Introduction
Colorectal cancer (CRC) originates from one colonic
epithelial cell due to an accumulation of genetic and epigenetic
changes that lead to malignancy (1). The incidence of CRC has a varied
distribution in the different geographical regions and a
significant difference has been observed between developed and
developing countries (2,3). A number of studies have shown show
that CRC is becoming increasingly common in Asian countries
(4,5,6).
According to the statistics issued by the Iranian Ministry of
Health, ~1,130 patients suffering from CRC succumbed in 2006, and
there was no significant difference between the mortality rates of
male and female patients (7). The
causes of CRC may include genetic and environmental factors. Based
on the observed molecular genetic alterations, it has been
indicated that CRC is a heterogeneous disease (8). For the development of more appropriate
diagnostic methods, there is a requirement for studies to identify
new molecular markers in the affected populations. Epidermal growth
factor (EGF) is one of the most important factors leading to the
development of tumors and plays a significant role in the
uncontrolled reproduction of cancerous cells (9). EGF is an important regulator of cell
survival, and in numerous types of cancer, including breast,
prostate, pancreas, colorectal, lung and head and neck, an increase
in EGF and ErbB, a member of the EGF receptor family, has
been demonstrated (10–12). The EGF gene is located on
chromosome 4q25–27 and produces various transcripts, the largest of
which has 24 exons and 23 introns (12,13).
Genetic factors play a key role in the susceptibility to diseases,
resistance against medications and interference in the interaction
with peripheral factors. Single nucleotide polymorphisms (SNPs) are
the most common genetic changes (14). As the polymorphisms of the
EGF gene affect the susceptibility to numerous types of
cancer, the majority of the case-control studies on EGF
genes have been conducted on exonic polymorphisms and untranslated
regions (UTRs) (12,15–17).
In the present study, the hypothesis that the noncoding
polymorphism, rs2298979, may be used to predict the susceptibility
to CRC was tested in the Iranian population.
Materials and methods
Study population
Genetic analysis was conducted on a population of
220 patients suffering from sporadic CRC and 220 normal individuals
who had been referred to the Research Institute for
Gastroenterology and Liver Diseases (RIGLD), Taleghani Hospital,
Shahid Beheshti University of Medical Sciences (Tehran, Iran).
Patients with a family history of hereditary non-polyposis CRC
(HNPCC) and familial adenomatous polyposis (FAP) were excluded from
this study. The patients and healthy individuals were all of
Iranian nationality. A colonoscopy was performed on all
participants in the patient and control groups, the medical
normality of the control group was confirmed and the histological
diagnosis of the pathologist was approved by the
gastroenterologist. The parameters of age, gender and cigarette
smoking status were controlled in the patient and control groups
and the stage of CRC was determined in the patient group. The tumor
stage was classified according to the tumor-node-metastasis (TNM)
classification of the Union for International Cancer Control (UICC;
Table I). This study was conducted
under the approval of the ethics committee of the Gastroenterology
and Liver Diseases Research Center, Shahid Beheshti University of
Medical Sciences (Tehran, Iran).
 | Table IDemographic characteristics of the
study population. |
Table I
Demographic characteristics of the
study population.
| Variable | Patients with
CRC | Controls |
|---|
| Age, years ±
mean | 43.12±15.366 | 59.17±13.615 |
| Gender, n (%) |
| Male | 124 (56.4) | 97 (44.1) |
| Female | 96 (43.6) | 123 (55.9) |
| Smoking, n (%) |
| Yes | 15 (6.8) | 16 (7.3) |
| No | 205 (93.2) | 204 (92.7) |
| Clinical stage, n
(%) |
| Stage I and II | 101 (45.9) | - |
| Stage III and
IV | 119 (54.1) | - |
DNA extraction
Subsequent to obtaining a letter of consent from
each individual, 5 ml peripheral blood was collected and stored at
4ºC in a bottle containing EDTA. Genomic DNA was extracted as soon
as possible following sampling using the standard salting out
method (18). The quality of the
extracted DNA was then assessed using a NanoDrop spectrophotometer
(NanoDrop Technologies, Inc., Wilmington, DE, USA).
EGF rs2298979 gene polymorphism
genotyping
Two specific primers were designed. The polymorphism
was determined using the polymerase chain reaction-restriction
fragment length polymorphism (PCR-RFLP) method. The characteristics
and sequence of the primers are shown in Table II. The 768-bp DNA fragment that was
located in the intron region of the EGF gene was amplified using
the specific primers (Table III).
The PCR products were digested by PciI endonuclease enzyme
(recognition sequence A/CATGT) for 6 h at 37ºC. In order to observe
the digested fragments, the RFLP solution was separated on a 3%
agarose gel and stained with ethidium bromide.
 | Table IIPrimer sequence and resulting fragment
length for the rs2298979 PCR. |
Table II
Primer sequence and resulting fragment
length for the rs2298979 PCR.
| Primer no. | Direction | Primer sequence | % GC | Result (bp) |
|---|
| 1 | Forward |
5′-CATACAATAAACACTCGATAAGCC-3′ | 37.5 | |
| 2 | Reverse |
5′-ACCTCCAACCAACCATACTACC-3′ | 50 | 768 |
 | Table IIITechnical data for the rs2298979
polymorphism detection method. |
Table III
Technical data for the rs2298979
polymorphism detection method.
| Factor | Value |
|---|
| PCR reaction
condition, no. of cycles |
| 94ºC for 5 min | 1 |
| 94ºC for 45 sec | 30 |
| 65ºC for 40 sec | - |
| 72ºC for 45 sec | - |
| 72ºC for 5 min | 1 |
| Master mix, μl |
| 10× PCR buffer | 2.50 |
| MgCl2 (50
mM) | 0.75 |
| Primer-forward (10
mM) | 1.00 |
| Primer-reverse (10
mM) | 1.00 |
| dNTP (10 mM) | 0.50 |
| Taq polymerase (5
U/μl) | 0.50 |
| Water | 17.75 |
| Restriction
enzyme | PciI |
| Restriction enzyme
time, h | 6 |
| Restriction pattern
length, bp |
| A | 768 |
| G | 508+260 |
| Agarose gel
concentration, % | 3 |
Sequencing
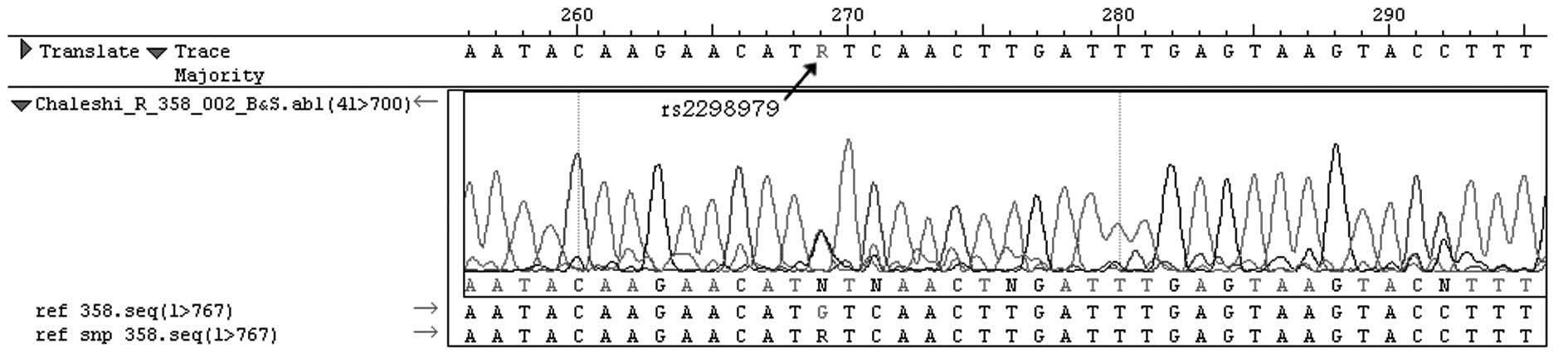
To confirm the RFLP procedure, 10% of the PCR
products were sequenced using the ABI PRISM 3130xL Genetic Analyzer
(Applied Biosystems®, Invitrogen Life Technologies,
Carlsbad, CA, USA) and the chain termination method (Fig. 1).
Statistical analysis
A Pearson χ2 test and Student's t-test
were used to calculate the P-value, with P<0.05 considered to
indicate a statistically significant difference. The data were
analyzed using SPSS statistical software version 13 (SPSS, Inc.,
Chicago, IL, USA).
Results
Following enzymatic digestion, it was revealed that
the size of the PCR products for the A/A genotype fragment was 768
bp in length, while the three fragments for the A/G genotype were
768, 508 and 260 bp, respectively. The genotype frequency
percentages of the rs2298970 polymorphism for the patients with CRC
were 28.6% for the A/A homozygote genotype and 71.4% for the A/G
genotype. Moreover, the genotype frequency percentages of the
controls were 16.4% for the A/A genotype and 83.6% for the A/G
genotype. In the entire patient and control group population, the
G/G genotype was not observed. Further details and frequency
percentages of the A and G alleles for the patient and control
groups are shown in Table IV.
According to the results of the present study, there was a
significant correlation between the A/G genotype of the rs2298979
polymorphism and a decreased risk of CRC compared with the control
group [odds ratio (OR), 0.488; 95% confidence interval (CI),
0.307–0.774; P=0.002]. However, no significant correlation was
observed among the genotypes of the patients in terms of the stage
parameters (Table V).
 | Table IVColorectal cancer risks correlated
with rs2298979 SNP that were examined in the present study. |
Table IV
Colorectal cancer risks correlated
with rs2298979 SNP that were examined in the present study.
| | | | OR (95% CI) | |
|---|
| | | |
| |
|---|
| Genetics | Patients, n (%) | Controls, n (%) | P-value | Unadjusted | Adjusted | P-value |
|---|
| Genotype |
| A/A | 63 (28.6) | 36 (16.4) | - | 1.00 (Reference) | 1.00 (Reference) | - |
| A/G | 157 (71.4) | 184 (83.6) | 0.002 | 0.488
(0.307–0.774) | 0.449
(0.263–0.767) | 0.003 |
| Alleles |
| A | 283 (64.3) | 256 (58.2) | - | 1.00 (Reference) | - | - |
| G | 157 (35.7) | 184 (41.8) | 0.062 | 0.772
(0.588–1.013) | - | - |
 | Table VTumor-stage specific distribution of
EGF rs2298979 genotypes among patients with CRC. |
Table V
Tumor-stage specific distribution of
EGF rs2298979 genotypes among patients with CRC.
| Genotype | Stage I, n (%) | Stage II, n
(%) | Stage III, n
(%) | Stage IV, n
(%) | P-value |
|---|
| A/A | 29 (64.4) | 38 (67.9) | 49 (75.4) | 41 (75.9) | 0.483 |
| A/G | 16 (35.6) | 18 (32.1) | 16 (24.6) | 13 (24.1) | - |
Discussion
EGF has a significant role in cell
proliferation, differentiation and tumorigenesis of epithelial
tissues (19). The action of a cell
continuing along its intended survival or death pathway is affected
by changes in the environmental signals. One of the most important
signals from the periphery of the cell inducing cell survival is
that of EGF, which attaches to the cell receptor (ErbB
receptor family) and stimulates intercellular pathways. EGF
signaling pathways are regulated by the concentration of EGF
present (11). It has been
demonstrated that the level of EGF in the plasma is significantly
correlated with CRC (20).
Polymorphisms of the EGF gene have also been shown to be
correlated with several other types of cancer. Early studies were
conducted to investigate the +61A/G polymorphism in the 5′UTR of
the EGF gene. The results revealed that the polymorphism was
correlated with cancer in individuals carrying the G allele of the
+61A/G polymorphism of the EGF gene (12,15,16).
To the best of our knowledge the correlation between the rs2298979
polymorphism of the EGF gene and sporadic CRC has been
studied for the first time in the present study. The rs2298979
polymorphism is located in intron 1 of the EGF gene. In the
present study, it was observed that the carriers of the A/G
genotype in the patient group were less susceptible to CRC in
comparison with the carriers of the hereditary A/A genotype. The
mechanism of this correlation is unknown. The region where the SNP
is located may be part of the regulatory sequence (21). The majority of studies on human
genome sequences focus on protein coding regions (exons) (22). The protein coding regions in
combination with the UTRs account for only 2% of the human genome.
It has been revealed that the non-coding regions of the genome are
effective in development, natural physiology and pathogenic
processes (22). In addition, it
has been demonstrated that there are SNPs present in intron regions
that have a significant correlation with cancer and different types
of diseases. In a study by Millar et al(23), it was revealed that the +1169A/T
polymorphism located within intron 4 of the growth hormone I (GHI)
gene was significantly correlated with the decrease in the
circulation of growth hormone and the risk of cancer of the large
intestine (23). Furthermore, Li
et al(24) investigated the
affect of two polymorphisms, +45G15G (T/G) and +276 (G/T), located
in exon 1 and intron 4 of the adiponectin gene, respectively, in
patients suffering from polycystic ovary syndrome (PCOS). No
significant correlation was revealed between the disease and the
+45G15G (T/G) polymorphism. By contrast, there was a statistically
significant correlation between the +276 (G/T) polymorphism,
located in intron 4, and PCOS disease (P=0.0126) (24). In conclusion, the present study
demonstrates that the carriers of the A/G genotype among patients
with CRC are less susceptible to the risk of cancer in comparison
with the carriers of A/A genotype. Further studies on the intron
regions of the EGF gene, particularly the rs2298979
polymorphism, are required in other populations.
Acknowledgements
The authors would like to thank all patients who
participated in the study, which was conducted with the support of
the Gastroenterology and Liver Diseases Research Center, Shahid
Beheshti University of Medical Science (Tehran, Iran).
References
|
1
|
Janakiram NB and Rao CV: Molecular markers
and targets for colorectal cancer prevention. Acta Pharmacol Sin.
29:1–20. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Haggar FA and Boushey RP: Colorectal
cancer epidemiology: incidence, mortality, survival, and risk
factors. Clin Colon Rectal Surg. 22:191–197. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chan AO, Soliman AS, Zhang Q, et al:
Differing DNA methylation patterns and gene mutation frequencies in
colorectal carcinomas from Middle Eastern countries. Clin Cancer
Res. 11:8281–8287. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yee YK, Tan VP, Chan P, Hung IF, Pang R
and Wong BC: Epidemiology of colorectal cancer in Asia. J
Gastroenterol Hepatol. 24:1810–1816. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
6
|
Cheung DY, Kim TH, Kim CW, et al: The
anatomical distribution of colorectal cancer in Korea: evaluation
of the incidence of proximal and distal lesions and synchronous
adenomas. Intern Med. 47:1649–1654. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Islamic Republic of Iran Ministry of
Health and Medical Education, Office of Deputy Minister for Health
Center for Disease Control, Cancer Office. Iranian Annual National
Cancer Registration Report. 2005–2006. Mar;2007.
|
|
8
|
Fahy B and Bold RJ: Epidemiology and
molecular genetics of colorectal cancer. Surg Oncol. 7:115–123.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lin L, Li G, Zhang Z, et al: Association
of epidermal growth factor +61 A/G polymorphism in Chinese patients
with colon cancer. Genet Test Mol Biomarkers. 16:1142–1145.
2012.
|
|
10
|
Wilson KJ, Gilmore JL, Foley J, Lemmon MA
and Riese DJ II: Functional selectivity of EGF family peptide
growth factors: implications for cancer. Pharmacol Ther. 122:1–8.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Henson ES and Gibson SB: Surviving cell
death through epidermal growth factor (EGF) signal transduction
pathways: implications for cancer therapy. Cell Signal.
18:2089–2097. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li TF, Ren KW and Liu PF: Meta-analysis of
epidermal growth factor polymorphisms and cancer risk: involving
9,779 cases and 15,932 controls. DNA Cell Biol. 31:568–574.
2011.PubMed/NCBI
|
|
13
|
Normanno N, De Luca A, Bianco C, et al:
Epidermal growth factor receptor (EGFR) signaling in cancer. Gene.
366:2–16. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li LC, Chui RM, Sasaki M, et al: A single
nucleotide polymorphism in the E-cadherin gene promoter alters
transcriptional activities. Cancer Res. 60:873–876. 2000.PubMed/NCBI
|
|
15
|
Zhang Y-M, Cao C and Liang K: Genetic
polymorphism of epidermal growth factor 61A>G and cancer risk: a
meta-analysis. Cancer Epidemiol. 34:150–156. 2010. View Article : Google Scholar
|
|
16
|
Xu W, Li Y, Wang X, et al: Association
between EGF promoter polymorphisms and cancer risk: a
meta-analysis. Med Oncol. 27:1389–1397. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tan D, Xu J, Li Y and Lai R: Association
between +61G polymorphism of the EGF gene and glioma risk in
different ethnicities: a meta-analysis. Tohoku J Exp Med.
222:229–235. 2010.
|
|
18
|
Miller SA, Dykes DD and Polesky HF: A
simple salting out procedure for extracting DNA from human
nucleated cells. Nucleic Acids Res. 16:12151988. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kang HG, Choi JE, Lee WK, et al: +61A>G
polymorphism in the EGF gene does not increase the risk of lung
cancer. Respirology. 12:902–905. 2007.
|
|
20
|
Spindler KL, Nielsen JN, Ornskov D,
Brandslund I and Jakobsen A: Epidermal growth factor (EGF) A61G
polymorphism and EGF gene expression in normal colon tissue from
patients with colorectal cancer. Acta Oncol. 46:1113–1117. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Werner T: Functional in silico analysis of
non-coding SNPs. Bioinformatics for Geneticists. Barnes MR and Gray
IC: John Wiley and Sons, Ltd; Chichester: pp. 273–287. 2003
|
|
22
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar
|
|
23
|
Millar DS, Horan M, Chuzhanova NA and
Cooper DN: Characterisation of a functional intronic polymorphism
in the human growth hormone (GH1) gene. Hum Genomics. 4:289–301.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li L, Yun JH, Lee JH, Song S, Choi BC and
Baek KH: Association study of +45G15G(T/G) and +276(G/T)
polymorphisms in the adiponectin gene in patients with polycystic
ovary syndrome. Int J Mol Med. 27:283–287. 2011.
|