Introduction
microRNAs (miRNAs/miRs) are non-coding RNA
molecules, 21–23 nucleotides long, that regulate gene expression at
the post-transcriptional level (1–3). miRNA
expression profiling analyses have revealed a global downregulation
of mature miRNA levels in primary human tumours relative to normal
tissues (4,5). Thus, miRNAs may function as tumour
suppressors or oncogenes, and dysregulated miRNA expression may
contribute to tumour cell metastasis.
Metastasis is a complex, multi-step and dynamic
biological event, and a critical process in the metastatic cascade
is the epithelial-to-mesenchymal transition (EMT). EMT is regulated
by a variety of signalling pathways that originate from the stroma
that surrounds cancer cells; these pathways are activated by
cytokines, including transforming growth factor-β (TGFβ),
hepatocyte growth factor, platelet-derived growth factor, epidermal
growth factor and integrin engagement, all of which converge at the
level of key transcription factors, ZEB, SNAIL and TWIST (6). Recently, the mechanisms behind EMT
have been elucidated, and the pathways involved in epithelial
marker downregulation and the corresponding mesenchymal marker
upregulation have been better characterised (7). One area of significant progress has
been the identification of the critical roles of miRNAs in these
processes.
The concept of miRNAs as powerful regulators of EMT
has transformed the conventional narrative of carcinoma
progression, and several miRNAs have now been described as crucial
EMT regulators. These miRNAs dynamically affect the balance between
EMT and the reverse process, termed mesenchymal-to-epithelial
transition (MET) (8). The
overexpression of miR-7 has been shown to partially reverse EMT to
MET in gastric cancer by targeting the insulin-like growth factor-1
receptor (IGF1R) (9). TGFβ appears
to play a dominant role by directly activating the ZEB, SNAIL and
TWIST transcription factors; the master regulators of EMT (6). A number of studies profiling miRNA
expression have been conducted to identify candidate miRNAs with
possible roles in TGFβ-induced EMT (10–14).
Several miRNAs have been shown to directly target families of EMT
transcription factors, including the miR-200 family; the members of
which target ZEB1 and ZEB2 (15).
Certain signalling pathways are also involved in EMT. Du et
al identified that miR-34a downregulation promoted EMT by
targeting the Notch signalling pathway in tubular epithelial cells
(16).
Recently, Xu et al identified that the
miR-503 expression level was downregulated in endometrial cancer
(EEC) cells, and that a relatively high miR-503 level in EEC
tissues indicates a longer EEC patient survival time (17). In addition, our previous study
revealed that miR-503 expression was downregulated in gastric
carcinoma using gene chip analysis (18). Using the Miranda prediction tool,
(http://www.microrna.org), it was predicted that
miR-503 may be upstream of EMT regulator proteins, including Notch
and IGF1R (19). The results
indicate that miR-503 may be an oncomiR and a regulator of EMT.
Therefore, further systemic delineation of miR-503 expression and
its function in gastric cancer is required.
Materials and methods
Human tissue specimens and cell
lines
Fresh tissues, consisting of 45 samples of human
gastric cancer (22 patients with metastasis and 23 patients without
metastasis) and 31 samples of adjacent normal mucosal tissues, were
collected from 76 patients who underwent surgery at the Second
Affiliated Hospital of Chongqing Medical University (Chongqing,
China) between 2012 and 2013. The present study complies with the
regulations of the Ministry of Health, the ‘biomedical research
involving human ethics review (tentative)’ and the Declaration of
Helsinki on the Ethical Principles for Medical Research Involving
Human Subjects. All patients provided informed consent according to
the protocols approved by the Institutional Review Board of the
Second Affiliated Hospital of Chongqing Medical University.
AGS and N87 cell lines were obtained from the
American Type Culture Collection (Manassas, VA, USA). MGC80-3,
SGC-7901, HGC-27, GES-1 and MKN-45 cell lines were purchased from
the Type Culture Collection of the Chinese Academy of Sciences
(Shanghai, China). The cell lines were cultured in RPMI 1640 medium
(Hyclone, Thermo Scientific, Waltham, MA, USA) supplemented with
10% fetal bovine serum (FBS), and were maintained at 37°C and 5%
CO2 in an incubator.
Primers, RNA isolation and miRNA
detection
miR-503 and U6 primers were purchased from Takara
Bio (Otsu, Japan). Total miRNA was extracted from cultured cells
and human tissue specimens with RNAiso for Small RNA (Takara Bio),
according to the manufacturer’s instructions. PolyA tails were
added to miR-503 and U6 with the miRNA Reaction Buffer Mix (Takara
Bio) and cDNA was synthesised from 5 ng total RNA using the miRNA
PrimeScriptRT Enzyme Mix (Takara Bio). Quantitative polymerase
chain reaction (qPCR) was run on a CFX96™ Real-Time PCR Detection
System (Bio-Rad, Hercules, CA, USA) with SYBR® Premix Ex
Taq™ II (Takara Bio). The qPCR conditions were as follows: 95°C for
30 sec, followed by 40 cycles of 95°C for 5 sec, then 60°C for 30
sec. The data were normalised against the U6snRNA. Subsequent to
amplification, a melting curve analysis was performed to ensure the
specificity of the products.
Oligonucleotide transfection
miR-503 and cont-miR (control-miR) mimics were
synthesised by Sangon Biotechnology, Co., Ltd. (Shanghai, China),
and mimic cotransfections were performed with Lipofectamine 2000
(Invitrogen Life Technologies, Carlsbad, CA, USA). At 24-h
post-transfection, the cells were plated for proliferation,
migration and invasion assays. The cells were harvested for RNA and
protein analyses at 48 h post-transfection.
Cell viability and clonability
assays
The transfected cells were seeded into 96-well
plates at a density of 1×104 cells/well. MTT solution
(20 μl of 5 mg/ml) was added to the cultures (200-μl volumes) prior
to a 4-h incubation at 37°C. Following removal of the culture
medium, the remaining crystals were dissolved in dimethylsulfoxide,
and the absorbance at 570 nm was measured. For colony formation
assays, the cells were seeded at a low density (1,000 cells/plate)
and allowed to grow until visible colonies appeared. The cells were
then stained with Giemsa, and the colonies were counted.
Migration and invasion assays
Cytoselect 24-well cell migration and invasion assay
kits (Cell Biolabs, Inc., San Diego, CA, USA) were used for the
migration and invasion assays, according to the manufacturer’s
instructions. AGS and MKN45 cell lines transfected with miR-503
mimics or control miR were harvested 72 h after transfection and
resuspended in serum-free Opti-minimum essential medium. The cells
(10×104 per 500 μl serum-free media) were added to the
upper chambers, and the lower chambers were filled with 750 μl
media with 10% FBS. The cells were incubated for 16 h at 37°C and
5% CO2 in a tissue culture incubator. After 16 h, the
non-migrated/non-invading cells were removed from the upper sides
of the Transwell membrane filter inserts with cotton-tipped swabs.
Migrated/invaded cells on the lower sides of the inserts were
stained, and the absorbance was read at 560 nm according to the
manufacturer’s instructions.
Antibodies
The anti-fibronectin antibody was obtained from
Santa Cruz Biotechnology, Inc.,(Santa Cruz, CA, USA). Antibodies
against vimentin, E-cadherin and N-cadherin were purchased from
Abcam (Cambridge, UK). Antibodies against SNAIL and GAPDH were
purchased from BD Biosciences (Franklin Lakes, NJ, USA).
Horseradish peroxidase (HRP)-conjugated goat anti-mouse and
anti-rabbit immunoglobulin G (IgG) were purchased from Santa Cruz
Biotechnology, Inc.
Immunoblotting
Total protein was extracted from the transfected
cells with radio-immunoprecipitation assay lysis buffer (Beyotime
Institute of Biotechnology, Shanghai, China), according to the
manufacturer’s instructions. After the whole-cell protein extracts
were quantified with a bicinchoninic acid protein assay, equivalent
amounts of cell lysates were resolved using 10% SDS-polyacrylamide
gel electrophoresis and transferred onto a polyvinylidene fluoride
membrane, which was then blocked in 5% skimmed milk in
Tris-buffered saline Tween 20 for 1 h at 4°C. The blots were then
incubated with the primary antibodies: mouse monoclonal antibody to
fibronectin, mouse monoclonal antibody to E-cadherin, rabbit
polyclonal antibody to N-cadherin, rabbit polyclonal antibody to
SNAIL, mouse monoclonal antibody to GAPDH. Following subsequent
incubations with HRP-conjugated goat anti-mouse and anti-rabbit
immunoglobulin G (IgG) secondary antibodies, the protein bands were
visualised with an enhanced chemiluminescence reagent (Millipore,
Billerica, MA, USA). The following antibody dilutions were used:
Anti-fibronectin, 1:200; anti-vimentin, anti-E-cadherin and
anti-N-cadherin, 1:1200; anti-SNAIL and anti-GAPDH, 1:500; and
HRP-conjugated IgG, 1:7000.
Statistical analysis
SPSS 13.0 software was used for the statistical
analysis (SPSS, Inc., Chicago, IL, USA). Data are presented as the
mean ± standard deviation. Group comparisons were performed with
Student’s t-test, and P<0.05 was considered to indicate a
significant difference.
Results
miR-503 expression is downregulated in
gastric cancer cell lines
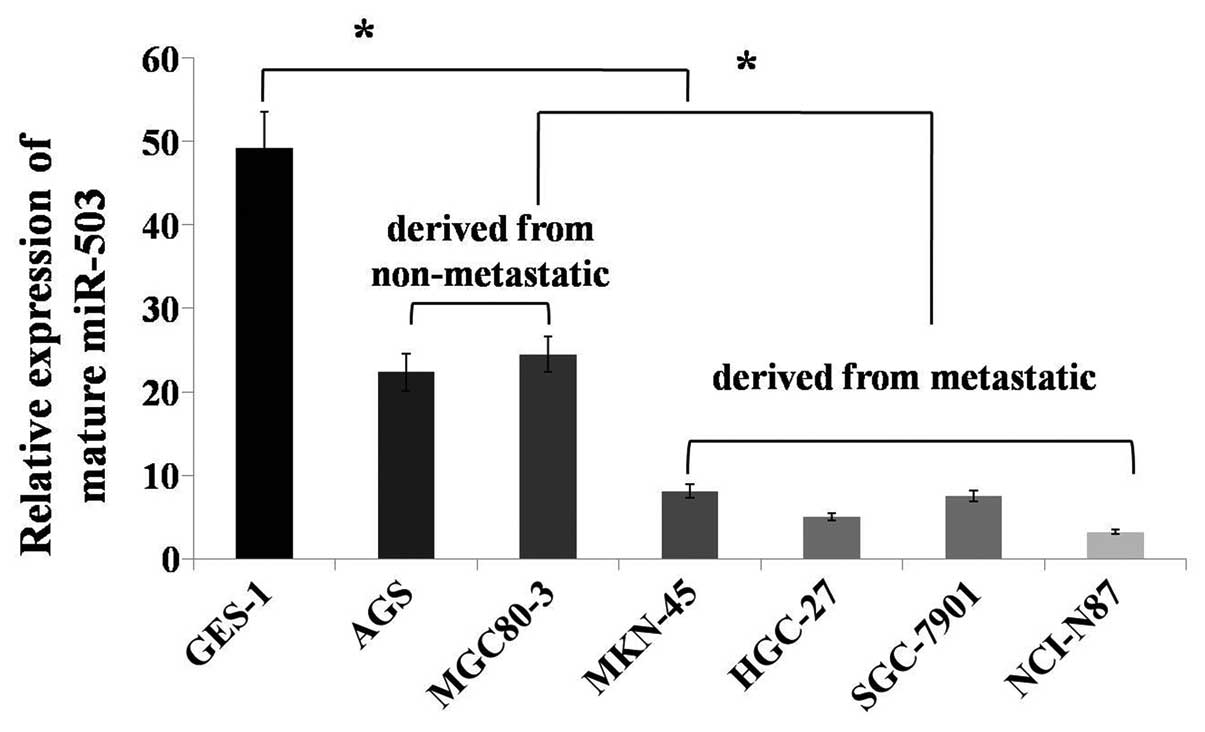
To investigate the miR-503 expression levels in
human gastric cancer cell lines, expression was monitored in
several cancer cell lines (SGC7901, HGC27, AGS, MKN45, NCI-N87 and
MGC80-3) and a normal gastric mucosa cell line (GES1). miR-503
expression was reduced in the gastric cancer cell lines compared
with the normal gastric mucosa cells. The AGS and MGC80-3 cell
lines were derived from non-metastatic tissues, and the SGC7901,
HGC27, MKN45 and NCI-N87 cell lines were derived from metastatic
tissues. miR-503 expression was upregulated in the gastric cancer
cell lines that were derived from non-metastatic tissues compared
with those derived from metastatic tissues (Fig. 1). Thus, miR-503 may play an
important role in gastric cancer. To further understand the role of
miR-503, its expression was detected in gastric cancer tissues.
miR-503 expression is downregulated in
gastric cancer tissues, and decreased miR-503 expression is
associated with gastric cancer metastasis
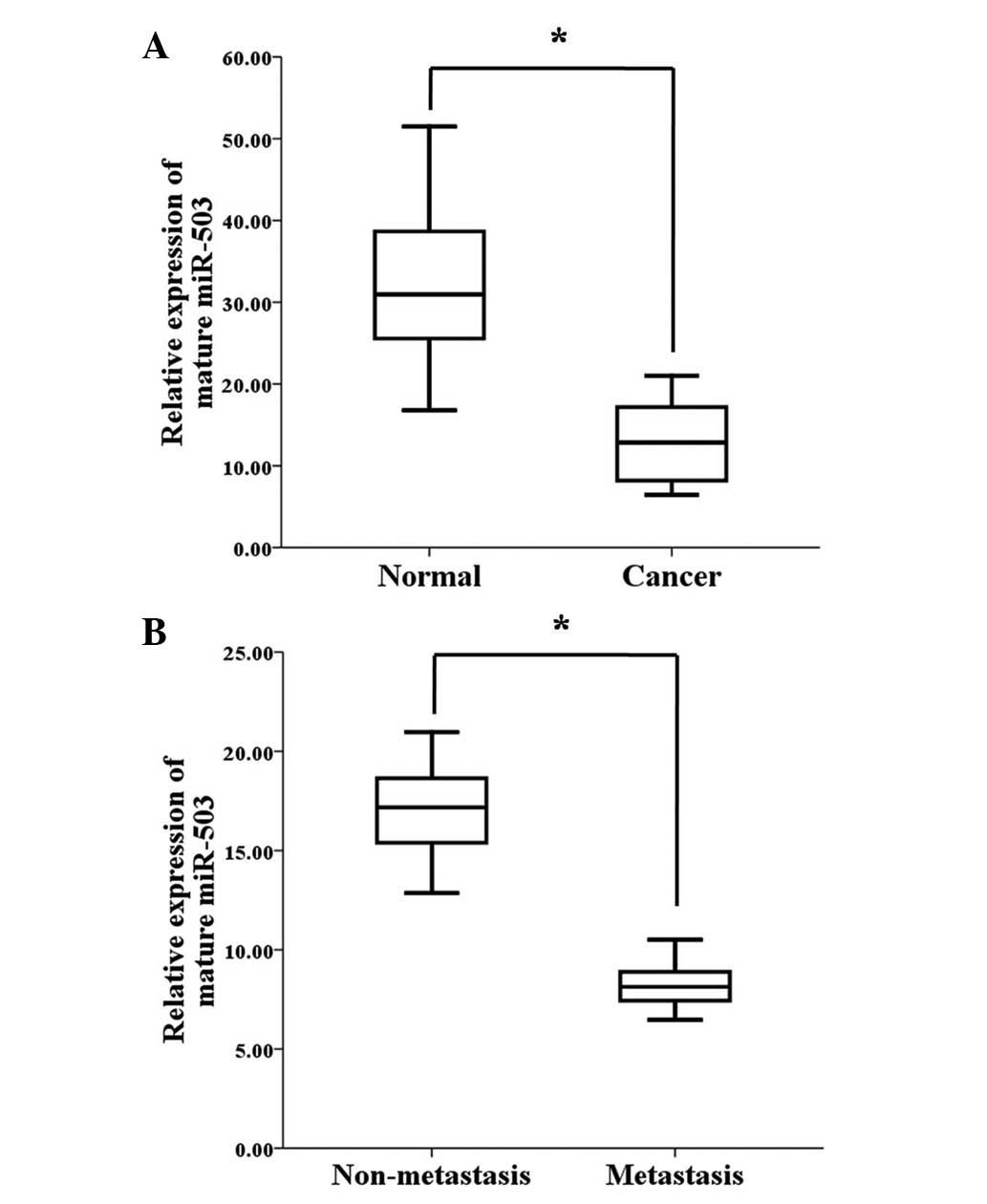
To explore the role of miR-503 in human gastric
cancer development, its expression levels were detected in 45 human
gastric cancer tissue samples and 31 adjacent normal mucosa tissue
samples. According to the qPCR analysis, the miR-503 expression
levels were significantly reduced in the tumour tissues compared
with the adjacent normal mucosa tissues (Fig. 2A). To determine whether miR-503
expression is associated with gastric cancer metastasis, the
miR-503 expression levels were examined in 45 archived primary
gastric tumours. These tumours were divided into two groups; the
tumours in one group were resected from 22 patients with lymph node
or distant organ metastases, and the tumours in the other group
were resected from 23 patients without metastases. According to the
qPCR analysis, the miR-503 expression levels were significantly
lower in the patients with metastasis when compared with the
patients without metastasis (Fig.
2B). These results demonstrate that miR-503 may be a critical
miRNA in gastric cancer progression, and that decreased miR-503
expression is associated with gastric cancer metastasis.
miR-503 inhibits gastric cancer cell
migration, invasion and proliferation
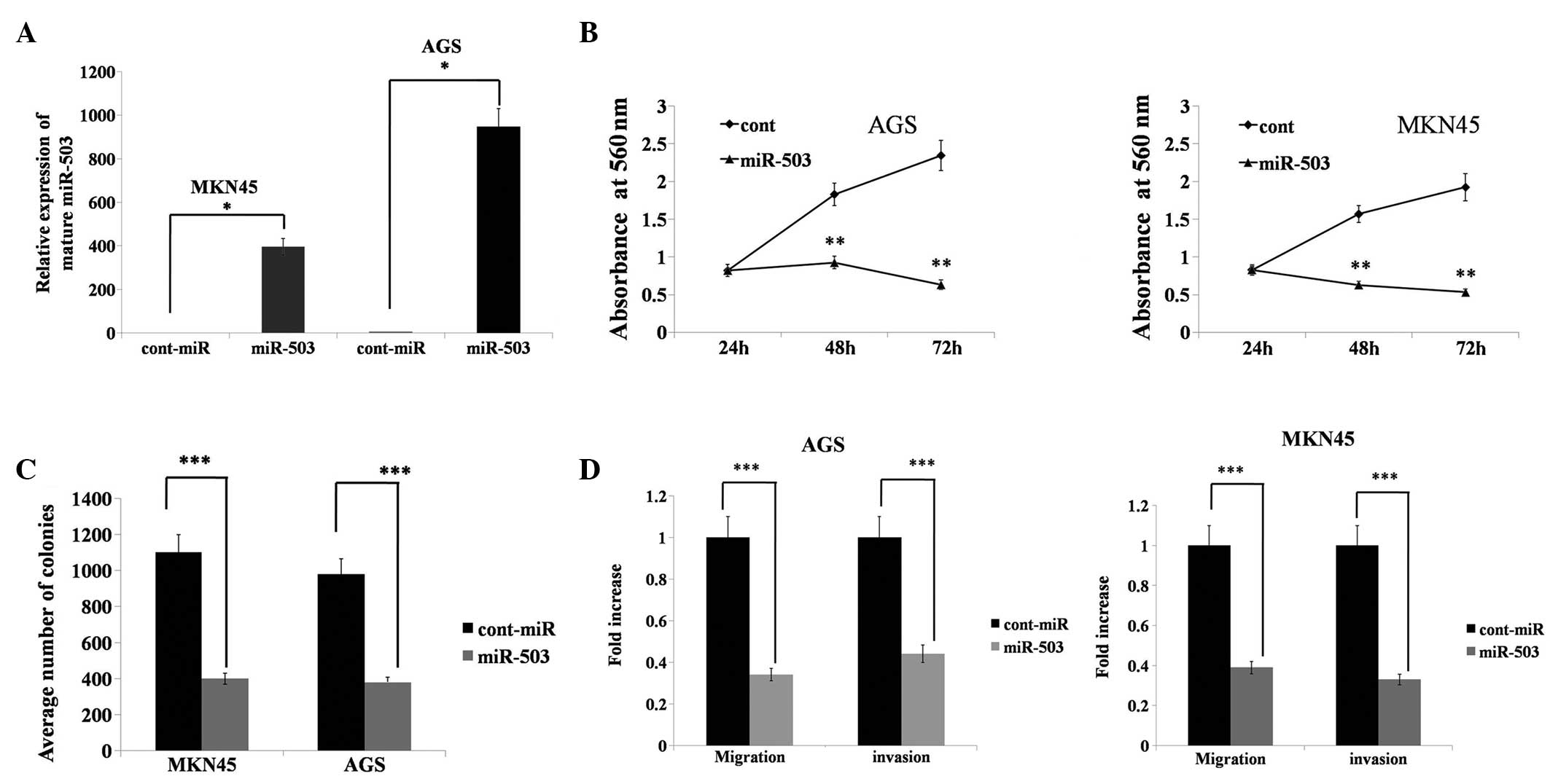
To determine the functional significance of miR-503
overexpression in gastric cancer, the AGS and MKN45 gastric cancer
cell lines were transfected with miR-503 mimics. miR-503 was
significantly overexpressed in the AGS and MKN45 cells lines
following miR-503 mimic transfection compared with the
cont-miR-transfected cells (Fig.
3A). Forced miR-503 expression significantly decreased cell
proliferation relative to cont-miR expression (Fig. 3B). miR-503-transfected cells also
exhibited a reduced colony-forming ability, as the number of foci
in the miR-503-expressing cells was reduced when compared with the
cont-miR-transfected cells (Fig.
3C). Transwell migration and Matrigel invasion assays
demonstrated that miR-503 significantly reduced the migration and
invasion capacities of the AGS and MKN45 cells (Fig. 3D). These results demonstrated that
miR-503 acts as a tumour suppressor gene in gastric cancer.
miR-503 promotes an epithelial phenotype
in gastric cancer
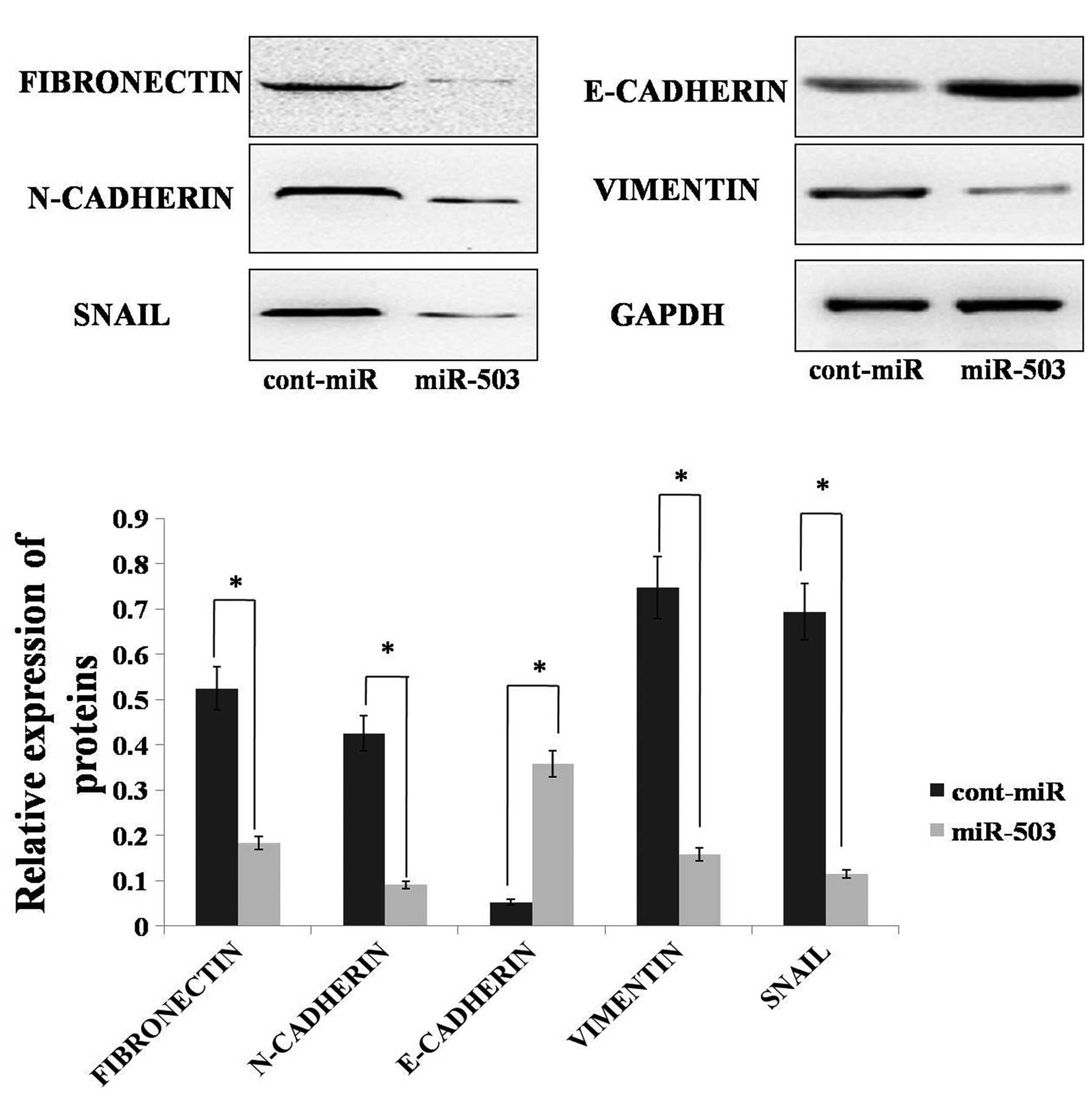
As miR-503 can inhibit gastric cancer cell migration
and invasion, we hypothesised that miR-503 could inhibit EMT in
gastric cancer. To determine if molecular changes typical of
reduced EMT occurred in miR-503-expressing cells, the expression of
mesenchymal markers, including fibronectin, vimentin, N-cadherin,
SNAIL and the epithelial marker, E-cadherin, was examined in the
AGS cell line. Immunoblot analysis showed that the expression
levels of fibronectin, vimentin and SNAIL were decreased in the AGS
cell line with forced miR-503 expression. The N-cadherin protein
level was also decreased in the AGS cell line with forced miR-503
expression. Furthermore, forced miR-503 expression increased
E-cadherin expression in the AGS cells, whereas the control
transfected cells remained E-cadherin-negative (Fig. 4). Altogether, these results
indicated that miR-503 could inhibit EMT in gastric cancer
cells.
Discussion
miR-503 is expressed differently in various types of
cancer; in certain types, miR-503 expression is upregulated. The
miR microarray technique has been used to demonstrate that miR-503
is upregulated in human retinoblastoma tissues (20). In addition, miR-503 expression has
been shown to be upregulated in human parathyroid carcinomas
(21). Additionally, high miR-503
expression has been significantly associated with shorter overall
survival in patients with adrenocortical carcinoma (22). However, all of these results were
obtained with the miR microarray technique and were not confirmed
by qPCR. A concrete mechanism to explain why miR-503 is upregulated
in certain cancers and the identity of the target gene of miR-503
remains unknown.
In other types of cancer, miR-503 expression is
downregulated. In oral cancer, miR-503 has been found to be
downregulated, according to the miR microarray technique (23). In addition, miR-503 has been
detected in non-metastatic prostate cancer xenografts, but not in
metastatic grafts (24). Jiang
et al identified the target gene of miR-503 and delineated
its function. The study revealed that miR-503 could silence cyclin
D1 (CCND1), a well-known proto-oncogene that is implicated in a
variety of cancer types. Thus, miR-503 could reduce S-phase cell
populations and cause cell growth inhibition (25). Xu et al also identified that
miR-503 directly targeted CCND1, and that abnormal miR-503
suppression led to elevated CCND1 levels, which may promote EEC
carcinogenesis and progression (17). Furthermore, miR-503 was identified
as a cell cycle regulator with involvement in cell adhesion,
migration and angiogenesis processes (26). This finding indicates that miR-503
may be a putative tumour suppressor. Zhou et al identified
the reason behind the downregulation of miR-503 expression in human
hepatocellular carcinomas (HCC) as namely due to the epigenetic
modulation of its promoter (27)
The results fully validated the finding that miR-503 was
downregulated in certain types of cancers and elucidated the reason
for the downregulation of miR-503 and the identity of the miR-503
target gene.
In the present study, miR-503 expression was found
to be reduced in gastric cancer cell lines compared with normal
gastric mucosa cell lines. Additionally, the miR-503 expression
levels were significantly reduced in tumour tissues compared with
the adjacent normal mucosa tissues. Meanwhile, the miR-503
expression levels in metastatic patients were significantly lower
than those in non-metastatic patients. The results showed that
miR-503 expression was correlated with gastric carcinoma
progression. Several studies have agreed with these results. A
study by Zhou and Wang identified that miR-503 was significantly
downregulated in HCCLM3, an HCC cell line with a strong metastatic
potential, when compared with MHCC97-L, an HCC cell line with a
lower metastatic potential. Decreased miR-503 expression was
identified in HCC cell lines with high metastatic abilities.
Meanwhile, overexpressed miR-503 inhibited the proliferation and
metastasis of HCCLM3 cells in vitro (28). In the present study, forced miR-503
expression was found to significantly decrease cell migration,
invasion and proliferation relative to cont-miR expression. The
reason why forced miR-503 expression inhibits gastric cancer cell
migration, invasion and proliferation is not fully understood. This
finding could be the result of several factors. First, one target
of miR-503 is cyclin D1, which can promote cancer cell growth.
Thus, when miR-503 is overexpressed in cancer cells, cyclin D1
expression is downregulated and cell growth is subsequently
inhibited. Second, increased miR-503 expression impairs endothelial
cell networking capacities, indicating an anti-angiogenic role for
this particular miR (26). Zhou
et al also identified that miR-503 directly targets and
negatively regulates the expression of vascular endothelial growth
factor (VEGF)-A and fibroblast growth factor (FGF)2. As VEGF-A and
FGF2 are important angiogenic factors, miR-503 can suppress tumour
growth by inhibiting angiogenesis (27).
In gastric cancers, EMT is known to be associated
with a migratory phenotype. Kurashige et al identified that
miR-200b regulates cell proliferation, invasion and migration by
directly targeting ZEB2 in gastric carcinomas (29). Zhang et al revealed that
miR-27 promotes human gastric cancer cell metastasis by inducing
EMT (30). However, it remains
unknown whether miR-503 can inhibit EMT in human gastric cancers.
We predicted that Notch and IGF1R, which are EMT regulators, may be
target genes of miR-503. In the present study, it was identified
that miR-503 overexpression could inhibit EMT in gastric cancer
cells. Therefore, these results show that miR-503 may act as an
important regulator of EMT in gastric cancer. In a forthcoming
study, we will determine whether Notch and IGF1R are target genes
of miR-503 with luciferase reporter assays, and the miR-503
regulatory mechanism will be studied further by xenograft
experiments.
In conclusion, the results of the present study have
identified miR-503 as a novel tumour suppressor gene in gastric
cancer. Furthermore, the data have demonstrated that miR-503 can
curb gastric cancer cell metastasis by inhibiting EMT. These
results indicate potential applications for miR-503 in gastric
cancer therapy.
Acknowledgements
This study was supported by a grant from the Science
and Technology Committee, Chongqin (no. 201302).
References
|
1
|
Rana TM: Illuminating the silence:
understanding the structure and function of small RNAs. Nat Rev Mol
Cell Biol. 8:23–36. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Valencia-Sanchez MA, Liu J, Hannon GJ and
Parker R: Control of translation and mRNA degradation by miRNAs and
siRNAs. Genes Dev. 20:515–524. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pillai RS, Bhattacharyya SN and Filipowicz
W: Repression of protein synthesis by miRNAs: how many mechanisms?
Trends Cell Biol. 17:118–126. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lu J, Getz G, Miska EA, et al: MicroRNA
expression profiles classify human cancers. Nature. 435:834–838.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Thomson JM, Newman M, Parker JS,
Morin-Kensicki EM, Wright T and Hammond SM: Extensive
post-transcriptional regulation of microRNAs and its implications
for cancer. Genes Dev. 20:2202–2207. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Thiery JP, Acloque H, Huang RY, et al:
Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gibbons DL, Lin W, Creighton CJ, et al:
Contextual extracellular cues promote tumor cell EMT and metastasis
by regulating miR-200 family expression. Genes Dev. 23:2140–2151.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhao X, Dou W, He L, et al: MicroRNA-7
functions as an anti-metastatic microRNA in gastric cancer by
targeting insulin-like growth factor-1 receptor. Oncogene.
32:1363–1372. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kong W, Yang H, He L, et al: MicroRNA-155
is regulated by the transforming growth factor beta/Smad pathway
and contributes to epithelial cell plasticity by targeting RhoA.
Mol Cell Biol. 28:6773–6784. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Korpal M, Lee ES, Hu G and Kang Y: The
miR-200 family inhibits epithelial-mesenchymal transition and
cancer cell migration by direct targeting of E-cadherin
transcriptional repressors ZEB1 and ZEB2. J Biol Chem.
283:14910–14914. 2008. View Article : Google Scholar
|
|
12
|
Gebeshuber CA, Zatloukal K and Martinez J:
miR-29a suppresses tristetraprolin, which is a regulator of
epithelial polarity and metastasis. EMBO Rep. 10:400–405. 2009.
View Article : Google Scholar
|
|
13
|
Cottonham CL, Kaneko S and Xu L: miR-21
and miR-31 converge on TIAM1 to regulate migration and invasion of
colon carcinoma cells. J Biol Chem. 285:35293–35302. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Eades G, Yao Y, Yang M, Zhang Y, Chumsri S
and Zhou Q: miR-200a regulates SIRT1 expression and epithelial to
mesenchymal transition (EMT)-like transformation in mammary
epithelial cells. J Biol Chem. 286:25992–26002. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gregory PA, Bert AG, Paterson EL, et al:
The miR-200 family and miR-205 regulate epithelial to mesenchymal
transition by targeting ZEB1 and SIP1. Nat Cell Biol. 10:593–601.
2008. View
Article : Google Scholar
|
|
16
|
Du R, Sun W, Xia L, et al: Hypoxia-induced
down-regulation of microRNA-34a promotes EMT by targeting the Notch
signaling pathway in tubular epithelial cells. PLoS One.
7:e307712012. View Article : Google Scholar
|
|
17
|
Xu YY, Wu HJ, Ma HD, Xu LP, Huo Y and Yin
LR: MicroRNA-503 suppresses proliferation and cell cycle
progression of endometrioid endometrial cancer via negatively
regulating cyclin D1. FEBS J. 280:3768–3779. 2013. View Article : Google Scholar
|
|
18
|
Luo HC, Zhang ZZ, Zhang X, Ning B, Guo JJ,
Nie N, Liu B and Wu XL: microRNA expression signature in gastric
cancer. Chin J Cancer Res. 21:74–80. 2009. View Article : Google Scholar
|
|
19
|
Zhang Y, Chen X, Lian H, et al:
MicroRNA-503 acts as a tumor suppressor in glioblastoma for
multiple antitumor effects by targeting IGF-1R. Oncol Rep.
31:1445–1452. 2014.PubMed/NCBI
|
|
20
|
Zhao JJ, Yang J, Lin J, et al:
Identification of miRNAs associated with tumorigenesis of
retinoblastoma by miRNA microarray analysis. Childs Nerv Syst.
25:13–20. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Corbetta S, Vaira V, Guarnieri V, et al:
Differential expression of microRNAs in human parathyroid
carcinomas compared with normal parathyroid tissue. Endocr Relat
Cancer. 17:135–146. 2010. View Article : Google Scholar
|
|
22
|
Özata DM, Caramuta S, Velázquez-Fernández
D, et al: The role of microRNA deregulation in the pathogenesis of
adrenocortical carcinoma. Endocr Relat Cancer. 18:643–655.
2011.PubMed/NCBI
|
|
23
|
Lu YC, Chen YJ, Wang HM, et al: Oncogenic
function and early detection potential of miRNA-10b in oral cancer
as identified by microRNA profiling. Cancer Prev Res (Phila).
5:665–674. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Watahiki A, Wang Y, Morris J, et al:
MicroRNAs associated with metastatic prostate cancer. PLoS One.
6:e249502011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jiang Q, Feng MG and Mo YY: Systematic
validation of predicted microRNAs for cyclin D1. BMC Cancer.
9:1942009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Caporali A, Meloni M, Völlenkle C, et al:
Deregulation of microRNA-503 contributes to diabetes
mellitus-induced impairment of endothelial function and reparative
angiogenesis after limb ischemia. Circulation. 123:282–291. 2011.
View Article : Google Scholar
|
|
27
|
Zhou B, Ma R, Si W, et al: MicroRNA-503
targets FGF2 and VEGFA and inhibits tumor angiogenesis and growth.
Cancer Lett. 333:159–169. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou J and Wang W: Analysis of microRNA
expression profiling identifies microRNA-503 regulates metastatic
function in hepatocellular cancer cell. J Surg Oncol. 104:278–283.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kurashige J, Kamohara H, Watanabe M, et
al: MicroRNA-200b regulates cell proliferation, invasion, and
migration by directly targeting ZEB2 in gastric carcinoma. Ann Surg
Oncol. 19(Suppl 3): S656–S664. 2012. View Article : Google Scholar
|
|
30
|
Zhang Z, Liu S, Shi R and Zhao G: miR-27
promotes human gastric cancer cell metastasis by inducing
epithelial-to-mesenchymal transition. Cancer Genet. 204:486–491.
2011. View Article : Google Scholar : PubMed/NCBI
|