilot study of circulating miRNAs as potential biomarkers of
early stage breast cancerPLoS
One5e13735201010.1371/journal.pone.001373521060830
|
|
13.
|
L GuoY LiuY BaiY SunF XiaoY GuoGene
expression profiling of drug-resistant small cell lung cancer cells
by combining microRNA and cDNA expression analysisEur J
Cancer4616921702201010.1016/j.ejca.2010.02.04320371173
|
|
14.
|
T VergoulisIS VlachosP AlexiouTarBase 6.0:
capturing the exponential growth of miRNA targets with experimental
supportNucleic Acids Res40Database
issueD222D229201210.1093/nar/gkr116122135297
|
|
15.
|
T XuY ZhuY XiongYY GeJP YunSM
ZhuangMicroRNA-195 suppresses tumorigenicity and regulates G1/S
transition of human hepatocellular carcinoma
cellsHepatology50113121200910.1002/hep.2291919441017
|
|
16.
|
W YeQ LvCK WongThe effect of central loops
in miRNA:MRE duplexes on the efficiency of miRNA-mediated gene
regulationPLoS
One3e1719200810.1371/journal.pone.000171918320040
|
|
17.
|
L LiuL ChenY XuR LiX DumicroRNA-195
promotes apoptosis and suppresses tumorigenicity of human
colorectal cancer cellsBiochem Biophys Res
Commun400236240201010.1016/j.bbrc.2010.08.04620727858
|
|
18.
|
Y MurakamiT YasudaK SaigoComprehensive
analysis of microRNA expression patterns in hepatocellular
carcinoma and non-tumorous
tissuesOncogene2525372345200610.1038/sj.onc.120928316331254
|
|
19.
|
Y LinJ WuH ChenCyclin-dependent kinase 4
is a novel target in micoRNA-195-mediated cell cycle arrest in
bladder cancer cellsFEBS
Lett586442447201210.1016/j.febslet.2012.01.02722289176
|
|
20.
|
C OneyamaJ IkedaD OkuzakiMicroRNA-mediated
downregulation of mTOR/FGFR3 controls tumor growth induced by
Src-related oncogenic
pathwaysOncogene3034893501201110.1038/onc.2011.6321383697
|
|
21.
|
M HafnerM LandthalerL
BurgerTranscriptome-wide identification of RNA-binding protein and
microRNA target sites by
PAR-CLIPCell141129141201010.1016/j.cell.2010.03.00920371350
|
|
22.
|
M DoghmanA El WakilB CardinaudRegulation
of insulin-like growth factor-mammalian target of rapamycin
signaling by microRNA in childhood adrenocortical tumorsCancer
Res7046664675201010.1158/0008-5472.CAN-09-397020484036
|
|
23.
|
LM GuoY PuZ HanMicroRNA-9 inhibits ovarian
cancer cell growth through regulation of NF-kappaB1FEBS
J27655375546200910.1111/j.1742-4658.2009.07237.x19702828
|
|
24.
|
L MaJ YoungH PrabhalamiR-9, a
MYC/MYCN-activated microRNA, regulates E-cadherin and cancer
metastasisNat Cell Biol12247256201020173740
|
|
25.
|
A GrimsonKK FarhWK JohnstonP
Garrett-EngeleLP LimDP BartelMicroRNA targeting specificity in
mammals: determinants beyond seed pairingMol
Cell2791105200710.1016/j.molcel.2007.06.01717612493
|
|
26.
|
ST LiffersJB MundingM VogtMicroRNA-148a is
down-regulated in human pancreatic ductal adenocarcinomas and
regulates cell survival by targeting CDC25BLab
Invest9114721479201110.1038/labinvest.2011.9921709669
|
|
27.
|
B ZhengL LiangC WangMicroRNA-148a
suppresses tumor cell invasion and metastasis by downregulating
ROCK1 in gastric cancerClin Cancer
Res1775747583201110.1158/1078-0432.CCR-11-171421994419
|
|
28.
|
R CuestaA Martínez-SánchezF
GebauermiR-181a regulates cap-dependent translation of p27(kip1)
mRNA in myeloid cellsMol Cell
Biol2928412851200910.1128/MCB.01971-0819273599
|
|
29.
|
W ZhuX ShanT WangY ShuP LiumiR-181b
modulates multidrug resistance by targeting BCL2 in human cancer
cell linesInt J Cancer12725202529201010.1002/ijc.2526020162574
|
|
30.
|
KH ShinSD BaeHS HongRH KimMK KangNH
ParkmiR-181a shows tumor suppressive effect against oral squamous
cell carcinoma cells by downregulating K-rasBiochem Biophys Res
Commun404896902201110.1016/j.bbrc.2010.12.05521167132
|
|
31.
|
J JiT YamashitaA BudhuIdentification of
microRNA-181 by genome- wide screening as a critical player in
EpCAM-positive hepatic cancer stem
cellsHepatology50472480200910.1002/hep.2298919585654
|
|
32.
|
X ZhangY NieY DuJ CaoB ShenY
LiMicroRNA-181a promotes gastric cancer by negatively regulating
tumor suppressor KLF6Tumour
Biol33921928201210.1007/s13277-012-0414-322581522
|
|
33.
|
T ChiyomaruH EnokidaS TataranomiR-145 and
miR-133a function as tumour suppressors and directly regulate FSCN1
expression in bladder cancerBr J
Cancer102883891201010.1038/sj.bjc.660557020160723
|
|
34.
|
M KanoN SekiN KikkawamiR-145, miR-133a and
miR-133b: Tumor-suppressive miRNAs target FSCN1 in esophageal
squamous cell carcinomaInt J
Cancer12728042814201010.1002/ijc.2528421351259
|
|
35.
|
J ZhangQ SunZ ZhangS GeZG HanWT ChenLoss
of microRNA- 143/145 disturbs cellular growth and apoptosis of
human epithelial cancers by impairing the MDM2-p53 feedback
loopOncogene2810381046201222330136
|
|
36.
|
Y AkaoY NakagawaT NaoeMicroRNA-143 and
-145 in colon cancerDNA Cell
Biol26311320200710.1089/dna.2006.055017504027
|
|
37.
|
M OsakiF TakeshitaY SugimotoMicroRNA-143
regulates human osteosarcoma metastasis by regulating matrix
metalloprotease-13 expressionMol
Ther1911231130201110.1038/mt.2011.5321427707
|
|
38.
|
PM BorralhoBT KrenRE CastroIB da SilvaCJ
SteerCM RodriguesMicroRNA-143 reduces viability and increases
sensitivity to 5-fluorouracil in HCT116 human colorectal cancer
cellsFEBS
J27666896700200910.1111/j.1742-4658.2009.07383.x19843160
|
|
39.
|
X ChenX GuoH ZhangRole of miR-143
targeting KRAS in colorectal
tumorigenesisOncogene2813851392200910.1038/onc.2008.47419137007
|
|
40.
|
H HeynM EngelmannS SchreekMicroRNA miR-335
is crucial for the BRCA1 regulatory cascade in breast cancer
developmentInt J Cancer12927972806201110.1002/ijc.2596221618216
|
|
41.
|
SF TavazoieC AlarcónT OskarssonEndogenous
human microRNAs that suppress breast cancer
metastasisNature451147152200810.1038/nature0648718185580
|
|
42.
|
YN LeeS BrandalP NoelKIT signaling
regulates MITF expression through miRNAs in normal and malignant
mast cell
proliferationBlood11736293640201110.1182/blood-2010-07-29354821273305
|
|
43.
|
SK PatnaikE KannistoS
YendamuriOverexpression of microRNA miR-30a or miR-191 in A549 lung
cancer or BEAS-2B normal lung cell lines does not alter
phenotypePLoS One5e9219201010.1371/journal.pone.000921920169152
|
|
44.
|
J KimDM CoffeyCJ CreightonZ YuSM HawkinsMM
MatzukHigh-grade serous ovarian cancer arises from fallopian tube
in a mouse modelProc Natl Acad Sci
USA10939213926201210.1073/pnas.111713510922331912
|
|
45.
|
P Puerta-GilR García-BaqueroAY JiamiR-143,
miR-222 and miR-452 are useful as tumor stratification and
noninvasive diagnostic biomarkers for bladder cancerAm J
Pathol18018081815201210.1016/j.ajpath.2012.01.03422426337
|
|
46.
|
MA CortezJW WelshGA CalinCirculating
microRNAs as noninvasive biomarkers in breast cancerRecent Results
Cancer Res195151161201210.1007/978-3-642-28160-0_1322527502
|
|
47.
|
JT MendellEN OlsonMicroRNAs in stress
signaling and human
diseaseCell14811721187201210.1016/j.cell.2012.02.00522424228
|
|
48.
|
E Varkonyi-GasicRP HellensQuantitative
stem-loop RT-PCR for detection of microRNAsMethods Mol
Biol744145157201110.1007/978-1-61779-123-9_1021533691
|
|
49.
|
T SaitoP SaetromMicroRNAs - targeting and
target predictionN
Biotechnol27243249201010.1016/j.nbt.2010.02.01620219708
|
|
50.
|
ME PeterTargeting of mRNAs by multiple
miRNAs: the next
stepOncogene2921612164201010.1038/onc.2010.5920190803
|
|
51.
|
S WuS HuangJ DingMultiple microRNAs
modulate p21Cip1/Waf1 expression by directly targeting its 3′
untranslated regionOncogene2923022308201020190813
|
|
52.
|
DW ThomsonCP BrackenGJ GoodallExperimental
strategies for microRNA target identificationNucleic Acids
Res3968456853201110.1093/nar/gkr33021652644
|
|
53.
|
TA FaraziJI SpitzerP MorozovT TuschlmiRNAs
in human cancerJ Pathol223102115201110.1002/path.2806
|
|
54.
|
SK ShenoudaSK AlahariMicroRNA function in
cancer: oncogene or a tumor suppressor?Cancer Metastasis
Rev28369378200910.1007/s10555-009-9188-520012925
|
|
55.
|
H ZhangX CaiY WangH TangD TongF
JimicroRNA-143, down-regulated in osteosarcoma, promotes apoptosis
and suppresses tumorigenicity by targeting Bcl-2Oncol
Rep2413631369201020878132
|
|
56.
|
L FanQ WuX XingY WeiZ ShaoMicroRNA-145
targets vascular endothelial growth factor and inhibits invasion
and metastasis of osteosarcoma cellsActa Biochim Biophys Sin
(Shanghai)44407414201210.1093/abbs/gms01922472569
|
|
57.
|
D LiX LiuL LinMicroRNA-99a inhibits
hepatocellular carcinoma growth and correlates with prognosis of
patients with hepatocellular carcinomaJ Biol
Chem2863667736685201110.1074/jbc.M111.27056121878637
|
|
58.
|
D SunYS LeeA MalhotramiR-99 family of
MicroRNAs suppresses the expression of prostate-specific antigen
and prostate cancer cell proliferationCancer
Res7113131324201110.1158/0008-5472.CAN-10-103121212412
|
|
59.
|
DM ÖzataS CaramutaD Velázquez-FernándezThe
role of microRNA deregulation in the pathogenesis of adrenocortical
carcinomaEndocr Relat Cancer18643655201121859927
|
|
60.
|
R MahnLC HeukampS RogenhoferA von
RueckerSC MüllerJ EllingerCirculating microRNAs (miRNA) in serum of
patients with prostate
cancerUrology771265.e916201110.1016/j.urology.2011.01.02021539977
|
|
61.
|
WC ChoEpigenetic alteration of microRNAs
in feces of colorectal cancer and its clinical significanceExpert
Rev Mol Diagn11691694201110.1586/erm.11.5721902530
|
|
62.
|
KB JonesZ SalahS Del MaremiRNA signatures
associate with pathogenesis and progression of osteosarcomaCancer
Res7218651877201210.1158/0008-5472.CAN-11-266322350417
|
|
63.
|
NM WhiteTT BaoJ GrigullmiRNA profiling for
clear cell renal cell carcinoma: biomarker discovery and
identification of potential controls and consequences of miRNA
dysregulationJ
Urol18610771083201110.1016/j.juro.2011.04.11021784468
|
|
64.
|
MM VickersJ BarI
Gorn-HondermannStage-dependent differential expression of microRNAs
in colorectal cancer: potential role as markers of metastatic
diseaseClin Exp
Metastasis29123132201210.1007/s10585-011-9435-322120473
|
|
65.
|
M ShuY ZhouW ZhuMicroRNA 335 is required
for differentiation of malignant glioma cells induced by activation
of cAMP/protein kinase A pathwayMol
Pharmacol81292298201210.1124/mol.111.07616622172575
|