Introduction
The hedgehog signaling pathway is critical for it’s
role in normal cell differentiation and embryonic development, as
well as in the pathological processes that drive cancer formation
(1–3). The ligands of sonic hedgehog (Shh)
bind to the transmembrane receptor, Patched (ptch) 1 and 2,
to relieve the suppression of the transmembrane protein, Smoothened
(Smo). This subsequently triggers the nuclear translocation
of various transcription factors to activate downstream target
genes (2,4). In various types of cancer, including
ovarian (5), lung (6,7),
breast (8), prostate (9), endometrial (10), skin (11) and gastrointestinal (12–14),
aberrant activation of Smo genes and loss of function
mutations in the ptch gene relieve the suppression of the
Smo protein and trigger full-length Gli1 translocation into the
nucleus, prompting excessive activation of downstream genes,
including c-myc and vascular endothelial growth factor (VEGF). It
has also been demonstrated that inhibition of the Shh pathway by a
Smo inhibitor, such as cyclopamine, slows or prevents the growth of
tumor tissues (15–17).
In the case of gastric cancer cells, excessive Shh
signaling activities are well known to affect cancer cell
proliferation, migration and invasion, and overexpression of Shh
was identified in intestinal metaplasia and stomach adenomas
(18). In in vitro studies,
the Shh pathway and downstream genes/proteins are highly involved
in the proliferation and migration of various gastric cancer cell
lines, including MKN1/7/45/74, MKN45 and AGS cells (19,20).
However, the exact mechanisms defining how the Shh pathway
regulates gastric tumorigenesis remains elusive.
In the present study, via the application of
cyclopamine, the Shh signaling pathway was inhibited in the human
gastric cancer cell line, AGS, and the effect on cell
proliferation, migration and invasion was evaluated. Furthermore,
it was demonstrated that the molecular and cellular expression of
key Shh signaling pathway-associated factors, Gli1 and CXCR4, were
markedly downregulated by cyclopamine in AGS cells.
Materials and methods
Cell culture and treatment
Human gastric cancer cell line AGS was obtained from
American Type Culture Collection (ATCC CRL-1739) and were
maintained in RPMI-1640 medium supplemented with 10% fetal bovine
serum (Invitrogen Life Technologies, Carlsbad, CA, USA) and 100
U/ml penicillin/streptomycin. The cells were cultured either with
cyclopamine (5–100 μM; Calbiochem, La Jolla, CA, USA) or without
cyclopamine for 24, 48 or 72 h.
Cell proliferation assay
Cells were plated at a concentration of
2.5×104 cells/ml of culture medium in 96-well plates for
24 and 72 h. Following the defined culture periods, an MTT assay
(Sigma, St. Louis, MO, USA) was applied according to the
manufacturer’s instructions to calculate the volume of viable cells
(21).
Apoptosis assay
Following in vitro culture for 24 h, the
gastric cancer cells, a total amount of 1×106, were
collected in a binding buffer (10 mM HEPES/NaOH, 140 mM NaCl, 2.5
mM CaCl2) after washing with phosphate-buffered saline
(PBS; 3×10 min). Fluorescence-activated cell sorting analysis for
apoptosis was conducted using an Annexin V-FITC/7-AAD kit according
to the manufacturer’s instructions (Beckman Coulter, Miami, FL,
USA). The mixture was incubated for 10 min in a dark room at room
temperature and the stained cells were immediately analyzed using a
flow cytometer (Cell Lab Quanta SC; Beckman Coulter) to determine
the percentage of apoptotic cells.
Invasion assay
Cancer cell migration/invasion was performed by a
quantitative cell migration assay (ECM500; Chemicon, Temecula, CA,
USA) according to the manufacturer’s instructions. Warm Knockout
DMEM (Sigma) in the amount of 200 μl was applied to the
extracellular matrix (ECM) layer to hydrate for 2 h at room
temperature. AGS cells were then dislodged by trypsinization (0.25%
trypsin; Sigma) and dispersed into a homogeneous single-cell
suspension at the concentration of 5×105 cells/ml,
followed by washing and resuspension in Knockout DMEM. Then, cell
suspension of 200 μl was allowed to adhere to the surface at 37°C
for 60 min. The migration mediums containing cyclopamine were then
put into the bottom chamber. Following 24 h of incubation at 37°C,
5% CO2 in air, the cells in the upper chamber were
stained for 20 min, and dissolved in 10% acetic acid and the
optical density (OD) was read at 560 nm on a standard reader.
Quantitative polymerase chain reaction
(qPCR)
A TRIzol reagent (Roche) was used to isolate total
RNA from 5×106 cells according to the manufacturer’s
instructions. First-strand cDNA synthesis and amplification was
conducted using an MBI Revert Aid First Strand cDNA Synthesis kit
(MBI Fermentas, Amherst NY, USA). The qPCR was performed using an
iQ5 Multicolor Real-Time PCR Detection system (Bio-Rad, Hercules,
CA, USA). The cycle threshold values were read from the ABI 7000
software. The primers were: Forward, 5′-TCCTTTGGGGTCCAGCCTTG-3′ and
reverse, 5′-ATGCCTGTGGAGTTGGGGCT-3′ for Gli1; forward,
5′-TCAGTCTGGACCGCTACCTG-3′ and reverse, 5′-CCACCCACAAGTCATTGGGG-3′
for CXCR4; and forward, 5′-AGGTCGGAGTCAACGGATTTG-3′ and reverse,
5′-GTGATGGCATGGACTGTGGT-3′ for GAPDH.
Western blot analysis
RIPA buffer (50 mM Tris, 150 mM NaCl, 1% Triton
X-100, 0.1% sodium dodecyl sulfate and 1% Na-deoxycholate; pH 7.4)
supplemented with protease inhibitor was used to collect the cell
suspension for the western blot analysis and a Bio-Rad protein
assay (Bio-Rad) was used to calculate the total protein
concentrations. Briefly, the protein lysates were resolved by
sodium dodecyl sulfate-polyacrylamide gel electrophoresis and
transferred onto nitrocellulose membranes (Hybond™-P; Amersham
Biosciences, Piscataway, NJ, USA). The membrane was blocked using
0.2% Tween-20 and 5% non-fat dry milk in PBS. The lysates were
incubated with a primary antibodies: GLI-1 rabbit polyclonal anti
human IgG (H-300, Santa Cruz Biotechnology, Inc., Santa Cruz, CA,
USA) and CXCR-4 rabbit polyclonal IgG anti-human (H-118, Santa Cruz
Biotechnology, Inc.) and a horseradish peroxidase-labeled rabbit
IgG secondary antibody (Santa Cruz Biotechnology, Inc.) and
detected using X-ray film.
Statistical analysis
Data were calculated in triplicate and expressed as
the mean ± standard error of the mean. Comparisons were made using
either student’s t-test or one-way analysis of variance post hoc
tests. P<0.05 was considered to indicate a statistically
significant result.
Results
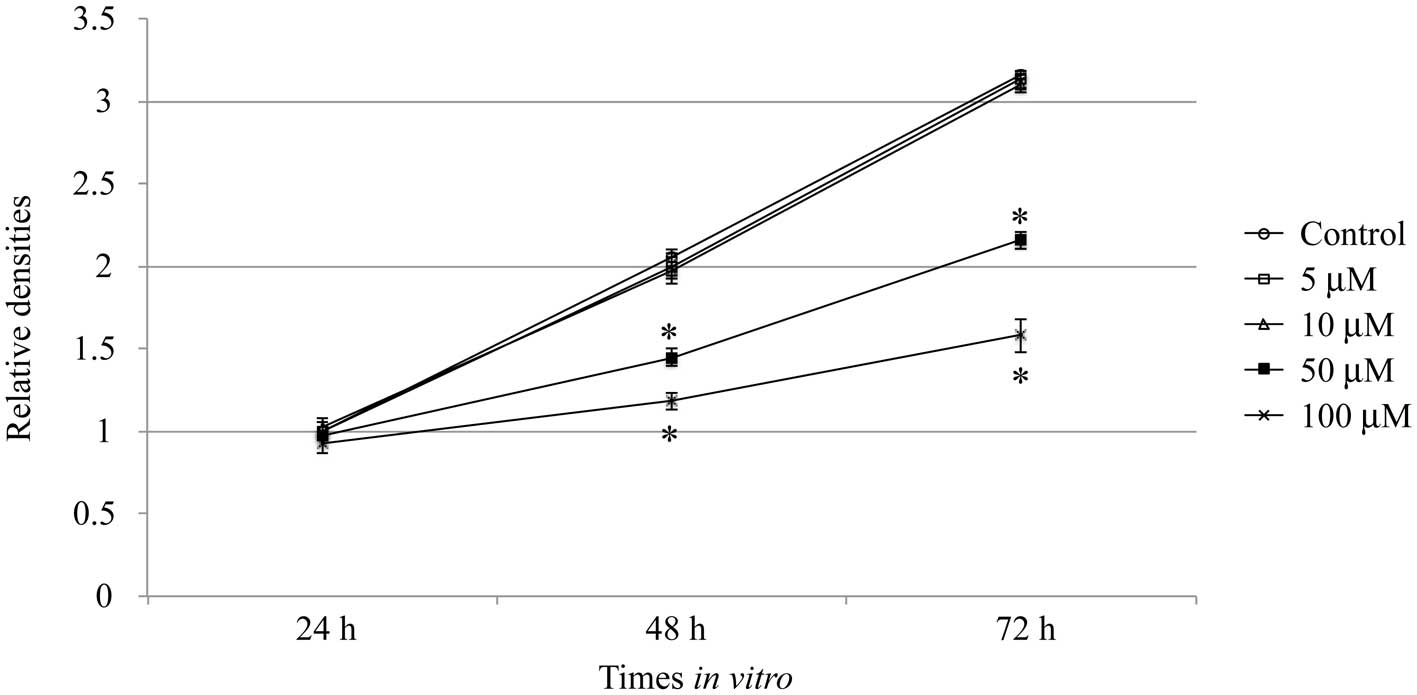
Inhibition of gastric cancer cell
proliferation by cyclopamine
AGS cells were cultured with or without cyclopamine
for 24, 48 and 72 h, and the effect of cyclopamine on cell
proliferation was measured (Fig.
1). The results demonstrated that when AGS cells were treated
with 5 or 10 μM of cyclopamine for 24, 48 or 72 h, the
proliferation densities were unaffected, as compared with the
control conditions (P>0.05). This indicated that the application
of cyclopamine at lower concentrations did not alter the cell
proliferation rate. However, while AGS cells that were treated with
50 or 100 μM cyclopamine for 48 or 72 h, respectively, cell
proliferation was significantly inhibited, indicating that a higher
concentration of cyclopamine inhibited the growth of AGS cells in a
dose-dependent manner (P<0.05).
Induction of apoptosis in gastric cancer
cells by cyclopamine
Secondly, the effects of cyclopamine on the AGS
cells were examined. The cells were either untreated (control) or
treated with cyclopamine (50 or 100 μM) for 24 or 48 h, followed by
annexin V staining. The results demonstrated that high
concentrations of cyclopamine (50 or 100 μM) induced significant
apoptosis in AGS cells (Table
I).
 | Table ICyclopamine induces apoptosis in
gastric cancer cells. |
Table I
Cyclopamine induces apoptosis in
gastric cancer cells.
| Parameter | Control | 50 μM | 100 μM |
|---|
| Rate of apoptosis, 24
h | 1.52±0.51 | 15.25±2.11a | 22.55±1.94a |
| Rate of apoptosis, 48
h | 3.15±0.63 | 24.32±2.37a | 30.12±2.33a |
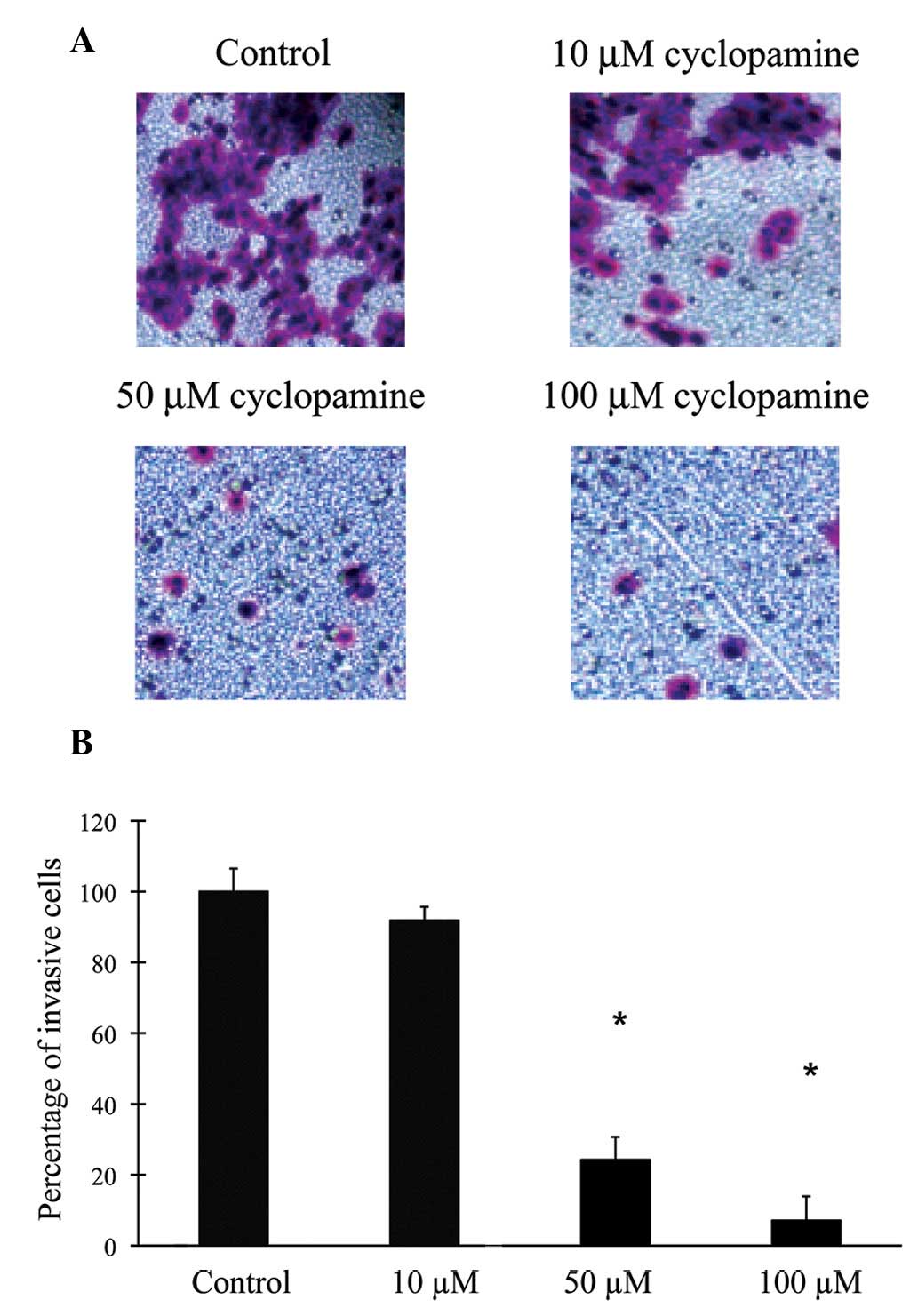
Inhibition of cell invasion in gastric
cancer cells by cyclopamine
A characteristic feature of gastric cancer cells is
their aggressive ability to filtrate and invade a reconstituted
basement membrane. The effect of cyclopamine on the cellular
invasion of human gastric cancer cells was assessed in the present
study. The cancer cells were either untreated (control) or treated
with cyclopamine at concentrations of 10, 50 and 100 μM, and
maintained in the culture medium for 24 h (Fig. 2). When treated with 10 μM of
cyclopamine, AGS cells demonstrated a similar rate of invasion, as
compared with that of the control condition (P>0.05). However,
with higher concentrations of cyclopamine (50 and 100 μM), the
baseline invasions were significantly inhibited. This response was
dose-dependent as the greater the concentration of cyclopamine was,
the higher the degree of inhibition it induced on cancer cell
migration (P<0.05).
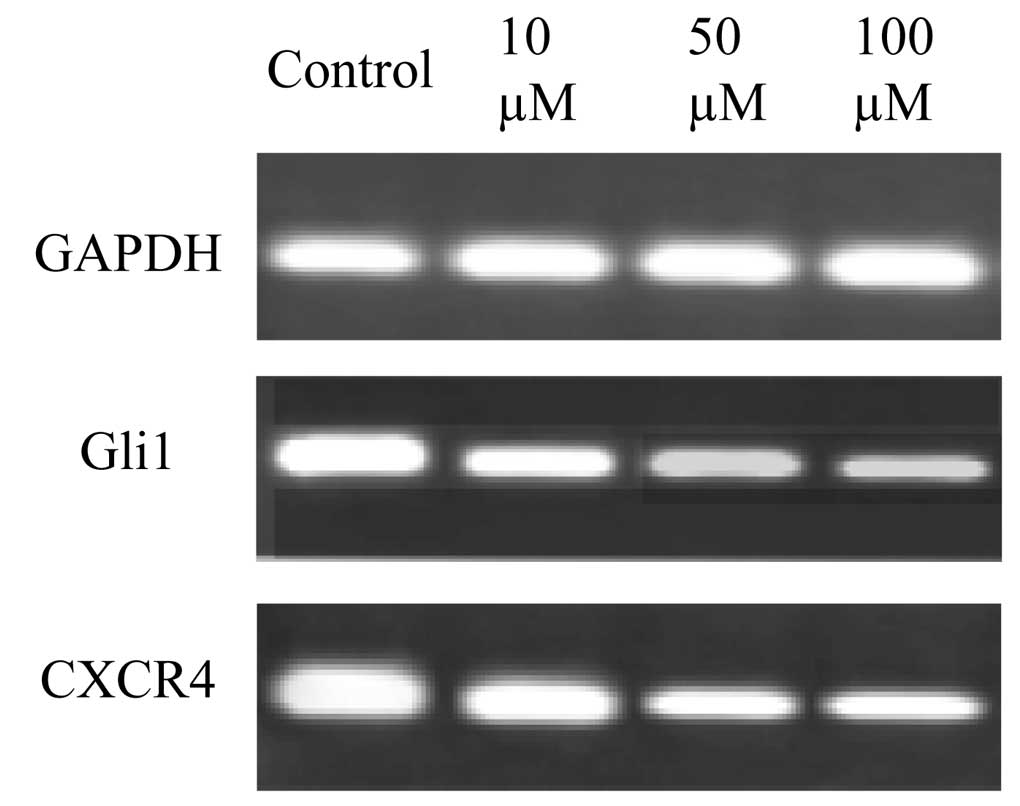
Downregulation of Shh-associated factors
by cyclopamine in gastric cancer cells
The effects of cyclopamine on gene regulation in AGS
cells are demonstrated in Fig. 3.
AGS cells were treated with 10, 50 and 100 μM cyclopamine for 24 h.
This identified that the higher concentrations of cyclopamine (50
and 100 μM) markedly downregulated the gene expression of Gli1 and
CXCR4 in the gastric cancer cells.
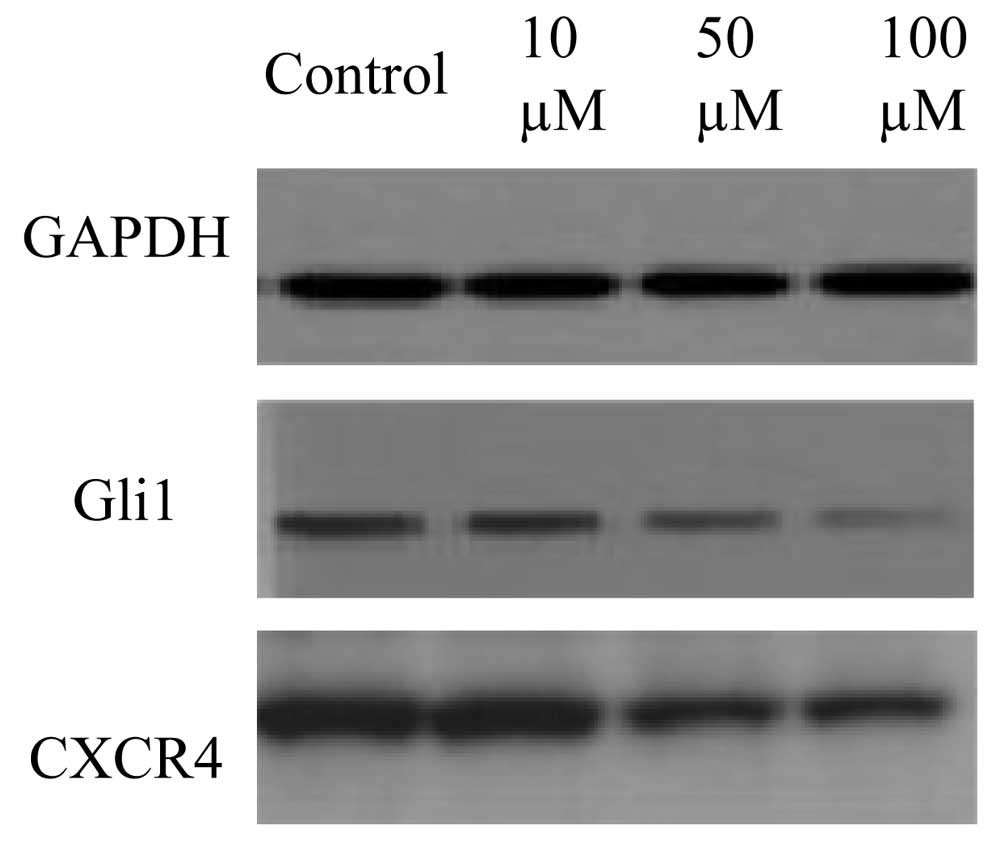
Cyclopamine downregulated Shh-associated
proteins in AGS cells
The effects of cyclopamine on Shh-related protein
expression in AGS cells are presented in Fig. 4. The results were consistent with
the gene expression results, as higher concentrations of
cyclopamine (50 and 100 μM) downregulated the protein expression of
Gli1 and CXCR4 in the gastric cancer cells.
Discussion
The Shh signaling pathway is important in cell
differentiation and maturation (1–3,22).
However, aberrant activation of the Shh pathway results in the
proliferation of various cancer cell types, including lung,
pancreatic and gastric (5,8,23–25).
While the mechanisms of the Shh signaling pathway in
promoting gastric tumor formation remain elusive, and the
downstream targeting genes continue to be largely unknown, recent
studies have indicated that various key factors, including Gil1 and
CXCR4, are closely associated with these pathological processes.
These studies identified that the chemokine receptor, CXCR4 and its
cognate ligand, CXCL12 were expressed in cancerous tissues and
possibly modulated the migration and invasion of tumors in
prostate, endometrial and breast cancer (26–29).
The in vivo and in vitro studies have identified that
CXCR4 was expressed in gastric carcinoma and gastric cancer cell
lines, and correlated with the late developmental stages of lymph
node cancer (30).
In the present study, it was demonstrated that,
following the inhibition of the Shh pathway through the application
of cyclopamine, the proliferation rates and migration capacities in
gastric cancer cells were significantly reduced in response to high
concentrations of the compound. In addition, it was revealed that
the gene and protein expression levels of Gli1 and CXCR4 were
consistently downregulated in the gastric cancer cells when high
concentrations of cyclopamine were applied. These results were
consistent with previous studies that demonstrated that Gli1 and
CXCR4 contributed to tumorigenesis in types of cancer other than
gastric (23,31,32).
In conclusion, the results of the present study provide invaluable
insights into the mechanisms of Shh signaling for the regulation of
gastric cancer cell growth in vitro and these data may
ultimately facilitate the development of novel therapeutic targets
for the treatment gastric of cancer in human patients.
References
|
1
|
Hooper JE and Scott MP: Communicating with
Hedgehogs. Nat Rev Mol Cell Biol. 6:306–317. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ingham PW and McMahon AP: Hedgehog
signaling in animal development: paradigms and principles. Genes
Dev. 15:3059–3087. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pasca di Magliano M and Hebrok M: Hedgehog
signalling in cancer formation and maintenance. Nat Rev Cancer.
3:903–911. 2003.PubMed/NCBI
|
|
4
|
Bale AE and Yu KP: The hedgehog pathway
and basal cell carcinomas. Hum Mol Genet. 10:757–762. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liao X, Siu MK, Au CW, et al: Aberrant
activation of hedgehog signaling pathway in ovarian cancers: effect
on prognosis, cell invasion and differentiation. Carcinogenesis.
30:131–140. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Watkins DN, Berman DM, Burkholder SG, Wang
B, Beachy PA and Baylin SB: Hedgehog signalling within airway
epithelial progenitors and in small-cell lung cancer. Nature.
422:313–317. 2003. View Article : Google Scholar
|
|
7
|
Gialmanidis IP, Bravou V, Amanetopoulou
SG, Varakis J, Kourea H and Papadaki H: Overexpression of hedgehog
pathway molecules and FOXM1 in non-small cell lung carcinomas. Lung
Cancer. 66:64–74. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
ten Haaf A, Bektas N, von Serenyi S, et
al: Expression of the glioma-associated oncogene homolog (GLI) 1 in
human breast cancer is associated with unfavourable overall
survival. BMC Cancer. 9:2982009.PubMed/NCBI
|
|
9
|
Karhadkar SS, Bova GS, Abdallah N, et al:
Hedgehog signalling in prostate regeneration, neoplasia and
metastasis. Nature. 431:707–712. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Feng YZ, Shiozawa T, Miyamoto T, et al:
Overexpression of hedgehog signaling molecules and its involvement
in the proliferation of endometrial carcinoma cells. Clin Cancer
Res. 13:1389–1398. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Daya-Grosjean L and Couvé-Privat S: Sonic
hedgehog signaling in basal cell carcinomas. Cancer Lett.
225:181–192. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Berman DM, Karhadkar SS, Maitra A, et al:
Widespread requirement for Hedgehog ligand stimulation in growth of
digestive tract tumours. Nature. 425:846–851. 2003. View Article : Google Scholar
|
|
13
|
Mori Y, Okumura T, Tsunoda S, Sakai Y and
Shimada Y: Gli-1 expression is associated with lymph node
metastasis and tumor progression in esophageal squamous cell
carcinoma. Oncology. 70:378–389. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Qualtrough D, Buda A, Gaffield W, Williams
AC and Paraskeva C: Hedgehog signalling in colorectal tumour cells:
induction of apoptosis with cyclopamine treatment. Int J Cancer.
110:831–837. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen JK, Taipale J, Cooper MK and Beachy
PA: Inhibition of Hedgehog signaling by direct binding of
cyclopamine to Smoothened. Genes Dev. 16:2743–2748. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen JK, Taipale J, Young KE, Maiti T and
Beachy PA: Small molecule modulation of Smoothened activity. Proc
Natl Acad Sci USA. 99:14071–14076. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lin TL and Matsui W: Hedgehog pathway as a
drug target: Smoothened inhibitors in development. Onco Targets
Ther. 5:47–58. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lee SY, Han HS, Lee KY, et al: Sonic
hedgehog expression in gastric cancer and gastric adenoma. Oncol
Rep. 17:1051–1055. 2007.PubMed/NCBI
|
|
19
|
Ohta M, Tateishi K, Kanai F, et al:
p53-Independent negative regulation of p21/cyclin-dependent
kinase-interacting protein 1 by the sonic
hedgehog-glioma-associated oncogene 1 pathway in gastric carcinoma
cells. Cancer Res. 65:10822–10829. 2005. View Article : Google Scholar
|
|
20
|
Fukaya M, Isohata N, Ohta H, et al:
Hedgehog signal activation in gastric pit cell and in diffuse-type
gastric cancer. Gastroenterology. 131:14–29. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fan XG, Kelleher D, Fan XJ, Xia HX and
Keeling PW: Helicobacter pylori increases proliferation of gastric
epithelial cells. Gut. 38:19–22. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
McMahon AP, Ingham PW and Tabin CJ:
Developmental roles and clinical significance of hedgehog
signaling. Curr Top Dev Biol. 53:1–114. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yoo YA, Kang MH, Kim JS and Oh SC: Sonic
hedgehog signaling promotes motility and invasiveness of gastric
cancer cells through TGF-beta-mediated activation of the ALK5-Smad
3 pathway. Carcinogenesis. 29:480–490. 2008. View Article : Google Scholar
|
|
24
|
Nagai S, Nakamura M, Yanai K, et al: Gli1
contributes to the invasiveness of pancreatic cancer through matrix
metalloproteinase-9 activation. Cancer Sci. 99:1377–1384. 2008.
View Article : Google Scholar
|
|
25
|
Feldmann G, Dhara S, Fendrich V, et al:
Blockade of hedgehog signaling inhibits pancreatic cancer invasion
and metastases: a new paradigm for combination therapy in solid
cancers. Cancer Res. 67:2187–2196. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Raman D, Baugher PJ, Thu YM and Richmond
A: Role of chemokines in tumor growth. Cancer Lett. 256:137–165.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Salvucci O, Bouchard A, Baccarelli A, et
al: The role of CXCR4 receptor expression in breast cancer: a large
tissue microarray study. Breast Cancer Res Treat. 97:275–283. 2006.
View Article : Google Scholar
|
|
28
|
Kodama J, Hasengaowa, Seki N, Kusumoto T
and Hiramatsu Y: Expression of the CXCR4 and CCR7 chemokine
receptors in human endometrial cancer. Eur J Gynaecol Oncol.
28:370–375. 2007.PubMed/NCBI
|
|
29
|
Engl T, Relja B, Marian D, et al: CXCR4
chemokine receptor mediates prostate tumor cell adhesion through
alpha5 and beta3 integrins. Neoplasia. 8:290–301. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lee HJ, Kim SW, Kim HY, et al: Chemokine
receptor CXCR4 expression, function, and clinical implications in
gastric cancer. Int J Oncol. 34:473–480. 2009.PubMed/NCBI
|
|
31
|
Yoon JW, Gilbertson R, Iannaccone S,
Iannaccone P and Walterhouse D: Defining a role for Sonic hedgehog
pathway activation in desmoplastic medulloblastoma by identifying
GLI1 target genes. Int J Cancer. 124:109–119. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Katoh M: Integrative genomic analyses of
CXCR4: transcriptional regulation of CXCR4 based on TGFbeta, Nodal,
Activin signaling and POU5F1, FOXA2, FOXC2, FOXH1, SOX17, and GFI1
transcription factors. Int J Oncol. 36:415–420. 2010.
|