Introduction
Heterogeneous ribonuclear protein C (hnRNPC) is an
RNA-binding protein located in the nuclei of normal cells; however,
it is also distributed in the cytoplasm of tumor cells (1). It is thought to be a prognostic marker
in tumors (2,3). hnRNPC has two isoforms, C2 and C1,
coded by a single gene and generated by alternative splicing of the
same transcript. The difference between the two isoforms is that C2
has an additional 13 amino acid insert after
Ser107(4). hnRNPC plays
multiple roles in post-transcriptional regulation, including
alternative splicing (5), nuclear
retention and export (6), stability
(7,8) and translation (3,9,10).
Several studies have shown that hnRNPC is overexpressed in tumors,
including hepatocellular carcinoma and breast cancer (2,11).
When its expression is repressed, tumor growth is suppressed and
occasionally inhibited (12,13).
Another important characteristic of tumors is
pleomorphism, including multinucleation, particularly in high grade
tumors (14,15). In humans, the vast majority of
normal cells are mononuclear except a few specific types of cells,
including hepatocytes (16).
Although multinucleation is a normal phenomenon in adult liver with
age, pathogens, including virus infection and carcinogens, are
indispensible elements to accelerate this process (17–19).
Multinucleation is the result of a change or disorder in gene
regulation whether for normal cell development progression or for
disease (16,20,21).
Among these genes, Aurora B is essential to chromosome segregation
and cytokinesis. It is an important component of the chromosomal
passenger complex and plays multiple roles in cell division such as
mitotic spindle assembly, kinetochore assembly, regulation of
mitotic checkpoints, chromosome compaction in anaphase and
regulation of cleavage furrow ingression (20–22).
During these processes, Aurora B is located at the midbody in late
anaphase and cytokinesis to recruit substrates that are necessary
for cytokinesis and exerts enzymatic activity to complete
cytokinesis (23–26). Upregulation of Aurora B and its
repression lead to cytokinesis failure and induced multinucleation
(27–29).
In this study, we found that hnRNPC2 is correlated
with multinucleation in hepatocellular carcinoma SMMC-7721 cells.
Further investigation revealed that hnRNPC2 induced multinucleation
by repressing the expression of Aurora B.
Materials and methods
Materials
The eukaryotic translational initiating factor 4E
(eIF4E) antibody and protein A/G-agarose were purchased from
Bioworld (Uitgeest, The Netherlands). The Aurora B antibody and
hnRNPC2 antibody were purchased from Epitomics (Burlingame, CA,
USA). TRIzol, Lipofectamine 2000 and RPMI-1640 were purchased from
Invitrogen Life Technologies (Carlsbad, CA, USA). The PrimeScript™
reverse transcription-polymerase chain reaction (RT-PCR) kit was
purchased from Takara Bio, Inc. (Shiga, Japan). Taq Platinum DNA
polymerase was purchased from Tiangen (Beijing, China). pEGFP-C1
was purchased from Clontech Laboratories (Mountain View, CA, USA).
Primer synthesis and DNA sequencing were performed by SunnyBio.
(Shanghai, China). siRNA was supplied by Genepharma (Shanghai,
China). Propidium iodide (PI) was purchased from Beyotime
(Jiangsu,China). 4,6-diamino-2-phenyl indole (DAPI) was purchased
from Sigma (St. Louis, MO, USA). The cell counting kit (CCK)-8 was
purchased from Dojindo (Kumamoto, Japan). iQ™ SYBR®-Green supermix
was purchased from Bio-Rad (Hercules, CA, USA). SMMC-7721 cells,
HL-7702 cells, A549 cells and BT549 cells were from the cell bank
of the Chinese Academy of Sciences.
The study was approved by the Ethics Committee of
the Institute of Biochemistry and Cell Biology, Shanghai Institutes
for Biological Sciences, Chinese Academy of Sciences, Shanghai,
China.
RNA extraction, cDNA synthesis and
expressional vector construction
SMMC-7721 cells (60 mm dish) were lysed by 1 ml
TRIzol following 3 washes with phosphate-buffered saline (PBS) to
extract the total RNA, following the manufacturer’s instructions.
cDNA synthesis was performed using the PrimeScript RT-PCR kit,
according to the manufacturer’s instructions and DNA amplification
was performed by Taq Platinum DNA polymerase with primers as
followed: hnRNPC (NM_001077442),
5′-ACCTCGAGACACGATGGCCAGCAACGTT-3′, 5′-CAG
AATTCGCTTAAGAGTCATCCTCGCC-3′. The amplified hnRNPC cDNA fragment
was T-A cloned into a pMD18-T vector. DNA sequencing was used to
obtain the hnRNPC2 gene, which was inserted into the pEGFP-C1
vector between the restriction sites XhoI and
EcoRI.
Cell culture, DNA transfection and cell
screening
Cells were cultured in RPMI-1640 medium plus 10%
newborn bovine serum (full medium). Cells were seeded in a 24-well
plate (1.5×105 cells per well) 24 h before transfection.
For transfection, 1.0 μg plasmid was used per well.
Transfection was performed according to the manufacturer’s
instructions. The transfectants were screened by full medium plus
800 μg/ml geneticin for 7 days and cultured in full medium
plus 400 μg/ml geneticin for a further 14 days. All cell
colonies displaying green fluorescence were obtained under a
fluorescent microscope and cultured together for proliferation.
Cell counting
Cell counting was performed according to the
instructions of the CCK-8 kit in a 96-well plate.
RNA interference
RNA interference was performed according the
instructions of Lipofectamine 2000. For each 35 mm dish 600 pmol
siRNA was used. The siRNA-Aurora B sequence was according to the
literature (30). At 72 h
post-transfection, cells were detected by flow cytometry and
western blotting.
Western blotting
Western blotting was performed according to the
literature (31).
PI staining and flow cytometry
Following 3 washes with PBS (pH 7.2), cells cultured
on cover slips or digested by 0.25% trypsin were fixed with
ice-cold 1.25% paraformaldehyde for 30 min [this step is only for
green fluorescent protein (GFP) or GFP fusion protein expressed
cells and their control cells]. Then, the fixed cells were washed
twice with PBS and fixed with ice-cold 75% ethanol for 2 h on ice.
Prior to staining with 5 μg/ml PI, cells were digested by 30
μg/ml RNase A at 37°C for 30 min. Finally, cells were
observed under a fluorescence microscope or detected by flow
cytometry.
Immunofluorescence staining and laser
scanning confocal microscopy
Cells were seeded on cover slips in a 24-well plate
24 h before transfection. At 72 h post-transfection, cover slips
with cells adhered to the surface were washed with PBS and fixed
with 4% paraformaldehyde for 40 min at room temperature. Then,
cells were permeabilized with 1% Triton X-100 for 5–10 min at room
temperature and blocked by 5% skimmed milk for 1 h at 37°C. Next,
cover slips were incubated with the primary antibody for 12 h then
the secondary antibody labeled with rhodamine for 8 h at 4°C. After
staining with 1 μg/ml DAPI in methanol for 4 min at room
temperature, cover slips were sealed with antifade mounting medium.
These stained cells were observed under a Leica TCS-SP laser
scanning confocal microscope.
mRNA-protein co-immunoprecipitation and
protein-protein co-immunoprecipitation
mRNA-protein co-immunoprecipitation (co-IP) was
performed according to the protocol (32). Protein-protein co-IP was performed
according to the above protocol with certain modifications: 6
μg/ml RNases and 4 U/ml DNase were substituted for the RNase
inhibitor and the extract was incubated at 37°C for 30 min to
digest DNA and RNA. Following co-IP, the harvested protein A/G
agarose was mixed with sodium dodecyl sulphate (SDS) loading buffer
and incubated at 50°C for 30 min. It was then centrifuged at 4000
rpm for 5 min and the supernate was used for immunoblot
analysis.
Real-time quantitative PCR
cDNAs were synthesized as mentioned above. The
real-time PCR reaction procedure was performed as follows: 95°C for
2 min; cycle: 95°C for 20 sec, 55°C for 30 sec and 72°C for 30 sec;
annealing from 65°C to 95°C with 0.5°C progressive increases. The
primers used in this study were: Aurora B (NM_004217):
5′-ATAGCAGTGGGACACCCGACAT-3′ and 5′-GGGACTTGAAGAGGACCTTGAGC-3′;
p190-B (NM_001030055): 5′-ATTTGACCTCCTGAGCACTT-3′ and
5′-TGTAGGCTTCATCCTCCATA-3′; glyceraldehyde 3-phosphate
dehydrogenase (GAPDH, NM_002046): 5′-CCTGTTCGACAGTCAGCCGCATC-3′ and
5′-CGACCA AATCCGTTGACTCCGACC-3′.
Statistical analysis
Each experiment was repeated at least three times
and data were analyzed by analysis of variance test. P<0.01 was
considered to indicate a statistically significant difference.
Results
Overexpression of hnRNPC2 induced
multinucleation in human hepatocellular carcinoma cells
To reveal whether there was a positive correlation
between hnRNPC2 and multinucleation, we constructed a eukaryotic
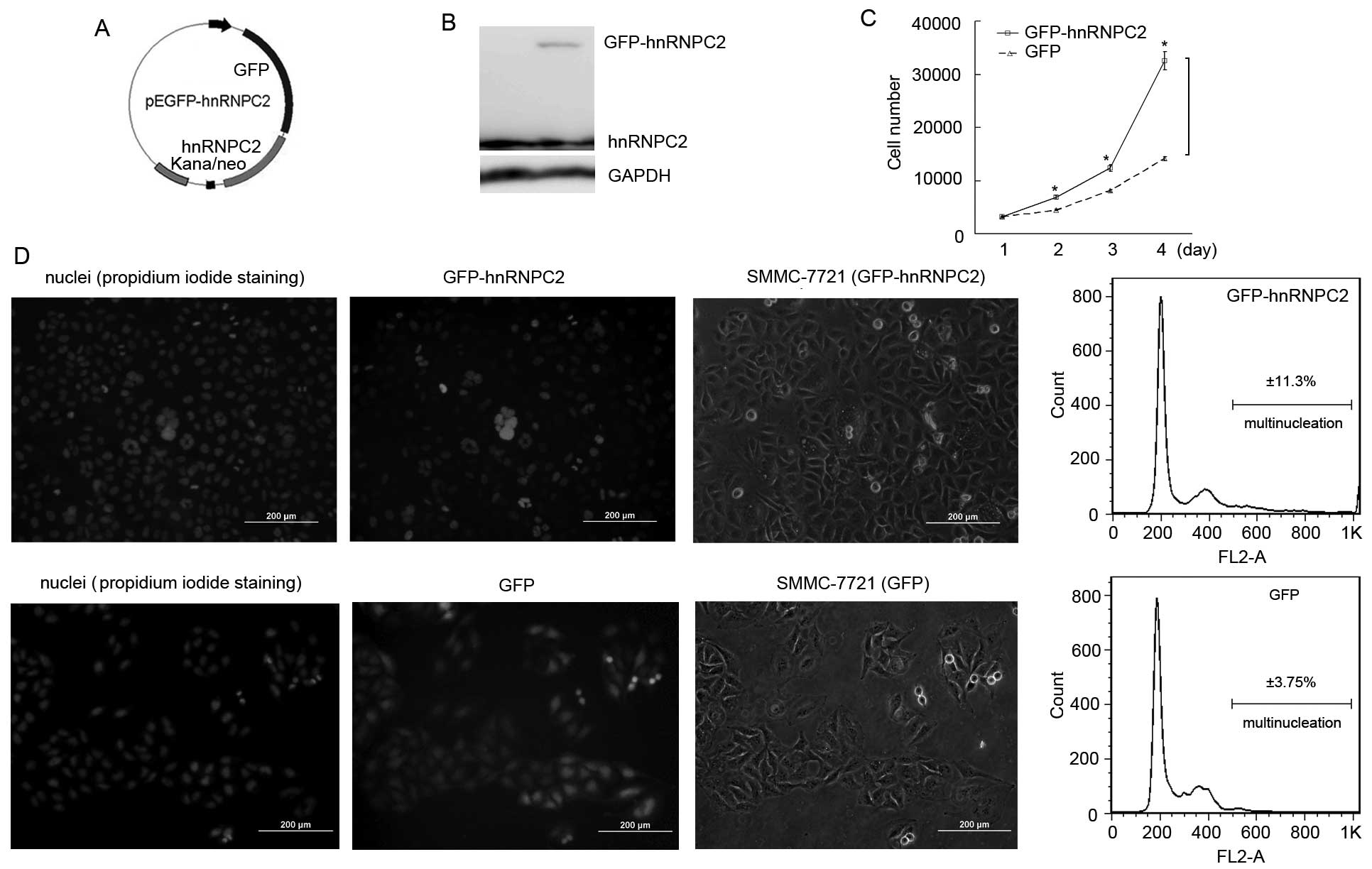
expressional vector pEGFP-hnRNPC2 (Fig.
1A) and transfected it into hepatocellular carcinoma SMMC-7721
cells. Western blot results revealed that GFP-hnRNPC2 is expressed
at 48 h post-transfection (Fig.
1B). Following screening by geneticin, GFP-hnRNPC2-expressed
cell colonies were all obtained under fluorescent microscope and
mixed into one culture. Then, cell proliferative rate tests were
carried out and the results revealed that the exogenous
hnRNPC2-expressed cells (hnRNPC2 overexpression) accelerated their
proliferation (Fig. 1C). Notably,
under the fluorescent microscope, we found that a number of those
cells showed more than two nuclei, glowing green fluorescence, in a
cell with an expanded cytoplasm (Fig.
1D). To evaluate the number of cells with multinucleation as a
result of overexpressed hnRNPC2, fluorescent microscopy and flow
cytometry were used. As Fig. 1D
shows, hnRNPC2-overexpressing SMMC-7721 cells possessed more
multinucleated cells; nearly 11.3% cells were induced to
multinucleation, while the control cells showed 3.8% multinucleated
cells. Furthermore, when pEGFP-hnRNPC2 was transfected into breast
cancer BT549 cells and noncancerous hepatocellular HL-7702 cells,
they showed similar results (data not shown). Collectively, these
results indicate that overexpression of hnRNPC2 is capable of
inducing multinucleation in hepatocellular carcinoma cells and this
effect may be universal.
Destiny of multinucleated cells induced
by overexpressed hnRNPC2
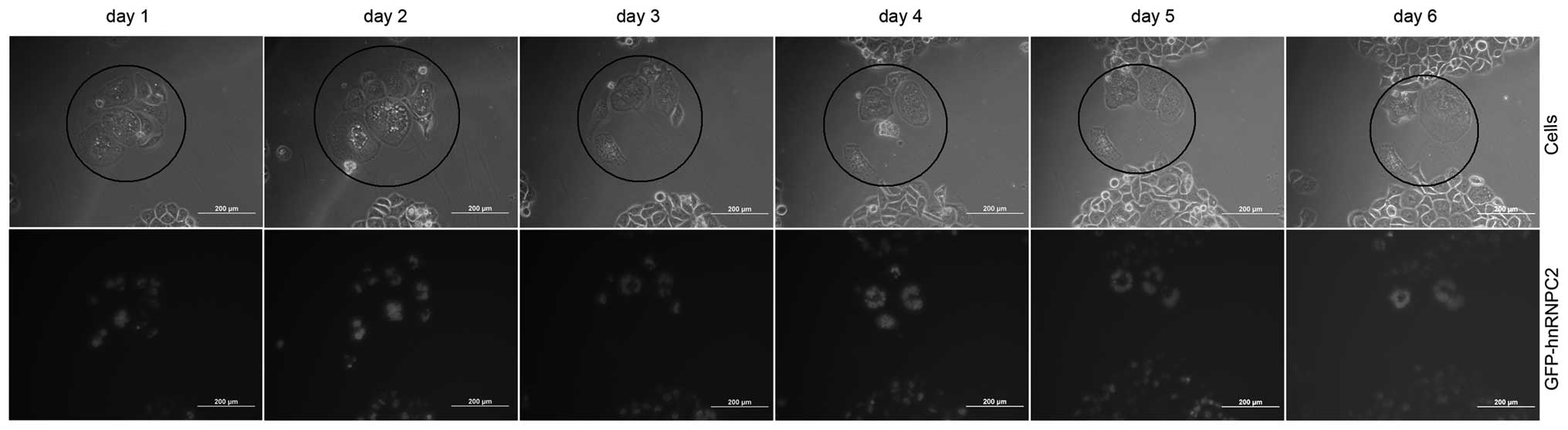
We demonstrated that multinucleated cells were
induced by the overexpression of hnRNPC2. To elucidate the destiny
of the induced multinucleated cells, we tracked the process of the
induced multinucleated cells’ progression every 24 h using a
fluorescent microscope. As Fig. 2
shows, the induced multinucleated cells lose the ability to divide
and they do not recover back to mononuclear cells. Instead, they
increase in nuclear number and undergo maximal expansion of their
cytoplasm. As time lapsed, they became giant multi-nucleated cells
and finally, due to an inability to divide, they died. In 8 groups
of tracking tests, all multinucleated cells died. We conclude that
the induced multinucleated cells lose the ability to divide and
therefore die.
Aurora B was repressed in
hnRNPC2-overexpressing cells
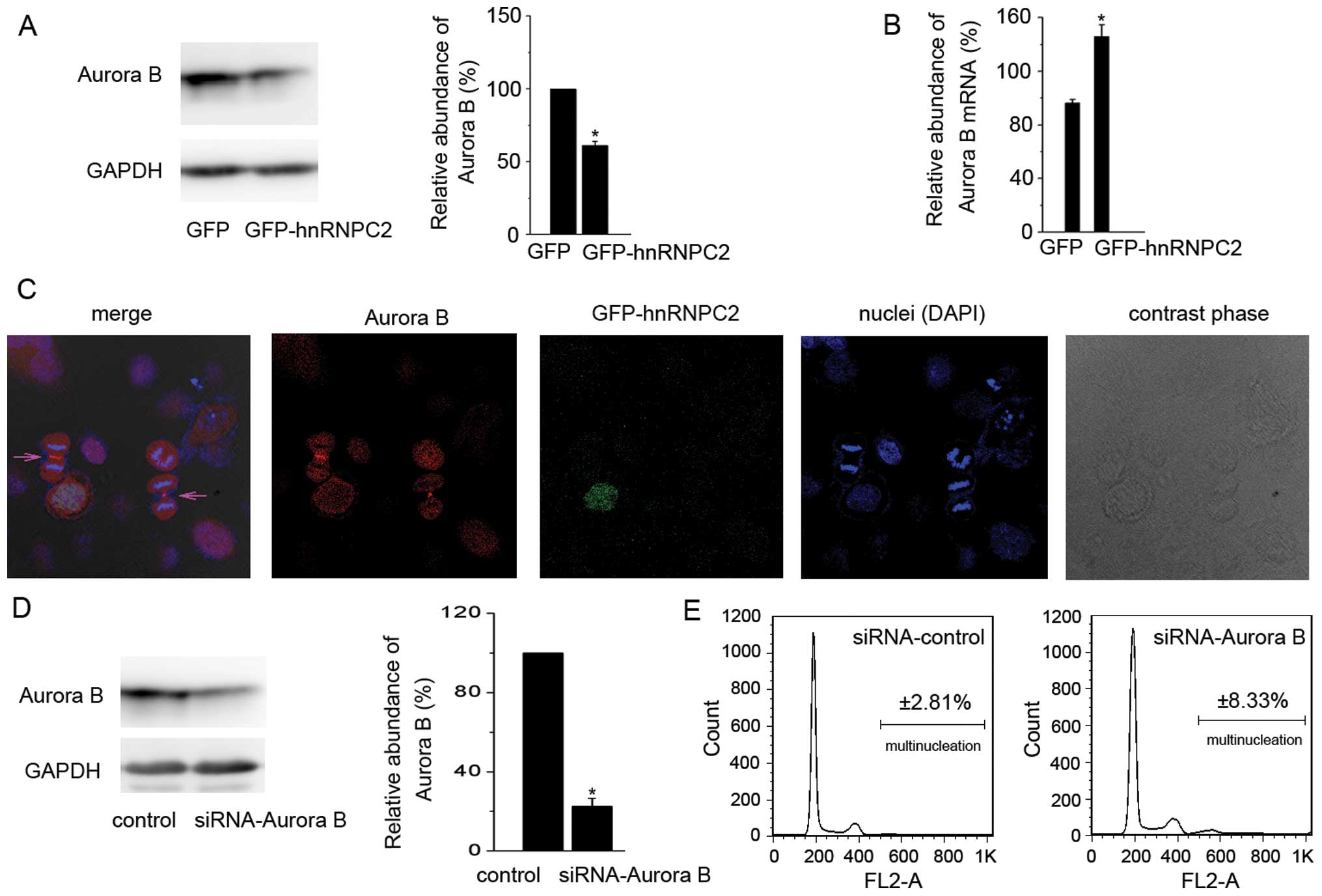
During this study, we found that the expression of
the Aurora B protein was repressed in hnRNPC2-overexpressing cells,
when compared to the control cells (Fig. 3A). Aurora B localizes at the midzone
in late anaphase and recruits and phosphorylates substrates that
are essential to complete cytokinesis (23–26).
To uncover the importance of Aurora B in cytokinesis of SMMC-7721
cells, immunofluorescent staining and confocal microscopy were
carried out together. As Fig. 3C
shows, Aurora B is located at the midbody during cytokinesis in
SMMC-7721 cells, while GFP-hnRNPC2-expressed multi-nucleated cells
lost the ability of cell division. In fact, we did not find an
induced multinucleated cell that was in cytokinesis in a series of
repeated experiments. Furthermore, we abolished the endogenous
Aurora B protein using RNA interference (Fig. 3D). As a result, the percentage of
multinucleation in Aurora B knockdown-SMMC-7721 cells increased to
8.3%, compared with 2.8% in the control cells (Fig. 3E). These results indicate that
Aurora B plays a vital role in cytokinesis and that overexpression
of hnRNPC2 induces multinucleation by repressing the Aurora B
protein.
hnRNPC2 repressed mRNA translation of
Aurora B by inhibiting eIF4E binding to its mRNA
We demonstrated that overexpression of hnRNPC2
induced multinucleation by repression of the Aurora B protein.
Next, we attempted to clarify how hnRNPC2 repressed the expression
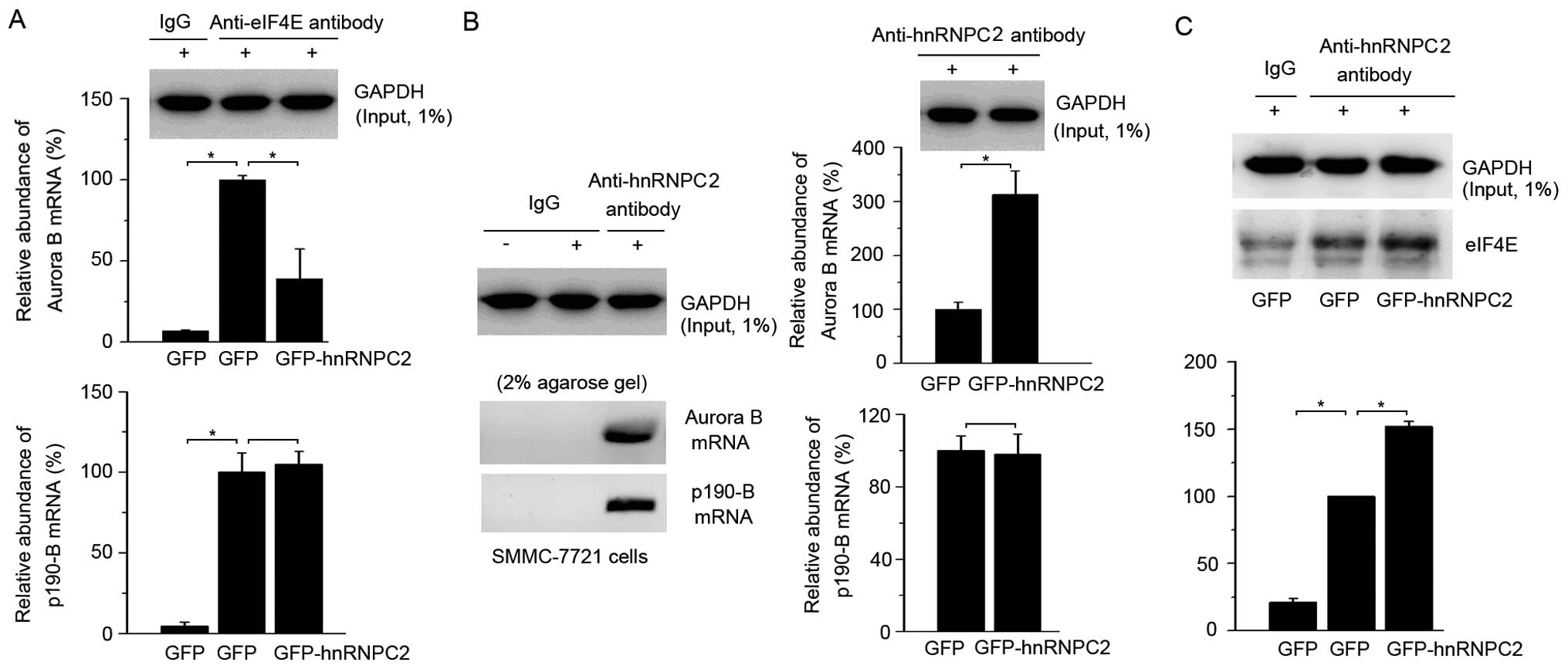
of Aurora B. First, we examined the Aurora B mRNA in cells. The
abundance of Aurora B mRNA increased in hnRNPC2-overexpressing
cells, which indicates that the repression of the Aurora B protein
is not caused by mRNA transcription but may instead be caused at
the translational level (Fig. 3B).
To obtain evidence for this hypothesis, we used mRNA-protein co-IP
method to detect whether mRNA translation was inhibited in
hnRNPC2-overexpressing cells. It is well-known that eIF4E is an
mRNA cap binding protein that is necessary for the initiation of
cap-dependent mRNA translation (3,33). The
mRNA-eIF4E co-IP results revealed that the Aurora B mRNA bound less
eIF4E while p190-B mRNA, as a control, did not change in
hnRNPC2-overexpressing cells, suggesting that the expression of the
Aurora B protein was specifically repressed by translational
initiation (Fig. 4A). Furthermore,
mRNA-hnRNPC2 co-IP results revealed that Aurora B mRNA specifically
bound with hnRNPC2 in SMMC-7721 cells and bound more in
hnRNPC2-overexpressing cells, while the relative abundance of
hnRNPC2 bound to p190-B mRNA changed little (Fig. 4B). To clarify how hnRNPC2 inhibited
eIF4E binding to Aurora B mRNA, protein-protein co-IP was carried
out. As expected, hnRNPC2 bound more eIF4E in
hnRNPC2-overexpressing cells (Fig.
4C). These results suggest that hnRNPC2 inhibits the binding of
eIF4E with Aurora B mRNA by binding with eIF4E, which represses
mRNA translational initiation and therefore results in the decrease
of the Aurora B protein.
Roles of hnRNPC2 in hepatocellular
carcinoma cells
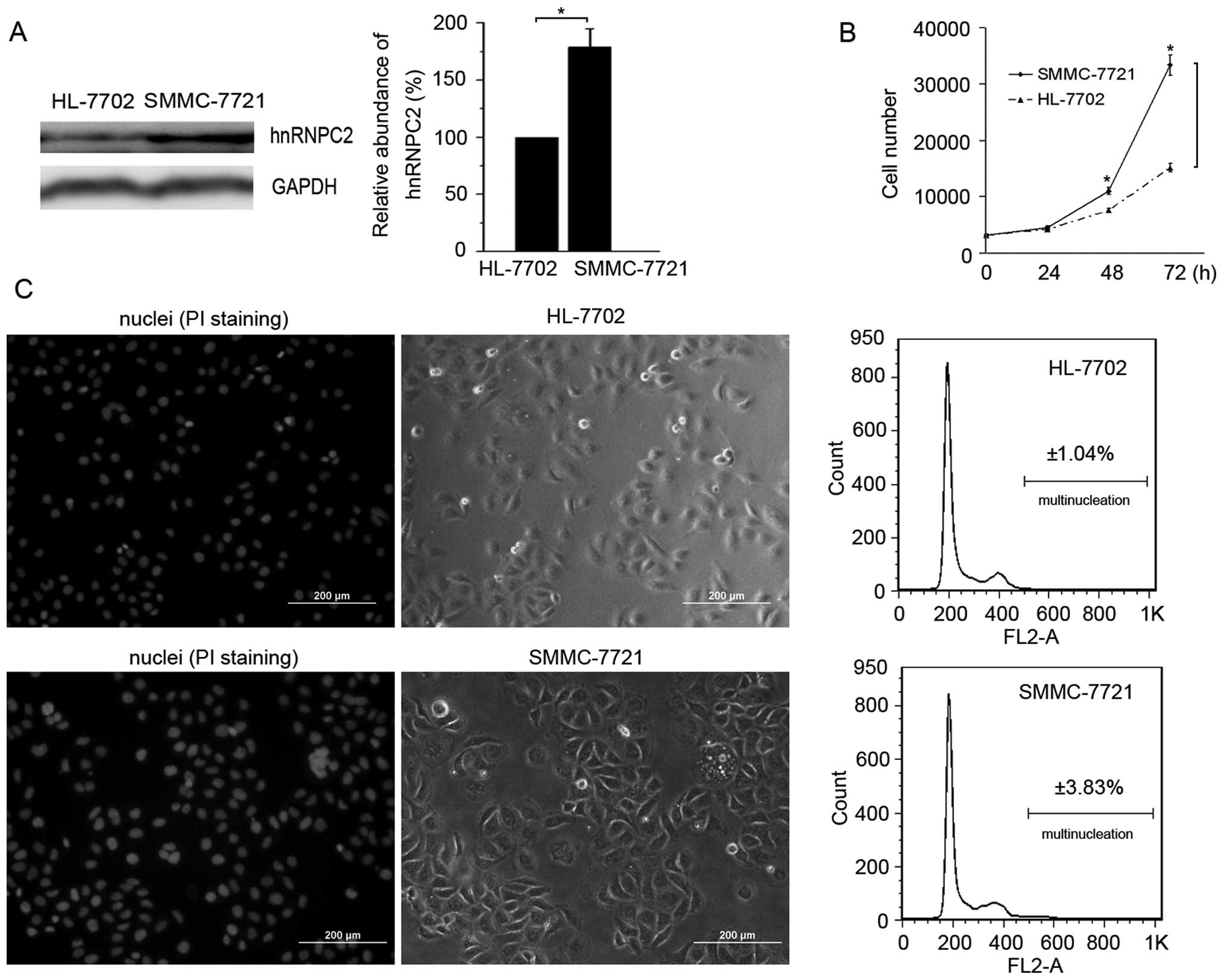
To explore the role of hnRNPC2 in the process of
hepatocellular carcinoma cell progression, we examined the
expression of hnRNPC2 in noncancerous hepatocellular HL-7702 cells
and hepatocellular carcinoma SMMC-7721 cells. Western blotting
results revealed that SMMC-7721 cells expressed more hnRNPC2 than
HL-7702 cells (Fig. 5A). Cell
proliferative rate tests demonstrated that the proliferation of
SMMC-7721 cells was much quicker than that of HL-7702 cells
(Fig. 5B). Moreover, fluorescent
microscopy and flow cytometry results revealed that the percentage
of multinucleated cells is three times larger in SMMC-7721 cells
when compared with that in HL-7702 cells (Fig. 5C). SMMC-7721 is a low-grade
malignant human hepatocellular cell line and hnRNPC and
multinucleation are related to tumor grade (2,11,34,35).
As mentioned above, we overexpressed hnRNPC2 in SMMC-7721 cells to
raise its tumor grade, resulting in an increase in multinucleation
(Fig. 1D). These results indicate
that the amount of multinucleation increased with increased
hnRNPC2, from noncancerous hepatocellular HL-7702 cells to
low-grade malignant hepatocellular SMMC-7721 cells and then to
SMMC-7721 cells expressing exogenous hnRNPC2, suggesting that
hnRNPC2 plays a role in hepatocellular carcinoma cell
progression.
Discussion
Multinucleation is an important characteristic in
tumor progression and correlates to tumor grade (35,36).
We found that overexpression of hnRNPC2 induced multinucleation in
hepatocellular carcinoma cells. From HL-7702 cells to SMMC-7721
cells and then to exogenous hnRNPC2 expressed SMMC-7721 cells, the
ratio of multinucleation increased positively with gradually
increased expression of hnRNPC2. In this process, Aurora B played
an important role to inhibit cytokinesis (23–26,37–40).
As indicated, deregulation of Aurora B, either downregulation or
upregulation, leads to cytokinesis failure in tumors (27–29,41).
Here, we observed that overexpression of hnRNPC2 in SMMC-7721 cells
repressed the expression of Aurora B, resulting in cytokinesis
failure and multinucleation appearance. This phenomena was also
induced by abolishing endogenous Aurora B protein by RNA
interference in SMMC-7721 cells, consistent with a previous report
(27). Thus, we conclude that
hnRNPC2 induces multinucleation by repressing the expression of the
Aurora B protein in hepatocellular carcinoma cells. In addition, we
found that multinucleation was an irreversible process. Once
mononuclear cells were transformed into multinucleated cells, they
lost the ability to divide. Instead, they accumulated more nuclei
and expanded their cytoplasm and thus became multi-nucleated giant
cells and eventually died. As elucidated above, Aurora B plays an
indispensible role in this process.
To uncover how hnRNPC2 repressed the expression of
the Aurora B protein, we first examined Aurora B mRNA and found
that it was increased in hnRNPC2-overexpressing cells. This may be
due to the function of hnRNPC2 that binds with AU-rich or U-rich
elements to stabilize mRNA (7,8). The
results indicate that the repression of the Aurora B protein has no
direct correlation with mRNA transcription and stability. We
studied eIF4E, which is fundamental to the initiation of protein
synthesis by binding with the 5′ terminal cap structure of mRNA
(3,33,42).
eIF4E is a presumptive oncogene and is frequently elevated in tumor
cells with an association with a poor prognosis (43,44).
In our results, Aurora B mRNA bound more hnRNPC2 and less eIF4E in
hnRNPC2-overexpressing cells, suggesting that the repression of
Aurora B, caused by overexpressed hnRNPC2, contributed to
translational initiation inhibition. Furthermore, we found that
hnRNPC2 bound more eIF4E in hnRNPC2-overexpressing SMMC-7721 cells.
Thus, we postulate that hnRNPC2 inhibits eIF4E binding to Aurora B
mRNA by binding with eIF4E, which may then repress eIF4E activity.
Even so, there are still many details of the mechanism to be
elucidated. For example, it remains unclear whether hnRNPC2
directly binds with eIF4E and whether other translational
initiation factors participate in the process. These questions are
worth further investigation.
Repression of Aurora B by hnRNPC2-induced
multinucleation results in cell death and overexpression of hnRNPC2
accelerates the cell proliferative rate in SMMC-7721 cells.
Previous reports have shown that Aurora B is increased to promote
cell proliferation, while inhibition of its activity by specific
inhibitors reduces the cell proliferative rate in certain tumors,
which therefore was treated as a potent therapeutic target and
prognostic marker (45,46). However, in certain tumors the
expression of Aurora B also decreases with increased cell
proliferative rate (41). However,
in this study the overexpressed hnRNPC2 repressed Aurora B
expression in hepatocellular carcinoma cells; however, the cell
proliferative rate is still elevated, which indicates that hnRNPC2
is involved in other routes of gene regulation. In fact, hnRNPC is
also treated as a potent therapeutic target and prognostic marker
in tumors; its high expression in tumors indicates fast cell
proliferation, high infiltration and invasion, poor therapeutic
effect and high recurrence rate following surgery (2,3,11).
Therefore, there is more to understand about the full function of
hnRNPC2 in tumorigenesis and its progression.
In conclusion, we found that the expression of
hnRNPC2 is positively correlated with multinucleation and
proliferation in hepatocellular carcinoma cells. Since both are
characteristics of tumors and are positively correlated with tumor
grade, we propose that hnRNPC2 may be treated as a potential target
for hepatocellular carcinoma cell diagnosis and treatment.
Acknowledgements
The authors would like to thank Bo-Wei
Zhang for pEGFP-C1 vector, Wei-Qi Wang for BT549 cells and all
members of the Core Facility for Molecular Biology and Core
Facility for Cell Biology in Shanghai Institute of Biological
Sciences for technical support. This study was supported by grants
from the State Natural Science Foundation of China (no. 30970585
and 31170722) and a grant for open research teams of the State Key
Laboratory of Molecular Biology.
References
|
1
|
Christian KJ, Lang MA and Raffalli-Mathieu
F: Interaction of heterogeneous nuclear ribonucleoprotein C1/C2
with a novel cis-regulatory element within p53 mRNA as a response
to cytostatic drug treatment. Mol Pharmacol. 73:1558–1567. 2008.
View Article : Google Scholar
|
|
2
|
Sun W, Xing B, Sun Y, et al: Proteome
analysis of hepatocellular carcinoma by two-dimensional difference
gel electrophoresis. Mol Cell Proteomics. 6:1798–1808. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang J, Cho SJ, Shu L, et al:
Translational repression of p53 by RNPC1, a p53 target
overexpressed in lymphomas. Genes Dev. 25:1528–1543. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Merrill BM, Barnett SF, LeStourgeon WM and
Williams KR: Primary structure differences between proteins Cl and
C2 of HeLa 40S nuclear ribonucleoprotein particles. Nucleic Acids
Res. 17:8441–8449. 1989. View Article : Google Scholar
|
|
5
|
Izquierdo JM: Heterogeneous
ribonucleoprotein C displays a repressor activity mediated by
T-cell intracellular antigen-1-related/like protein to modulate Fas
exon 6 splicing through a mechanism involving Hu antigen R. Nucleic
Acids Res. 38:8001–8014. 2010. View Article : Google Scholar
|
|
6
|
Nakielny S and Dreyfuss G: The hnRNP C
proteins contain a nuclear retention sequence that can override
nuclear export signals. J Cell Biol. 134:1365–1373. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Velusamy T, Shetty P, Bhandary YP, Liu MC
and Shetty S: Posttranscriptional regulation of urokinase receptor
expression by heterogeneous nuclear ribonuclear protein C.
Biochemistry. 47:6508–6517. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shetty S: Regulation of urokinase receptor
mRNA stability by hnRNP C in lung epithelial cells. Mol Cell
Biochem. 272:107–118. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lee EK, Kim HH, Kuwano Y, et al: hnRNP C
promotes APP translation by competing with FMRP for APP mRNA
recruitment to P bodies. Nat Struct Mol Biol. 17:732–739. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kim JH, Paek KY, Choi K, et al:
Heterogeneous nuclear ribonucleoprotein C modulates translation of
c-myc mRNA in a cell cycle phase-dependent manner. Mol Cell Biol.
23:708–720. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Blume SW, Jackson NL, Frost AR, et al:
Northwestern profiling of potential translation-regulatory proteins
in human breast epithelial cells and malignant breast tissues:
evidence for pathological activation of the IGF1R IRES. Exp Mol
Pathol. 88:341–352. 2010. View Article : Google Scholar
|
|
12
|
Hossain MN, Fuji M, Miki K, Endoh M and
Ayusawa D: Downregulation of hnRNP C1/C2 by siRNA sensitizes HeLa
cells to various stresses. Mol Cell Biochem. 296:151–157. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Meng Z, Jackson NL, Choi H, King PH,
Emanuel PD and Blume SW: Alterations in RNA-binding activities of
IRES-regulatory proteins as a mechanism for physiological
variability and pathological dysregulation of IGF-IR translational
control in human breast tumor cells. J Cell Physiol. 217:172–183.
2008. View Article : Google Scholar
|
|
14
|
Ariizumi T, Ogose A, Kawashima H, Hotta T,
Umezu H and Endo N: Multinucleation followed by an acytokinetic
cell division in myxofibrosarcoma with giant cell proliferation. J
Exp Clin Cancer Res. 28:442009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Blanchard Z, Malik R, Mullins N, et al:
Geminin overexpression induces mammary tumors via suppressing
cytokinesis. Oncotarget. 2:1011–1027. 2011.PubMed/NCBI
|
|
16
|
Margall-Ducos G, Celton-Morizur S, Couton
D, Brégerie O and Desdouets C: Liver tetraploidization is
controlled by a new process of incomplete cytokinesis. J Cell Sci.
120(Pt 20): 3633–3639. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Toyoda H, Bregerie O, Vallet A, et al:
Changes to hepatocyte ploidy and binuclearity profiles during human
chronic viral hepatitis. Gut. 54:297–302. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Studach LL, Rakotomalala L, Wang WH, et
al: Polo-like kinase 1 inhibition suppresses hepatitis B virus X
protein-induced transformation in an in vitro model of liver cancer
progression. Hepatology. 50:414–423. 2009. View Article : Google Scholar
|
|
19
|
Hsieh SY, Huang SF, Yu MC, et al:
Stathmin1 overexpression associated with polyploidy, tumor-cell
invasion, early recurrence, and poor prognosis in human hepatoma.
Mol Carcinog. 49:476–487. 2010.PubMed/NCBI
|
|
20
|
Xu Z, Ogawa H, Vagnarelli P, et al:
INCENP-aurora B interactions modulate kinase activity and
chromosome passenger complex localization. J Cell Biol.
187:637–653. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Petsalaki E, Akoumianaki T, Black EJ,
Gillespie DA and Zachos G: Phosphorylation at serine 331 is
required for Aurora B activation. J Cell Biol. 195:449–466. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Emanuele MJ, Lan W, Jwa M, Miller SA, Chan
CS and Stukenberg PT: Aurora B kinase and protein phosphatase 1
have opposing roles in modulating kinetochore assembly. J Cell
Biol. 181:241–254. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yan J, Jin S, Li J and Zhan Q: Aurora B
interaction of centrosomal Nlp regulates cytokinesis. J Biol Chem.
285:40230–40239. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mora-Bermudez F, Gerlich D and Ellenberg
J: Maximal chromosome compaction occurs by axial shortening in
anaphase and depends on Aurora kinase. Nat Cell Biol. 9:822–831.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Miyauchi K, Zhu X, Foong C, Hosoya H and
Murata-Hori M: Aurora B kinase activity is required to prevent
polar cortical ingression during cytokinesis. Cell Cycle.
6:2549–2553. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Song SJ, Kim SJ, Song MS and Lim DS:
Aurora B-mediated phosphorylation of RASSF1A maintains proper
cytokinesis by recruiting Syntaxin16 to the midzone and midbody.
Cancer Res. 69:8540–8544. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Delaval B, Ferrand A, Conte N, et al:
Aurora B -TACC1 protein complex in cytokinesis. Oncogene.
23:4516–4522. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
You J, Li Q, Wu C, Kim J, Ottinger M and
Howley PM: Regulation of aurora B expression by the bromodomain
protein Brd4. Mol Cell Biol. 29:5094–5103. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Qi G, Ogawa I, Kudo Y, et al: Aurora-B
expression and its correlation with cell proliferation and
metastasis in oral cancer. Virchows Arch. 450:297–302. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hauf S, Cole RW, LaTerra S, et al: The
small molecule Hesperadin reveals a role for Aurora B in correcting
kinetochore-microtubule attachment and in maintaining the spindle
assembly checkpoint. J Cell Biol. 161:281–294. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang Y, Sun DQ and Liu DG: Tumor
suppression by RNA from C/EBPβ 3′UTR through the inhibition of
protein kinase Cε activity. PLoS One. 6:e165432011.
|
|
32
|
Peritz T, Zeng F, Kannanayakal TJ, et al:
Immunoprecipitation of mRNA-protein complexes. Nat Protoc.
1:577–580. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yanagiya A, Suyama E, Adachi H, et al:
Translational homeostasis via the mRNA cap-binding protein, eIF4E.
Mol Cell. May 10–2012.(Epub ahead of print).
|
|
34
|
Ding ZB, Shi YH, Zhou J, et al:
Association of autophagy defect with a malignant phenotype and poor
prognosis of hepatocellular carcinoma. Cancer Res. 68:9167–9175.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Couto SS, Griffey SM, Duarte PC and
Madewell BR: Feline vaccine-associated fibrosarcoma: morphologic
distinctions. Vet Pathol. 39:33–41. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ganem NJ, Storchova Z and Pellman D:
Tetraploidy, aneuploidy and cancer (Review). Curr Opin Genet Dev.
17:157–162. 2007. View Article : Google Scholar
|
|
37
|
Guse A, Mishima M and Glotzer M:
Phosphorylation of ZEN-4/MKLP1 by aurora B regulates completion of
cytokinesis. Curr Biol. 15:778–786. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Capalbo L, Montembault E, Takeda T, Bassi
ZI, Glover DM and D’Avino PP: The chromosomal passenger complex
controls the function of endosomal sorting complex required for
transport-III Snf7 proteins during cytokinesis. Open Biol.
2:1200702012. View Article : Google Scholar
|
|
39
|
Xu Z, Vagnarelli P, Ogawa H, Samejima K
and Earnshaw WC: Gradient of increasing Aurora B kinase activity is
required for cells to execute mitosis. J Biol Chem.
285:40163–40170. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Guse A, Mishima M and Glotzer M:
Phosphorylation of ZEN-4/MKLP1 by aurora B regulates completion of
cytokinesis. Curr Biol. 15:778–786. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Baldini E, Arlot-Bonnemains Y, Mottolese
M, et al: Deregulation of Aurora kinase gene expression in human
testicular germ cell tumours. Andrologia. 42:260–267. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Graham PL, Yanowitz JL, Penn JK, Deshpande
G and Schedl P: The translation initiation factor eIF4E regulates
the sex-specific expression of the master switch gene Sxl in
Drosophila melanogaster. PLoS Genet. 7:e10021852011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Nasr Z, Robert F, Porco JA Jr, Muller WJ
and Pelletier J: eIF4F suppression in breast cancer affects
maintenance and progression. Oncogene. Apr 9–2012.(Epub ahead of
print).
|
|
44
|
Ilic N, Utermark T, Widlund HR and Roberts
TM: PI3K-targeted therapy can be evaded by gene amplification along
the MYC-eukaryotic translation initiation factor 4E (eIF4E) axis.
Proc Natl Acad Sci U S A. 108:E699–708. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Portella G, Passaro C and Chieffi P:
Aurora B: a new prognostic marker and therapeutic target in cancer.
Curr Med Chem. 18:482–496. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Xie F, Lang Q, Zhou M, et al: The dietary
flavonoid luteolin inhibits Aurora B kinase activity and blocks
proliferation of cancer cells. Eur J Pharm Sci. 46:388–396. 2012.
View Article : Google Scholar : PubMed/NCBI
|