Introduction
Malignant tumors are treated with a combination of
therapeutic modalities, including surgery, radiotherapy,
chemotherapy, immunotherapy and hyperthermia therapy. Hyperthermia
is effective in killing tumor cells; moreover, it is capable of
increasing the sensitivity of tumor cells towards radiotherapy and
numerous antitumor drugs (1).
Therefore, hyperthermia is considered to be an effective and
promising tumor therapy (2).
Hyperthermia inhibits tumor cells through the five following
mechanisms: i) by inhibiting tumor cell respiration, thereby
increasing anaerobic glycolysis and increasing the acidity of the
environment, which may result in damage to the tumor cell membranes
and the release of lysosomal acid hydrolase; ii) by altering the
enzyme structure of the cytoplasm and the nucleus, thereby leading
to a metabolic disorder; iii) by inhibiting the synthesis of DNA,
RNA and the associated proteins, thereby leading to the aggregation
of denatured nuclear matrix proteins; iv) by impairing the
cytoskeleton; and v) by inhibiting anti-apoptotic proteases and/or
the activity of oncogenes, and enhancing pro-apoptotic proteases
and/or tumor suppressor gene activity, thereby leading to tumor
cell necrosis and/or the induction of apoptosis (3–6).
Hyperthermic temperatures between 40 and 45°C kill
tumor cells, mainly by inducing apoptosis (7,8). The
therapeutic temperature for treating an oral tumor is 43°C, which
is used to treat malignant oral tumors by inducing tumor cell
apoptosis. Hyperthermia-induced tumor cell apoptosis relies on
numerous approaches and regulation factors. It has been
demonstrated that hyperthermia is able to activate the release of
pro-apoptotic factors, such as cytochrome C,
apoptosis-inducing factor (AIF) and Smac/Diablo, from the
mitochondria, and activate downstream effectors to induce apoptosis
(9). In addition, hyperthermia has
been demonstrated to enhance Fas-L, tumor necrosis factor-α (TNF-α)
and TNF-related apoptosis-inducing ligand (TRAIL) expression, and
to increase tumor cell sensitivity towards their receptors, which
ultimately induces apoptosis through the death receptor pathway
(10–12). Furthermore, it has been indicated
that hyperthermia induces intracellular reactive oxygen species
(ROS) production and the activation of downstream effectors, which
leads to apoptosis (13,14). The treatment also promotes the
influx of extracellular Ca2+ and/or the damage of the
cytoplasmic calcium pool, thereby increasing intracellular
Ca2+ concentrations and activating
Ca2+-related enzymes, thus resulting in apoptosis
(15,16). Moreover, it has been demonstrated
that hyperthermia is able to activate or upregulate the gene or
protein expression of p53, Bax, Bak and caspase family members,
again leading to apoptosis (9,17–20).
During hyperthermia-induced tumor cell apoptosis, anti-apoptotic
protective factors are produced by the tumor cells for the
maintenance of self-survival and for protecting the cell from
damage. For example, heat shock proteins (HSPs) are produced under
stress to maintain cell survival (21). It has been demonstrated that the
expression of HSPs is increased significantly following
hyperthermia (22–25), which may inhibit apoptosis.
Additionally, it has been demonstrated that Bcl-2 and Mcl-1 are
also able to protect tumor cells from hyperthermic damage (26,27).
However, the mechanism of hyperthermia-induced tumor cell apoptosis
has not been fully elucidated.
In our previous study, we demonstrated that
hyperthermia-induced Tca8113 cell apoptosis involved changes in the
expression and function of multiple proteins (28). In the present study, a
hyperthermia-induced apoptosis model was established using the
Tca8113 cell line derived from a human tongue squamous cell
carcinoma. Cell lysates were subjected to fluorescent differential
display two-dimensional (2D) gel electrophoresis 2, 6, 8, 12 and 24
h after hyperthermia treatment. A total of 107 proteins were
detected that exhibited different expression levels in the
hyperthermia-treated cells compared with in the controls, using
matrix-associated laser desorption/ionization (MALDI)-time of
flight (TOF) or MALDI-TOF/TOF mass spectrometry analysis. This
method allowed us to obtain the peptide mass fingerprint, and 57
proteins were identified by searching the SwissProt and NCBInr
databases. Following the analysis of the protein profiles, the
results indicated that hyperthermia-induced Tca8113 cell apoptosis
is controlled by multiple factors, including time and regulatory
proteins.
Materials and methods
Materials
The human tongue squamous cell carcinoma cell line,
Tca8113, was purchased from Shanghai Cell Bank (Chinese Academy of
Sciences, Shanghai, China). The CyDye Difference Gel
Electrophoresis (DIGE) fluorescent Cy2, Cy3 and Cy5 were purchased
from GE Healthcare (Uppsala, Sweden). The heat shock 70 kDa protein
(HSP70; D69), stathmin 1 and Lamin A/C polyclonal antibodies were
purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA).
Cofilin 1 (5) sc-53934 mouse
monoclonal IgG2b was purchased from Santa Cruz
Biotechnology, Inc. (Santa Cruz, CA, USA). The
anti-phosphoglycerate mutase 1 (PGAM1) antibody (rabbit polyclonal
to PGAM1; ab96622) and the anti-Δ(3,5)-Δ(2,4)-dienoyl-CoA isomerase mitochondrial
(ECH1p) antibody (rabbit polyclonal to ECH1p; ab90645) were
purchased from Abcam (Cambridge, UK).
Cell culture and hyperthermia
treatment
The Tca8113 cells were cultured in RPMI-1640 medium
containing 10% fetal bovine serum (with 1×105 U/l
penicillin and 1×102 mg/l streptomycin) at 37°C with 5%
CO2. Cells at 80–90% confluence were treated in a
temperature-controlled water bath for 40 min at 43°C, and then
cultured under normal conditions for 2, 6, 8, 12 and 24 h. The
cells solely cultured under normal conditions were used as the
control.
Sample preparation and quantitation
The Tca8113 cells (2×107) were collected
and centrifuged at 228 × g for 5 min. The subsequent cell pellets
were washed with phosphate-buffered saline (PBS) and then
centrifuged at 228 × g for 5 min. This was repeated three times.
The cell pellets were resuspended in 250 μl DIGE lysis
buffer containing 7 M urea, 2 M thiourea, 65 mM Tris, 4% CHAPS,
0.2% IPG buffer and a protease inhibitor cocktail (Roche
Diagnostics GmbH, Mannheim, Germany), which was sonicated briefly
on ice. The cell lysates were centrifuged at 15,600 × g for 20 min
at 4°C. The supernatant was collected as the cell lysate, and the
protein concentration was measured with the Bio-Rad Protein Assay
(Bio-Rad Laboratories, Hercules, CA, USA) and adjusted to the same
concentration for each sample.
2D-DIGE assay
Electrophoresis was conducted as described
previously (29), and was performed
with a model E-600 electrophoresis system (Amersham, Piscataway,
NJ, USA). The pH of the protein samples was set at 8.0–9.0, and the
samples were diluted to 5 μg/μl. All samples were
mixed to equal amounts and aliquoted at 50 μg/10μl,
which was then used as the internal standard. A total of 50
μg sample was labeled with 400 pmol/μl Cy3 and Cy5,
respectively. The internal standard was labeled with 400
pmol/μl Cy2. The three types of labeled samples were mixed
evenly and the proteins were separated by isoelectric focusing
electrophoresis (IFE), with a pH gradient of 3.0–10.0 on a 13-cm
non-linear strip. The IFE conditions were as follows: 30 V for 12
h, 500 V for 1 h, 1,000 V for 1 h, 8,000 V for 8 h and 500 V for 4
h. Following isoelectric focusing, IPG strips were placed in
equilibrium buffer and the second electrophoresis stage [12.5%
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
(SDS-PAGE)] was subsequently conducted. The SDS-PAGE conditions
were 15 mA for 20 min and 30 mA until the bromophenol blue front
reached the bottom of the gel.
Gel scanning and software analysis
The gel pattern was scanned with a Typhoon scanner
(GE Healthcare Biosciences, Pittsburgh, PA, USA) at wavelengths of
488, 532 and 633 nm for Cy2, Cy3 and Cy5, respectively, and
optimized by ImageQuant software (GE Healthcare Biosciences).
Differential points were analyzed by DeCyder™ 2D software, version
6.5 (GE Healthcare).
Protein identification
The gels were stained with Coomassie brilliant blue
R350 (Amersham). The differential points were removed and the stain
was removed in NH4HCO3/30% ACN buffer, prior
to being digested in trypsin for 20 h. Peptides were extracted,
desalted with a ZipTip (Millipore Corporation, Billerica, MA, USA)
and analyzed by a 4800 Plus MALDI TOF/TOF™ analyzer (Applied
Biosystems, Foster City, CA, USA). The differentially expressed
proteins were identified by software analysis, as well as SwissProt
and NCBInr database searches.
Western blot analysis
Equal amounts of protein were separated by 10%
SDS-PAGE and electroblotted onto Immobilon-P membranes (Millipore
Corporation). The membranes were incubated with the primary
antibody at 4°C overnight and then incubated with horseradish
peroxidase-conjugated secondary antibody at room temperature for 1
h. Following washing with 1X Tris-buffered saline with 0.1%
Tween-20 (TBST), the membranes were visualized by chemiluminescence
(Pierce, Biotechnology, Inc., Rockford, IL, USA). The relative
protein content is the ratio of the test protein gray scale value
in contrast with the α-tubulin gray scale value, measured by ImageJ
software (version 2.1.4.6; National Institutes of Health, Bethesda,
MD, USA).
Statistical analysis
Differences in the protein expression profile were
analyzed by DeCyder 2D software, version 6.5, and a Student’s
t-test was applied. Differences among the 6 groups were compared
using a one-way ANOVA. P<0.01 was considered to indicate a
statistically significant difference.
Results
2D-DIGE gel pattern and analysis of
differential protein points
The Tca8113 cells were incubated for 40 min in a
water bath at 43°C, and subsequently transferred to normal culture
conditions for 2, 6, 8, 12 and 24 h. Protein samples were labeled
with Cy2, Cy3 and Cy5, which were separated by 2D-DIGE, and the
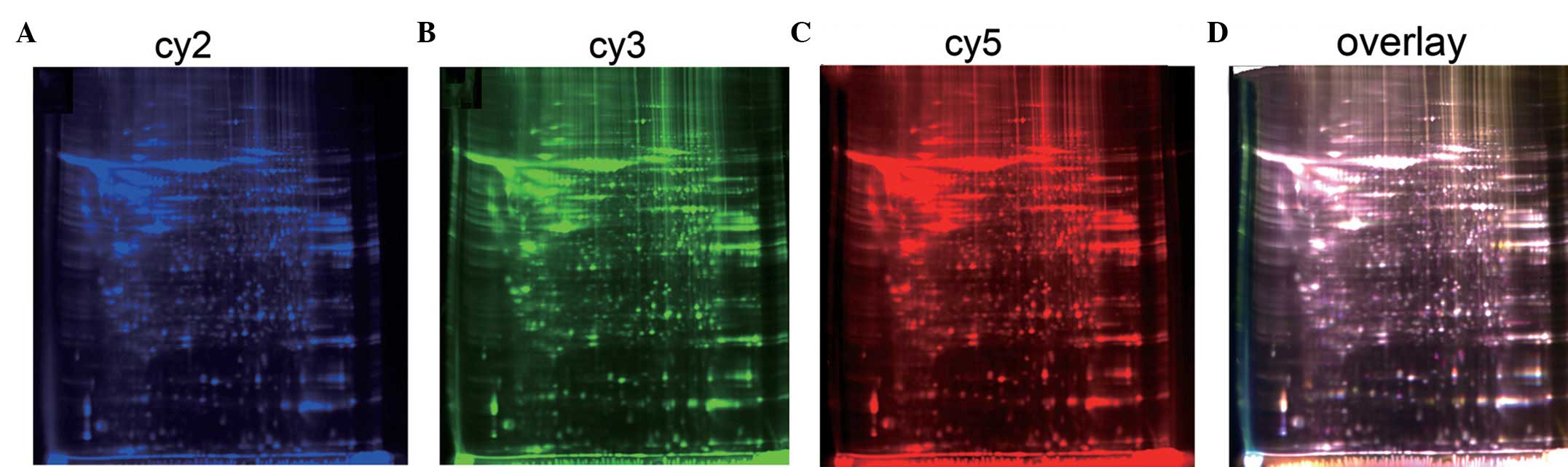
respective blue, green and red gel images were captured for each
dye (Fig. 1). The gel images were
analyzed by DeCyder 2D software, version 6.5, and the data were
compared by a one-way statistical analysis, which identified 107
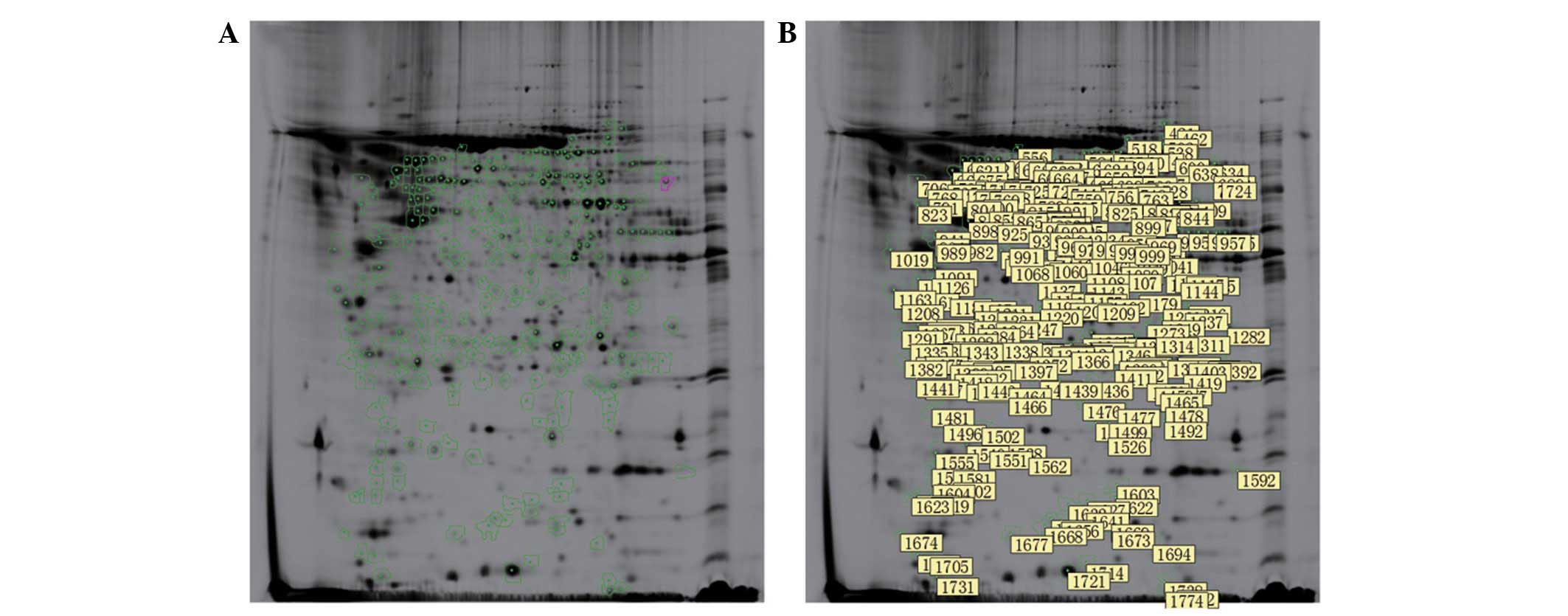
differentially expressed proteins (P=0.000; Fig. 2).
Differential protein point identification
and dynamic expression analysis
A total of 107 differential protein points were
detected by tandem mass spectroscopy and analyzed using the
SwissProt and NCBInr databases, which identified 57 types of
proteins (Tables I and II). The early period was considered to be
within 8 h of hyperthermic treatment, while 8–24 h post-treatment
was considered to be the middle to late period. Protein expression
levels that were higher in the hyperthermia-treated cells compared
with the controls were considered to be upregulated, while proteins
with lower expression levels were considered to be downregulated.
The dynamic expression changes in 57 proteins, at 2, 6, 8, 12 and
24 h after hyperthermic treatment, were characterized. In total, 44
proteins were demonstrated to be upregulated and 13 proteins were
shown to be downregulated.
 | Table I.Upregulation of 44 proteins during
the 24 h following hyperthermia. |
Table I.
Upregulation of 44 proteins during
the 24 h following hyperthermia.
| Upregulated
continuously | Upregulated
early | Upregulated at a
middle to late period |
|---|
| α-enolase | BolA-like protein
2 |
Peroxiredoxin-6 |
| Actin, cytoplasmic
2 | Cofilin-1 | Phosphoglycerate
mutase 1 |
| Crystal structure
of Ufc1, the Ufm1-conjugating enzyme 1, chain A | Deoxyuridine
5′-triphosphate nucleotidohydrolase, mitochondrial | Human eukaryotic
translation initiation factor 1A, chain A |
| F-actin-capping
protein subunit β | Glutathione
S-transferase P | T-complex protein 1
subunit ζ |
| 26S proteasome
non-ATPase regulatory subunit 14 | Far upstream
element-binding protein 1 | Splicing factor
U2AF 65 kDa subunit |
|
Glyceraldehyde-3-phosphate
dehydrogenase |
Peroxiredoxin-2 | Scavenger
mRNA-decapping enzyme DcpS |
| Heat shock protein
β-1 | Pyruvate kinase
isozymes M1/M2 | Keratin, type II
cytoskeletal 8 |
| Inorganic
pyrophosphatase | Tubulin β
chain | 60 kDa heat shock
protein, mitochondrial |
| Heterogeneous
nuclear ribonucleoprotein H | | 3-hydroxyacyl-CoA
dehydrogenase type-2 isoform 1 |
| Mitotic checkpoint
protein BUB3 | | |
| Phosphoglycerate
kinase 1 | | |
| hCG15971, isoform
CRA_b | | |
| Protein
disulfide-isomerase A3 | | |
| Proteasome
activator complex subunit 1 | | |
|
Peroxiredoxin-4 | | |
| Serum albumin | | |
| Flavin
reductase | | |
| Galactokinase | | |
| Prohibitin | | |
| Protein 4.1,
isoform 3 | | |
|
Pyridoxine-5′-phosphate oxidase | | |
| ATP synthase
subunit d, mitochondrial | | |
| Heat shock cognate
71 kDa protein | | |
| Heat shock 70 kDa
protein 1 | | |
| Keratin, type I
cytoskeletal 10 | | |
| Keratin, type II
cytoskeletal 7 | | |
| Keratin, type II
cytoskeletal 1 | | |
 | Table II.Downregulation of 13 proteins during
the 24 h following hyperthermia. |
Table II.
Downregulation of 13 proteins during
the 24 h following hyperthermia.
| Downregulated
continuously | Downregulated
early | Downregulated at a
middle to late period |
|---|
| ATP synthase
subunit α, mitochondrial | Argininosuccinate
synthase, mitochondrial | Hydroxysteroid
(17-β) dehydrogenase 10 |
| Crystal structure
of the human eukaryotic translation initiation factor 5A, chain
A |
D-3-phosphoglycerate dehydrogenase | Lamin-A/C |
|
Fructose-bisphosphate aldolase A |
Δ(3,5)-Δ(2,4)-dienoyl-coenzyme A
isomerase, mitochondrial | |
| Vimentin | Eukaryotic
translation initiation factor 5A-1 | |
| Stathmin
1/oncoprotein 18 |
Proliferation-associated protein 2G4 | |
| Mitochondrial
succinyl-CoA:3-ketoacid-coenzyme A transferase 1 | |
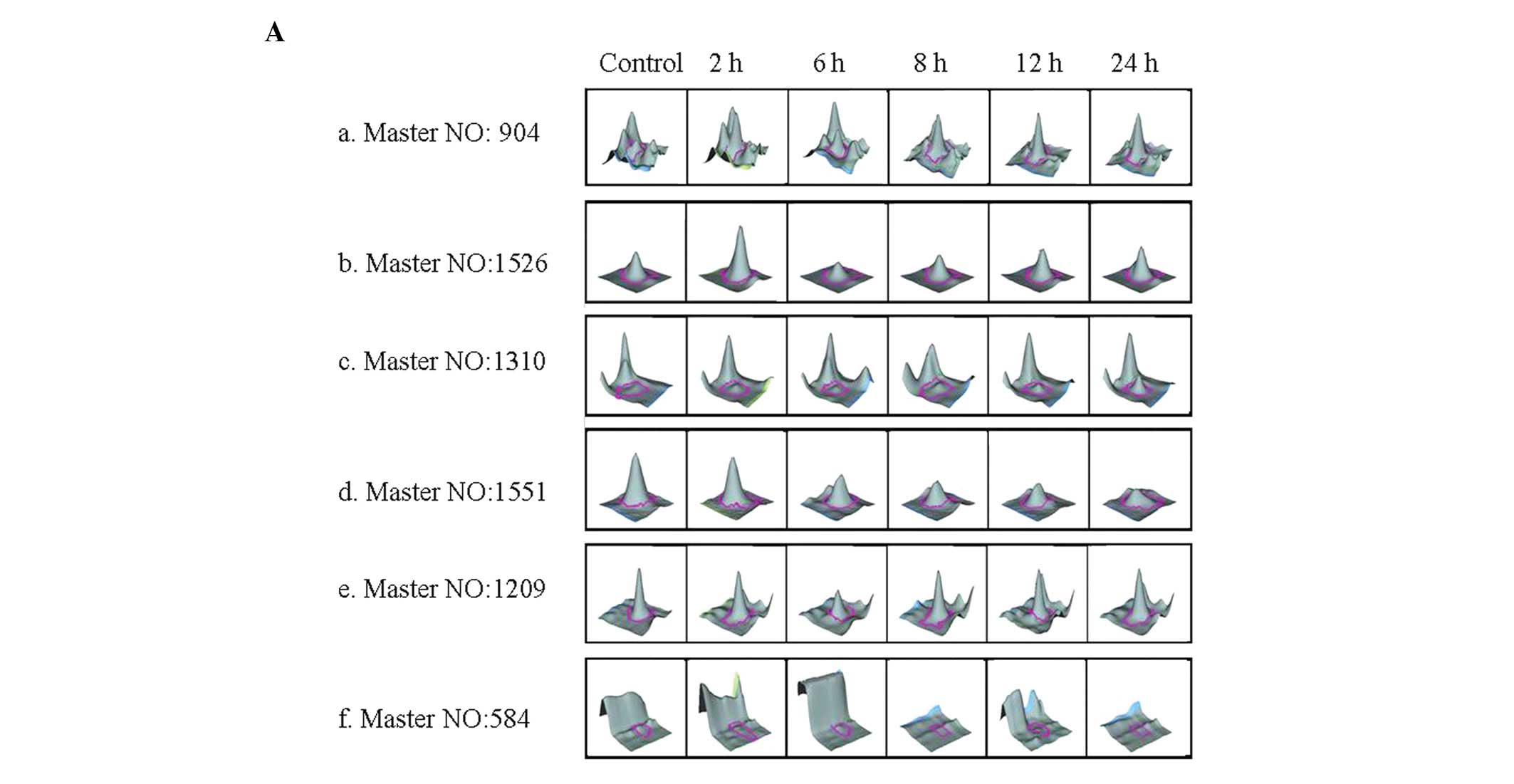
Among the 44 upregulated proteins, 27 were
upregulated continuously, 8 were upregulated during the early time
period and 9 were upregulated during the middle to late time period
(Table I; Fig. 3Aa–c, a representative figure of the
protein expression changes during the three time periods). Among
the 13 downregulated proteins, 5 were downregulated continuously, 6
were downregulated during the early time period and two were
downregulated during the middle to late time period (Table I; Fig.
3Ad–f).
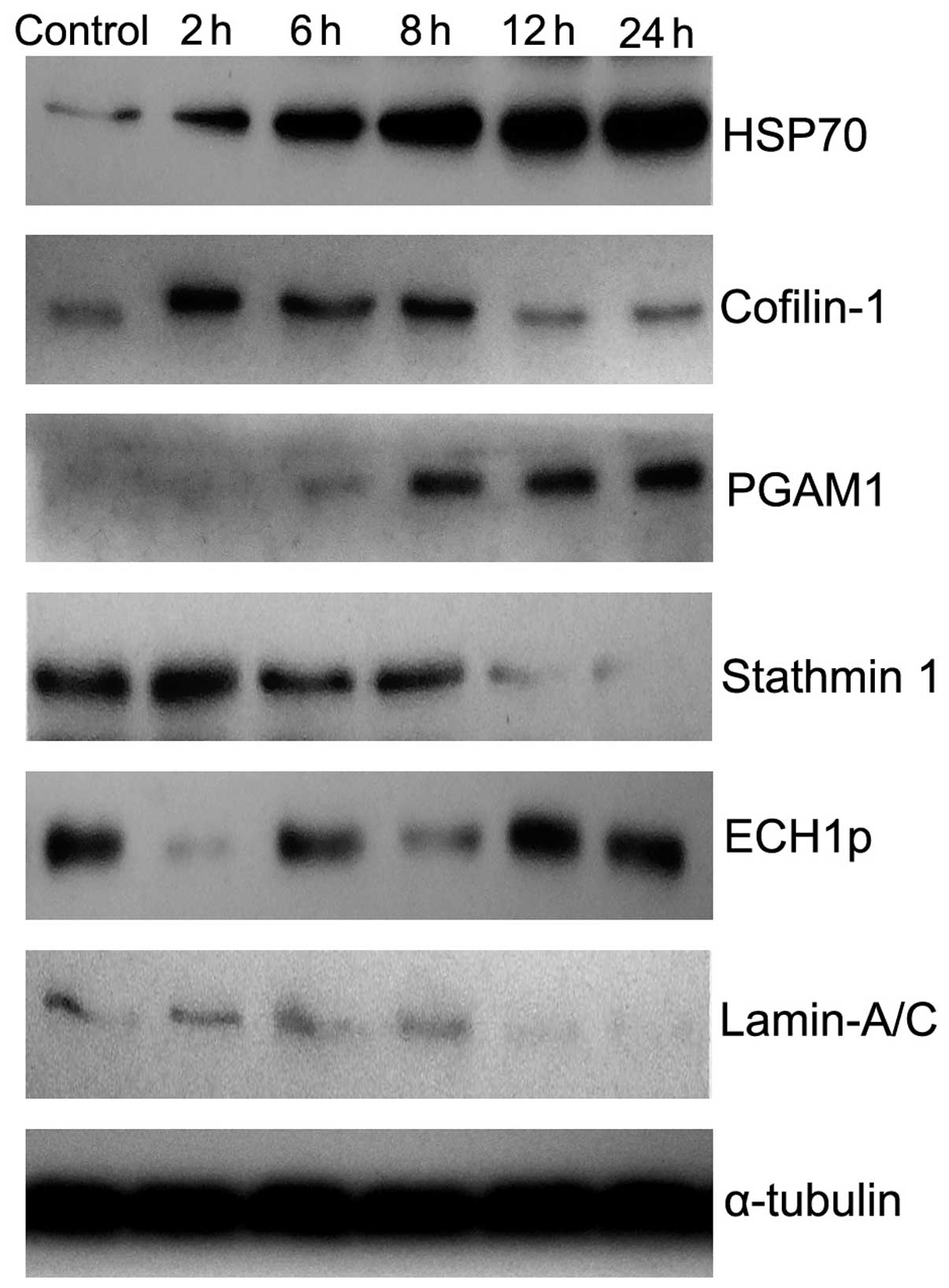
These protein expression profiles were further
confirmed by western blot analysis (Fig. 4). The relative content of these
proteins, following heating for 0, 2, 6, 8, 12 and 24 h, are shown
in Table III and Fig. 3B (P= 0.000). Therefore, the results
demonstrated that protein expression changed in a dynamic manner
during hyperthermia-induced Tca8113 cell apoptosis.
 | Table III.The relative content of
representative proteins following heating for different time
periods. |
Table III.
The relative content of
representative proteins following heating for different time
periods.
| Master no. | 0 h | 2 h | 6 h | 8 h | 12 h | 24 h |
|---|
| 904 | 0.07±0.02 | 0.19±0.05 | 0.69±0.11 | 0.78±0.11 | 0.80±0.15 | 0.99±0.20 |
| 1526 | 0.11±0.03 | 0.73±0.12 | 0.57±0.11 | 0.55±0.13 | 0.21±0.09 | 0.24±0.08 |
| 1310 | 0.02±0.01 | 0.02±0.01 | 0.09±0.02 | 0.54±0.17 | 0.57±0.20 | 0.62±0.23 |
| 1551 | 0.52±0.14 | 0.72±0.19 | 0.31±0.10 | 0.33±0.09 | 0.10±0.02 | 0.08±0.03 |
| 1029 | 0.68±0.17 | 0.07±0.02 | 0.32±0.09 | 0.22±0.07 | 0.56±0.11 | 0.59±0.09 |
| 584 | 0.10±0.02 | 0.13±0.03 | 0.11±0.03 | 0.10±0.03 | 0.03±0.01 | 0.02±0.01 |
Discussion
The present study investigated protein expression
differences in Tca8113 cells following hyperthermia, utilizing
fluorescent differential display 2D gel electrophoresis. Changes in
expression level were observed in a total of 107 proteins (Fig. 2B), and 57 of these proteins were
identified by mass spectrometry analysis. The expression of these
proteins varied over different time periods following hyperthermia
treatment (Tables I and II) according to the following profiles: 27
were upregulated and 5 were downregulated continuously; 8 were
upregulated and 6 were downregulated during the early time period;
and 9 were upregulated and 2 were downregulated during the middle
to late time period. These results demonstrated that the protein
types and expression levels were each upregulated and downregulated
during hyperthermia-induced Tca8113 cell apoptosis. These findings
suggest that hyperthermia-induced Tca8113 cell apoptosis is
controlled by multiple factors, which include time and regulatory
proteins.
Overall, the proteins were grouped into five
functional categories. The first functional category consisted of
energy metabolism-related proteins. This category included the
following proteins: Glyceraldehyde-3-phosphate dehydrogenase
(GAPDH); α-enolase; inorganic pyrophosphatase; galactokinase;
flavin reductase; phosphoglycerate kinase 1;
pyridoxine-5′-phosphate oxidase; ATP synthase subunit d,
mitochondrial; 26S proteasome non-ATPase regulatory subunit 14;
protein disulfide-isomerase A3 (PDIA3); glutathione S-transferase
P; PGAM1; 3-hydroxyacyl-CoA dehydrogenase type-2 isoform 1; crystal
structure of Ufc1, the Ufm1-conjugating enzyme 1, chain A; pyruvate
kinase isozymes M1/M2 (PKM1/2); and peroxiredoxin-4, -6 and -2. The
levels of these energy metabolism-related proteins increased
following hyperthermic induction. By contrast, the levels of the
following energy metabolism-related proteins decreased: ATP
synthase subunit α, mitochondrial; fructose-bisphosphate aldolase A
(ALDOA); mitochondrial succinyl-CoA:3-ketoacid-coenzyme A
transferase 1; D-3-phosphoglycerate dehydrogenase;
argininosuccinate synthase, mitochondrial; Δ(3,5)-Δ(2,4)-dienoyl-CoA isomerase, mitochondrial
(ECH1p); and hydroxysteroid (17-β) dehydrogenase 10 (HSD17B10). The
changes in the expression levels of metabolism-related proteins may
lead to impaired tumor cell substance metabolism and energy
metabolism, which subsequently affect cell survival. Moreover, it
has been demonstrated that certain proteins involved in the
regulation of cell apoptosis (with the exception of metabolic
proteins, such as GAPDH) may be induced to undergo nuclear
translocation by ROS, and that increased GAPDH levels are a
necessary step for apoptosis (30).
For example, PKM2 initiates cell apoptosis by nuclear translocation
through a caspase-independent pathway (31). Furthermore, argininosuccinate
synthase levels correlate with Bcl-2 levels, and drug-induced cell
apoptosis may be enhanced by downregulating argininosuccinate
synthase expression (32). In
addition, PGAM1 is overexpressed in multiple types of tumors and is
involved in tumor formation; it has therefore been suggested that
liver cancer cell apoptosis may be induced by the inhibition of
PGAM1 (33). Moreover, the
increased expression of peroxiredoxin II facilitates tumor cell
survival (34), while peroxiredoxin
IV inhibits radiation-induced tumor cell apoptosis (35). In the present study, the up- or
downregulation of these aforementioned proteases after different
time periods following hyperthermic treatment may have been
involved in the regulation of apoptosis.
Cytoskeleton-related proteins are another of the
five functional categories of proteins. This category includes:
Actin, cytoplasmic 2; F-actin-capping protein subunit β; cofilin-1;
tubulin β chain; keratin, type I cytoskeletal 10 (CK-10); keratin,
type II cytoskeletal 1, 7 and 8 (CK-1,7 and 8); protein 4.1,
isoform 3; and far upstream element-binding protein 1 (FBP1). Such
proteins were upregulated after different time periods following
hyperthermic treatment. The following three cytoskeleton-related
proteins were downregulated: Lamin-A/C; vimentin and stathmin 1
(oncoprotein 18). These cytoskeletal proteins are involved in
formation of the cell cytoskeleton (involving actin, microtubules,
intermediate filaments and the microbeam network) in order to
maintain cellular integrity, which is associated with tumorigenesis
and the regulation of apoptosis (36–40).
Changes in the expression levels of cytoskeleton-related proteins
indicate damage to the cytoskeleton, which consequently affects the
cell morphology and function. Cytoskeletal proteins have also been
demonstrated to be involved in the regulation of apoptosis. Kouzu
et al revealed that CK8 is involved in inhibiting several
types of drug-induced apoptosis (41). Additionally, inhibiting far upstream
element binding protein 1 (FBP1) has been shown to increase
hepatocellular carcinoma (HCC) sensitivity towards apoptotic
stimulation, which therefore inhibits cell proliferation (42). Furthermore, stathmin has been
demonstrated to be downregulated upon the induction of
hyperthermia, which also inhibits cell proliferation (43) and results in stathmin
phosphorylation and dysfunctional microtubule assembly. Overall,
these changes have been demonstrated to induce Jurkat cell
apoptosis (44). The present study
results indicated that within 24 h of hyperthermic induction,
changes in the expression levels of a variety of cytoskeletal
proteins became evident. In particular, changes were observed in
the expression levels of stathmin and vimentin, which are proteins
associated with tumor growth, while the tumor cell invasion and
tumorigenesis dramatically decreased. Therefore, the results
indicated that different cytoskeletal proteins are involved in the
regulation of apoptosis after different time periods following
hyperthermia.
Another of the functional protein categories was
that of chaperones. This category included the following proteins:
HSP70-1; heat shock cognate 71 kDa protein (HSPA8); HSP β-1
(HSPB1/HSP27); PDIA3; T-complex protein 1 subunit ζ; and 60 kDa
HSP, mitochondrial (HSPD1). Such proteins were upregulated
following the induction of hyperthermia. The main function of the
chaperones is to maintain cell survival; HSP family members
regulate mitochondrial pathway-mediated apoptosis and the death
receptor-mediated apoptotic pathway. By blocking the
hyperthermia-induced apoptotic pathway, HSPs are able to
consequently inhibit apoptosis (22–25,28).
In addition, T-complex protein 1 subunit ζ participates in the
assembly of the cytoskeletal proteins actin and tubulin. Moreover,
protein disulfide-isomerase A3 is located in the endoplasmic
reticulum lumen and acts as a component of the
calnexin/calreticulin chaperone complex. Therefore, this protein
plays a significant role in calcium homeostasis and regulates free
Ca2+, which is able to activate related enzymes and
promote apoptosis (15,16). The present study results
demonstrated that HSPs are upregulated within 24 h following
hyperthermia, which may result in the regulation of apoptosis
through different apoptotic signaling pathways. As PDIA3 and
T-complex protein 1 subunit ζ were also upregulated following
hyperthermia, we therefore hypothesized that these two proteins may
participate in the regulation of hyperthermia-induced
apoptosis.
Transcription factors and protein-synthesis related
proteins comprise another of the functional protein categories.
This particular category included the following proteins: Scavenger
mRNA-decapping enzyme DcpS; crystal structure of Ufc1, the
Ufm1-conjugating enzyme 1, chain A; heterogeneous nuclear
ribonucleoprotein H; proteasome activator complex subunit 1;
deoxyuridine 5′-triphosphate nucleotidohydrolase, mitochondrial
(dUTPase); human eukaryotic translation initiation factor 1A
(eIF1A), chain A; and splicing factor U2AF 65 kDa subunit (U2AF65).
These proteins were upregulated after different time periods
following hyperthermic treatment. By contrast, eIF5A-1 and the
crystal structure of the human eIF5A, chain A were downregulated.
It has been confirmed that certain transcription factors and
protein synthesis-related proteins are involved in the regulation
of apoptosis. For example, the overexpression of heterogeneous
nuclear ribonucleoprotein H (hnRNP H) has been shown to partially
counteract apoptosis induced by etoposide, and also to block
mammalian STE20-like protein kinase 2 (MST2; proapoptotic MST2
kinase)-mediated apoptosis in cancer cells (45). In addition, eIF5A is able to
regulate p53-dependent apoptosis by regulating p53 activity in
COS-7 cells (46), while splicing
factor U2AF65 subunit participates in Fas-mediated apoptotic
regulation (47). Overall, these
proteins are up- and downregulated following the induction of
hyperthermia, and thus may contribute to hyperthermia-induced
apoptotic regulation.
The final functional protein category comprised the
cell division- and proliferation-related proteins, including
prohibitin, mitotic checkpoint protein BUB3 and BolA-like protein
2, which were upregulated following hyperthermia treatment.
However, one of the proteins in this category,
proliferation-associated protein 2G4, was downregulated. Prohibitin
is a multifunctional protein that is involved in the following
processes: i) the regulation of cell signaling, apoptosis and
survival (48); ii) the regulation
of cell cycle progression and function as an anti-proliferative
protein that is considered to be a tumor suppressor; and iii) the
inhibition of cell division and tumor growth (49). The present study results indicated
that prohibitin is upregulated continuously following hyperthermia
treatment. As a tumor suppressor, this protein may participate in
the promotion of hyperthermia-induced Tca8113 cell apoptosis.
The present study demonstrated that the expression
levels of 57 proteins were dramatically regulated in Tca8113 cells
within 24 h of hyperthermia treatment. The altered proteins were
grouped within the following functional classes: energy
metabolism-related enzymes, cytoskeleton-related proteins,
chaperones, transcription factors and protein synthesis-related
proteins and cell division- and proliferation-related proteins.
However, no changes were detected in the expression levels of the
hyperthermia-induced apoptosis-related proteins that had been
previously identified to exhibit such changes, such as p53, Bax,
Bak, caspases, cytochrome C (9,17–20),
apoptosis-inducing factor (AIF), Bcl-2 and Mcl-1, which may have
been due to the low expression of such proteins. Moreover, certain
proteins were not detectable by 2D-DIGE. The present study found
that the following pro-apoptotic proteins were upregulated: GAPDH,
PKM2, eIF5A, prohibitin and mitotic checkpoint protein BUB3. In
addition, certain anti-apoptotic proteins were downregulated,
including stathmin, vimentin and argininosuccinate synthase.
Notably, certain anti-apoptotic proteins were upregulated,
including FBP1, hnRNP H, the peroxiredoxins, the HSPs and PGAM1.
The expression of 57 proteins was either upregulated or
downregulated within 24 h of hyperthermic treatment. These findings
indicate that hyperthermia-induced Tca8113 cell apoptosis is
controlled by multiple factors, which include time and regulatory
proteins; however, the underlying mechanisms for these changes
require further investigation.
Acknowledgements
The study was supported in part by the
National Natural Science Foundation of China (grant no. 81160325),
the Ministry of Education Program for New Century Excellent Talents
in University (grant no. NCET-07-0388) and the Project of the
Department of Science and Technology of Yunnan Province (grant no.
2009CC021).
References
|
1.
|
Hildebrandt B and Wust P: The biologic
rationale of hyperthermia. Cancer Treat Res. 134:171–184.
2007.PubMed/NCBI
|
|
2.
|
van der Zee J: Heating the patient: a
promising approach? Ann Oncol. 13:1173–1184. 2002.PubMed/NCBI
|
|
3.
|
Yonezawa M, Otsuka T, Matsui N, et al:
Hyperthermia induces apoptosis in malignant fibrous histiocytoma
cells in vitro. Int J Cancer. 66:347–351. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Dewhirst MW, Vujaskovic Z, Jones E and
Thrall D: Re-setting the biologic rationale for thermal therapy.
Int J Hyperthermia. 21:779–790. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Basile A, Biziato D, Sherbet GV, Comi P
and Cajone F: Hyperthermia inhibits cell proliferation and induces
apoptosis: relative signaling status of P53, S100A4, and Notch in
heat sensitive and resistant cell lines. J Cell Biochem.
103:212–220. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Milleron RS and Bratton SB: ‘Heated’
debates in apoptosis. Cell Mol Life Sci. 64:2329–2333. 2007.
|
|
7.
|
Harmon BV, Takano YS, Winterford CM and
Gobé GC: The role of apoptosis in the response of cells and tumours
to mild hyperthermia. Int J Radiat Biol. 59:489–501. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Moriyama-Gonda N, Igawa M, Shiina H,
Urakami S, Shigeno K and Terashima M: Modulation of heat-induced
cell death in PC-3 prostate cancer cells by the antioxidant
inhibitor diethyldithiocarbamate. BJU Int. 90:317–325. 2002.
View Article : Google Scholar
|
|
9.
|
Shelton SN, Dillard CD and Robertson JD:
Activation of caspase-9, but not caspase-2 or caspase-8, is
essential for heat-induced apoptosis in Jurkat cells. J Biol Chem.
285:40525–40533. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Yoo J, Kim HR and Lee YJ: Hyperthermia
enhances tumour necrosis factor-related apoptosis-inducing ligand
(TRAIL)-induced apoptosis in human cancer cells. Int J
Hyperthermia. 22:713–728. 2006. View Article : Google Scholar
|
|
11.
|
Moulin M, Dumontet C and Arrigo AP:
Sensitization of chronic lymphocytic leukemia cells to
TRAIL-induced apoptosis by hyperthermia. Cancer Lett. 250:117–127.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Tran S, Meinander A, Holmström T, et al:
Heat stress down-regulates FLIP and sensitizes cells to Fas
receptor-mediated apoptosis. Cell Death Differ. 10:1137–1147. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Zhao QL, Fujiwara Y and Kondo T: Mechanism
of cell death induction by nitroxide and hyperthermia. Free Radic
Biol Med. 40:1131–1143. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Katschinski DM, Boos K, Schindler SG and
Fandrey J: Pivotal role of reactive oxygen species as intracellular
mediators of hyperthermia-induced apoptosis. J Biol Chem.
275:21094–21098. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Ahmed K, Zhao QL, Matsuya Y, et al:
Enhancement of macrosphelide-induced apoptosis by mild
hyperthermia. Int J Hyperthermia. 23:353–361. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Hashimoto T, Shibata M, Ito Y, Nakao KI,
Sasaki S and Otsuki Y: Elevated levels of intracellular
Ca2+ and apoptosis in human lung cancer cells given
heat-shock. Int J Hyperthermia. 19:178–192. 2003.
|
|
17.
|
Klostergaard J, Leroux ME, Auzenne E, et
al: Hyperthermia engages the intrinsic apoptotic pathway by
enhancing upstream caspase activation to overcome apoptotic
resistance in MCF-7 breast adenocarcinoma cells. J Cell Biochem.
98:356–369. 2006. View Article : Google Scholar
|
|
18.
|
Pagliari LJ, Kuwana T, Bonzon C, et al:
The multidomain proapoptotic molecules Bax and Bak are directly
activated by heat. Proc Natl Acad Sci USA. 102:17975–17980. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Shellman YG, Howe WR, Miller LA, et al:
Hyperthermia induces endoplasmic reticulum-mediated apoptosis in
melanoma and non-melanoma skin cancer cells. J Invest Dermatol.
128:949–956. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Bonzon C, Bouchier-Hayes L, Pagliari LJ,
Green DR and Newmeyer DD: Caspase-2-induced apoptosis requires bid
cleavage: a physiological role for bid in heat shock-induced death.
Mol Biol Cell. 17:2150–2157. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Beere HM: Death versus survival:
functional interaction between the apoptotic and stress-inducible
heat shock protein pathways. J Clin Invest. 115:2633–2639. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Banerjee Mustafi S, Chakraborty PK, Dey RS
and Raha S: Heat stress upregulates chaperone heat shock protein 70
and antioxidant manganese superoxide dismutase through reactive
oxygen species (ROS), p38MAPK, and Akt. Cell Stress Chaperones.
14:579–589. 2009.
|
|
23.
|
Shackley DC, Haylett A, Whitehurst C, et
al: Comparison of the cellular molecular stress responses after
treatments used in bladder cancer. BJU Int. 90:924–932. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Li GC and Calderwood SK: Hyperthermia
classic article commentary: ‘Re-induction of hsp70 synthesis: an
assay for thermotolerance’ by Gloria C. Li and Johnson Y. Mak,
International Journal of Hyperthermia 1989;5:389–403. Int J
Hyperthermia. 25:258–261. 2009.
|
|
25.
|
Watanabe N, Tsuji N, Akiyama S, et al:
Induction of heat shock protein 72 synthesis by endogenous tumor
necrosis factor via enhancement of the heat shock element-binding
activity of heat shock factor 1. Eur J Immunol. 27:2830–2834. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Strasser A and Anderson RL: Bcl-2 and
thermotolerance cooperate in cell survival. Cell Growth Differ.
6:799–805. 1995.PubMed/NCBI
|
|
27.
|
Stankiewicz AR, Livingstone AM, Mohseni N
and Mosser DD: Regulation of heat-induced apoptosis by Mcl-1
degradation and its inhibition by Hsp70. Cell Death Differ.
16:638–647. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Jiang W, Bian L, Ma LJ, Tang RZ, Xun S and
He YW: Hyperthermia-induced apoptosis in Tca8113 cells is inhibited
by heat shock protein 27 through blocking phospholipid scramblase 3
phosphorylation. Int J Hyperthermia. 26:523–537. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Tannu NS and Hemby SE: Two-dimensional
fluorescence difference gel electrophoresis for comparative
proteomics profiling. Nat Protoc. 1:1732–1742. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Hara MR, Agrawal N, Kim SF, et al:
S-nitrosylated GAPDH initiates apoptotic cell death by nuclear
translocation following Siah1 binding. Nat Cell Biol. 7:665–674.
2005. View
Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Steták A, Veress R, Ovádi J, Csermely P,
Kéri G and Ullrich A: Nuclear translocation of the tumor marker
pyruvate kinase M2 induces programmed cell death. Cancer Res.
67:1602–1608. 2007.PubMed/NCBI
|
|
32.
|
Goodwin BL, Solomonson LP and Eichler DC:
Argininosuccinate synthase expression is required to maintain
nitric oxide production and cell viability in aortic endothelial
cells. J Biol Chem. 279:18353–18360. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Ren F, Wu H, Lei Y, et al: Quantitative
proteomics identification of phosphoglycerate mutase 1 as a novel
therapeutic target in hepatocellular carcinoma. Mol Cancer.
9:812010. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Lee KW, Lee DJ, Lee JY, Kang DH, Kwon J
and Kang SW: Peroxiredoxin II restrains DNA damage-induced death in
cancer cells by positively regulating JNK-dependent DNA repair. J
Biol Chem. 286:8394–8404. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Park JJ, Chang HW, Jeong EJ, et al:
Peroxiredoxin IV protects cells from radiation-induced apoptosis in
head-and-neck squamous cell carcinoma. Int J Radiat Oncol Biol
Phys. 73:1196–1202. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Lundkvist A, Reichenbach A, Betsholtz C,
Carmeliet P, Wolburg H and Pekny M: Under stress, the absence of
intermediate filaments from Müller cells in the retina has
structural and functional consequences. J Cell Sci. 117:3481–3488.
2004.PubMed/NCBI
|
|
37.
|
Lau AT and Chiu JF: The possible role of
cytokeratin 8 in cadmium-induced adaptation and carcinogenesis.
Cancer Res. 67:2107–2113. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Bhat KM and Setaluri V:
Microtubule-associated proteins as targets in cancer chemotherapy.
Clin Cancer Res. 13:2849–2854. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Prasad S, Soldatenkov Va, Srinivasarao G
and Dritschilo A: Intermediate filament proteins during
carcinogenesis and apoptosis (Review). Int J Oncol. 14:563–570.
1999.PubMed/NCBI
|
|
40.
|
Rana S, Maples PB, Senzer N and Nemunaitis
J: Stathmin 1: a novel therapeutic target for anticancer activity.
Expert Rev Anticancer Ther. 8:1461–1470. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Kouzu Y, Uzawa K, Koike H, et al:
Overexpression of stathmin in oral squamous-cell carcinoma:
correlation with tumour progression and poor prognosis. Br J
Cancer. 94:717–723. 2006.PubMed/NCBI
|
|
42.
|
Nakamura K, Zhang X, Kuramitsu Y, et al:
Analysis on heat stress-induced hyperphosphorylation of stathmin at
serine 37 in Jurkat cells by means of two-dimensional gel
electrophoresis and tandem mass spectrometry. J Chromatogr A.
1106:181–189. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
43.
|
Cajone F and Sherbet GV: Stathmin is
involved in S100A4-mediated regulation of cell cycle progression.
Clin Exp Metastasis. 17:865–871. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
44.
|
Rabenhorst U, Beinoraviciute-Kellner R,
Brezniceanu ML, et al: Overexpression of the far upstream element
binding protein 1 in hepatocellular carcinoma is required for tumor
growth. Hepatology. 50:1121–1129. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45.
|
Rauch J, O’Neill E, Mack B, et al:
Heterogeneous nuclear ribonucleoprotein H blocks MST2-mediated
apoptosis in cancer cells by regulating A-Raf transcription. Cancer
Res. 70:1679–1688. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46.
|
Li AL, Li HY, Jin BF, et al: A novel eIF5A
complex functions as a regulator of p53 and p53-dependent
apoptosis. J Biol Chem. 279:49251–49258. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
47.
|
Izquierdo JM: Fas splicing regulation
during early apoptosis is linked to caspase-mediated cleavage of
U2AF65. Mol Biol Cell. 19:3299–3307. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
48.
|
Zhu B, Zhai J, Zhu H and Kyprianou N:
Prohibitin regulates TGF-beta induced apoptosis as a downstream
effector of Smad-dependent and -independent signaling. Prostate.
70:17–26. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49.
|
Wang S, Nath N, Adlam M and Chellappan S:
Prohibitin, a potential tumor suppressor, interacts with RB and
regulates E2F function. Oncogene. 18:3501–3510. 1999. View Article : Google Scholar : PubMed/NCBI
|