Introduction
The etiology and pathogenesis of gastric cancer
remains enigmatic. Cloning and functional studies of gastric
cancer-related genes are important for revealing the gene changes
and molecular mechanisms in gastric cancer. As in other
malignancies, the development of gastric cancer is a pathological
process of an accumulation of multigene, multistage mutations
(1–3). A high frequency of loss of
heterozygosity (LOH) has been reported on chromosome 1p35–36 for a
variety of tumors (4,5), indicating that tumor-related genes
exist that have not been cloned in this chromosomal region.
Igarashi et al identified a high frequency LOH in the
chromosome 1p35–36 region in gastric cancer (6). The downregulation of a newly
identified gene of the expressed sequence tag segment, MDSCBC11, in
the chromosome 1p35–36 region was detected in our previous
investigation using cDNA microarray analysis (7). In the present study, the
MDSCBC11-represented gene was cloned and its full cDNA sequence was
obtained. Gene expression was investigated in gastric carcinomas,
adjacent gastric mucosa and normal gastric mucosa.
Materials and methods
Northern blotting
Total RNA was extracted from normal human liver
tissue and the primer was designed according to the sequence of
MDSCBC11. RT-PCR was performed using isotope 32P-labeled
dCTP as the substrate as follows: 30 cycles of 95°C for 5 min, 94°C
for 35 sec, 56°C for 35 sec and 72°C for 35 sec, and a final
extension at 72°C for 10 min. The PCR product, which was used as
the probe in the following study, was sequenced to confirm that it
was consistent with the sequence of MDSCBC11. A northern blot was
performed using multiple-tissue northern blots (MTN; Cat. no.
636803; Clontech Inc., Mountain View, CA, USA) with the purified
probe.
Rapid amplification of cDNA ends (RACE)
experiment to obtain the full-length cDNA of the
MDSCBC11-represented gene
Total RNA was extracted from normal fetal liver
tissue, and cDNA was obtained by a two-step RT-PCR. Following this,
5′ RACE and 3′ RACE procedures were performed following the
instructions of the SMART™ RACE cDNA Amplification kit (Cat. no.
634914; Clontech Inc.). The primers and amplification conditions
are shown in Table I. The PCR
products were sequenced. According to sequencing results, the
overlapped sequences of the 5′-RACE and 3′-RACE cDNA fragments were
removed and the full-length cDNA was obtained.
 | Table IPrimer sequences and amplification
conditions of the MDSCBC11 segment and RACE. |
Table I
Primer sequences and amplification
conditions of the MDSCBC11 segment and RACE.
| Primer | Sequence (5′-3′) | Annealing
temperature, °C | Product size, bp |
|---|
| MDSCBC11 |
| F |
GCGATGTAACACGAGAAAG | 55 | 392 |
| R |
GGAAATGGTGAAGGGAGAC | | |
| GAPDH |
| F |
AACTGTGGCGTGATGGCCGC | 58 | 500 |
| R |
GCAGGGACTCCCCAGCAGTG | | |
| RACE |
| F |
GCACATACCAAGGCCACCACACA | 57 | |
| R |
CAGGCATCACCCCGCTAAATCCC | | |
Bioinformatics analysis
The bioinformatics-associated software and website
(http://www.ncbi.nlm.nih.gov/mapview/)
were adopted to predict and analyze the structure and homology of
the MDSCBC11-represented gene.
Detection of ELCOX3 expression in gastric
cancer
Specimens from 46 patients with gastric cancer who
had not been administered radiotherapy, chemotherapy or other
anti-tumor treatments were collected at the First Affiliated
Hospital of the University of South China (Hengyang, Hunan, China).
The study was approved by the ethics committee of University of
South China (Hengyang, China). Written informed consent was
obtained from the patients. Three sections consisting of gastric
carcinoma, adjacent gastric mucosa at 1–2 cm from the cancer-foci
edge and normal gastric mucosa from the cancer-foci edge (>5 cm)
were isolated from each specimen within 30 min of being excised
from the body. Two to three pieces (~100 mg) for each section were
prepared and the tissue pieces were individually transferred to
Eppendorf microcentrifuge tubes for liquid nitrogen
cryopreservation subsequent to being rinsed with aseptic 0.1%
diethypyrocarbonate water. Among the 46 cases, 26 were male and 20
were female, with an age range of 26–77 years (mean, 56 years).
There were 11 cases of gastric fundus or cardia cancer, 17 of
gastric body cancer and 18 of gastric antrum cancer. There were 19
cases of well- or moderately-differentiated adenocarcinoma,
including mucinous adenocarcinoma, and 27 cases of
poorly-differentiated adenocarcinoma. Of the total cases, 18 were
Borrmann type I+II and 28 cases were type III+IV. There were 24
cases of TNM stages I+II and 22 cases of stages III+IV. Lymph node
metastasis was observed in 30 cases and was absent in 16. The
clinical data for all the cases were available and all specimens
were confirmed by histopathology.
RNA was extracted according to the manufacturer's
instructions for the EZNA Total RNA kit (Omega Corp., Guangzhou,
China) A two-step RT-PCR procedure was performed and primers were
designed according to the sequence of the target gene and the
reference glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene,
which were synthesized by Shanghai Invitrogen Biotechnology Co.,
Ltd. (Shanghai, China). The primer sequences and amplified fragment
lengths were as follows: ELCOX3 forward, 5′-CGCGATGTAACACGAGAAAG-3′
and reverse, 5′-TATTAGTTGGCGGATGAAGC-3′ (PCR product size, 500 bp);
and reference GAPDH gene forward, 5′-GTCAGTGGTGGACCTGACCT-3′ and
reverse, 5′-TGAGGAGGGGAGATTCAGTG-3′ (PCR product size, 400 bp).
Statistical analysis
All data are expressed as mean ± SD. Data were
analyzed using one-way ANOVA, t-test and Spearman's correlation
coefficient. P<0.05 was considered to indicate a statistically
significant difference.
Results
Size and tissue distribution of
MDSCBC11-represented gene transcript
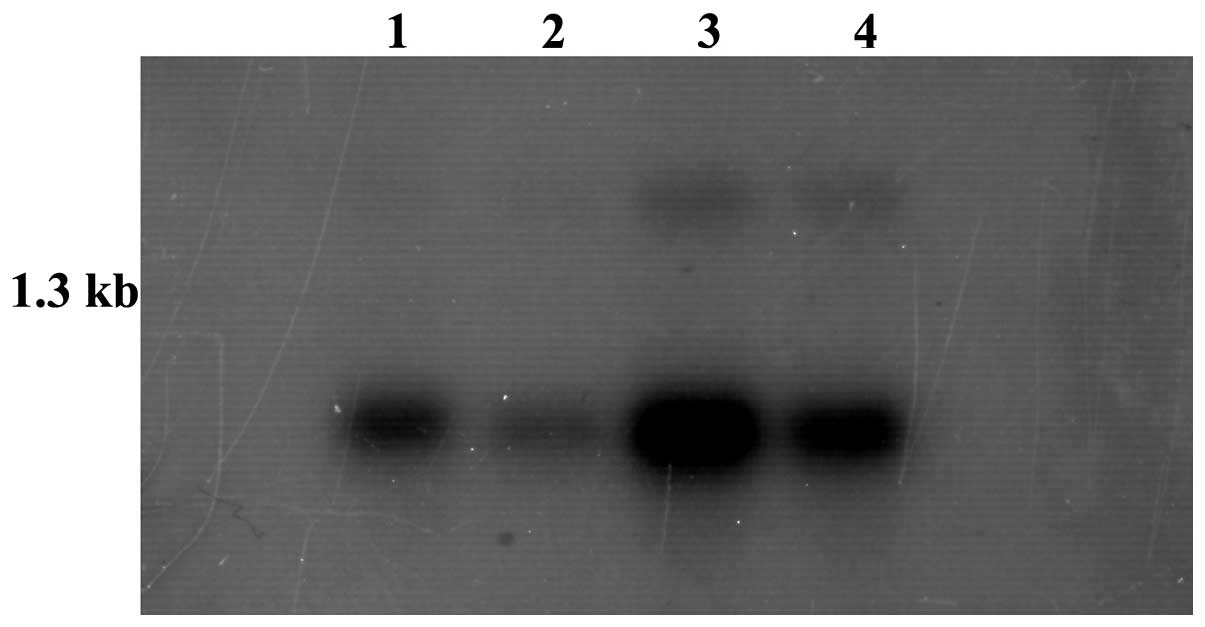
A northern blot of α-32P-dCTP-labeled
MDSCBC11 cDNA with MTN (Cat. no. 636803) revealed that the
MDSCBC11-represented gene was expressed in the liver, brain, kidney
and lungs. The kidney exhibited the highest expression level and
the lung exhibited the lowest expression level. There were two
transcripts in the kidney and liver, with a small transcript of 0.8
kb and a larger transcript of 1.5 kb (Fig. 1).
Cloning of the full-length cDNA of the
small transcript (0.8 kb)
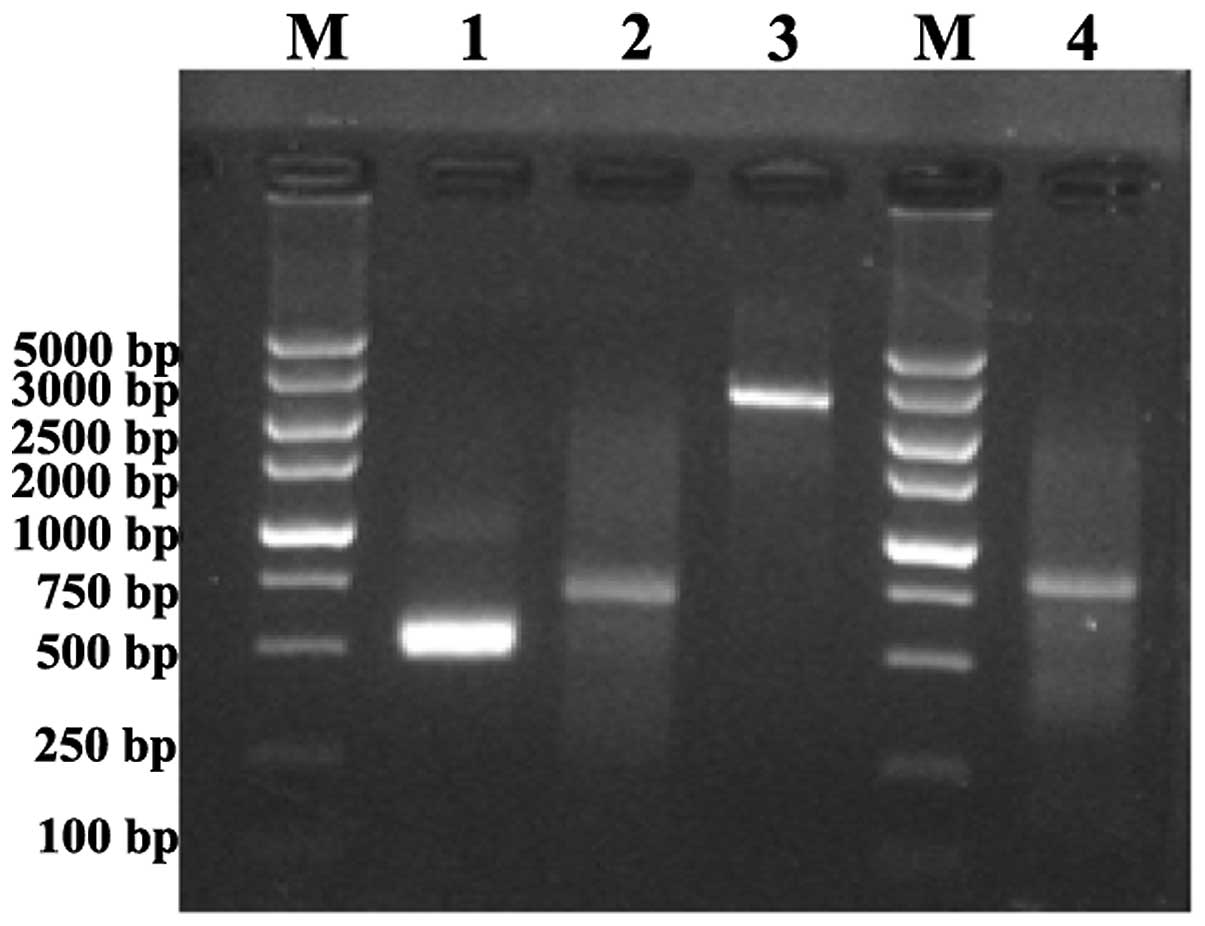
The distribution of the 5′ and 3′ end sequences were
acquired by RACE using RNA that was extracted from normal embryonic
liver tissue by the SMART RACE cDNA Amplification kit (Cat. no.
634914). An agarose gel electrophoretogram revealed that the
5′-RACE product was ~700 bp in size and the 3′-RACE product was
~500 bp in size (Fig. 2). The PCR
products were sent to Takara (Kyoto, Japan) for sequencing.
According to the sequencing results, the overlapped sequences of
5′-RACE and 3′-RACE cDNA fragments were removed and a full-length
cDNA of 822 bp was obtained.
The full-length cDNA of the small transcript
sequence is shown in Fig. 3.
Bioinformatics analysis of ELCOX3
The results of the BLAST analysis (blast.ncbi.nlm.nih.gov/Blast.cgi) for the
MDSCBC11 cDNA indicated that the cDNA of the MDSCBC11 small
transcript had 99% homology with the human cytochrome c
oxidase subunit III (COX3) gene in the mitochondria and was
therefore named ELCOX. The ELCOX3 and COX3 genes were blasted and
it was observed that the cDNA sequence of COX3 at the 635th C
changed to the T of ELCOX3, which resulted in an amino acid codon
change from UCA to UUA and an amino acid change at residue 212 from
serine to leucine. The results of the chromosome location analysis
demonstrated that the ELCOX3 gene was located inside the
mitochondria.
Using the open reading frame (ORF) finder server of
NCBI (http://www.ncbi.nlm.nih.gov/gorf/gorf.html), the
sequence had a complete ORF, which encoded 261 amino acids from the
8th to 793rd bases. The results of the analysis by ExPASy and NCBI
BLAST indicated that the ELCOX3 encoded protein had 99% homology
with the human COX3 gene and that no CpG islands or introns were
detected in the 5′ untranslated regions. The encoded protein was a
weak acidic protein with a isoelectric point of 6.78 and a
molecular weight (MW) of 29.97 kDa. The domain prediction results
revealed that the ELCOX3 encoded protein was a type of COX,
polychain transmembrane protein and telomerase, which exists in
eukaryotes and the majority of bacteria.
Expression level of ELCOX3 in gastric
cancer
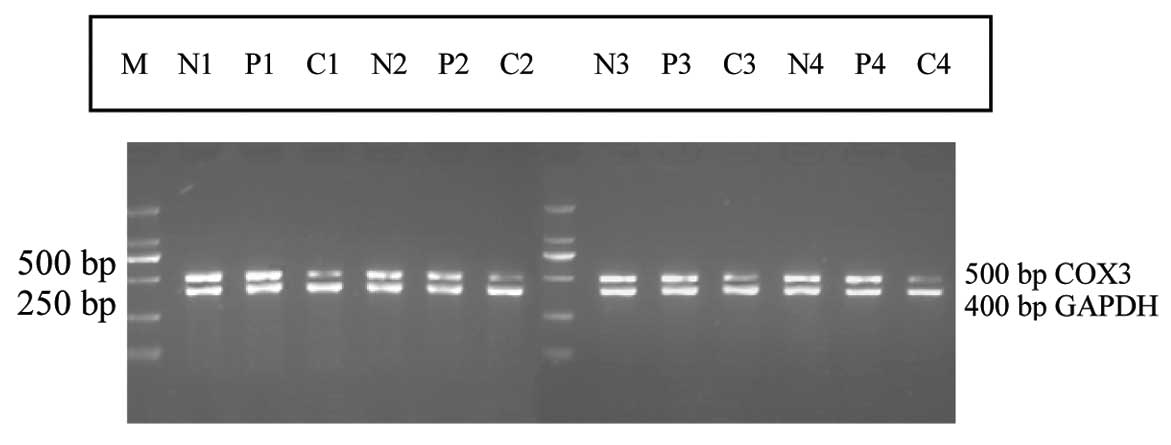
From the RT-PCR detection results, the size of the
ELCOX3 gene product in gastric mucosa was recorded as 500 bp, while
the internal reference of the GAPDH gene product was 400 bp
(Fig. 4). The expression level of
ELCOX3 in the gastric carcinoma samples was lower than in the
adjacent gastric mucosa and normal gastric mucosa samples, with a
downregulation of 23.91% (11/46 cases). The optical density ratio
analysis revealed that the relative expression values of ELCOX3
mRNA in the gastric carcinomas, adjacent gastric mucosa and normal
gastric mucosa were 0.7012±0.1920, 1.1128±0.1605 and 1.1356±0.1537,
respectively. The expression level in the gastric carcinomas was
significantly lower than that in the corresponding normal gastric
mucosa (P=0.016; P<0.05), while there was no significant
difference between the corresponding adjacent gastric mucosa and
the normal gastric mucosa (P=0.812; P>0.05).
The analysis of the correlation between the
clinicopathological parameters of the gastric cancer cases and
ELCOX3 expression in the gastric carcinomas demonstrated that the
expression of ELCOX3 mRNA in the gastric carcinomas was not
correlated with gender, age, tumor size, Borrman classification,
differentiation degree, invasion depth or TNM stage (P>0.05). No
significant correlation was identified between the downregulation
of ELCOX3 mRNA in primary gastric carcinoma and lymph node
metastasis (Spearman's correlation coefficient, r=0.088; P=0.559;
P>0.05).
Discussion
Gastric cancer is the result of the interaction of
genetic, environmental and other factors, involving changes in the
expression and regulation of a large number of genes. In the
present study, MDSCBC11 was selected from the
differentially-expressed genes that are associated with gastric
cancer by cDNA microarray (7) to
perform RACE, and 5′ and 3′ end products were acquired. A
full-length cDNA of 822 bp was obtained. This gene had no intron
and its ORF was located at the 8th to 793rd bases, which encoded
261 amino acids and had a MW of 29.97 kDa. BLAST analysis indicated
that ELCOX3 had 99% homology with the COX3 gene. MTN revealed that
the MDSCBC11-represented gene had two transcripts, a small
transcript of 0.8 kb and a larger transcript of 1.5 kb. The
full-length cDNA of the 0.8 kb transcript was obtained in the
present study and the 1.5 kb transcript, which may be a new gene
that has not been cloned, remains to be investigated.
The animal mitochondrial genome (mtDNA) contains 13
protein genes, including the three subunits of COX, subunits I, II
and III (COX1, 2 and 3, respectively), and Cytb, ATP6 and ATP8.
These genes are significant components for the inner mitochondrial
membrane respiratory chain. The homology of COX1, 2 and 3 is ~80%.
Therefore, the cloning and analysis of these genes remains an
effective way to investigate the phylogeny and characterization of
distant relatives (8,9). Subunits I, II and III of COX are
encoded in the mitochondrial genome of eukaryotes and are
evolutionarily conserved from bacteria to humans (10). These three subunits constitute the
catalytic core of mitochondrial oxidase, as well as the catalytic
core of all bacteria aa3 type COX (11–14).
Subunit III is more conservative than subunits I and II and cannot
transfer electrons directly since it contains no metal centers, but
is able to pump protons through cytochrome oxidase. The effect of
cytochrome oxidase decreases when the level of subunit III is
reduced. However, the specific mechanism is unclear (15–17).
In the present study, the stop codon of human ELCOX3 was shown to
be T, as in Anopheles quadrimaculatus, Anopheles
gambiae and Penaeus monodon (tiger prawns), and a polyA
tail is required to be added to codon T as the final stop codon
(18–20). There were certain differences
between the human COX3 and ELCOX3 sequences. With the exception of
codons 1–7, there was one base change; the 635th C changed to a T.
The ELCOX3-represented protein was 99% homologous with the human
COX3 protein, with only the 212th serine changed to leucine.
mt-COX is a rate-limiting enzyme in cell respiration
chain transmission. mt-COX cooperates with cytochrome c and
plays a significant role in cell mitochondria apoptosis.
mt-COX-encoded gene mutations or expression changes may induce
biological characteristics and functional changes in its
corresponding protein, which thus makes the cell abnormal. mt-COX
gene mutation is associated with the development of tumors
(21–25), and COX plays a specific role in
tumor development, mainly through an increase in reactive oxygen
species in mitochondria oxidative phosphorylation.
Semi-quantitative RT-PCR introduced an internal
reference as a contrast. The scanning density ratio of the target
gene and internal reference gene as the relative expression of the
target gene may not only confirm the integrity of RNA extraction
and the success of RT-PCR, but also provides the target gene with a
quantitative criteria (26,27). In the present study, ELCOX3 mRNA
expression in gastric carcinomas, corresponding adjacent gastric
mucosa and distal normal gastric mucosa was detected by RT-PCR. The
results demonstrated that the expression level in the gastric
carcinoma samples was significantly lower than that in the
corresponding normal gastric mucosa (P<0.05), while there was no
significant difference between the adjacent gastric mucosa and the
corresponding normal gastric mucosa (P>0.05). Compared with the
corresponding normal gastric mucosa, the expression levels of
ELCOX3 mRNA in the gastric carcinomas were downregulated at a rate
of 23.91% (11/46 cases). The down regulation of the ELCOX3 mRNA was
not correlated with lymph node metastasis. Therefore, ELCOX3 mRNA
downregulation may be an early event during the course of gastric
cancer and may be associated with the pathogenesis of human gastric
cancer.
In summary, the full cDNA sequence of the small
transcript of the MDSCBC11-represented gene was identified to be
822 bp in size and was named ELCOX3 due to the homology with the
COX3 gene in the human mitochondria. The downregulation of ELCOX3
gene expression was shown to be associated with the development of
human gastric carcinomas.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (grant nos. 30772116 and 81172576), the
Hengyang Natural Science Foundation of Hunan Province (grant no.
10JJ8006) and the Foundation of the Construct Program of the Key
Discipline in Hunan Province (no. 2011-76).
Abbreviations:
|
COX3
|
cytochrome c oxidase III
|
References
|
1
|
Jaiswal BS, Kljavin NM, Stawiski EW, Chan
E, Parikh C, et al: Oncogenic ERBB3 mutations in human cancers.
Cancer Cell. 13:603–617. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Otani K, Li X, Arakawa T, Chan FK and Yu
J: Epigenetic-mediated tumor suppressor genes as diagnostic or
prognostic biomarkers in gastric cancer. Expert Rev Mol Diagn.
13:445–455. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yoon K, Lee S, Han TS, Moon SY, Yun SM, et
al: Comprehensive genome- and transcriptome-wide analyses of
mutations associated with microsatellite instability in Korean
gastric cancers. Genome Res. 23:1109–1117. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang Q, Diskin S, Rappaport E, Attiyeh E,
Mosse Y, Shue D, Seiser E, Jagannathan J, Shusterman S, Bansal M,
Khazi D, Winter C, Okawa E, Grant G, Cnaan A, Zhao H, Cheung NK,
Gerald W, London W, Matthay KK, Brodeur GM and Maris JM:
Integrative genomics identifies distinct molecular classes of
neuroblastoma and shows that multiple genes are targeted by
regional alterations in DNA copy number. Cancer Res. 66:6050–6062.
2006. View Article : Google Scholar
|
|
5
|
Janoueix-Lerosey I, Novikov E, Monteiro M,
Gruel N, Schleiermacher G, Loriod B, Schleiermacher G, Loriod B,
Nguyen C and Delattre O: Gene expression profiling of 1p35–36 genes
in neuroblastoma. Oncogene. 23:5912–5922. 2004.
|
|
6
|
Igarashi J, Nimura Y, Fujimori M, Mihara
M, Adachi W, Kageyama H and Nakagawara A: Allelic loss of the
region of chromosome 1p35-pter is associated with progression of
human gastric carcinoma. Jpn J Cancer Res. 91:797–801. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xie HL, Li ZY, Gan RL, Li XJ, Zhang QL,
Hui M and Zhou XT: Differential gene and protein expression in
primary gastric carcinomas and their lymph node metastases as
revealed by combined cDNA microarray and tissue microarray
analysis. J Dig Dis. 11:167–175. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Baden KN, Murray J, Capaldi RA and
Guillemin K: Early developmental pathology due to cytochrome
c oxidase deficiency is revealed by a new zebrafish model. J
Biol Chem. 282:34839–34849. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fontanesi F, Soto IC and Barrientos A:
Cytochrome c oxidase biogenesis: new levels of regulation.
IUBMB Life. 60:557–568. 2008.
|
|
10
|
Fontanesi F, Soto IC, Horn D and
Barrientos A: Assembly of mitochondrial cytochrome
c-oxidase, a complicated and highly regulated cellular
process. Am J Physiol Cell Physiol. 291:C1129–C1147.
2006.PubMed/NCBI
|
|
11
|
Saraste M: Oxidative phosphorylation at
the fin de siécle. Science. 283:1488–1493. 1999.
|
|
12
|
Muramoto K, Ohta K, Shinzawa-Itoh K, Kanda
K, Taniguchi M, Nabekura H, Yamashita E, Tsukihara T and Yoshikawa
S: Bovine cytochrome c oxidase structures enable O2
reduction with minimization of reactive oxygens and provide a
proton-pumping gate. Proc Natl Acad Sci USA. 107:7740–7745.
2010.PubMed/NCBI
|
|
13
|
Hino T, Matsumoto Y, Nagano S, Sugimoto H,
Fukumori Y, Murata T, Iwata S and Shiro Y: Structural basis of
biological N2O generation by bacterial nitric oxide
reductase. Science. 330:1666–1670. 2010.
|
|
14
|
Svensson-Ek M, Abramson J, Larsson G,
Törnroth S, Brzezinski P and Iwata S: The X-ray crystal structures
of wild-type and EQ(I–286) mutant cytochrome c oxidases from
Rhodobacter sphaeroides. J Mol Biol. 321:329–339.
2002.PubMed/NCBI
|
|
15
|
Nguyen XT, Pabarue HA, Geyer RR, Shroyer
LA, Estey LA, Parilo MS, Wilson KS and Prochaska LJ: Biochemical
and biophysical properties of purified phospholipid vesicles
containing bovine heart cytochrome c oxidase. Protein Expr
Purif. 26:122–130. 2002. View Article : Google Scholar
|
|
16
|
Soto IC, Fontanesi F, Valledor M, Horn D,
Singh R and Barrientos A: Synthesis of cytochrome c oxidase
subunit 1 is translationally downregulated in the absence of
functional F1F0-ATP synthase. Biochim Biophys Acta. 1793:1776–1786.
2009.PubMed/NCBI
|
|
17
|
Hosler JP: The influence of subunit III of
cytochrome c oxidase on the D pathway, the proton exit
pathway and mechanism-based inactivation in subunit I. Biochim
Biophys Acta. 1655:332–339. 2004.PubMed/NCBI
|
|
18
|
Mitchell SE, Cockburn AF and Seawright JA:
The mitochondrial genome of Anopheles quadrimaculatus
species A: complete nucleotide sequence and gene organization.
Genome. 36:1058–1073. 1993.PubMed/NCBI
|
|
19
|
Beard CB, Hamm DM and Collins FH: The
mitochondrial genome of the mosquito Anopheles gambiae: DNA
sequence, genome organization, and comparisons with mitochondrial
sequences of other insects. Insect Mol Biol. 2:103–124.
1993.PubMed/NCBI
|
|
20
|
Wilson K, Cahill V, Ballment E and Benzie
J: The complete sequence of the mitochondrial genome of the
crustacean Penaeus monodon: are malacostracan crustaceans
more closely related to insects than to branchiopods? Mol Biol
Evol. 17:863–874. 2000.PubMed/NCBI
|
|
21
|
Ray AM, Zuhlke KA, Levin AM, Douglas JA,
Cooney KA and Petros JA: Sequence variation in the mitochondrial
gene cytochrome c oxidase subunit I and prostate cancer in
African American men. Prostate. 69:956–960. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Athar M, Chaudhury NK, Hussain ME and
Varshney R: Hoechst 33342 induced reactive oxygen species and
impaired expression of cytochrome c oxidase subunit 1
leading to cell death in irradiated human cancer cells. Mol Cell
Biochem. 352:281–292. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Payne CM, Holubec H, Bernstein C,
Bernstein H, Dvorak K, Green SB, Wilson M, Dall'Agnol M, Dvorakova
B, Warneke J and Garewal H: Crypt-restricted loss and decreased
protein expression of cytochrome C oxidase subunit I as
potential hypothesis-driven biomarkers of colon cancer risk. Cancer
Epidemiol Biomarkers Prev. 14:2066–2075. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bernstein C, Facista A, Nguyen H, Zaitlin
B, Hassounah N, Loustaunau C, Payne CM, Banerjee B, Goldschmid S,
Tsikitis VL, Krouse R and Bernstein H: Cancer and age related
colonic crypt deficiencies in cytochrome c oxidase I. World
J Gastrointest Oncol. 2:429–442. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gutierrez-Gonzalez L, Graham TA,
Rodriguez-Justo M, Leedham SJ, Novelli MR, Gay LJ, Ventayol-Garcia
T, Green A, Mitchell I, Stoker DL, Preston SL, Bamba S, Yamada E,
Kishi Y, Harrison R, Jankowski JA, Wright NA and McDonald SA: The
clonal origins of dysplasia from intestinal metaplasia in the human
stomach. Gastroenterology. 140:1251–1260. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ding Y, Zhang H, Zhong M, Zhou Z, Zhuang
Z, Yin H, Wang X and Zhu Z: Clinical significance of the uPA system
in gastric cancer with peritoneal metastasis. Eur J Med Res.
18:282013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li C, Wang L, Su J, Zhang R, Fu L and Zhou
Y: mRNA expression and hypermethylation of tumor suppressor genes
apoptosis protease activating factor-1 and death-associated protein
kinase in oral squamous cell carcinoma. Oncol Lett. 6:280–286.
2013.PubMed/NCBI
|