Introduction
Biliary tract cancers (BTCs) are a heterogeneous
group of tumors arising from the epithelial cells of the intra- and
extra-hepatic bile ducts and gallbladder (1,2).
Histologically, the majority of BTCs are adenocarcinomas and have a
poor prognosis. The majority of BTC patients exhibit an
unresectable disease at the time of diagnosis due to the advanced
cancer stage. Although patients rarely have identical risk factors,
it is clear that the disorders that cause chronic inflammation of
the biliary tract, including primary sclerosing cholangitis,
gallstones and bile duct stones, are associated with an increased
incidence of BTC.
Little is known about the molecular pathogenesis of
BTC (1,2). Although alterations in a number of
cancer-associated genes, including p53 and KRAS, have
been identified as potential risk factors, the frequency of these
alterations is low. Interleukin 6 (IL-6), an inflammatory cytokine,
appears to have a more definite role in the pathogenesis of BTC.
The activation of EGFR, ERBB2 and HGF has also been reported in BTC
(1,2).
Homozygous deletions have been useful in the
positional cloning of a number of tumor suppressor genes. Using
high resolution single nucleotide polymorphism (SNP) arrays, we
previously detected novel regions of homozygous deletions and
identified potential tumor suppressor genes in human cancers
(3,4). In the present study, DNA copy number
aberrations in human BTC cell lines were investigated using SNP
arrays to identify the genes potentially involved in BTC. It was
observed that a novel homozygous deletion at the chromosomal region
7p21.3 occurred in a BTC cell line and that the plant homeodomain
(PHD) finger protein 14 (PHF14) gene, which lies within the
19p13.2 chromosomal region, was homozygously deleted. The present
study also further examined whether defective PHF14
expression has a functional role in BTC cells.
Materials and methods
Cell lines
The following eight human BTC cell lines were
studied: TFK1, HuCCT1, OCUG1, NOZ, OZ, SSP25, HuH28, and TKKK.
These cell lines were obtained from the Health Science Research
Resources Bank (Osaka, Japan) and the American Type Culture
Collection (Manassas, VA, USA). The cells were maintained in
Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf
serum. This study was approved by the Ethics Committee of Kyoto
Prefectural University of Medicine, Kyoto, Japan.
SNP array analysis
DNA copy number changes were analyzed using the
GeneChip Mapping 250K Sty array (Affymetrix, Santa Clara, CA, USA)
according to the manufacturer’s instructions, as previously
described (3–5). Briefly, 250 ng genomic DNA was
digested with a restriction enzyme, then ligated to an adaptor and
amplified by PCR. The amplified products were fragmented,
biotinylated and hybridized to the microarrays. Hybridization was
detected by incubation with a streptavidin-phycoerythrin conjugate
and scanning of the array. Following the appropriate normalization
of the mean array intensities, signal ratios were calculated
between the BTC cell lines and the anonymous normal references.
Copy numbers were then inferred from the observed signal ratios
based on the hidden Markov model using Copy Number Analyzer for
Affymetrix GeneChip mapping arrays (CNAG) software (available at
http://www.genome.umin.jp).
PCR analysis
Conventional PCR was performed using Ex Taq DNA
polymerase (Takara, Otsu, Japan) according to the manufacturer’s
instructions. Genomic DNA and mRNA were quantified using the
real-time fluorescence detection method, as described previously
(5). The primers that were used for
the PCR are shown in Table I. The
endogenous controls for the mRNA and genomic DNA levels were
GAPDH and long interspersed nuclear element-1 (LINE-1),
respectively.
 | Table IPrimers used for PCR. |
Table I
Primers used for PCR.
| PHF14 | STS-marker | Forward primer | Reverse primer |
|---|
| Exon 4 | |
5′-TTGGAAATGCATATAATAATGTTTAAG-3′ |
5′-AGCCACAGTCAGCCATTTCT-3′ |
| Exon 5 | |
5′-TTCTTTTTCTTTTGTGATTTTATGTGA-3′ |
5′-AGGGAAGTCAAAGGCAGACA-3′ |
| Exon 6 | |
5′-TGTTTGTTTTGTGTGTGGGAAT-3′ |
5′-GCCAGGTAAACTAACAAGTAAAACC-3′ |
| Exon 7 | |
5′-TGGAAATAAGTTTGCTTTGAGAA-3′ |
5′-TGTTTTCTGAACGTCTGACTAGC-3′ |
| Exon 17 | |
5′-TGTCAGTGTTCTAAATATTTGTTTTGT-3′ |
5′-GGTGTACTGGTTAAAATGTTGGTTC-3′ |
| Exon 18 | |
5′-CAGATGCAGTTAAAATCTGTCAA-3′ |
5′-AAACTTTTAAAGGTCCAGCTTTTG-3′ |
| Genomic DNA | SWSS2137 |
5′-GACAGGCTCAGATATTTC-3′ |
5′-CAACCATCTGTTGTCTTC-3′ |
| mRNA | |
5′-AGCAACTATCACCAGAAGCACA-3′ |
5′-TTTTCCTGAATTTGAATCATGC-3′ |
Immunoblotting
Immunoblots were prepared according to previously
published methodology (5). Cell
lysates (20 μg protein per sample) were separated via
SDS-polyacrylamide gel electrophoresis using 10% acrylamide gels.
The anti-PHF14 rabbit polyclonal antibody and the anti-β-actin
mouse monoclonal antibody were purchased from Sigma-Aldrich (Tokyo,
Japan). The anti-PHF14 and anti-β-actin antibodies were used for
immunoblotting at dilutions of 1:400 and 1:5,000, respectively. The
anti-mouse or anti-rabbit IgG (Amersham, Tokyo, Japan) used for
secondary immunodetection was diluted to 1:5,000. Antibody binding
was detected using an ECL system (Amersham).
RNA interference (RNAi)
To knock down PHF14 expression in the cells,
two small interfering RNA (siRNA) duplex oligoribonucleotides
targeting PHF14 [PHF14 Stealth Select RNAi™ siRNA HSS114491
(siRNAb) and HSS114492 (siRNAc)] and negative control siRNA
duplexes were purchased from Invitrogen (Carlsbad, CA, USA). The
siRNAs were delivered into OCUG1 cells using Lipofectamine RNAiMAX
(Invitrogen), according to the manufacturer’s instructions. The
cell viability was assessed by measuring
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
(Nacalai Tesque, Kyoto, Japan) dye absorbance (MTT assay),
according to the manufacturer’s instructions, at 24, 48 and 72 h
after siRNA transfection.
Statistical analysis
Differences between the groups were evaluated using
the Student’s t-test. The statistical analyses were performed on
SPSS 15.0 software (SPSS Inc., Chicago, IL, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
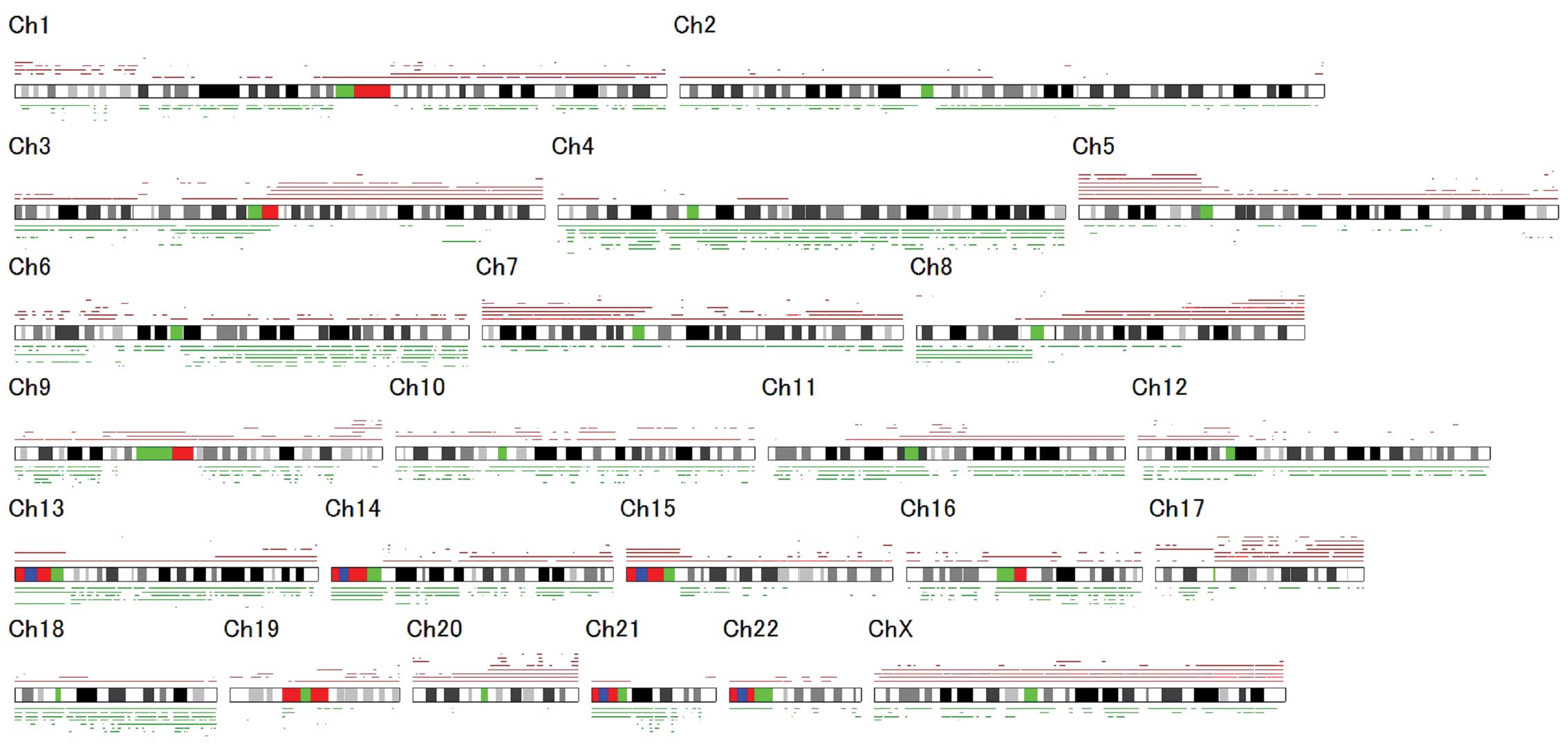
Overview of genomic changes in BTC cell
lines
To identify the genes involved in BTC, eight BTC
cell lines were screened for DNA copy number aberrations using SNP
array analysis. The genetic changes that were detected are shown in
Fig. 1. Chromosomal regions
frequently involved in the gain of DNA were identified at 5p and
17q (seven cases, 88%), as well as 8q (six cases, 75%). The
chromosomal regions most frequently associated with DNA loss were
identified at 4p and 4q (seven cases, 88%) and 6q (six cases, 75%).
The homozygous deletions and chromosomal amplifications are shown
in Table II. SNP array analyses
successfully identified chromosomal amplification regions
containing known oncogenes, including KRAS (12p12.1) and
ERBB2 (17q12), as well as chromosomal homozygous deletion
regions containing known tumor suppressor genes, including
FHIT (3p14.2), CDKN2A (9p21), CDKN2B (9p21)
and WWOX (16q23.1; Table
II). Of these chromosomal regions, the homozygous deletion at
7p21.3 became the focus for further investigation as it was a novel
alteration in BTC.
 | Table IIChromosomal regions that were
amplified or homozygously deleted in BTC cell lines. |
Table II
Chromosomal regions that were
amplified or homozygously deleted in BTC cell lines.
| DNA copy number | Chromosomal
region | Cell line | Known oncogene or
tumor suppressor gene | Number of genes |
|---|
| Amplification | 12p11.1-q11 | NOZ | | 1 |
| 12p12.1 | NOZ | KRAS | 8 |
| 12q12 | TKKK | | 1 |
| 17q12 | TKKK | ERBB2 | 35 |
| 22q11.2 | TKKK | | 20 |
| Homozygous
deletion | 3p14.2a | TFK1, HuCCT1,
OCUGI | FHIT | 1 |
| 5q12 | OCUG1 | | 1 |
| 6q16.3-q21 | OZ | | 2 |
| 7p21.3 | OZ | | 5 |
| 9p21a | TFK1, OZ | CDKN2A,
CDKN2B | 3 |
| 16q23.1 | NOZ | WWOX | 1 |
| 20p12.1a | TFK1, OCUGI | | 1 |
| 21q21.3 | TKKK | | 1 |
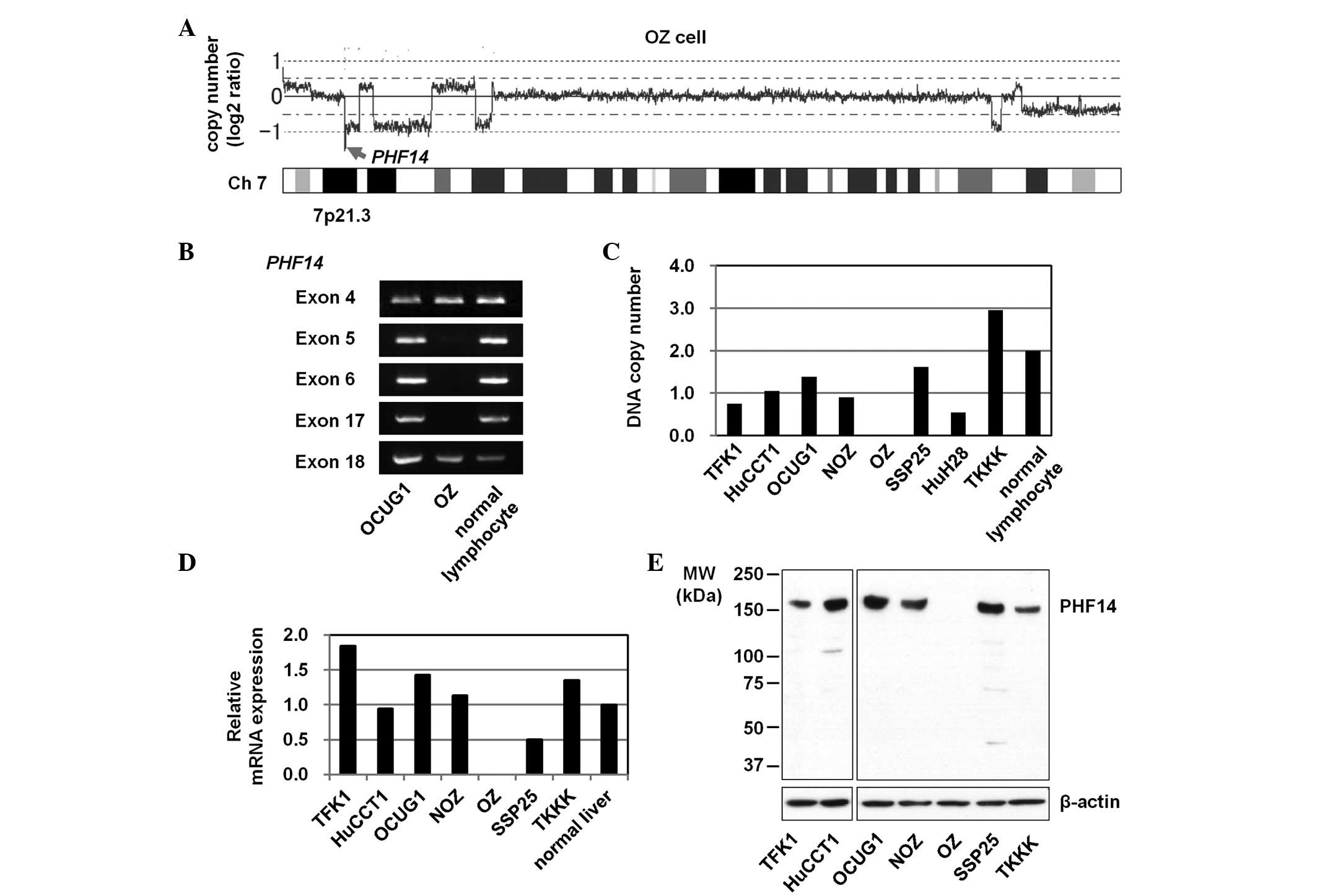
Identification of homozygous PHF14 gene
deletion
Among the eight cell lines screened, the OZ cell
line (6) exhibited a homozygous
deletion at chromosomal region 7p21.3 (Fig. 2A). It was estimated that the region
of deletion included five genes. Further validation experiments
using genomic PCR revealed a homozygous deletion of a single gene,
PHF14. The extent of the homozygous deletion was narrowed
down to a location between exons 5 and 17 of the PHF14 gene
(Fig. 2B).
Copy number and expression of PHF14 gene
in BTC cell lines
The DNA copy numbers and expression levels of the
PHF14 gene in the BTC cell lines and control normal
lymphocytes or liver (Fig. 2C–E)
were then analyzed. Real-time quantitative reverse transcription
(RT)-PCR and immunoblot analyses did not detect PHF14 mRNA
or protein expression, respectively (Fig. 2D and E), thus demonstrating the
absence of the PHF14 gene from the OZ cell line.
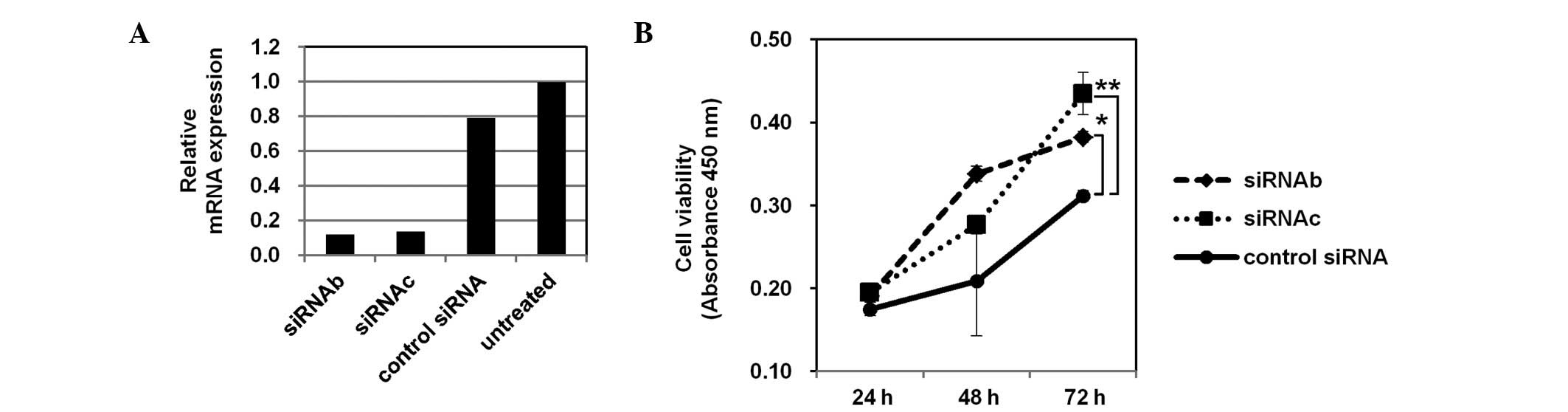
Enhanced growth of BTC cells by
PHF14-knockdown
To determine whether the defective expression of
PHF14 had a functional role in the BTC cells, PHF14
expression was knocked down with two independent siRNA molecules
(siRNAb and siRNAc) in OCUG1 cells (Fig. 3A). The PHF14-knockdown led to
an upregulation of cell growth, as determined via the MTT assay 72
h after the transfection with siRNAb and siRNAc (Fig. 3B). These observations suggest that
the defective expression of PHF14 may promote the
proliferation of BTC cells.
Discussion
In the present study, a novel homozygous deletion at
chromosomal region 7p21.3 was identified in the OZ cell line, a
human BTC cell line that was established from the ascites of a
patient with mucin-secreting BTC in the hepatic hilus (6). Subsequent detailed analyses revealed
that the homozygous deletion was located between exons 5 and 17 of
the PHF14 gene. Moreover, the present data suggest that the
defective expression of PHF14 may promote the proliferation
of the BTC cells.
Based on the amino acid sequence homology,
PHF14 is considered to be a PHD finger protein. The PHD
finger protein is known to be involved in chromatin-mediated
transcriptional regulation (7–9). The
PHD finger domain recognizes the methylation status of histone
lysine residues, including histone H3 trimethylated at lysine 4,
which is associated with an ‘open’ chromatin structure and
transcriptional activation. Mutations, deletions and chromosomal
translocation in the genes encoding PHD finger proteins, such as
the tumor suppressor ING1, have been associated with various types
of cancer (8). A mutation in
PHF14 was previously identified in a colon cancer cell line
(10). However, the function of
PHF14 has remained unknown. Phf14, a mouse homologue of PHF14, was
identified as a novel transcriptional factor that acts as a
negative regulator of platelet-derived growth factor receptor-α
(PDGFRα) expression in mouse mesenchymal cells (11). Furthermore, Phf14-null mice
exhibited interstitial pulmonary hyperplasia. Mesenchymal
fibroblasts derived from the Phf14-null mice showed an
increased proliferation rate, accompanied by the enhanced
expression of PDGFRα (11). The
increased growth of Phf14−/− mesenchymal cells
supports the present observation that the knockdown of PHF14
enhances the growth of BTC cells. Although the mechanisms by which
PHF14 functions in tumors remain to be elucidated, the
present data suggest that alterations in the expression of
PHF14 may be involved in the tumorigenesis of BTC.
References
|
1
|
Koti RS and Davidson BR: Malignant biliary
diseases. Sherlock’s Diseases of the Liver and Biliary System.
Dooley JS, Lok ASF, Burroughs AK and Heathcote EJ: 12th edition.
Wiley-Blackwell; Oxford: pp. 294–311. 2011
|
|
2
|
Goodman ZD, Terracciano LM and Wee A:
Tumours and tumour-like lesions of the liver. MacSween’s Pathology
of the Liver. Burt A, Portmann B and Ferrell L: 6th edition.
Churchill Livingstone; Philadelphia: pp. 761–851. 2011
|
|
3
|
Zen K, Yasui K, Gen Y, et al: Defective
expression of polarity protein PAR-3 gene (PARD3) in
esophageal squamous cell carcinoma. Oncogene. 28:2910–2918. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Endo M, Yasui K, Zen Y, et al: Alterations
of the SWI/SNF chromatin remodelling subunit-BRG1 and BRM in
hepatocellular carcinoma. Liver Int. 33:105–117. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zen K, Yasui K, Nakajima T, et al: ERK5 is
a target for gene amplification at 17p11 and promotes cell growth
in hepatocellular carcinoma by regulating mitotic entry. Genes
Chromosomes Cancer. 48:109–120. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Homma S, Nagamori S, Fujise K, et al:
Human bile duct carcinoma cell line producing abundant mucin in
vitro. Gastroenterol Jpn. 22:474–479. 1987.PubMed/NCBI
|
|
7
|
Aasland R, Gibson TJ and Stewart AF: The
PHD finger: implications for chromatin-mediated transcriptional
regulation. Trends Biochem Sci. 20:56–59. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Baker LA, Allis CD and Wang GG: PHD
fingers in human diseases: disorders arising from misinterpreting
epigenetic marks. Mutat Res. 647:3–12. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Saiga S, Möller B, Watanabe-Taneda A, Abe
M, Weijers D and Komeda Y: Control of embryonic meristem initiation
in Arabidopsis by PHD-finger protein complexes. Development.
139:1391–1398. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ivanov I, Lo KC, Hawthorn L, Cowell JK and
Ionov Y: Identifying candidate colon cancer tumor suppressor genes
using inhibition of nonsense-mediated mRNA decay in colon cancer
cells. Oncogene. 26:2873–2884. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kitagawa M, Takebe A, Ono Y, Imai T, Nakao
K, Nishikawa S and Era T: Phf14, a novel regulator of mesenchyme
growth via platelet-derived growth factor (PDGF) receptor-α. J Biol
Chem. 287:27983–27996. 2012.PubMed/NCBI
|