Introduction
Cholangiocarcinoma (CCA) is a malignancy arising
from the biliary tract epithelium (1). CCA is rare, with an annual incidence
of 2–3 cases per 100,000 individuals in the Western population
(2). However, it is the second most
common primary hepatic malignancy, with recent epidemiological
studies suggesting a progressively increasing incidence in Western
countries (3–5). Anatomically, CCA is classified as
intrahepatic (IH-CCA) or extrahepatic (EH-CCA). IH-CCA arises from
the intrahepatic ducts, which extend from the periphery of the
liver to the second-order bile ducts within the liver (6). EH-CCA is observed and defined in three
different growth patterns: periductal infiltrating, papillary or
intraductal and mass forming (7).
The high mortality rate of CCA is mainly due to its aggressive
behavior. Indeed, the majority of these tumors are diagnosed at a
late stage of disease progression, precluding surgical therapies
(8,9). Furthermore, CCA is characterized by a
marked resistance to chemotherapy (10,11).
Several drugs have been tested in phase II studies in unresectable
CCA (5-fluorouracil, gemcitabine methasulfon-m-anisidide,
cisplatin, rifampicin, mitomycin C and paclitaxel) with partial
response rates below 9% and average survival shorter than 12 months
(8). Therefore, studies for the
identification of key factors that play a critical role in tumor
chemosensitivity/resistance for the selection of patients with the
highest likelihood of responding to these therapies are urgently
required.
Polycomb group (PcG) proteins are epigenetic
chromatin modifiers involved in cancer development and their roles
are now being evaluated in numerous human malignancies (12). PcG proteins are essential for cancer
stem cell (CSC) self-renewal. PcG members are organized in two main
protein complexes: Polycomb repressive complex 1 and 2 (PRC1, PRC2)
(13). PRC2 is required in the
initial stage of silencing, through the histone H3 Lys 27
trimethylation (14) which
contributes to the recruitment of PRC1. PRC1 is required for stable
maintenance of the initiated PcG gene silencing, through histone
H2A ubiquitylation (14), on
specific target loci. Enhancer of zeste homolog 2 (EZH2) is the
catalytic subunit of PRC2; while B-cell-specific Moloney murine
leukemia virus integration site 1 (BMI-1) contributes to the
ubiquitin E3 ligase activity in PRC1. EZH2 is overexpressed in
poorly differentiated CCA (15).
Several genetic and epigenetic factors may be involved in the
deregulation and modulation of key signaling pathways in tumor
aggressiveness and chemoresistance. The abnormal expression of EZH2
is involved in the tumorigenic processes and is regarded as a
potential marker of aggressive types of cancer with poor prognoses
(16,17). Previous studies have demonstrated
that EZH2 contributes to the epigenetic silencing of several target
genes that control cell growth and proliferation, including
E-cadherin, Rb and p16 (18). The
overexpression of EZH2 may induce hypermethylation of the promoter
of the p16 gene, reducing the expression of p16, which is a key
step in the multistep cholangiocarcinogenesis from hepatolithiasis
to intraepithelial neoplasia (19).
A recent study described 26 single nucleotide
polymorphisms (SNPs) in the EZH2 locus, including SNPs correlated
with lung cancer risk (20).
Genetic analysis of germinal variants may be an easy-to-perform
prognostic tool, readily transferable into the clinic. Therefore,
the study of candidate polymorphisms of EZH2 as biomarkers of
clinical outcome may provide effective prognostic markers in CCA
patients. The aim of the current study was to evaluate a
correlation between EZH2 SNPs and clinical outcome in CCA
patients.
Materials and methods
Patients
A total of 75 patients with histologically confirmed
unresectable biliary tract cancer were enrolled in this
retrospective pharmacogenetic single-center study at the Department
of Oncology of Carrara Civic Hospital (Carrara, Italy), between
February 2004 and November 2010. CCA patients were treated upfront
with intravenous or intra-arterial cisplatin (Platinol®,
Bristol-Myers Squibb, Roma, Italy) and epirubicin
(Pharmorubicin®, Pfizer Italia S.R.L., Latina, Italy)
and oral capecitabine (Xeloda®, Roche S.p.A, Milano,
Italy; ECX regimen). This study was approved by the ethics review
board of Carrara Hospital. Informed consent was obtained from all
the patients.
DNA isolation
Genomic DNA was extracted from peripheral venous
blood samples (5 ml) from an antecubital vein of 75 CCA patients
and stored anonymously at −20°C in the Laboratory of VU University
Medical Center, Department of Medical Oncology (Amsterdam, The
Netherlands). Genomic DNA was isolated through the QIAamp DNA mini
kit (Qiagen, Venlo, The Netherlands). The purity and quantity of
DNA obtained were measured by spectrophotometer
NanoDrop®-1000-Detector (NanoDrop-Technologies,
Wilmington, DE, USA). The absorbance was read at 260 and 280 nm and
the contamination by proteins was estimated through the calculation
of 260/280 ratio.
In silico analysis
A total of 26 previously described EZH2 SNPs were
functionally tested by the appropriate software. SNPs were screened
through in silico characterization based on functional
relevance [missense mutation, transcription factor binding (TFB),
miRNA binding]. In particular, the PROMO3.0 (http://alggen.lsi.upc.es/cgibin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3)
in which human factors and binding sites were considered, with a
maximum matrix dissimilarity rate of 15 (21,22),
GeneCard (http://www.genecards.org/) and
MicroSNiper (http://cbdb.nimh.nih.gov/microsniper) software were
used (23).
SNP genotyping
EZH2 SNPs g.148525904C>G (rs2302427),
g.148519011C>T (rs6464926), g.148517456T>G (rs17171119) and
g.148505302 C>T (rs887569) were analyzed with real-time PCR.
Applied Biosystems SNP genotyping assays were used for reactions.
The PCR assays were performed using 20 ng of genomic DNA diluted in
5.94 μl DNase RNase free water, 6.25 μl of TaqMan Universal Master
Mix (Applied Biosystems, Foster City, CA, USA) with AmpliTaq Gold
and 0.3125 μl of the assay mix (specific primers and probe) in 12.5
μl total volume. The allelic content of each sample in the plate
was determined by reading the generated fluorescence.
Statistical analysis
All SNPs were examined for Hardy-Weinberg
equilibrium (24). Overall survival
(OS) and time to progression (TTP) curves were obtained through the
Kaplan-Meier method and the log rank test was used to compare the
survival distributions. P<0.05 was considered to indicate a
statistically significant result. The Cox regression model was used
to test the effect of g.148505302 C>T SNP and prognostic factors
on OS.
Results
Patient characteristics and
responses
Patient characteristics are summarized in Table I. The median ECOG performance status
and median Ca19.9 level at diagnosis were 1 and 204 IU/ml,
respectively. Of the 74 evaluable patients, 3 complete responses
(CR) were observed (4.1%), while a partial response (PR) was
observed in 10 of 74 evaluable patients (13.5%), stable disease
(SD) in 34 patients (45.9%) and progressive disease (PD) in 27
patients (36.5%). A median follow-up of 42.3 months revealed that
the median OS was 14.9 months (8.2–21.6) and the 1-year survival
rate was 56%.
 | Table IPatient characteristics. |
Table I
Patient characteristics.
| Characteristics | No. of patients |
|---|
| General |
| Total | 75 |
| Evaluable
disease | 74 |
| Median age (range),
years | 62.3 (26–80) |
| Gender |
| Male | 44 |
| Female | 31 |
| ECOG |
| 0 | 35 |
| 1 | 30 |
| 2 | 10 |
| Tumor diagnosis |
| Intrahepatic
cholangiocarcinoma | 56 |
| Gallbaldder
carcinoma | 11 |
| Common bile
duct | 8 |
| Sites of
metastases |
| Liver | 60 |
| Involvement
<50% | 40 |
| Involvement
>50% | 20 |
| Lymph nodes | 10 |
| Peritoneum | 7 |
| Local disease
recurrence | 6 |
| Other | 4 |
| Median Ca19.9
level (range, IU/ml) | 204 (0–11400) |
In silico characterization of g.148505302
C>T, g.148525904C>G, g.148519011C>T and
g.148517456T>G
Among the 26 SNPs in the EZH2 locus described by
Yoon et al(20), only 1 SNP
(g.148525904C>G) is located on an exon (exon 6) and is
responsible for an amino acid change (histidine/aspartate).
Conversely, 25 SNPs are located in EZH2 non-coding regions and they
may affect EZH2 expression by affecting miRNA, transcription
regulator binding or mRNA splicing. However, no SNPs affecting
miRNA binding or splicing were identified, while the PROMO 3.0
software detected noteworthy correlations between the allelic
variants of 3 of these EZH2 SNPs and TFB sites (Fig. 1). In g.148505302 C>T, the T
allele creates a binding site for the peroxisome
proliferator-activated receptor (PPAR)-α/retinoid-X-receptor
(RXR)-α. The C allele of g.148519011C>T creates a binding site
for E2F-1 and the G allele of g.148517456T>G creates a binding
site for Pax-5 and p53. All these TFs are expressed in CCA cell
lines (25–28).
SNP genotyping
The 4 SNPs selected (Table II) were successfully evaluated in
all available DNA samples. The g.148505302 C>T,
g.148519011C>T and g.148525904C>G SNPs were in Hardy-Weinberg
equilibrium. Conversely, the g.148517456T>G SNP did not follow
the Hardy-Weinberg equilibrium. However, all SNPs had frequencies
comparable to those observed in Caucasian populations reported in
Pubmed Reference-SNP (RefSNP).
 | Table IIPosition and functional
characteristics of the investigated SNPs. |
Table II
Position and functional
characteristics of the investigated SNPs.
| SNP | Position | Change | Comments |
|---|
|
g.148525904C>G | Exon 6 |
C:Histidine/G:Aspartate | |
|
g.148519011C>T | Intron 8 | C: E2F-1 TFB | TF expressed in CCA
(23) |
|
g.148517456T>G | Intron 8 | G: Pax-5 and p53
TFB | TF expressed in CCA
(24) |
| g.148505302
C>T | Intron 19 | A:PPAR-α/RXR-α
TFB | TF expressed in CCA
(25) |
EZH2 SNP correlation with clinical
outcome
No significant correlations were identified between
the g.148519011C>T, g.148525904C>G and g.148517456T>G SNPs
and clinical outcome, while the g.148505302 C>T (rs887569) SNP
had a significant association with OS. In particular, the patients
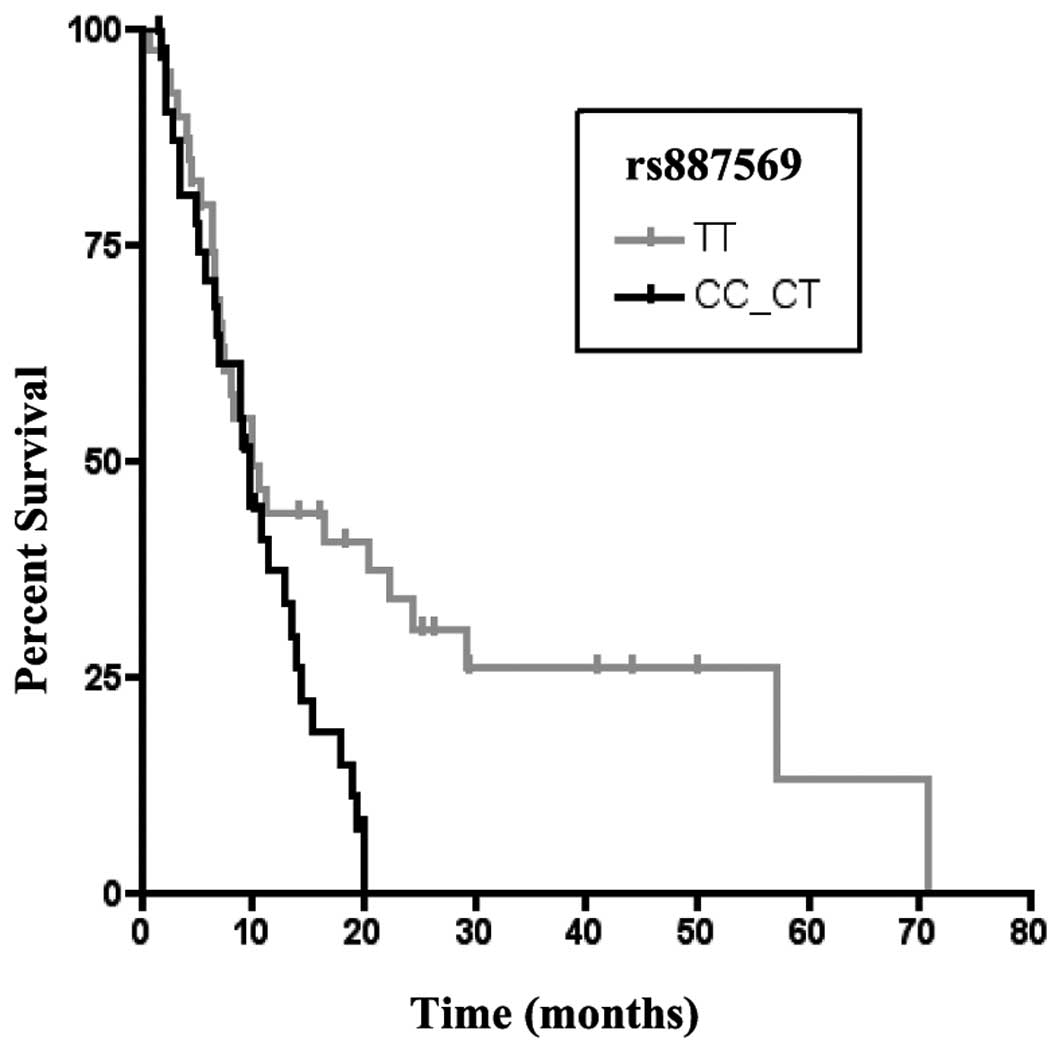
harboring the TT genotype had a significantly longer OS (TT vs.
CT-CC, P=0.036; Fig. 2). Moreover,
the TT genotype revealed a trend-like correlation with OS (P=0.075)
in multivariate analysis.
Discussion
The present study revealed that the g.148505302 T
allele is associated with a longer OS in CCA patients. The
g.148505302 C>T SNP is located in intron 19 of the EZH2 gene
(20). Through in silico
analysis it was revealed that the T allele creates a binding site
for the PPAR-α/RXR-α heterodimer. The PPARs were identified in 1990
by Issemann and Green (29). PPARs
are ligand-activated transcription factors that directly influence
the transcription of target genes (30). Three related PPAR isotypes have been
identified (PPAR-α, -β/δ and -γ) (31). PPAR-α binds to DNA as a
heterodimeric complex with RXR-α (32). This complex binds to a specific
sequence in regulatory regions of target genes, known as peroxisome
proliferator response element (PPRE), with two copies of a
hexameric nucleotide sequence, TGACCT-like (32). Several studies have suggested
differential mechanism underlying the role of PPAR-α in cancer,
including key roles in modulation of cell-cycle genes, cell
proliferation and cellular apoptosis (33). In accordance with these hypotheses,
the g.148505302 T allele, which binds the PPAR-α/RXR-α complex, may
trigger mechanisms involved in apoptosis and cell proliferation
inhibition by downregulation of the EZH2 oncogene. EZH2 is a
transcriptional repressor involved in cell proliferation (19). Overexpression of EZH2 is associated
with aggressive and metastatic disease in various types of cancer
(34), including CCA. In
particular, EZH2 is not expressed in the cholangiocytes or
hepatocytes of livers without tumors, but is overexpressed in
poorly differentiated carcinoma (35). Therefore, EZH2 expression may be a
predictor of the biological aggressiveness and poor prognosis in
CCA (36). Cell culture studies
have confirmed the expression of EZH2 mRNA in CCA cells, but not in
normal cells (35). These studies
have also demonstrated that when EZH2 is decreased by
suberoylanilide hydroxamic acid (SAHA; a histone deacetylase
inhibitor) treatment, the tumor suppressors p16, E-cadherin and p21
are activated (35). Previous
studies have shown that EZH2 expression had a stepwise increase in
aggressive and invasive CCA (36,37).
Since EZH2 drives CSC self-renewal and is associated with poor
prognosis in most malignancies, it is conceivable that these cells
contribute to the maintenance of the tumoral mass and are
implicated in CCA chemoresistance.
EZH2 expression and activity may be affected by
functional polymorphisms. SNP genotyping is particularly attractive
for tumors detected in the advanced stages, including CCA, since it
is an easy-to-perform analysis. This analysis may be performed with
a small volume of biological fluids (e.g. 200 μl of blood
specimens) in less than 24 h.
The current study demonstrates how a candidate EZH2
SNP may be a novel biomarker correlated with clinical outcome in
CCA patients. Due to the relatively small sample size and
retrospective design, further studies are required in order to
validate the prognostic role of this SNP in CCA. Our in
silico prediction should also be extended by appropriate
molecular analyses, which go beyond the scope of the present
analysis. In addition, the possibility that the g.148505302 C>T
SNP is in linkage disequilibrium with other polymorphic variants,
which may be responsible for the prognostic significance of this
marker, cannot be ruled out.
In conclusion, to the best of our knowledge, this is
the first study to show an EZH2 SNP having a significant impact on
CCA outcome, possibly through its role in the PPAR-α/RXR-α complex
interaction with EZH2. If these results are confirmed by larger
prospective studies, this EZH2 polymorphism may be useful for
predicting the clinical outcome in CCA patients.
Acknowledgements
This study was partially supported by grants from
the Netherlands Organization for Scientific Research (NWO) to Dr
Elisa Giovannetti and from the Ministero dell’Istruzione,
dell’Universita’ e Ricerca (PRIN 2008) to Dr Elisa Giovannetti and
Professor Romano Danesi.
References
|
1
|
Parkin DM, Ohshima H, Srivatanakul P and
Vatanasapt V: Cholangiocarcinoma: epidemiology, mechanisms of
carcinogenesis and prevention. Cancer Epidemiol Biomarkers Prev.
2:537–544. 1993.PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
3
|
Welzel TM, Graubard BI, El-Serag HB, Shaib
YH, Hsing AW, Davila JA and McGlynn KA: Risk factors for
intrahepatic and extrahepatic cholangiocarcinoma in the United
States: a population-based case-control study. Clin Gastroenterol
Hepatol. 5:1221–1228. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shaib YH, El-Serag HB, Nooka AK, Thomas M,
Brown TD, Patt YZ and Hassan MM: Risk factors for intrahepatic and
extrahepatic cholangiocarcinoma: a hospital-based case-control
study. Am J Gastroenterol. 102:1016–1021. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Albores-Saavedra J, Murakata L, Krueger JE
and Henson DE: Noninvasive and minimally invasive papillary
carcinomas of the extrahepatic bile ducts. Cancer. 89:508–515.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Farges O and Fuks D: Clinical presentation
and management of intrahepatic cholangiocarcinoma. Gastroenterol
Clin Biol. 34:191–199. 2010. View Article : Google Scholar
|
|
7
|
Lim JH and Park CK: Pathology of
cholangiocarcinoma. Abdom Imaging. 29:540–547. 2004.
|
|
8
|
Gatto M and Alvaro D: New insights on
cholangiocarcinoma. World J Gastrointest Oncol. 2:136–145. 2010.
View Article : Google Scholar
|
|
9
|
Leelawat K, Narong S, Wannaprasert J and
Ratanashu-ek T: Prospective study of MMP7 serum levels in the
diagnosis of cholangiocarcinoma. World J Gastroenterol.
16:4697–4703. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Seehofer D, Kamphues C and Neuhaus P:
Management of bile duct tumors. Expert Opin Pharmacother.
9:2843–2856. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
de Groen PC, Gores GJ, LaRusso NF,
Gunderson LL and Nagorney DM: Biliary tract cancers. N Engl J Med.
341:1368–1378. 1999.
|
|
12
|
Valk-Lingbeek ME, Bruggeman SW and van
Lohuizen M: Stem cells and cancer; the polycomb connection. Cell.
118:409–418. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lund AH and van Lohuizen M: Polycomb
complexes and silencing mechanisms. Curr Opin Cell Biol.
16:239–246. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mathews LA, Crea F and Farrar WL:
Epigenetic gene regulation in stem cells and correlation to cancer.
Differentiation. 78:1–17. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu DC and Yang ZL: Overexpression of EZH2
and loss of expression of PTEN is associated with invasion,
metastasis and poor progression of gallbladder adenocarcinoma.
Pathol Res Pract. 207:472–478. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Varambally S, Dhanasekaran SM, Zhou M,
Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt
RG, Otte AP, Rubin MA and Chinnaiyan AM: The polycomb group protein
EZH2 is involved in progression of prostate cancer. Nature.
419:624–629. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kleer CG, Cao Q, Varambally S, Shen R, Ota
I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, Sabel MS,
Livant D, Weiss SJ, Rubin MA and Chinnaiyan AM: EZH2 is a marker of
aggressive breast cancer and promotes neoplastic transformation of
breast epithelial cells. Proc Natl Acad Sci USA. 100:11606–11611.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jacobs JJ, Kieboom K, Marino S, DePinho RA
and van Lohuizen M: The oncogene and Polycomb-group gene bmi-1
regulates cell proliferation and senescence through the ink4a
locus. Nature. 397:164–168. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sasaki M, Yamaguchi J, Itatsu K, Ikeda H
and Nakanuma Y: Over-expression of polycomb group protein EZH2
relates to decreased expression of p16 INK4a in
cholangiocarcinogenesis in hepatolithiasis. J Pathol. 215:175–183.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yoon KA, Gil HJ, Han J, Park J and Lee JS:
Genetic polymorphisms in the polycomb group gene EZH2 and the risk
of lung cancer. J Thorac Oncol. 5:10–16. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Messeguer X, Escudero R, Farré D, Núñez O,
Martinez J and Albà MM: PROMO: detection of known transcription
regulatory elements using species-tailored searches.
Bioinformatics. 18:333–334. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Farré D, Roset R, Huerta M, Adsuara JE,
Rosello L, Albà MM and Messeguer X: Identification of patterns in
biological sequences at the ALGGEN server: PROMO and MALGEN.
Nucleic Acids Res. 31:3651–3653. 2003.PubMed/NCBI
|
|
23
|
Barenboim M, Zoltick BJ, Guo Y and
Weinberger DR: MicroSNiPer: a web tool for prediction of SNP
effects on putative microRNA targets. Hum Mutat. 31:1223–1232.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rodriguez S, Gaunt TR and Day IN:
Hardy-Weinberg equilibrium testing of biological ascertainment for
Mendelian randomization studies. Am J Epidemiol. 169:505–514. 2009.
View Article : Google Scholar
|
|
25
|
Yoon JH, Higuchi H, Werneburg NW, Kaufmann
SH and Gores GJ: Bile acids induce cyclooxygenase-2 expression via
the epidermal growth factor receptor in a human cholangiocarcinoma
cell line. Gastroenterology. 122:985–993. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kim EM, Kim JS, Choi MH, Hong ST and Bae
YM: Effects of excretory/secretory products from Clonorchis
sinensis and the carcinogen dimethylnitrosamine on the
proliferation and cell cycle modulation of human epithelial HEK293T
cells. Korean J Parasitol. 46:127–132. 2008.PubMed/NCBI
|
|
27
|
Zheng T, Wang J, Chen X, Meng X, Song X,
Lu Z, Jiang H and Liu L: Disruption of p73-MDM2 binding synergizes
with gemcitabine to induce apoptosis in HuCCT1 cholangiocarcinoma
cell line with p53 mutation. Tumour Biol. 31:287–295. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Han C, Lim K, Xu L, Li G and Wu T:
Regulation of Wnt/beta-catenin pathway by cPLA2alpha and PPARdelta.
J Cell Biochem. 105:534–545. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Issemann I and Green S: Activation of a
member of the steroid hormone receptor superfamily by peroxisome
proliferators. Nature. 347:645–650. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Evans RM: The steroid and thyroid hormone
receptor superfamily. Science. 240:889–895. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Youssef J and Badr M: Enhanced
hepatocarcinogenicity due to agonists of peroxisome
prolifer-atoractivated receptors in senescent rats: role of
peroxisome proliferation, cell proliferation and apoptosis.
ScientificWorldJournal. 2:1491–1500. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kliewer SA, Umesono K, Noonan DJ, Heyman
RA and Evans RM: Convergence of 9-cis retinoic acid and peroxisome
proliferator signalling pathways through heterodimer formation of
their receptors. Nature. 358:771–774. 1992. View Article : Google Scholar
|
|
33
|
Tugwood JD, Issemann I, Anderson RG,
Bundell KR, McPheat WL and Green S: The mouse peroxisome
proliferator activated receptor recognizes a response element in
the 5′ flanking sequence of the rat acyl CoA oxidase gene. EMBO J.
11:433–439. 1992.PubMed/NCBI
|
|
34
|
Chase A and Cross NC: Aberrations of EZH2
in cancer. Clin Cancer Res. 17:2613–2618. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yamaguchi J, Sasaki M, Sato Y, Itatsu K,
Harada K, Zen Y, Ikeda H, Nimura Y, Nagino M and Nakanuma Y:
Histone deacetylase inhibitor (SAHA) and repression of EZH2
synergistically inhibit proliferation of gallbladder carcinoma.
Cancer Sci. 101:355–362. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sasaki M, Ikeda H, Itatsu K, Yamaguchi J,
Sawada S, Minato H, Ohta T and Nakanuma Y: The overexpression of
polycomb group proteins Bmi1 and EZH2 is associated with the
progression and aggressive biological behavior of hepatocellular
carcinoma. Lab Invest. 88:873–882. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Nakanuma Y, Sasaki M, Sato Y, Ren X, Ikeda
H and Harada K: Multistep carcinogenesis of perihilar
cholangiocarcinoma arising in the intrahepatic large bile ducts.
World J Hepatol. 1:35–42. 2009. View Article : Google Scholar : PubMed/NCBI
|