Introduction
Human males absent on the first (hMOF), also known
as Moz-Ybf2/Sas3-Sas2-Tip60 1 (MYST1), is the human ortholog of the
Drosophila MOF protein. The protein contains a chromodomain
and acetyl-CoA binding motif, which is one of the key components of
the male specific lethal (dMSL) complex (1). Biochemical purifications have revealed
that hMOF forms at least two distinct multiprotein complexes in
mammalian cells. The hMSL complex is responsible for the majority
of histone H4 acetylation at lysine (K) 16 (2,3), while
the hNSL (non-specific lethal) complex reveals a broad substrate
specificity, which is able to acetylate histone H4 at K5, 8 and 16
(4). Although the functions of the
two complexes in human cells are unclear, their ability to
acetylate histone H4 at K16 indicates the significance of H4K16
acetylation in cells. It has been reported that the depletion of
hMOF leads to a global reduction in histone H4K16 acetylation in
human cells, in addition to genomic instability, spontaneous
chromosomal aberrations, cell cycle defects, reduced transcription
of certain genes, defective DNA damage repair and early embryonic
lethality (4–9). This suggests a critical role for hMOF
in fundamental processes, including gene transcription, cell
proliferation, differentiation and the DNA repair response.
Ovarian cancer is the most common cause of
cancer-related mortality from gynecological tumors (10). Due to difficulties in early
detection, the majority of ovarian cancers are diagnosed at
advanced stages. Studies have identified various biomarkers for
diagnosing and predicting the prognosis of ovarian cancer,
including the potential tumor hypoxic marker, hypoxia inducible
factor-1α (HIF1α), and its regulated genes, vascular endothelial
growth factor (VEGF), carbonate anhydrate IX (CA9) and
stanniocalcin 1 (STC1) (11–13).
However, the markers are not specific or sensitive enough to
accurately predict the survival of patients with ovarian cancer
(14,15). Studies have indicated that
epigenetic alterations play a significant role in carcinogenesis
and more recent studies have involved the use of global histone
modifications as predictors of cancer recurrence in various tumor
entities (16,22–24).
Although the role of hMOF and its corresponding modification in
transcription regulation is not completely understood, the abnormal
expression of H4K16 has been identified in a number of primary
cancer tissues. The expressional behavior of hMOF varies in
different primary cancers. Frequent downregulation of hMOF
expression has been identified in primary breast cancer and
medulloblastoma (17). In contrast,
hMOF was demonstrated to be overexpressed in non-small cell lung
carcinoma tissues (25). In a
previous study, we also showed that hMOF gene expression levels
were frequently downregulated in renal cell carcinoma (RCC)
(21). hMOF protein expression
correlates with histone H4K16 acetylation. The previous
observations strongly suggest that the histone acetyltransferase
(HAT), hMOF, and the corresponding histone H4K16 acetylation may be
involved in tumorigenesis. However, little is known with regard to
the role of hMOF and its corresponding modification in ovarian
carcinomas. The present study examined hMOF mRNA and protein
expression levels in primary ovarian carcinomas using quantitative
polymerase chain reaction (qPCR), western blotting and
immunohistochemistry. Furthermore, the mRNA expression levels of
the hMOF-regulated non-protein coding human leukocyte antigen (HLA)
complex P5 (HCP5) were examined in ovarian cancer tissues.
Materials and methods
Antibodies and tissue collection
Anti-H4K16Ac (H9164) and anti-MYST1 (SAB4503328)
polyclonal antibodies were purchased from Sigma-Aldrich (St Louis,
MO, USA). Rabbit polyclonal anti-GAPDH and anti-hMOF were raised
against bacterially-expressed proteins (Jilin University,
Changchun, Jilin, China). Human clinical ovarian cancer and normal
tissues were collected from patients with primary ovarian cancer,
who underwent radical ovarian tumor surgery at The First Clinical
Hospital of Jilin University (Changchun, Jilin, China) between
January 2010 and July 2012. Approval for the study was obtained
from the Ethics Committee of The First Clinical Hospital of Jilin
University and all patients provided their informed consent. All
the tissues that were removed during the surgery were frozen
immediately in liquid nitrogen and stored at −80°C. Patient medical
records, including tumor staging, pathological diagnosis and
surgical records, were reviewed. No patients were administered
chemotherapy or radiotherapy prior to surgery.
PCR
Total RNA from ovarian cancer and normal tissues
were isolated using TRIzol® LS Reagent (Invitrogen,
Carlsbad, CA, USA). A total of 1 μg RNA from each sample was
used as a template to produce cDNA using the PrimeScript 1st Strand
cDNA Synthesis kit (Takara, Shiga, Japan). hMOF, CA9, VEGF, HIF1α,
hSTC1 and GAPDH mRNA levels were analyzed by PCR using the C1000™
Thermal Cycler (Bio-Rad, Hercules, CA, USA) and by qPCR using Real
Time PCR Detector Chromo 4 (Bio-Rad). All PCR reactions were
performed under the following conditions: An initial denaturation
step at 95°C for 3 min, followed by 35 cycles of denaturation at
95°C for 30 sec, annealing at 60°C for 30 sec and extension at 72°C
for 30 sec. The primer sets that were used for the PCR were: GAPDH
forward, 5′-ATCACTGCCACCCAGAAGAC-3′ and reverse,
5′-ATGAGGTCCACCACCCTGTT-3′, yielding a 460 bp product; hMOF
forward, 5′-GGC TGGACGAGTGGGTAGACAA-3′ and reverse, 5′-TGGTGA
TCGCCTCATGCTCCTT-3′, yielding a 227 bp product; CA9 for wa rd, 5′-
G CAG GAG GAT TCCCCCT TG-3′ and reverse,
5′-GGAGCCTCAACAGTAGGTAGAT-3′, yielding a 185 bp product; VEGF
forward, 5′-GAACTT TCTGCTGTCTTGGGTGCAT-3′ and reverse, 5′-GGTCTG
CATTCACATTTGTTGTGCTG-3′, with a 392 bp product; HIF1α forward,
5′-GCACAGGCCACATTCACG-3′ and reverse,
5′-TGAAGATTCAACCGGTTTAAGGA-3′, yielding a 520 bp product; hSTC1
forward, 5′-CAC ACCCACGAGCTGACTTC-3′ and reverse, 5′-TTATGCACT
CTCATGGGATGTGCG-3′, yielding a 130 bp product; and HCP5 forward,
5′-GACTCTCCTACTGGTGCTTGGT-3′ and reverse,
5′-CACTGCCTGGTGAGCCTGTT-3′, yielding a 240 bp product.
Western blotting
Ovarian cancer or normal tissue samples (200 mg)
were homogenized with liquid nitrogen and solubilized in 200
μl cold PBS containing 1.0% Nonidet P-40, 0.5%
Na-deoxycholate, 0.1% SDS, 0.05 mM PMSF and a protease inhibitor
cocktail. The homogenate was swirled and kept on ice for 30 min.
Whole cell extracts were prepared by sonication (Scientz-IID;
Ningbo Scientz Biotechnology Co., Ltd., Ningbo, China) for 10 sec
with 50% duty cycle and centrifugation at 13,400 x g for 30 min.
The amount of total proteins in the resulting supernatant was
measured according to the instructions from the Bio-Rad Protein
Assay kit (500-0201). Equal total amounts of the denatured proteins
were separated using 12% SDS polyacrylamide gel electrophoresis
(SDS-PAGE). The specific proteins were detected by immunoblotting
using hMOF, H4K16Ac and GAPDH polyclonal antibodies.
Immunohistochemical staining
Formalin-fixed and paraffin-embedded ovarian cancer
tissue blocks were obtained from The First Clinical Hospital of
Jilin University. The tissue blocks were sectioned and
deparaffinized in xylene and rehydrated through a graded ethanol
series. The tissue slides were then subjected to antigen retrieval
by boiling in 0.01 M sodium citrate buffer (pH 6) in a microwave
oven for 10 min. Endogenous peroxidase activity was blocked by
incubation for 10 min in 3% hydrogen peroxide in methanol. Finally,
the reactions were detected using the DAB detection kit (Dako,
Carpinteria, CA, USA). Anti-MYST1 (SAB4503328) and acetylated H4K16
polyclonal antibodies (H9164) were used at a 1:500 dilution. The
MYST1 protein expression status and the histone H4K16 acetylation
levels were estimated in a four-step scale (19).
RNAi treatment and DNA microarray
HeLa cells were cultured in 6-well tissue culture
plates (∼2×105 cells/well) in DMEM medium (Sigma)
containing 5% glucose and 10% fetal bovine serum. The cells were
transfected with 20 nM hMOF siRNA (D-014800, Dharmacon, Lafayette,
CO, USA) and non-targeting siRNA (D-001206, Dharmacon). At 24 h
post-transfection, the cells were split into new 6-well plates for
immunoblotting, PCR and DNA microarray analysis. Subsequent to 24
h, the cells were harvested and lysed. Whole cell extracts were
prepared from two wells of a 6 well plate by adding 4 × SDS sample
buffer, and total RNA was isolated from one well of a 6 well plate
using TRIzol® LS Reagent (Invitrogen). In addition,
cells from one well of the 6-well plate were rinsed twice with warm
PBS and harvested. The cells were then stored in an RNA hold
solution (ER501-01; Beijing Transgen Biotech Co., Ltd., Beijing,
China) and sent to OneArray by Phalanx Biotech Group (Belmont, CA,
USA) for DNA microarray analysis.
Chromatin immunoprecipitation (ChIP)
One or two 10-cm dishes containing ∼1×107
HeLa cells grown to ∼80% confluence were used for each ChIP. The
cells were cross-linked with 5 ml 1% formaldehyde in PBS for 15 min
at room temperature, followed by incubation with 125 mM glycine for
5 min. In order to shear the DNA to lengths of 200–1,000 base
pairs, the cell lysates were sonicated using a Scientz-IID for 5×60
sec, with a one sec interval between each round, at a setting of
45% duty, level 2. Equal amounts of sonicated chromatin from each
sample were incubated at 4°C overnight with 5–10 μg of
antibodies against H4K16Ac (H9164, Sigma) or hMOF rabbit polyclonal
antibodies. Total rabbit IgG (sc-2027, Santa Cruz Biotechnology,
Inc., Santa Cruz, CA, USA) and pre-immune serum were used as
controls. Following 24 h of incubation, 50 μl protein A
agarose containing salmon sperm DNA (10 μg) and BSA (25
μg; 50% Slurry) were added to the mixture and further
incubated for 2.5 h at 4°C to collect the agarose/antibody/protein
complex. The protein A agarose/antibody/protein complex was washed
for 5 min on a rotating platform with 1 ml buffer according to the
order of Low Salt- (0.1% SDS, 1% Triton-100, 2 mM EDTA, 20 mM Tris,
pH 8.0 and 150 mM NaCl) High Salt- (0.1% SDS, 1% Triton-100, 2 mM
EDTA, 20 mM Tris, pH 8.0 and 500 mM NaCl) LiCl- (0.25 M LiCl, 1%
NP-40, 1% NaDOC, 1 mM EDTA and 10 mM Tris, pH 8.0) TE- (10 mM Tris,
pH 8.0 and 1 mM EDTA) TE (10 mM Tris, pH 8.0 and 1 mM EDTA).
Finally, the washed beads were eluted using a 480 μl elution
buffer containing 0.1 M NaHCO3 and 1% SDS. DNA was
extracted by phenol/chloroform and precipitated using ethanol.
Amplification of 1–2 μl immunoprecipitated or 100-fold
diluted input DNA was performed using Real Time PCR Detector Chromo
4 (Bio-Rad). Each experiment was performed independently, 2–3
times. All ChIP signals were normalized to the total input. The
primer sets of the HCP5 promoter (−142 to +50) were forward,
5′-TCCACCTTTCCCAACCTGTGTC-3′ and reverse,
5′-GGACTCCATGACCCGCAACC-3′.
Statistical analysis
The gene expression signals on the 2% agarose gel
and western blot images were scanned and quantified with Quantity
One Basic software (Bio-Rad). The differences in the expression of
genes and proteins between ovarian cancer and normal tissues were
statistically analyzed. Statistical analysis was completed with
SPSS 17.0 (SPSS, Inc., Chicago IL, USA). Statistical comparisons
were analyzed using the student’s t-test. P<0.05 was considered
to indicate a statistically significant difference.
Results
A reduction in hMOF mRNA expression
levels is observed in ovarian cancer tissues
In order to identify the expression levels of
several cancer-related genes in the pathogenesis of primary ovarian
cancer, the present study examined the mRNA levels of hMOF, CA9,
VEGF, HIF1α and hSTC1 in 47 patients with pathologically-diagnosed
ovarian cancer using PCR. Fig. 1A
displays a section of the PCR results (n=30). Compared with the
contralateral normal ovarian tissues or the normal ovarian tissues
from an alternative patient, the mRNA expression levels of hMOF,
CA9, VEGF, HIF1α and hSTC1 in the ovarian cancer tissues revealed
varying behaviors on the DNA agarose gel. Among them, the hMOF gene
expression levels showed a decreasing tendency in the ovarian
cancer tissues. The quantified mRNA levels are shown in Fig. 1C. The gene expression levels of hMOF
and HIF1α were markedly decreased in the ovarian cancer tissues
compared with the normal tissues (P<0.01 and P<0.05,
respectively). However, no significant differences were observed in
VEGF, CA9 and hSTC1 expression. An analysis of mRNA expression in
47 samples revealed the downregulation of hMOF mRNA in 81% (38/47)
of patients, whereas only 13% (6/47) showed upregulation and 6%
(3/47) showed no change (Fig. 1B).
In contrast with hMOF, the expression levels of VEGF were
upregulated in more than half of the cases (27/47, 57%), while only
17% (8/47) of patients exhibited downregulation and 26% (12/47)
exhibited no change. Although HIF1α mRNA expression was
downregulated in 40% (19/47) of patients, the majority of the cases
did not change (26/47, 55%). Almost half of the patients (23/47,
49%) revealed no changes in the mRNA expression levels of hSTC1.
The percentage change between the upregulation and downregulation
was 30% (14/47) and 21% (10/47), respectively. There were no
significant changes in the number of cases of varying CA9
expression. The percentage changes in the upregulation,
downregulation or no change in CA9 mRNA expression were 38%, 34%
and 28%, respectively.
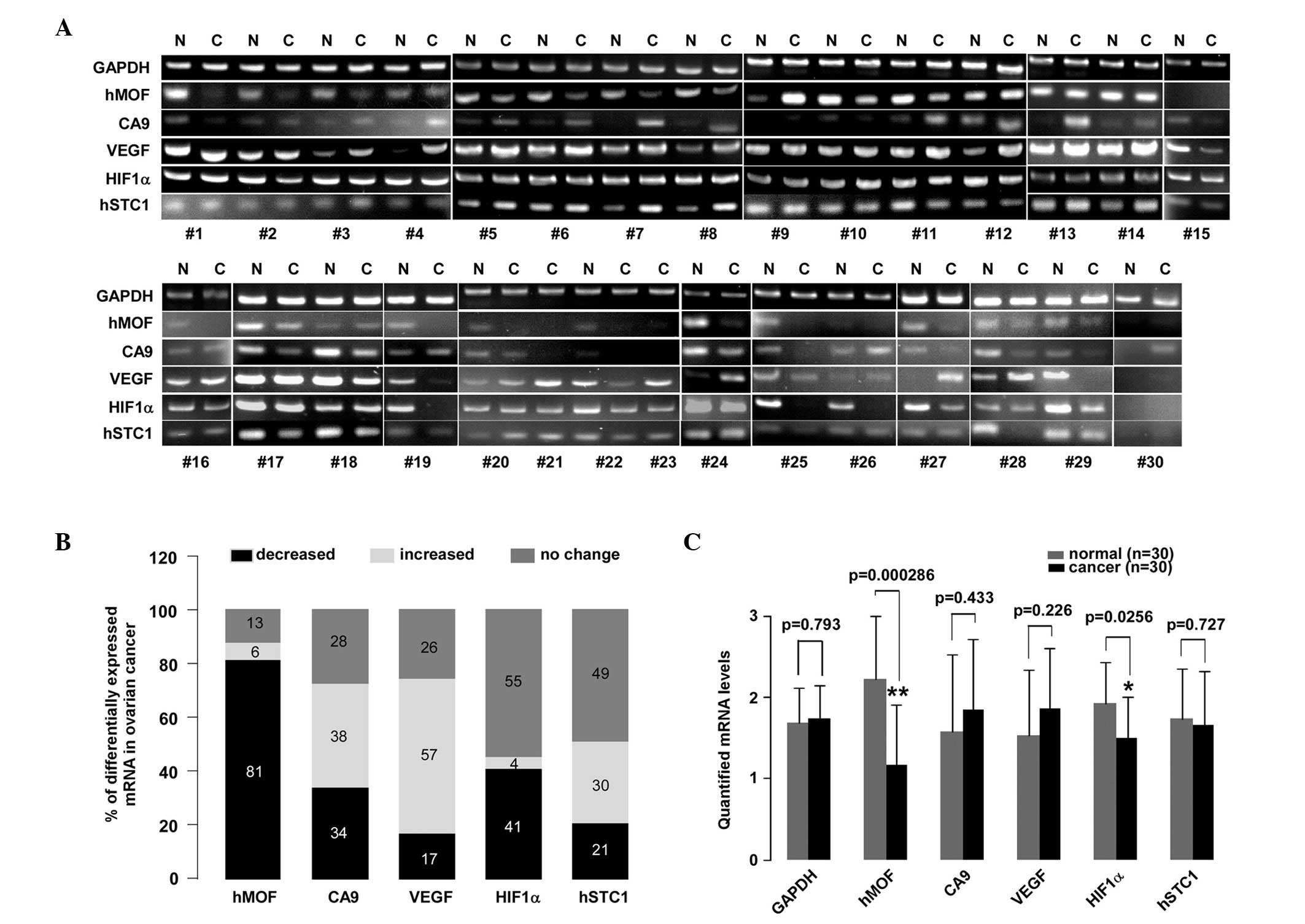 | Figure 1.A reduction in hMOF mRNA levels is
observed in human ovarian cancer. (A) PCR analysis of 47 clinical
ovarian cancer tissues. Total RNA was isolated from the tissues
using TRIzol. The PCR assay was performed to detect the mRNA
expression levels of hMOF, CA9, VEGF, HIF1α and hSTC1 in clinical
ovarian cancer and normal ovarian tissues. The PCR products were
then separated by electrophoresis on a 2% agarose gel. The DNA
fragments were visualized and photographed under ultraviolet light
with ethidium bromide. The mRNA levels from 37 ovarian cancer
tissues were compared with corresponding contralateral ovarian
normal tissues. However, 10 clinical ovarian cancer tissues were
missing contralateral ovarian normal tissues and were compared with
non-corresponding normal ovarian tissues. (B) Summarization of the
PCR results. The 100% stacked column charts were used to compare
the case numbers of differentially-expressed mRNAs in the ovarian
cancer tissues. The total case numbers of differentially-expressed
mRNAs (increased, decreased and no change) in the ovarian cancer
tissues is equal to 100%. (C) Statistical analysis of quantified
mRNA levels between the ovarian cancer and normal tissues. The mRNA
expression signals shown in (A) were quantified by densitometry
using Quantity One Basic Software. The significant difference is
expressed as *P<0.05, **P<0.01. hMOF,
human males absent on the first; PCR, polymerase chain reaction;
CA9, carbonate anhydrate IX; VEGF, vascular endothelial growth
factor; HIF1α, hypoxia-inducible factor-1α; hSTC1, human
stanniocalcin 1; N, normal tissue; C, cancer tissue. |
Frequent downregulation of hMOF in
ovarian cancer tissues is confirmed using qPCR
The results of the PCR analysis clearly revealed a
downregulation of hMOF gene expression in ovarian cancer. To
further validate the frequent downregulation of hMOF mRNA
expression in primary ovarian cancer, an additional 57 clinically
diagnosed ovarian cancer tissues and 15 normal tissues were used in
this experiment (8/15 cases were contralateral normal ovarian
tissues). The expression levels of hMOF were measured using qPCR.
As shown in Fig. 2A, the analysis
of mRNA expression in the 57 samples revealed a significant
(>2-fold decreased) downregulation of hMOF mRNA in 65% (37/57)
of patients, whereas 11% (6/57) of patients showed significant
(>2-fold increased) upregulation of hMOF. In addition, a
<2-fold reduction of hMOF was observed in 11% (6/57) of
patients. In contrast, 12% (7/57) of patients showed a <2-fold
elevation of hMOF. The significant differences in hMOF expression
between ovarian cancer and normal tissues were analyzed using
Student’s t-test. As shown in Fig.
2B, the hMOF expression levels were significantly reduced in
ovarian cancer (P<0.01). In order to determine whether the
reduction of hMOF expression resulted in decreased hMOF protein
levels, four randomly selected ovarian cancer tissues and matched
contralateral normal ovarian tissues were used (Fig. 2B). As shown in the lower panel,
aliquots of whole cell extract from the tissues were analyzed by
western blotting using the indicated antibodies. As expected, the
level of hMOF protein expression was decreased in the ovarian
cancer tissues compared with the matched normal tissues.
Simultaneously, the acetylation status of histone H4K16 was also
significantly reduced or lost in the ovarian cancer samples. To
further confirm this result, immunohistochemical staining was
performed for hMOF and histone H4K16 acetylation in the
formalin-fixed, paraffin-embedded tissue sections from 20 patients.
However, the immunohistochemical staining of certain cases failed.
Therefore, no immunohistochemistry analysis data are presented. As
an example, high or low immunohistochemical staining for hMOF and
H4K16Ac in ovarian cancer is shown in Fig. 2D.
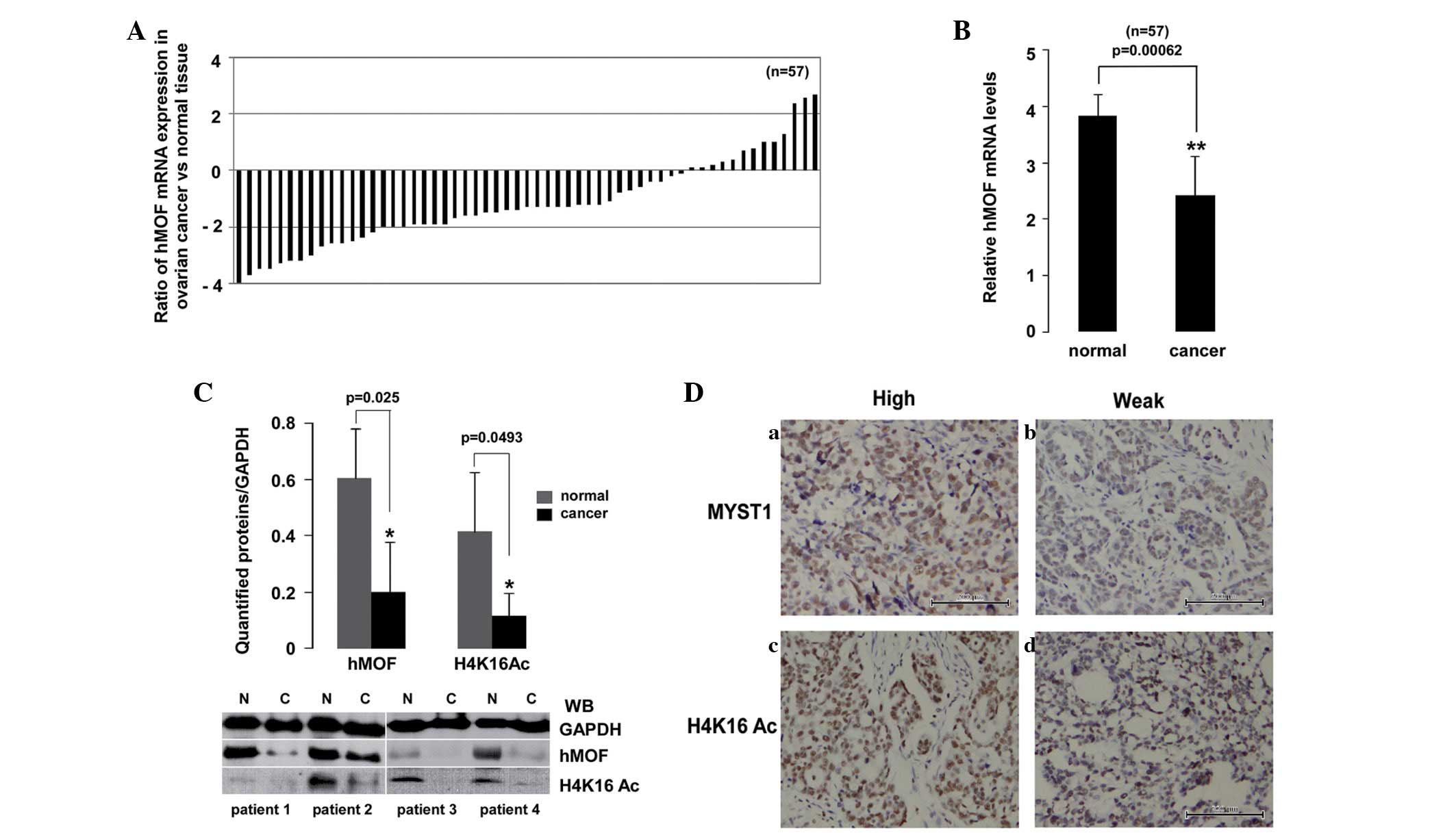 | Figure 2.Downregulation of hMOF mRNA expression
in ovarian cancer tissues confirmed using qPCR and immunoblotting.
(A) Expression patterns of hMOF in clinical ovarian cancer tissues.
Total RNA was isolated from 57 clinical ovarian cancer and 15
normal tissues (8/15 cases were the contralateral normal tissues).
The relative mRNA expression levels of hMOF were analyzed using
qPCR. Expression is displayed as a ratio of hMOF gene expression in
the ovarian cancer versus normal tissues. Each bar is the log2
value of the ratio of hMOF expression levels between the ovarian
cancer and normal tissues. Bar value >1 represents >2-fold
increase, whereas bar value <1, represents >2-fold decrease.
(B) Statistical analysis of the qPCR data. Each bar represents the
mean of three independent experiments. The significant difference
is expressed as **P<0.01. (C) Four randomly selected,
pathologically diagnosed ovarian cancer and contralateral normal
tissues from the same patients were used. A whole cell extract was
prepared from the tissues and equivalent total protein amounts were
subjected to SDS-PAGE in 12% gels. Proteins were detected by
western blotting with anti-hMOF, H4K16Ac and GAPDH antibodies
(lower panel). Western blotting images were quantified using
Quantity One software and normalized by GAPDH levels. The
significant difference is expressed as *P<0.05. (D)
Immunohistochemical staining for (a and b) hMOF and (c and d)
H4K16Ac in ovarian cancer tissues (×200). (a and c) High or (b and
d) low hMYST1 and H4K16Ac expression levels in ovarian cancer
tissues. hMOF, human males absent on the first; qPCR, quantitative
polymerase chain reaction; H4K16, histone H4 lysine 16; MYST1,
Moz-Ybf2/Sas3-Sas2-Tip60 1; N, normal tissue; C, cancer tissue. |
HCP5, an hMOF-regulated gene, is also
frequently downregulated in ovarian cancer tissues
In order to confirm that hMOF was downregulated in
ovarian cancer, hMOF target genes were screened from gene
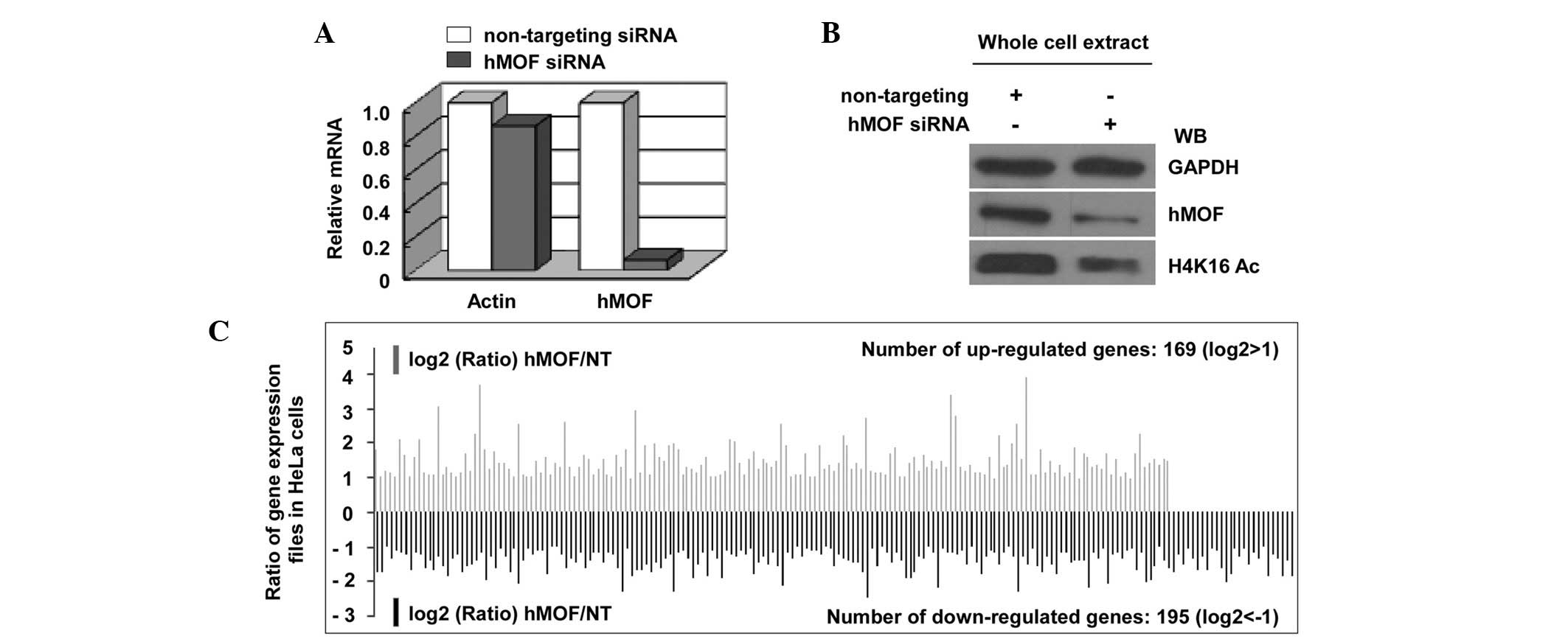
expression profiles in hMOF siRNA knockdown HeLa cells. The effect
of siRNA interference was confirmed using qPCR and immunoblotting
(Fig. 3A and B). As a result of the
DNA microarray analysis by the Phalanx Human OneArray™ (HOA 5.2:
Phalanx Biotech Group, Inc.), a total of 364 genes were shown to be
differentially-expressed between the hMOF siRNA knockdown and
non-targeting (NT) siRNA-treated HeLa cells, of which 169 genes
were upregulated and 195 genes were downregulated by >2-fold
(Fig. 3C; Table I). HCP5 was confirmed as one of the
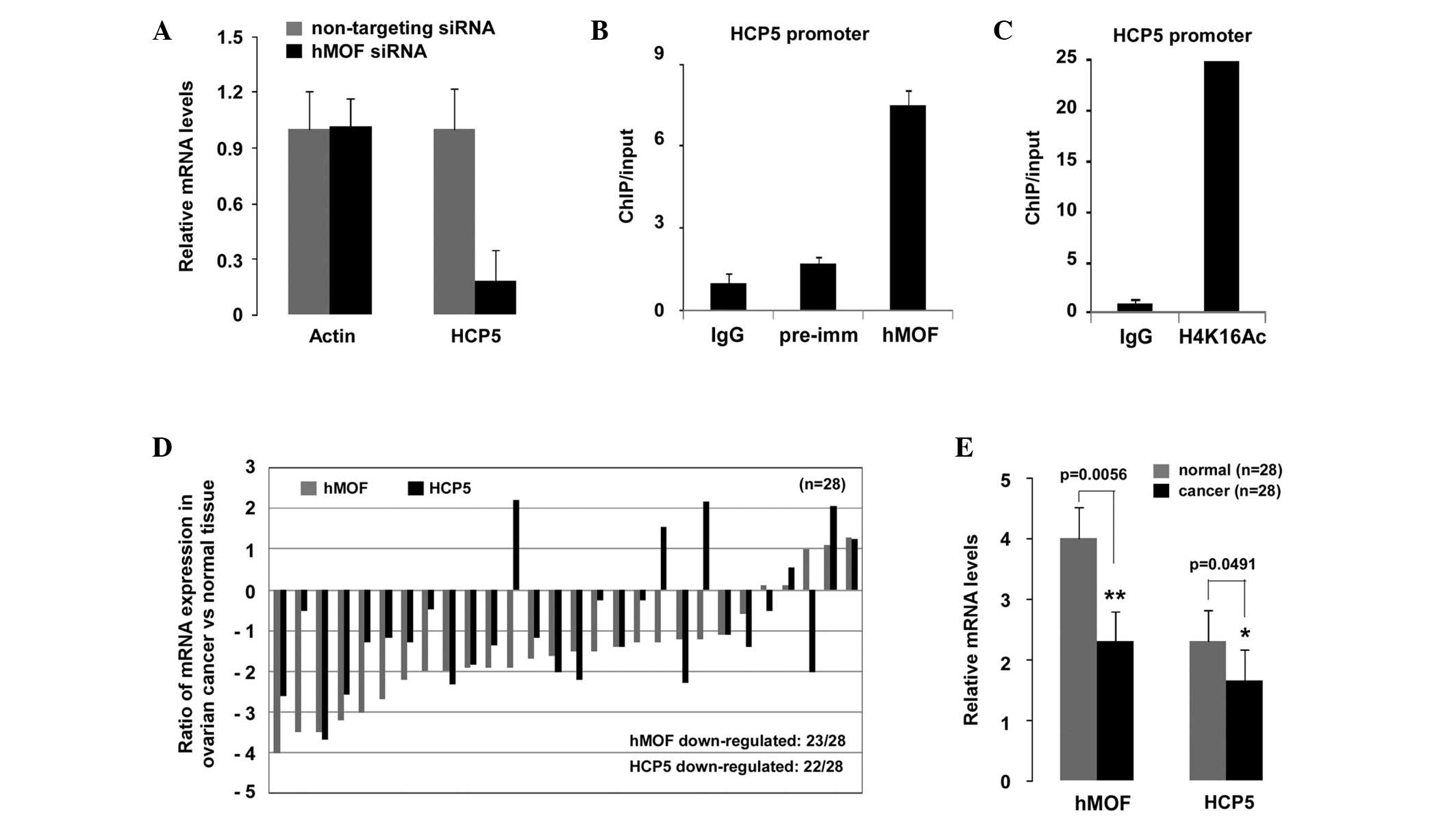
hMOF downregulated genes by qPCR and ChIP. As shown in Fig. 4A, the relative mRNA levels of HCP5
in the hMOF siRNA knockdown HeLa cells were markedly decreased
compared with those of the NT siRNA-treated cells. To determine
whether hMOF was recruited to the HCP5 promoter, ChIP experiments
were performed using chromatin from NT siRNA or hMOF siRNA
knockdown HeLa cells. As expected, hMOF-specific and histone
H4K16Ac antibodies precipitated a DNA fragment encompassing the
HCP5 promoter (Fig. 4B and C),
suggesting that HCP5 was an hMOF target gene. To further
investigate whether the mRNA expression levels of HCP5 were also
affected in hMOF-downregulated human ovarian cancer tissues, 28
clinically diagnosed ovarian cancer tissues were selected. The
total mRNA expression levels of hMOF and HCP5 were observed to be
significantly decreased compared with those of the normal tissues
(P<0.01 and P<0.05, respectively; Fig. 4E). The results are shown in Fig. 4D, where 23 of the 28 patients showed
a downregulation of hMOF, among which, 20 (87%) also exhibited a
reduction in HCP5 expression.
 | Table I.Downregulated gene expression profiles
in hMOF siRNA knockdown HeLa cells. |
Table I.
Downregulated gene expression profiles
in hMOF siRNA knockdown HeLa cells.
| Gene symbol | Gene ID No. | Gene description | Fold-change |
|---|
| ANKRD33B | NM_001164440 | Ankyrin repeat domain
33B | −3.11 |
| PSMB 10 | NM_002801 | Proteasome subunit, β
type, 10 | −3.18 |
| ANKRD2 | NM_020349 | Ankyrin repeat domain
2 | −3.48 |
| CARS2 | NM_024537 | Cysteinyl-tRNA
synthetase 2 | −3.09 |
| RHEBL1 | NM_144593 | Ras homolog enriched
in brain like 1 | −4.61 |
| STK36 | NM_015690 | Serine/threonine
kinase 36 | −3.64 |
| SEMA7A | NM_003612 | Semaphorin 7A, GPI
membrane anchor | −3.35 |
| CXCL10 | NM_001565 | Chemokine (C-X-C
motif) ligand 10 | −3.84 |
| ACSL5 | NM_016234 | Acyl-CoA synthetase
long-chain family member 5 | −3.04 |
| SAA1 | NM_000331 | Serum amyloid A1 | −3.65 |
| HCP5 | NR_040662 | HLA complex P5
(non-protein coding) | −3.19 |
| DHX58 | NR_024119 | DEXH (asp-Glu-X-His)
box polypeptide 58 | −3.21 |
| IFI27 | NM_005532 | Interferon,
α-inducible protein 27 | −3.85 |
Discussion
Histone acetylation, as one of the
best-characterized epigenetic modifications, is controlled by HATs
and histone deacetylases (HDACs). The balance between histone
acetylation and deacetylation serves as a key epigenetic mechanism
for gene expression, DNA repair, developmental processes and
tumorigenesis (5–7). Thus, an imbalance may lead to abnormal
cell functions and cancer. MYST1, also known as hMOF, a member of
the MYST family of HATs and an epigenetic marker of active genes,
is responsible for histone H4K16 acetylation in human cells
(2,3). Studies have demonstrated that hMOF
participates in a number of biological processes, including gene
transcription, cell proliferation, differentiation and the DNA
repair response (4–9). The fact that hMOF is involved in these
critical cellular functions suggests that hMOF may play a
significant role in tumorigenesis. Although little is known about
the mechanism of hMOF in tumor development and progression, hMOF
expression in clinical cancer tissues has been reported by several
studies. A frequent downregulation of hMOF in primary breast
carcinomas, renal cell carcinoma and medulloblastomas has been
identified and the reduction in hMOF protein expression has been
shown to correlate with H4K16 acetylation in those tumors (17,21).
In addition, the analysis of tissue microarray slides has revealed
low or absent histone H4K16 acetylation in the majority of breast
cancer tissues (16). However, the
expression of hMOF in non-small cell lung carcinoma tissues has
been shown to be frequently elevated (25). The present study investigated the
expression of the HAT, hMOF, and its corresponding H4K16
acetylation in a series of primary ovarian cancer tissues by qPCR,
western blotting and immunohistochemistry. The results revealed
that hMOF mRNA or protein expression was frequently downregulated
in human ovarian cancer (>75%), and that hMOF protein expression
was correlated with histone H4K16 acetylation in parallel.
Furthermore, the hMOF-regulated gene, HCP5, was also found to be
downregulated in ovarian cancer tissues. Therefore, hMOF may have a
significant role in primary ovarian carcinoma tumorigenesis.
HLA is the human version of the major
histocompatibility complex (MHC). The genes in this complex are
categorized into classes I, II and III. MHC or HLA modulates the
efficacy of cytotoxic immune responses through the presentation of
tumor antigens (19). The
modulation of tumor antigen-specific immune responses by an
abnormal expression of HLA-class I and II molecules has been
identified in a variety of carcinomas, including ovarian cancer
(20). HCP5 is localized within the
MHC class I region. Although the function of HCP5 is not well
known, the sequence of the gene is associated with the human
endogenous retroviruses, HERV-L and HERV-16 (18). In the present study, the expression
of hMOF was frequently downregulated in the clinical ovarian cancer
tissues (Figs. 1 and 2) and, more notably, HCP5 was presented in
gene expression profiles as a hMOF-downregulated gene. Together,
this suggests that HCP5, as a hMOF-target gene, may also be
downregulated in ovarian cancer. This prediction was confirmed
using qPCR. HCP5 gene expression was downregulated in >87% of
patients with a decreased hMOF level. Although the functional
mechanism between hMOF and HCP5 is unclear, a reduction in hMOF may
lead to defective expression of its target genes.
In summary, frequent downregulation of hMOF and a
loss of H4K16 acetylation are observed in ovarian cancer tissues.
Although a large series of clinical cases and analyses of overall
survival are required for further investigations, the molecular
mechanism that links a loss of hMOF expression to ovarian cancer
will be a promising area for further research. Combined with the
results from previous studies, the present study concluded that the
abnormal expression of hMOF in tumors may be a common feature,
suggesting that hMOF may be a novel epigenetic biomarker for tumor
diagnosis.
Abbreviations:
|
HAT
|
histone acetyltransferase;
|
|
MYST
|
Moz-Ybf2/Sas3-Sas2-Tip60;
|
|
HLA
|
human leukocyte antigen;
|
|
HCP5
|
HLA complex P5;
|
|
RT-PCR
|
reverse transcription-polymerase chain
reaction;
|
|
qPCR
|
quantitative PCR.
|
Acknowledgements
This study was supported by the
National Natural Science Foundation of China (no. 31070668) and the
Research Fund for the Doctoral Program of Higher Education of China
(no. 20110061110020).
References
|
1.
|
Hilfiker A, Hilfiker-Kleiner D, Pannuti A
and Lucchesi JC: mof, a putative acetyl transferase gene related to
the Tip60 and MOZ human genes and to the SAS genes of yeast, is
required for dosage compensation in Drosophila. EMBO J.
16:2054–2060. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Smith ER, Cayrou C, Huang R, Lane WS, Côtê
J and Lucchesi JC: A human protein complex homologous to the
Drosophila MSL complex is responsible for the majority of histone
H4 acetylation at lysine 16. Mol Cell Biol. 25:9175–9188. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Mendjan S, Taipale M, Kind J, Holz H,
Gebhardt P, Schelder M, Vermeulen M, Buscaino A, Duncan K, Mueller
J, Wilm M, Stunnenberg HG, Saumweber H and Akhtar A: Nuclear pore
components are involved in the transcriptional regulation of dosage
compensation in Drosophila. Mol Cell. 21:811–823. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Cai Y, Jin J, Swanson SK, Cole MD, Choi
SH, Florens L, Washburn MP, Conaway JW and Conaway RC: Subunit
composition and substrate specificity of a MOF-containing histone
acetyltransferase distinct from the male-specific lethal (MSL)
complex. J Biol Chem. 285:4268–4272. 2010. View Article : Google Scholar
|
|
5.
|
Carrozza MJ, Utley RT, Workman JL and Côté
J: The diverse functions of histone acetyltransferase complexes.
Trends Genet. 19:321–329. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Gupta A, Guerin-Peyrou TG, Sharma GG, Park
C, Agarwal M, Ganju RK, Pandita S, Choi K, Sukumar S, Pandita RK,
Ludwig T and Pandita TK: The mammalian ortholog of Drosophla
MOF that acetylates histone H4 lysine 16 is essential for
embryogenesis and oncogenesis. Mol Cell Biol. 28:397–409. 2008.
|
|
7.
|
Sharma GG, So S, Gupta A, Kumar R, Cayrou
C, Avvakumov N, Bhadra U, Pandita RK, Porteus MH, Chen DJ, Cote J
and Pandita TK: MOF and histone H4 acetylation at lysine 16 are
critical for DNA damage response and double-strand break repair.
Mol Cell Biol. 30:3582–3595. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Rea S, Xouri G and Akhtar A: Males absent
on the first (MOF): from flies to humans. Oncogene. 26:5385–5394.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Taipale M, Rea S, Richter K, Vilar A,
Lichter P, Imhof A and Akhtar A: hMOF histone acetyltransferase is
required for histone H4 lysine 16 acetylation in mammalian cells.
Mol Cell Biol. 25:6798–6810. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar
|
|
11.
|
Schwab LP, Peacock DL, Majumdar D, Ingels
JF, Jensen LC, Smish KD, Cushing RC and Seagroves TN:
Hypoxia-inducible factor 1α promotes primary tumor growth and
tumor-initiating cell activity in breast cancer. Breast Cancer Res.
14:R62012.
|
|
12.
|
McDonald PC, Winum JY, Supuran CT and
Dedhar S: Recent developments in targeting carbonic anhydrase IX
for cancer therapeutics. Oncotarget. 3:84–97. 2012.PubMed/NCBI
|
|
13.
|
Law AY and Wong CK: Stanniocalcin-2 is a
HIF-1 target gene that promotes cell proliferation in hypoxia. Exp
Cell Res. 316:466–476. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Choschzick M, Oosterwijk E, Müller V,
Woelber L, Simon R, Moch H and Tennstedt P: Overexpression of
carbonic anhydrase IX (CAIX) is an independent unfavorable
prognostic marker in endometrioid ovarian cancer. Virchows Arch.
459:193–200. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Seeber LM, Horrée N, Vooijs MA, Heintz AP,
van der Wall E, Verheijen RH and van Diest PJ: The role of hypoxia
inducible factor-1alpha in gynecological cancer. Crit Rev Oncol
Hematol. 78:173–184. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Elsheikh SE, Green AR, Rakha EA, Powe DG,
Ahmed RA, Collins HM, Soria D, Garibaldi JM, Paish CE, Ammar AA,
Grainge MJ, Ball GR, Abdelghany MK, Martinez-Pomares L, Heery DM
and Ellis IO: Global histone modifications in breast cancer
correlate with tumor phenotypes, prognostic factors, and patient
outcome. Cancer Res. 69:3802–3809. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Pfister S, Rea S, Taipale M, Mendrzyk F,
Straub B, Ittrich C, Thuerigen O, Sinn HP, Akhtar A and Lichter P:
The histone acetyltransferase hMOF is frequently downregulated in
primary breast carcinoma and medulloblastoma and constitutes a
biomarker for clinical outcome in medulloblastoma. Int J Cancer.
122:1207–1213. 2008. View Article : Google Scholar
|
|
18.
|
Kulski JK and Dawkins RL: The P5 multicopy
gene family in the MHC is related in sequence to human endogenous
retroviruses HERV-L and HERV-16. Immunogenetics. 49:404–412. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Salih HR and Nüssler V: Commentary: Immune
escape versus tumor tolerance: how do tumors evade immune
surveillance? Eur J Med Res. 6:323–332. 2001.PubMed/NCBI
|
|
20.
|
Kübler K, Arndt PF, Wardelmann E, Landwehr
C, Krebs D, Kuhn W and van der Ven K: Genetic alterations of
HLA-class II in ovarian cancer. Int J Cancer. 123:1350–1356.
2008.PubMed/NCBI
|
|
21.
|
Wang Y, Zhang R, Wu D, Lu Z, Sun W, Cai Y,
Wang C and Jin J: Epigenetic change in kidney tumor: downregulation
of histone acetyltransferase MYST1 in human renal cell carcinoma. J
Exp Clin Cancer Res. 32:82013. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Seeber LM and van Diest PJ: Epigenetics in
ovarian cancer. Methods Mol Biol. 863:253–269. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Asadollahi R, Hyde CA and Zhong XY:
Epigenetics of ovarian cancer: from the lab to the clinic. Gynecol
Oncol. 118:81–87. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Balch C, Matei DE, Huang TH and Nephew KP:
Role of epigenomics in ovarian and endometrial cancers.
Epigenomics. 2:419–447. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Song JS, Chun SM, Lee JY, Kim DK, Kim YH
and Jang SJ: The histone acetyltransferase hMOF is overexpressed in
non-small cell lung carcinoma. Korean J Pathol. 45:386–396. 2011.
View Article : Google Scholar
|