Introduction
Thyroid cancer is the most common endocrine
malignancy with a high incidence in numerous regions of the world
(1,2). Thyroid cancer is histologically
classified into papillary thyroid cancer (PTC), follicular thyroid
cancer (FTC) and anaplastic thyroid cancer (ATC). Thyroid cancers
frequently harbor activating mutations in the mitogen-activated
protein kinase (MAPK) and phosphatidylinositol 3-kinase (PI3K)/Akt
signaling pathways, as represented by RET/PTC, RAS and
BRAF mutations in the former and by PIK3CA and
PTEN mutations in the latter (3,4). As a
significant mechanism for the tumorigenesis of thyroid cancer,
aberrant activation of these two important signaling pathways by
such mutations causes uncontrolled cell division, proliferation and
survival, leading to malignancy.
High-frequency somatic mutations of the
GNA11, matrix metalloproteinase (MMP)27, TRRAP and
GRM3 genes have been reported in uveal melanomas and
melanomas with various incidences (5–8). We
previously demonstrated that FGD1 was normally maintained,
hypomethylated and overexpressed by BRAF (V600E) in thyroid
cancer cells (9). GNA11 activates
the MAPK signaling pathway. Particularly frequent somatic mutations
of the GNA11 gene at codon 209 in exon 5 and codon 183 in
exon 4, resulting in mutant GNA11Q209L and GNA11R183C,
respectively, have been reported in uveal melanoma and blue nevi
(5). The GNA11 gene encodes
a G-protein α-subunit (Gα11) that mediates signals from
G-protein-coupled receptors (GPCRs) to the MAPK pathway. The normal
amino acid, glutamine, encoded by codon 209 of the GNA11
gene, lies within the RAS-like domain of GNA11 (corresponding to
residue 61 of Ras) and is essential for GTP hydrolysis. In members
of the RAS family, mutations at this site and at codon 12 cause the
loss of GTPase activity with constitutive activation of Ras. The
GNA11Q209L and GNA11R183C mutants have been demonstrated
to be able to transform 3T3 cells and form tumors in
immunocom-promised mice (5). MMPs
are proteolytic enzymes that degrade components of the
extracellular matrix and basement membranes. MMP abnormalities have
been associated with the metastasis of various types of cancer
(6,10–12).
In particular, mutations of the MMP27 gene have been
observed in melanoma. The majority of the mutations in this gene
have been identified in exons 1, 2, 3, 8 and 9 (6). FGD1 gene mutations have been
reported in Aarskog-Scott syndrome (AAS), or facio-digito-genital
dysplasia (13). At present, 20
different FGD1 gene mutations have been reported in this syndrome
(13). FGD1 is a Dbl family
member that has been shown to function as a CDC42-specific guanine
nucleotide exchange factor (GEF). It has also been demonstrated
that FGD1 expression is sufficient to cause tumorigenic
transformation of NIH3T3 fibroblasts (14). Two studies from the same group
reported that the TRRAP gene was recurrently mutated and
clustered in one amino acid position S722F (7). Furthermore, a frequent mutation of the
GRM3 gene has been reported, and the authors also noted that
the mutant selectively regulated the phosphorylation of MEK in the
activation of the MAPK signaling pathway, leading to the
anchorage-independent growth and migration of cells (8). The mutation status in the
GNA11, MMP27, FGD1, TRRAP and
GRM3 genes has not been studied in thyroid cancer. The
present study was conducted to investigate the mutational status of
these genes in thyroid cancer.
Materials and methods
Cell lines, tumor samples and DNA
extraction
A total of 89 samples, consisting of 12 thyroid
cancer cell lines and 77 thyroid tumor samples were used for the
mutational analysis of the GNA11 gene. For the MMP27
mutational analysis, 29 samples consisting of 12 thyroid cancer
cell lines and 17 ATC samples were used. The FGD1,
TRRAP and GRM3 genes were analyzed in 28 samples,
including 12 thyroid cancer cell lines and 16 PTC samples. The
thyroid cancer cell lines and tumor samples were used as described
previously and with institutional review board (IRB; The Johns
Hopkins University School of Medicine, Baltimore, MD, USA) approval
(15). The cell lines were
authenticated as described previously (16). With the exception of the FTC133
cells cultured in Dulbecco’s modified Eagle’s medium (DMEM)/Ham’s
F-12 medium, all tumor cell lines were cultured in RPMI-1640 medium
supplemented with 10% fetal bovine serum (FBS), streptomycin (100
μg/ml), penicillin (100 U/ml) and 2 mM glutamine. Genomic DNA from
the cell lines and tumors was isolated by standard
phenol-chloroform extraction using MaXtract high-density gel tubes
followed by ethanol precipitation procedures (Qiagen, Valencia, CA,
USA) (15).
PCR amplification and sequencing of the
GNA11, MMP27, FGD1, TRRAP and GRM3 genes
The primer sequences and PCR conditions for the
amplification of exons 4 and 5 of the GNA11 gene and exons
1, 2, 3, 8 and 9 of the MMP27 gene were as described
previously (5,6). For the mutational analysis of the
MMP27 gene, in addition to the above primers, additional
primers were used to shorten the amplicon size. These primers are
shown in Table I. The PCR
conditions for the amplification of the MMP27 with the
additional primers were as follows: One cycle of 94°C for 3 min;
ten cycles of 94°C for 30 sec, 67°C for 30 sec with a 1°C reduction
for each cycle and 72°C for 30 sec; thirty two cycles of 94°C for
30 sec, 57°C for 30 sec and 72°C for 30 sec; followed by 72°C for 7
min as a final extension; and 4°C as the storage temperature. The
primer sequences for the amplification of the FGD1 gene are
shown in Table I. The PCR reaction
conditions for the FGD1 amplification were as follows: After
an initial denaturation at 94°C for 2 min, the amplification was
performed at 94°C for 1 min, followed by annealing temperatures
(exons 11, 14 and 18, 55°C; exons 1, 2 and 16, 57°C; exons 5, 6, 7,
8, 13 and 17, 58°C; exons 3, 12 and 15, 60°C; and exons 4, 9 and
10, 62°C) for 1 min for 35 cycles, with a final extension at 72°C
for 7 min. The primer sequences and PCR conditions for the
amplification of exon 1 of the TRRAP gene and exons 1, 2, 3,
4 and 5 of the GRM3 gene were as described previously
(7,8). The PCR products were directly
sequenced using a BigDye Terminator v3.1 Cycle Sequencing ready
reaction kit (Applied Biosystems, Foster City, CA, USA). These
exons were investigated as they harbored the majority of the
reported mutations in these genes in human cancers. The GenBank
accession numbers were NM_002067.2 (GNA11), NM_022122
(MMP27), NM_004463 (FGD1), NM_003496 (TRRAP)
and NM_000840 (GRM3).
 | Table I.Primer sets used for PCR amplification
of the MMP27 and FGD1 genes. |
Table I.
Primer sets used for PCR amplification
of the MMP27 and FGD1 genes.
| Exon | Forward | Reverse |
|---|
| MMP27 | | |
| 1-1 |
GCCAATTCATGCCACGTCTCAC |
GAAAATATGCAACTGGCTCAGG |
| 1-2 |
GAACCGGCTTCAGCTGAAGAAAG |
CACATTCCTGCAAAAGAGTCCTG |
| 2-1 |
CCTGAGATGGAGATTTGCTCTC |
GGATTGACAGTGACTGGAAAAC |
| 2-2 |
TCTTTTTGGTCAGGCATATCTC | ATCATG
AAGACACCCAGGTGTG |
| 2-3 | CTCAACCAGT
TCTACTCTCTTG |
GTATGGCTACACCCTCCCT |
| 2-4 | TGAGATCATG
AAGACACCCAGG |
CATTCAATTGACTGAGCACTTC |
| FGD1 | | |
| 1 |
GGCTTGAGTCTCTGCAGTG |
AACAAGAACCCGCTCCCAGTAC |
| 2 |
AGTCCTAACTTTAACCCCAGTC |
CTGGCTAACTTCTCCCCTCCTC |
| 3 |
TTCACCATGTTAGCCAGGCTC |
GTATGAGCTTGACTGAGAGGC |
| 4 |
AGCCTGGGACAGGAAGGGATA |
AAAGGCGCTTCCAGGTTCTCC |
| 5 |
TATTAGGCTTAGAGTGGCATG |
ACTGCCTCCTTGAAACGCACC |
| 6 |
TCAGTCTCAAGACCAATGCTG |
GAAGTCTTGTGTACACCTCTG |
| 7 |
ACTGAGATGAAAGGTATCTGC |
TCAGATCTGGCTGCAGATGCC |
| 8 |
AAGCTGGAAGGAGCAGACTTG |
AGAGCTATTAGTGTGGAGAAG |
| 9 |
AGGCAGAGGTTGTGGTGAGCC |
GTGCCAGCCTCCTGTCAGATG |
| 10 |
ATCTGACAGGAGGCTGGCACC |
AGTACCAGGTCACTATGTGTG |
| 11 |
CAGTAAAGCTTCAGGGCAAG |
AGCCAGCATCTTTGTTCCTC |
| 12 |
TGTGTGTGTATGTGTGCAGAG |
TCTCTGGGCCTGGAATGCCTC |
| 13 |
AGGTGAAGAAGAGGGTCAAGC |
ACCATTCTGGTTAGCTGTGAG |
| 14 |
CTAGGGTATACGAAGGTGAGGC |
TAAAGGTCAGGTGGGCATTTGG |
| 15 |
ATGATAATCCAAGCGTTGGAG |
AGGGTACCCACTCTGCAGTGG |
| 16 |
TGCTGTGGGAGTTGGTATGCTG |
TGCTGTGGGAGTTGGTATGCTG |
| 17 |
TTCATCTGAGATAGGAATAGC |
TCTTCCAAGGCTCACCTTATC |
| 18 |
GCAAAACCAGTTAGAAGCTGGG |
TTCAAGTATTGACTGAGCTGGG |
Results
The strategy of the present study was to investigate
the gene exons that were the most likely to carry mutations. In
particular, exons 4 and 5 of the GNA11 gene were examined
for mutations since all of the known GNA11 mutations have
been reported in codons 209 and 183 of these exons. Exons 1, 2, 3,
8 and 9 of the MMP27 gene were selected for sequencing as
they have been shown to carry somatic mutations in melanomas. All
the exons of the FGD1 gene were analyzed for mutation, as
mutations in this gene have never been reported in human cancers.
Exon 1 of the TRRAP gene and exons 1, 2, 3, 4 and 5 of the
GRM3 gene were analyzed as these exons have also been
reported to harbor somatic mutations in melanoma.
The sequencing results showed no mutations in and
around the hot spot of codons 209 and 183 in the GNA11 gene
in 12 thyroid cancer cell lines and 46 thyroid cancer samples
(including 26 FTC and 20 ATC samples). No novel MMP27
somatic mutations were identified in 12 thyroid cancer cell lines
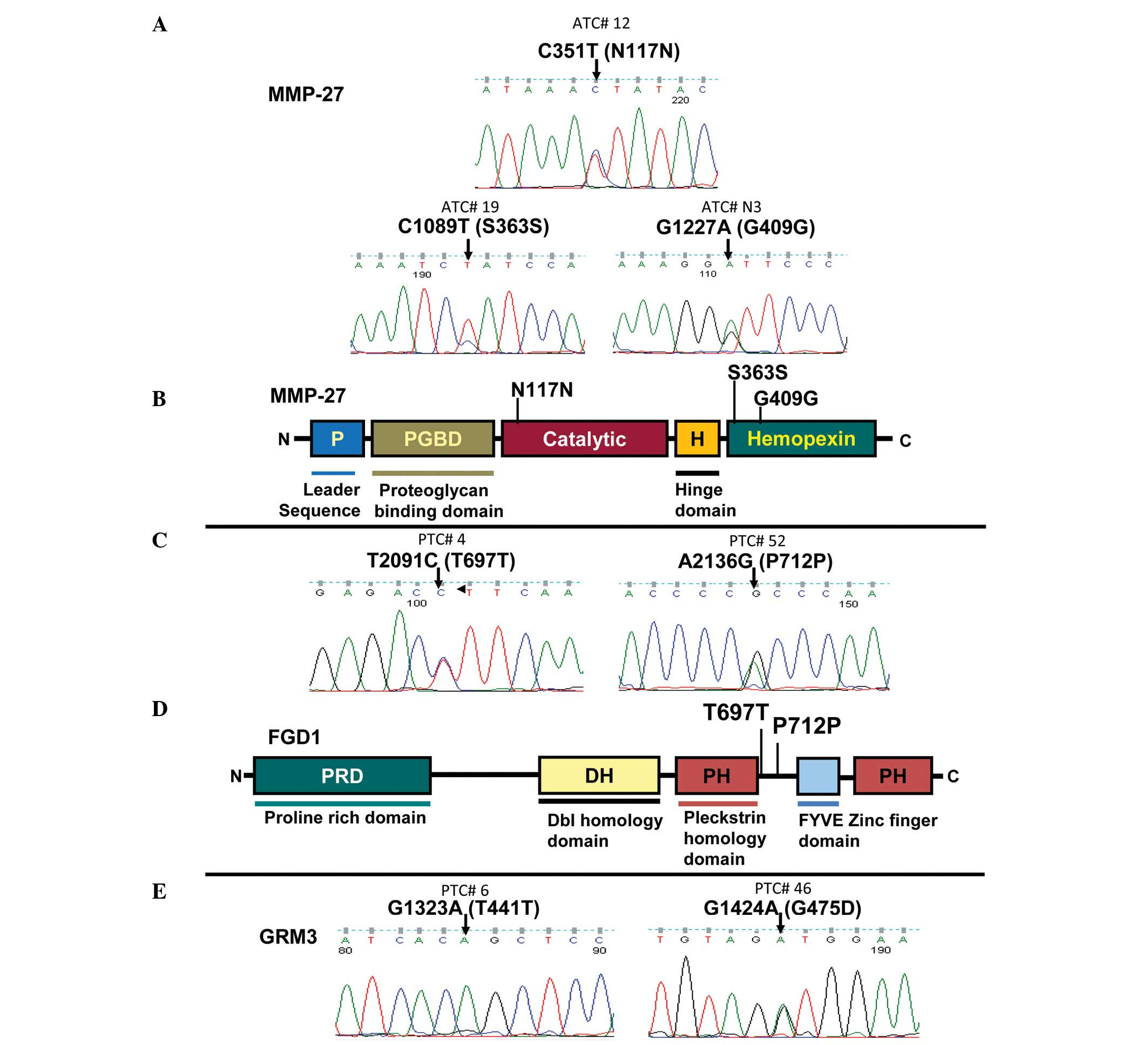
and 15 ATC tumor samples. As shown in Fig. 1A and B, an infrequent [1/17 (5.8%)]
C>T transition was observed at nucleotide position 351,
resulting in a codon change of AAC>AAT and amino acid N117N in
exon 3. In exon 8, an infrequent [1/17 (5.8%)] C>T transition
was observed at nucleotide position 1089, resulting in a codon
change of TCC>TCT and amino acid S363S. In exon 9, a frequent
[7/17 (41.2%)] G>A transition at nucleotide position 1227 was
also observed, resulting in a codon change of GGG>GGA and amino
acid G409G. The two N117N and S363S mutations were rare and novel
silent mutations that have not been previously reported in the SNP
database.
No FGD1 mutations were identified in 12
thyroid cancer cell lines. However, as illustrated in Fig. 1C and D, an infrequent [1/16 (6.3%)]
T>C mutation was observed at nucleotide position 2091, resulting
in a codon change of ACT>ACC and amino acid T697T. An infrequent
[1/16 (6.3%)] A>G was also observed at nucleotide position 2136,
resulting in a codon change of CCA>CCG and amino acid P712P.
These silent T697T (rs12011120) and P712P (rs1126744) mutations
were rare SNPs that have been reported in the SNP database
(http://www.ncbi.nlm.nih.gov/projects/SNP/).
Mutations were not identified in the TRRAP
gene in 12 thyroid cancer cell lines and 16 PTC tumor samples. No
GRM3 mutations were detected in 12 thyroid cancer cell
lines. A G>A mutation was observed at nucleotide position 1323,
resulting in a codon change of ACG>ACA and amino acid T441T in
all 12 thyroid cancer cell lines and 16 PTC samples. An infrequent
[1/16 (6.3%)] G>A mutation was also observed at nucleotide
position 1424 resulting in a codon change of GGT>GAT and amino
acid G475D (Fig. 1E). The T441T
mutation was a novel synonymous SNP that has not previously been
reported in the SNP database. G475D (rs17161026) was a
non-synonymous SNP that has previously been reported in the SNP
database (http://www.ncbi.nlm.nih.gov/projects/SNP/). Fig. 1 shows the mutations identified in
the present study, and a summary of the results is presented in
Table II.
 | Table II.Mutations of the MMP27,
FGD1 and GRM3 genes in thyroid cancers. |
Table II.
Mutations of the MMP27,
FGD1 and GRM3 genes in thyroid cancers.
| S. No. | Gene | Exon | Nucleotide | Codon | Amino acid | Type of
mutation |
|---|
| ATC# 12 | MMP27 | 3 | C351T | AAC-AAT | N117N | Silent |
| ATC# 19 | MMP27 | 8 | C1089T | TCC-TCT | S363S | Silent |
| ATC# N3 | MMP27 | 9 | G1227A | GGG-GGA | G409G | Silent |
| PTC# 4 | FGD1 | 14 | T2091C | ACT-ACC | T697T | Silent |
| PTC# 52 | FGD1 | 14 | A2136G | CCA-CCG | P712P | Silent |
| PTC# 6 | GRM3 | 3 | G1323A | ACG-ACA | T441T | Silent |
| PTC# 46 | GRM3 | 3 | G1424A | GGT-GAT | G475D | Missense |
Discussion
In the present study, several genes were analyzed
for the first time for possible mutations in thyroid cancer.
GNA11 mutations were analyzed in all types of thyroid cancer
(PTC, FTC and ATC) as they have been frequently identified in uveal
melanoma and are known to activate the MAPK signaling pathway
(5), which is one of the most
deregulated signaling pathways in thyroid cancer (3). However, no mutations were detected in
and around codons 209 and 183. These two hot spot codons were
selectively analyzed as GNA11 mutations have consistently been
identified only in these two residues (5). The MMPs are proteolytic enzymes that
degrade the components of the extracellular matrix and basement
membranes, which are associated with cancer metastasis (6,10–12).
ATC is the most aggressive type of thyroid cancer that is often
associated with deadly metastasis (17). Therefore ATC was particularly
analyzed for the mutation of the MMP27 gene. Two MMPs
have been reported to occasionally be mutated in melanomas. The
MMP27 gene was analyzed for mutation in ATC in the present
study as we had already previously analyzed the second gene,
MMP8, in thyroid cancer (18). Three uncommon mutations were
identified; C351T resulting in N117N, C1089T resulting in S363S and
G1227A resulting in G409G silent mutations. The silent mutations
are unlikely to be involved in thyroid carcinogenesis as these
mutations do not change the basic amino acids.
We previously revealed that FGD1 was normally
maintained, hypomethylated and overexpressed by BRAF (V600E)
in thyroid cancer cells and in turn observed that it was
hyper-methylated after ShRNA-mediated knockdown of BRAF
(V600E) in thyroid cancer cell lines (9). Based on these findings and the high
transforming and invasive potential of the FGD1 gene
(4), we considered there to be a
high possibility of identifying oncogenic mutations in FGD1.
All 18 exons of the gene were sequenced to be analyzed for
mutations, but only two silent mutations (T697T and P712P) were
detected. These mutations are unlikely to have a significant role
in PTC. No somatic missense mutations were identified in the
FGD1 gene.
The TRRAP gene has been reported to be
mutated in a particular codon, S722F (7). As GRM3 activates the MAPK signaling
pathway (8), the present study
investigated whether GRM3 is mutated in PTC samples, since
the majority of PTCs harbor genetic deregulation in the MAPK
signaling pathway. No mutations were detected in TRRAP, while two
SNPs (T441T and G475D) were identified in the GRM3 gene.
This suggests that TRRAP and GRM3 may not have
important roles in the pathogenesis of this type of thyroid
cancer.
In conclusion, the present findings suggested that
genetic alterations in the GNA11, MMP27, FGD1, TRRAP and
GRM3 genes may not be significant in the tumorigenesis of
thyroid cancer. It is not surprising that mutations in these genes
are not common in thyroid cancer since a number of the upstream
effectors involved in cellular transformation, growth and
metastasis, including EGFR, RET/PTC, ALK, RAS, BRAF, PTEN, PIK3CA,
PIK3CB and PDK1, are commonly genetically altered via mutations or
genetic amplifications that are able to independently activate the
MAPK or PI3K/Akt pathways in thyroid cancer (3,4,16,18,19).
Acknowledgements
The authors would like to thank Drs
N.E. Heldin, K.B. Ain, N. Onoda, M. Santoro, D. Wynford Thomas, G.
Brabant, A.P. Dackiw, R Schweppe and B Haugen for kindly providing
access to the cell lines used in the present study. The study was
supported by NIH R01 grant CA 134225.
References
|
1.
|
Leenhardt L, Grosclaude P and
Chérié-Challine L: Thyroid Cancer Committee: Increased incidence of
thyroid carcinoma in France: a true epidemic or thyroid nodule
management effects? Report from the French Thyroid Cancer
Committee. Thyroid. 14:1056–1060. 2004. View Article : Google Scholar
|
|
2.
|
Sprague BL, Warren Andersen S and
Trentham-Dietz A: Thyroid cancer incidence and socioeconomic
indicators of health care access. Cancer Causes Control.
19:585–593. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Xing M: BRAF mutation in thyroid cancer.
Endocr Relat Cancer. 12:245–262. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Liu Z, Hou P, Ji M, et al: Highly
prevalent genetic alterations in receptor tyrosine kinases and
phosphatidylinositol 3-kinase/akt and mitogen-activated protein
kinase pathways in anaplastic and follicular thyroid cancers. J
Clin Endocrinol Metab. 93:3106–3116. 2008. View Article : Google Scholar
|
|
5.
|
Van Raamsdonk CD, Griewank KG, Crosby MB,
et al: Mutations in GNA11 in uveal melanoma. N Engl J Med.
363:2191–2199. 2010.PubMed/NCBI
|
|
6.
|
Palavalli LH, Prickett TD, Wunderlich JR,
et al: Analysis of the matrix metalloproteinase family reveals that
MMP8 is often mutated in melanoma. Nat Genet. 41:518–520. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Wei X, Walia V, Lin JC, et al: Exome
sequencing identifies GRIN2A as frequently mutated in melanoma. Nat
Genet. 43:442–446. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Prickett TD, Wei X, Cardenas-Navia I, et
al: Exon capture analysis of G protein-coupled receptors identifies
activating mutations in GRM3 in melanoma. Nat Genet. 43:1119–1126.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Hou P, Liu D and Xing M: Genome-wide
alterations in gene methylation by the BRAF V600E mutation in
papillary thyroid cancer cells. Endocr Relat Cancer. 18:687–697.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Brinckerhoff CE and Matrisian LM: Matrix
metalloproteinases: a tail of a frog that became a prince. Nat Rev
Mol Cell Biol. 3:207–214. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Egeblad M and Werb Z: New functions for
the matrix metalloproteinases in cancer progression. Nat Rev
Cancer. 2:161–174. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Freije JM, Balbín M, Pendás AM, Sánchez
LM, Puente XS and López-Otín C: Matrix metalloproteinases and tumor
progression. Adv Exp Med Biol. 532:91–107. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Orrico A, Galli L, Faivre L, et al:
Aarskog-Scott syndrome: clinical update and report of nine novel
mutations of the FGD1 gene. Am J Med Genet A. 152A:313–318. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Whitehead IP, Abe K, Gorski JL and Der CJ:
CDC42 and FGD1 cause distinct signaling and transforming
activities. Mol Cell Biol. 18:4689–4697. 1998.PubMed/NCBI
|
|
15.
|
Murugan AK, Dong J, Xie J and Xing M: MEK1
mutations, but not ERK2 mutations, occur in melanomas and colon
carcinomas, but none in thyroid carcinomas. Cell Cycle.
8:2122–2124. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Murugan AK and Xing M: Anaplastic thyroid
cancers harbor novel oncogenic mutations of the ALK gene. Cancer
Res. 71:4403–4411. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Neff RL, Farrar WB, Kloos RT and Burman
KD: Anaplastic thyroid cancer. Endocrinol Metab Clin North Am.
37:525–538. 2008. View Article : Google Scholar
|
|
18.
|
Murugan AK, Dong J, Xie J and Xing M:
Uncommon GNAQ, MMP8, AKT3, EGFR, and PIK3R1 mutations in thyroid
cancers. Endocr Pathol. 22:97–102. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Mitsiades CS, Kotoula V, Poulaki V, et al:
Epidermal growth factor receptor as a therapeutic target in human
thyroid carcinoma: mutational and functional analysis. J Clin
Endocrinol Metab. 91:3662–3666. 2006. View Article : Google Scholar
|