Introduction
The use of in vitro cell lines is a widely
established approach to study the growth properties and regulatory
pathways of malignant tumors that are considered important for
tumor expansion and spread. This is extremely important in breast
cancer studies as a number of well-established cell lines have been
in use for several years.
Previously, the application of microarray analyses
has enabled clustering of several tumor types into specific groups.
These studies did not provide a ‘breast cancer specific’ cluster
and the MDA-MB-435 breast cancer cell line was out of range of
other breast cancer cell lines, appearing in the same cluster as
melanoma cell lines due to expression of specific melanocytic genes
(1). Further studies using protein
expression analyses clearly demonstrated that two MDA-MB-435
sublines (termed MDA-MB-435S and -HGF) expressed the melanocytic
genes melan A and tyrosinase (2),
but not typical breast cancer genes, mammaglobin, prolactin and
pS2. Rae et al(3) revealed
that a number of MDA-MB-435 cell lineages used in various
laboratories were genetically identical to the parental cell line
and also exhibited a melanocytic phenotype. Collectively, these
studies reported that the widely used breast cancer model cell line
MDA-MB-435 does not reflect a breast carcinoma cell line and may
originate from a metastasis from an undetected malignant
melanoma.
By contrast, other studies have reported clear
evidence that MDA-MB-435 cells produced milk lipid droplets
following differentiation (4). In
addition, the cell line has been found to express epithelial
markers, including pan-cytokeratin (CK), indicative of an
epithelial origin (5). Finally, an
extensive study by Sellappan et al(6) confirmed the epithelial characteristics
of those cells based on the production of typical milk lipid
droplets together with the expression of pan-cytokeratin,
epithelial membrane-antigen (EMA), β-casein and α-lactalbumin.
However, this cell line also revealed specific expression of the
melanoma markers tyrosinase and melan A. The authors hypothesized
that the MDA-MB-435 cells exhibited lineage instability and
classified them as a breast epithelial cell line that had undergone
lineage infidelity (6).
These observations are consistent with our own
previous study (7) in which 159
breast cancer samples were analyzed for melanocyte and epithelial
markers and CKs. These tumors were identified to contain a small
subpopulation of cells with high expression levels of Melan A.
However, the immunohistochemical identification of focal positivity
for Melan A in otherwise (clinically) clear-cut breast carcinoma
must not be interpreted as melanoma metastases. The association
between Melan A expression and differentiation grading indicates
that lineage infidelity correlates with a reduction in cellular
differentiation and therefore, Melan A may be an important marker
for reduction in tumor cell differentiation (7)
The current study is a comparative study of
MDA-MB-435 (directly obtained from the initial, unmodified cell
line) and additional commonly used breast cancer cell lines (MCF-7
and MDA-MB-231), aiming to extend our previous studies. The
expression of breast cancer and/or melanocytic marker profiles in
various growth stages of breast cancer cells was determined by
quantification of gene and protein expression techniques. Results
demonstrate that lineage infidelity of MDA-MB-435 cells is
associated with tumor cell density, indicating that the growth
properties of these cells have an effect on the gain of melanocytic
properties. The observation that gene and protein expression levels
change with cell density has been previously reported by this
research group. In these studies, breast cancer cells growing in
cultures with low density were demonstrated to exhibit a more
aggressive phenotype with elevated proteolytic activity and
invasiveness (8–10).
In the present study, MDA-MB-435 cells growing in
dense cultures were found to exhibit lower expression of the breast
cancer marker, mammaglobin and higher expression of the melanocyte
markers, Melan A and S100.
The results are consistent with the hypothesis that
MDA-MB-435 cells were initially of epithelial origin and therefore
suitable for the study of breast cancer. Furthermore, for the first
time, cell density was demonstrated to correlate with
differentiation state in breast cancer and additional evidence that
Melan A is a suitable marker for differentiation in breast cancer
is provided.
Materials and methods
Cell culture conditions
The breast cancer cell lines MCF-7, MDA-MB-231 (both
American Type Culture Collection, Manassas, VA, USA) and MDA-MB-435
(kindly donated by Dr Janet E. Price, Anderson Cancer Center,
Houston, TX, USA) used in this study reflect a stepwise increase in
malignant biological behavior on the basis of methodical
retransplantation studies, MDA-MB-435>MDA-MB-231>MCF-7
(11,12). These cell lines are commonly used
for breast cancer studies and therefore are well-defined in their
growth, invasive and metastatic characteristics.
Cells were cultured at 37°C in a humidified
atmosphere of 5% CO2 and 95% air. For comparative
analysis of melan A mRNA expression, the established melanoma cell
line A2058 was also included (American Type Culture Collection).
Cells were grown in MEM (Eagle’s) with Earle’s salts supplemented
with 20% heat inactivated fetal calf serum, 1% L-glutamine solution
(200 mM), 1% sodium pyruvate solution (100 mM), 1% non-essential
amino acids for MEM (100X solution) and 2% vitamins for MEM (100X
solution). The medium was changed every three days. For
subcultures, cells were harvested following brief treatment with
0.1% trypsin/EDTA solution and seeded at a dilution of 1:5. Cells
of 10 subsequent passages were used for studies.
Cells were grown to subconfluency (<50% of
confluence) or ∼90% confluence in 75-cm2 plastic culture
flasks containing 15 ml medium.
Immunocytochemistry
Immunocytochemistry was used to determine the
cellular pattern of pan-CK (clone Lu-5) and various CK isoforms
(KL-1 and CK-10, -14, -18, -19 and 20), the breast cancer marker,
mammaglobin and the melanocytic markers, S100 and melan-A.
Antibodies against the CKs were all purchased from Dako (Hamburg,
Germany). Monoclonal antibodies against melan A, mammaglobin and
S100 were obtained from Zytomed (Berlin, Germany).
Cells were grown to subconfluency or confluency on
silanized sterile glass slides (SuperFrost plus; Menzel,
Braunschweig, Germany). The medium was discarded, the cells rinsed
with Tris-buffer, fixed in methanol/acetone (2:1 v/v) for 2 min and
rinsed again. Following incubation with the specific polyclonal
primary antibodies for 30 min (37°C), rinsing with Tris-buffer and
application of the secondary antibody system
(Streptavidin-Biotin-Complex method; Dako), the resulting antibody
complexes were visualized with diaminobenzidine (Sigma-Aldrich,
Deisenhofen, Germany).
Quantitative RT-PCR
RNA was extracted from cells using the RNeasy
Protect Mini kit (Qiagen, Hilden, Germany) according to the
manufacturer’s instructions and reverse transcribed using oligo dT
primers in 20 μl final volume. All primers for the genes
tested were designed using Primer3 software (13) with a Tm optimum of ∼60°C
and a product length of 100–150 nt (Table I). Real time PCR was performed on an
I-Cycler (Bio-Rad, Hercules, CA, USA) using iQ Supermix (Bio-Rad)
supplemented with 10 nM fluorescein (Bio-Rad), 0.1X Sybr-Green I
(Sigma-Aldrich), 2.5 μl cDNA (5X diluted) and 3 pmol sense
and antisense primers in a final reaction volume of 25 μl.
Following an initial denaturation step of 3 min during which the
well factor was measured, 50 cycles of 15 sec at 95°C followed by
30 sec at 60°C were performed. Fluorescence was measured during the
annealing step in each cycle. Following amplification, melting
curves with 80 steps of 15 sec at 0.5°C increments were performed
to monitor amplicon identity. Amplification efficiency was assessed
for all primer sets utilized in separate reactions and primers with
efficiencies >94% were used. Expression data were normalized
against GAPDH and on RNA polymerase II (RPII) gene expression data
obtained in parallel. Results are expressed with standard errors
and statistical comparisons (unpaired two-tailed t-test) were
obtained using Qgene software (14). Expression changes were calculated
using the mean value of normalizations obtained using GAPDH and
RPII genes as references.
 | Table IPrimer sequences for mammaglobin and
melan A. |
Table I
Primer sequences for mammaglobin and
melan A.
| Gene | Sense | Antisense |
|---|
| Mammaglobin |
5′-TCCAAGACAATCAATCCACAAG-3′ |
5′-CAGTTCTGTGAGCCAAAGGTC-3′ |
| Melan A |
5′-ATCGGGACAGCAAAGTGTCTC-3′ |
5′-GAGTTTCTCATAAGCAGGTGGAG-3′ |
Statistical analysis
Statistical significance was assessed by comparing
mean ± SD values, which were normalized to the control group with
student’s t-test for independent groups. One-way analysis of
variance was used to test for statistical significance and when
significance was determined, Bonferroni’s multiple comparison test
was performed post hoc. P<0.05 was considered to indicate a
statistically significant difference. Statistical analysis was
performed using the Prism software (GraphPad, San Diego, CA,
USA).
Results
Expression pattern of CKs in breast
cancer cell lines of various levels of tumorigenicity and growth
density
Expression of CKs were analyzed to verify the
epithelial origin of the three human breast cancer cell lines,
MCF-7, MDA-MB-231 and MDA-MB-435, which constitute our in
vitro cell model. All cell lines have been previously reported
to originate from pleural effusions (12,15).
MCF-7 cells have been found to exhibit low invasive abilities and
are essentially non-metastatic. By contrast, injection of
MDA-MB-231 and -435 cells into the mammary fat pad of nude mice has
been demonstrated to result in tumor formation and distant
metastases in lungs and lymph nodes of specific mice (12), however, the extent of these events
varied between cell lines and growth velocity differed.
All three human breast cancer cell lines exhibited
the epithelial marker, pan-CK (clone Lu-5) and the low-molecular
weight CK, KL-1 (Fig. 1). However,
there were line-specific differences in the tested CK isoforms.
Similarly, MCF-7 revealed specific positive staining for CK-18, -19
and -20 and the MDA-MB-231 cells for CK-18 and -19. The MDA-MB-435
cells, however, did not react for CK18, -19 or -20. All three cell
lines were negative for CK-10 and -14 (squamous cell
differentiation markers; Table II).
These patterns were identified in the subconfluent and confluent
growth conditions investigated in the three cell lines.
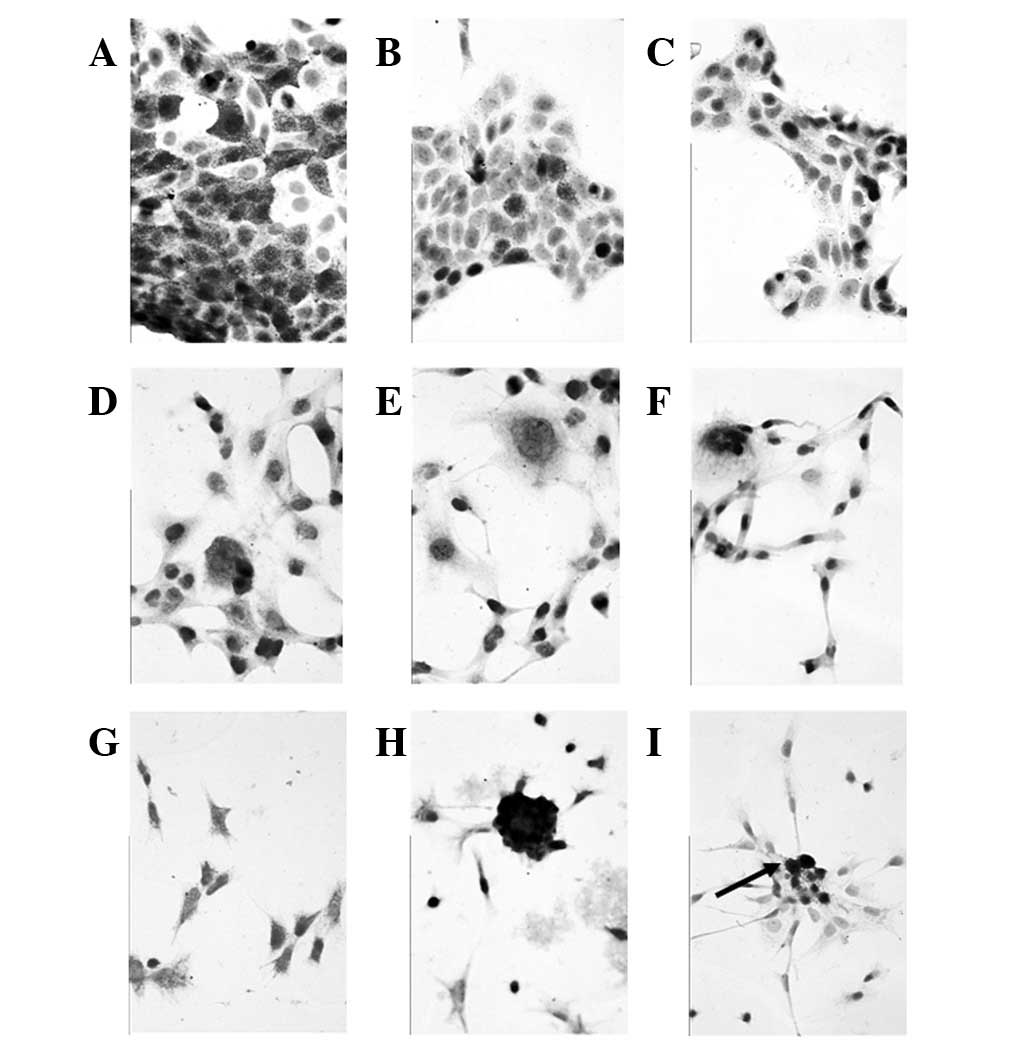 | Figure 1Immunocytochemical staining pattern
for cell-associated epithelial and melanocytic markers in (A–C)
MCF-7, (D–F) MDA-MB-231 and (G–I) MDA-MB-435 cells. Cytoplasmic
staining against (A,D,G) pan-CK, (B,E,H) mammaglobin and (C,F,I)
melan A. MCF-7 and MDA-MB-231 cells were negative for melan 1,
while MDA-MB-435 cells reveal a focal typical positive reaction for
this melanocytic marker, particularly in regions with accumulated,
‘denser’ tumor cells (magnification, ×400). CK, cytokeratin. |
 | Table IIImmunocytochemical staining for CKs
and mammaglobin in breast cancer cell lines. |
Table II
Immunocytochemical staining for CKs
and mammaglobin in breast cancer cell lines.
| Breast cancer cell
lines
|
|---|
| Target | MCF-7 | MDA-MB-231 | MDA-MB-435 |
|---|
| LMW-CK (KL-1) | + | + | + |
| CK-10 | 0 | 0 | 0 |
| CK-14 | 0 | 0 | 0 |
| CK-18 | + | + | 0 |
| CK-19 | + | + | 0 |
| CK-20 | + | 0 | 0 |
| Mammaglobin | + | + | + |
Expression pattern of mammaglobin in the
various cell lines
Immunohistochemistry was used to analyze mammaglobin
protein expression in the cell lines (Fig. 1). As identified for pan-CK, all cell
lines were positively stained (Table
II). No differences in expression were found between
subconfluent and confluent cells.
Expression of melanocytic expression
markers in the various cell lines
While CKs and mammaglobin protein were found in all
three cell lines, the melanocytic markers melan A and S100-protein
were negative in MCF-7 and MDA-MB-231 cells of subconfluent and
confluent growth status. However, MDA-MB-435 cells were negative
for these markers under subconfluent conditions only, while in
confluent cell cultures a focal specific cytoplasmic staining for
melan A and S100 was detected. Staining was observed to be higher
in areas where densely grown cells formed cluster-like aggregates
(Table III and Fig. 1).
 | Table IIIImmunocytochemical staining for
melanocytic markers in MDA-MB-435 cells. |
Table III
Immunocytochemical staining for
melanocytic markers in MDA-MB-435 cells.
| Cell confluency
|
|---|
| Markers | Subconfluent | Densely packed
(confluent) |
|---|
| Melan A | 0 | + |
| S100-protein | 0 | + |
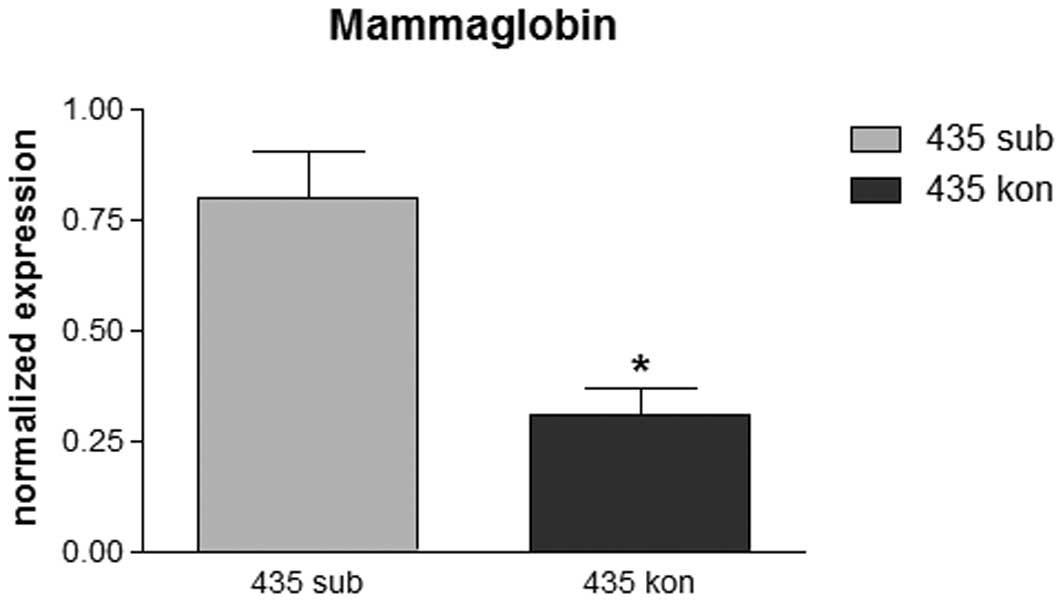
Quantitative mRNA expression of
mammaglobin in the cell lines
To verify the immunohistochemical protein pattern,
quantitative RT-PCR was performed to determine mRNA expression of
mammaglobin in various settings. While MCF-7 and MDA-MB-231 cells
revealed moderate expression of mammaglobin at various levels of
confluence without significant differences (data not shown),
MDA-MB-435 cells revealed statistically significant differences
between cell densities. The mRNA level for mammaglobin was ∼3-fold
higher in subconfluent than in confluent cells (Fig. 2).
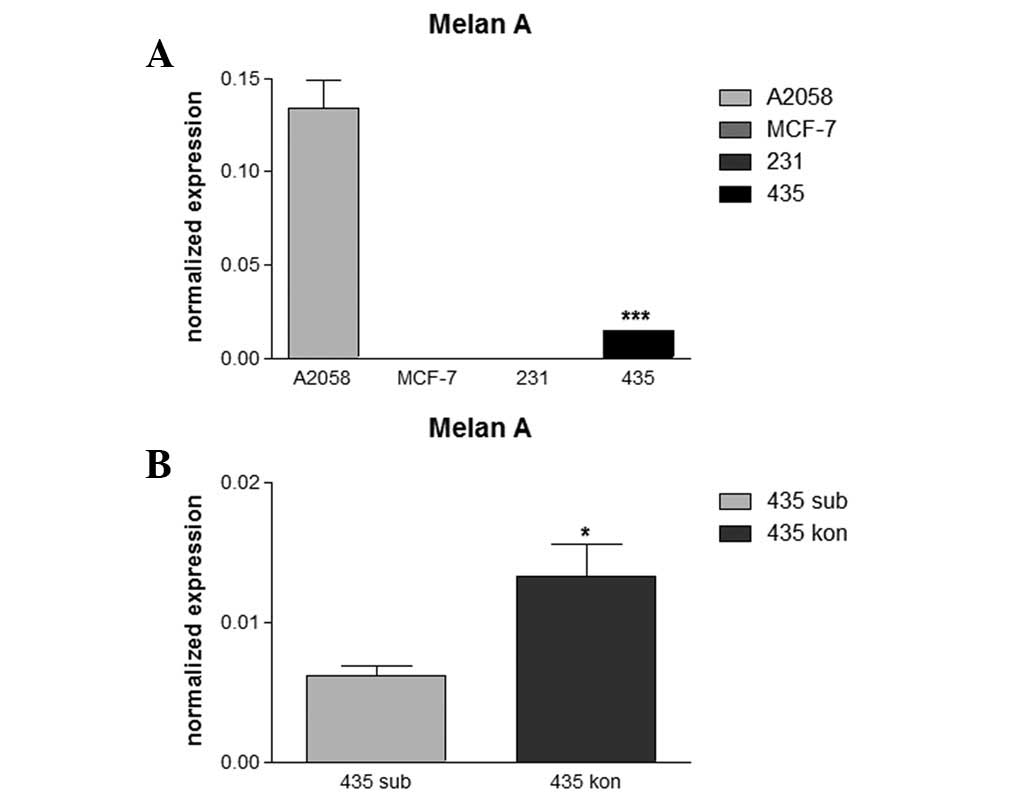
Quantitative mRNA expression of melan A
in various breast cancer cell lines
Analysis of melan A mRNA expression levels revealed
an absence of this melanocytic marker in MCF-7 and MDA-MB-231 cells
(Fig. 3A), consistent with
immunohistochemistry results. In MDA-MB-435 cells, a low level of
melan A expression was observed in subconfluent cell cultures which
was found to be significantly increased ∼2-fold in the dense
cultures (Fig. 3B). The melanocytic
cell line A2058 was used as control and revealed high expression
levels of melan A which exceeded levels in confluent MDA-MB-435
cells by >10-fold (Fig. 3A).
Expression of melan A mRNA was not detected in tthe MCF-7 and
MDA-MB-231 cell lines.
Discussion
In vitro cell lines as model systems are an
extremely broad application for cancer research. However, the
results obtained from using cell lines are markedly dependent on
prevention of contamination, correct attribution of a cell line to
the initial primary tumor and stability. The correct identification
of the breast cancer cell line, MDA MB-435, a widely used cell line
in breast cancer research remains an area of fierce debate. Similar
to several other breast cancer cell lines, MDA MB-435 cells
originate from the pleural effusion of a patient who succumbed to
mammary carcinoma (11,16).Establishing a permanent cell line
from a primary tumor is extremely difficult, while metastatic tumor
cells, particularly those that have detached from the surrounding
stroma and are freely floating in effusion fluids, are
significantly easier to handle. However, in rare instances, the
metastases may not occur from the predicted primary tumor and may
originate from an unknown synchronous malignant tumor that
systemically spreads more readily.
The development of molecular array techniques has
enabled high throughput analysis of thousands of genes and a number
of tumors and tumor cell lines have been molecularly screened to
analyze gene expression pattern. Ross et al(1) performed an extensive study on the
expression levels of various tumor cell lines. By this approach,
MDA-MB-435 cells were identified to express typical markers of
melanocytic origin. As a consequence, those authors hypothesized
that the MDA-MB-435 cells may have originated from an unknown
metastasizing malignant melanoma which occurred in parallel to a
(non-metastazing) breast cancer. In addition, two subsequent
studies on in vitro cells and tumor tissue obtained
following tumor cell transplantation into nude mice (2,3),
exclusively detected melanocytic markers in MDA-MB-435 cells and
transplanted tumor nodules.
However, other studies performed morphology analysis
and immunocytochemistry and demonstrated that MDA-MB-435 cells
expressed epithelial cell markers, in particular milk lipid
droplets (4) and CKs (5). In addition, a previous study by
Sellappan et al(6)
identified epithelial cell markers (CKs) and breast
epithelia-specific genes, including β-casein and α-lactalbumin, in
MDA MB-435 cells. In this study, authors observed expression of
melanocytic markers, tyrosinase and melan A, in those cells,
hypothesizing that lineage infidelity had occured during tumor
progression.
The results of the present study are consistent with
observations of Sellappan et al, demonstrating that
MDA-MB-435 cells coexpress breast epithelia-specific (CKs and
mammaglobin) and melanocytic (melan A and S100) markers. In
addition, the locally invasive, but non-metastasizing, estrogen
receptor-positive MCF-7 cell line and the invasive and moderately
metastasizing, estrogen receptor-negative MDA-MB-231 cell line were
screened for lineage instability. Neither of the cell lines
revealed a melanocytic phenotype.
To characterize the cell lines with respect to their
epithelial origin, the expression of various CKs was analyzed to
determine the type and degree of cellular differentiation and
maturation (17). By this approach,
the non-invasive MCF-7 cells were found to be ‘more differentiated’
than the invasive MDA-MB-231 cells, since the MCF-7 cells
synthesized various glandular CKs, including CK-20, which was not
identified in the MDA-MB-231 cells. The highly aggressive
MDA-MB-435 cell line was not found to express glandular or squamous
CKs and assumed to exhibit a significantly lower degree of
differentiation.
In addition, the expression patterns for epithelial
and melanocytic markers were compared in all three breast cancer
cell lines with respect to cell density and thereby the growth
status of the cells. MCF-7 and MDA-MB-231 cells revealed a stable
epithelial phenotype, however, MDA-MB-435 cells reacted differently
when subconfluent and confluent cells were compared. Subconfluent
cells were observed to have undergone epithelial differentiation,
however, in dense cultures, foci which had undergone melanocytic
differentiation were identified. This observation is consistent
with the tumor transplantation experiments by Sellapan et
al(6) in which small foci of
the tumor transplant were detected to express the melanocytic
marker HMB-45. In the present study, mammaglobin mRNA expression
was found to decrease, concomitant with a gradual increase of melan
A mRNA in fully confluent cells. These observations are consistent
with coexpression of the epithelial marker, EMA and the melanocytic
marker, HMB-45, reported in a previous study in tumor transplants
(6). These observations markedly
indicate a dual differentiation status of densely packed cells.
In summary, the present study clearly demonstrates
that MDA-MB-435 cells undergo a high level of lineage instability
leading to the gain of melanocytic lineage characteristics. This
instability is markedly dependent on the growth stage of cells and
occurs only in foci of densely growing tumor cells. There is clear
evidence that the MDA-MB-435 cells are of epithelial origin and
therefore no metastasis of an unknown malignant melanoma has been
hypothesized to be included in the initial pleural effusion where
the cells were obtained. In addition, lineage instability was not
found in the other breast cancer cell lines, MCF-7 and MDA-MB-231,
at subconfluent or confluent growth stages. Cell density was
observed to correlate with differentiation status in breast cancer
and is consistent with the hypothesis that Melan A is a suitable
marker for differentiation in breast cancer.
Finally, our observations provide a significant
evidence for the further use of MDA-MB-435 cells as breast cancer
cells in various model systems, in particular in those that
correlate molecular observations with tumor cell behaviour
(9). However, additional studies
must be performed to identify the molecular mechanism of this
lineage instability, including the microdis-section of melanocytic
versus epithelial MDA-MB-435 cells to understand the molecular
and/or genetic background of this unusual observation.
References
|
1
|
Ross DT, Scherf U, Eisen MB, et al:
Systematic variation in gene expression patterns in human cancer
cell lines. Nat Genet. 24:227–235. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ellison G, Klinowska T, Westwood RF,
Docter E, French T and Fox JC: Further evidence to support the
melanocytic origin of MDA-MB-435. Mol Pathol. 55:294–299. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rae JM, Ramus SJ, Waltham M, et al: Common
origins of MDA-MB-435 cells from various sources with those shown
to have melanoma properties. Clin Exp Metastasis. 21:543–552.
2004.PubMed/NCBI
|
|
4
|
You H, Yu W, Sanders BG and Kline K:
RRR-alpha-tocopheryl succinate induces MDA-MB-435 and MCF-7 human
breast cancer cells to undergo differentiation. Cell Growth Differ.
12:471–480. 2001.PubMed/NCBI
|
|
5
|
Bachmeier BE, Nerlich AG, Lichtinghagen R
and Sommerhoff CP: Matrix metalloproteinases (MMPs) in breast
cancer cell lines of different tumorigenicity. Anticancer Res.
21:3821–3828. 2001.PubMed/NCBI
|
|
6
|
Sellappan S, Grijalva R, Zhou X, et al:
Lineage infidelity of MDA-MB-435 cells: expression of melanocyte
proteins in a breast cancer cell line. Cancer Res. 64:3479–3485.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bachmeier BE, Nerlich AG, Mirisola V,
Jochum M and Pfeffer U: Lineage infidelity and expression of
melanocytic markers in human breast cancer. Int J Oncol.
33:1011–1015. 2008.PubMed/NCBI
|
|
8
|
Bachmeier BE, Albini A, Vene R, et al:
Cell density-dependent regulation of matrix metalloproteinase and
TIMP expression in differently tumorigenic breast cancer cell
lines. Exp Cell Res. 305:83–98. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bachmeier BE, Rohrbach H, De Waal J,
Jochum M and Nerlich AG: Enhanced expression and activation of
major matrix metalloproteinases in distinct topographic areas of
invasive breast carcinomas. Int J Oncol. 26:1203–1207.
2005.PubMed/NCBI
|
|
10
|
Bachmeier BE, Vene R, Iancu CM, et al:
Transcriptional control of cell density dependent regulation of
matrix metalloproteinase and TIMP expression in breast cancer cell
lines. Thromb Haemost. 93:761–769. 2005.PubMed/NCBI
|
|
11
|
Price JE, Polyzos A, Zhang RD and Daniels
LM: Tumorigenicity and metastasis of human breast carcinoma cell
lines in nude mice. Cancer Res. 50:717–721. 1990.PubMed/NCBI
|
|
12
|
Zhang RD, Fidler IJ and Price JE: Relative
malignant potential of human breast carcinoma cell lines
established from pleural effusions and a brain metastasis. Invasion
Metastasis. 11:204–215. 1991.
|
|
13
|
Rozen S and Skaletsky H: Primer3 on the
WWW for general users and for biologist programmers. Methods Mol
Biol. 132:365–386. 2000.PubMed/NCBI
|
|
14
|
Muller PY, Janovjak H, Miserez AR and
Dobbie Z: Processing of gene expression data generated by
quantitative real-time RT-PCR. Biotechniques. 32:1372–1379.
2002.PubMed/NCBI
|
|
15
|
Soule HD, Vazguez J, Long A, Albert S and
Brennan M: A human cell line from a pleural effusion derived from a
breast carcinoma. J Natl Cancer Inst. 51:1409–1416. 1973.PubMed/NCBI
|
|
16
|
Cailleau R, Olive M and Cruciger QV:
Long-term human breast carcinoma cell lines of metastatic origin:
preliminary characterization. In Vitro. 14:911–915. 1978.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Moll R: Molecular diversity of
cytokeratins: significance for cell and tumor differentiation. Acta
Histochem Suppl. 41:117–127. 1991.PubMed/NCBI
|